Abstract
In the burgeoning field of Plasmodium gene expression, there are—to borrow some famous words from a former U.S. Secretary of Defense—“known knowns, known unknowns, and unknown unknowns.” This is in itself an important achievement, since it is only in the past decade that facts have begun to move from the third category into the first. Nevertheless, much remains in the middle ground of known or suspected “unknowns.” It is clear that the malaria parasite controls vital virulence processes such as host cell invasion and cytoadherence at least partly via epigenetic mechanisms, so a proper understanding of epigenetic transcriptional control in this organism should have great clinical relevance. Plasmodium, however, is an obligate intracellular parasite: it operates not in a vacuum but rather in the complicated context of its metazoan hosts. Therefore, as valuable data about the parasite's basic epigenetic machinery begin to emerge, it becomes increasingly important to relate in vitro studies to the situation in vivo. This review will focus upon the challenge of understanding Plasmodium epigenetics in an integrated manner, in the human and insect hosts as well as the petri dish.
WHY DOES EPIGENETICS MATTER IN PLASMODIUM?
The malaria parasite has a complicated life cycle involving several functionally distinct forms living in two very different host species. It must be able to perform rapid transitions between morphological states and also long-term antigenic variation to sustain chronic infections in human hosts. To achieve all this, the parasite employs various types of regulation, including transcriptional and posttranscriptional regulation of gene expression, translational repression, and posttranslational protein modification. While several of these regulatory modes are suited to respond rapidly to host conditions during an acute infection or a life cycle transition, only the epigenetic control of gene expression can then maintain that “imprinted” pattern in later generations. (The term “epigenetic” will be used here to refer to the heritable marking of genetic material without alterations in the actual genetic code.) Epigenetic mechanisms of transcriptional control are thus likely to act in at least three major areas of Plasmodium biology.
The first area is the cascade of changing gene expression which occurs during asexual replication in the human bloodstream. This is the most studied stage of the life cycle and the one which causes all the clinical symptoms of malaria. Plasmodium falciparum, the main Plasmodium species responsible for human mortality, has an unusual, formulaic mode of gene expression during its 48-h developmental process inside the erythrocyte, implying tight and integrated genome-wide regulation of transcription (13, 62, 65). It was initially thought that the Plasmodium genus had a striking paucity of classical transcription factors because few could be found in the sequenced genome of P. falciparum (49), and epigenetics was postulated as an alternative primary mode of transcriptional regulation. More recently, a large family of “ApiAP2” transcription factors has been identified in apicomplexans; this family is related to the AP2 family in plants and may fill the transcription factor “gap” (6, 30, 56, 110). Nevertheless, epigenetic histone modifications are also known to be involved in the erythrocytic cycle, because treating parasites with drugs which inhibit histone modifiers can disrupt their normal development (15). It is also notable that one ApiAP2 factor associates with the heterochromatin protein HP1 and another, in Toxoplasma gondii, copurifies with a histone-modifying enzyme, suggesting that the two modes of transcriptional regulation may converge (41, 96).
The second area in which epigenetics is likely to be important is in controlling sexual and morphological differentiation during the rest of the life cycle. The parasite has distinct hepatocytic as well as erythrocytic forms in the mammalian host; it can form sexually differentiated gametocytes during the blood stage and these gametocytes mate to produce ookinetes and then sporozoites in the mosquito. Transcriptional profiling has demonstrated distinct profiles in all of these stages: the gametocyte (103), ookinete (90), oocyst sporozoite (59), salivary gland sporozoite (73), hepatocyte stage (95, 105), and erythrocyte stage (62). Additional shifts in gene expression probably occur as the sporozoite migrates through the skin and invades a hepatocyte (102), after which replication occurs many hundredfold, giving rise to unique metabolic requirements. Likewise, many genes must be up- or downregulated at the abrupt transition to the insect gut with its distinct temperature and metabolic profile. Studies in the rodent malaria species P. berghei have shown that the latter transition is regulated partly by posttranslational repression (72) and partly by a single AP2 transcription factor that activates many genes required for mosquito midgut invasion (110). It has also been shown that an AP2 factor is involved in sporozoite development (109) and that differentiation into gametocytes may be regulated by another translational repressor, PfPuf2 (36, 78). These are the only two life cycle transitions for which underlying mechanisms have begun to emerge and it is not yet known whether epigenetic regulation plays a major role in any life cycle transition. However, in the related apicomplexan Toxoplasma gondii, life cycle transitions can be induced by inhibiting conserved histone-modifying enzymes (T. gondii HDAC3 [TgHDAC3] and TgCARM1), suggesting that similar pathways may exist in the malaria parasite (12, 96).
Finally, epigenetics is centrally involved in the variant expression of gene families which determine virulence processes such as cytoadherence and variant erythrocyte invasion. The best-characterized family of antigen-encoding genes is the “var” family in P. falciparum. This gene family encodes ∼60 variants of P. falciparum erythrocyte membrane protein 1 (PfEMP1), the main surface adhesin expressed on infected erythrocytes. var genes are generally silenced, with only one or a few being expressed at any one time (16, 32, 40, 43, 76, 79, 80, 100), and their variant expression is clinically important, being associated with immune evasion and recurring waves of high parasitemia in chronically infected patients (93). The expressed var gene(s) does not undergo recombination into a defined expression site, as occurs in Trypanosoma brucei, but instead is marked with “activating” histone modifications and probably is located in a special perinuclear site that is permissive for transcription (33, 45, 67). Furthermore, var gene silencing is relaxed when either of the two class 3 histone deacetylase enzymes or “sirtuins” in the parasite is knocked out (33, 106). All these findings are classical hallmarks of epigenetic gene control.
A second example of variant expression in P. falciparum is the use of alternative invasion pathways. During the erythrocytic cycle, merozoites can invade new erythrocytes via binding between variantly expressed ligands and specific host receptors (19), and evidence is beginning to emerge that at least some genes encoding invasion proteins are regulated epigenetically, in a way similar to that for var genes (20, 58).
WHAT DO WE KNOW ABOUT THE EPIGENETIC MACHINERY IN PLASMODIUM?
Given the apparent importance of epigenetics in Plasmodium, considerable effort has been exerted in recent years to characterize the epigenetic machinery of the parasite. The proteins involved can be divided into “writers” of epigenetic histone marks, “readers” of these marks, and structural chromatin proteins involved in enforcing these marks (98). The proteins identified thus far in all three classes are analogous to those in model organisms, and they are almost entirely conserved across Plasmodium species as well as in T. gondii. The conservation of epigenetic machinery in this early-diverging phylum speaks to the sustained importance of epigenetic pathways in apicomplexans. It has also proved to be a boon for parasitologists, since epigenetics in model organisms is very well studied and many useful experimental tools have been developed.
The galaxy of histone modifications and histone-modifying proteins found in P. falciparum is the subject of several recent reviews (22, 53, 104). In brief, histone tails can be acetylated, methylated, and sumoylated as in higher eukaryotes (54, 55, 77), and the parasite accordingly possesses histone acetylases, deacetylases, methylases, and demethylases, several of which have been characterized (35, 37, 75).
Recently, data have also begun to emerge about a possible role for noncoding RNA (ncRNA) in Plasmodium genome regulation. In higher eukaryotes ncRNAs have well-established roles in the establishment of heterochromatin via RNA interference (RNAi) proteins, but this is a controversial subject for Plasmodium, since the genus does not possess a functional RNAi system (11). Nevertheless, long ncRNAs have been found associated with P. falciparum centromeres (64), and a genome-wide survey revealed both sense and antisense transcripts derived from telomeres and telomere-associated repeats (TAREs). These ncRNAs may be involved in telomeric silencing even in the absence of RNAi, perhaps by stalling polymerases or forming RNA duplexes which prevent translation or cause retention in the nucleus (89). The same study detected a plethora of antisense RNAs from subtelomeric var genes and these have been reported to associate physically with var-carrying chromatin. However, there is no evidence that antisense transcription actually correlates with var gene silencing (34, 91). Much remains to be discovered in this area but it does seem likely that Plasmodium somehow employs ncRNAs as well as histone modifications to enforce epigenetic silencing.
WHAT EXPERIMENTAL TOOLS HAVE BEEN USED TO STUDY PLASMODIUM EPIGENETICS?
The present literature on Plasmodium epigenetics derives largely from experiments carried out with P. falciparum, with supporting data from T. gondii. These two species are uniquely amenable to molecular study because both can be grown in human cell culture, whereas most other species of Plasmodium cannot. The experimental methods used fall into three main categories: drug studies, genetic manipulations of histone-modifying enzymes, and studies profiling chromatin states and gene expression, in either a directed or a genome-wide manner.
Drug studies.
Blood-stage Plasmodium can be killed with many drugs which inhibit histone-modifying enzymes. These include the histone acetylase inhibitors curcumin and anacardic acid (23, 24), the sirtuin inhibitor nicotinamide (88), and a multitude of known and novel inhibitors of type 1 and type 2 histone deacetylases (1, 2, 4, 28, 70, 83). These results suggest that histone modification is crucial for normal parasite development, and deacetylase inhibitors are therefore under investigation as antimalarial drugs (3). For functional studies, however, drug treatment tends to be a “blunt instrument” because a drug may affect several different enzyme classes and parasites may be killed by off-target effects. For example, both the P. falciparum sirtuins can be knocked out genetically, so nicotinamide may not actually kill parasites via sirtuin inhibition unless the two enzymes are highly functionally redundant. When establishing whether a particular drug actually does kill via disrupted histone modification, it is clearly important to identify the genes affected, and in this regard, recent microarray studies using either anacardic acid or apicidin (a histone deacetylase inhibitor) revealed profoundly deregulated transcription across the genome and throughout the developmental cycle, demonstrating that histone acetylases and deacetylases act very widely (15, 24).
Studies of histone-modifying enzymes.
A more targeted approach than a drug study is to make genetic knockouts of particular histone modifiers or to characterize recombinant versions of these enzymes. Both P. falciparum sirtuins have been knocked out, affecting very specific, partially overlapping sets of subtelomeric, variantly expressed genes (33, 106). Therefore, in contrast to the type 1 and 2 histone deacetylases, the sirtuins have strikingly restricted genomic targets; in fact, these knockouts may be possible only because antigenic variation is dispensable in vitro. One sirtuin, Sir2a, as well as the GCN5 acetyltransferase and the PRMT1 arginine methyltransferase have been expressed in vitro and shown to be active enzymes (35, 37, 46, 75), but the repertoire of histone modifiers in the Plasmodium genome is large, so many enzymes remain to be characterized.
Studies of epigenetic marks.
The third branch of research in this field is the analysis of epigenetic readouts: the state of chromatin as assessed by chromatin immunoprecipitation (ChIP) and the levels of gene expression as measured by reverse transcription-PCR (RT-PCR) or microarray analysis. Targeted ChIP studies have shown that “activating” and “silencing” histone marks correlate with patterns of gene expression in several virulence gene families of interest, including the var genes and certain invasion-related genes (17, 44, 58, 67). Several “ChIP on chip” studies have also been published recently, expanding our picture of chromatin modification in P. falciparum by analyzing ChIP results on whole-genome microarrays. In the first of these studies, the presence of the acetylase GCN5 and an activating histone mark, histone 3 lysine 9 acetylation (H3K9ac), was found to correlate with gene transcription, albeit weakly (25). A similar but stronger association has also been observed in T. gondii (51). Two subsequent studies found that by contrast, the silencing histone mark histone 3 lysine 9 trimethylation (H3K9me3) is tightly restricted to subtelomeric regions carrying variant gene families, whose members are indeed mostly silenced at any one time (68, 99). The distribution of this mark was disrupted when Sir2a was knocked out, but only within the var and rif families, upon which this sirtuin has been shown to have a transcriptional effect. Thus, certain histone marks correlate well with patterns of gene expression and have actually been proposed as tools for predicting the locations of genes (in the case of H3K9ac) or the identity of variant gene families (in the case of H3K9me3).
Analyses of physical chromatin characteristics, including nucleosomal occupancy (87, 108) and the distribution of heterochromatin protein 1 (HP1) (42, 84), have also been made. Nucleosomal occupancy is highest around telomeres and lowest in housekeeping genes but it is not more generally correlated with levels of transcription throughout the genome. P. falciparum HP1, meanwhile, appears to be a constituent of heterochromatin, just as it is in model organisms: it binds to H3K9me3 and is tightly associated with subtelomeric gene families, although in contrast to the case in other organisms, neither HP1 nor H3K9me3 are found at centromeres, suggesting that these may have a somewhat unusual structure in P. falciparum. Overall, genome-wide studies of epigenetic markers are clearly valuable in providing a comprehensive picture of the genome, measuring the extent to which features can be generalized, and defining the extent and nature of variant gene families. Such studies must, however, be followed up experimentally to establish direct links between epigenotypes and phenotypes, as has been done for the var genes. Even in this well-studied example, much still remains to be discovered about the mechanisms which actually link epigenetic marks to gene expression and to antigenic switching.
WHAT ASPECTS OF PLASMODIUM EPIGENETICS MIGHT WE BE MISSING?
Parasite diversity.
Our present understanding of Plasmodium epigenetics has a very limited scope in terms of parasite diversity, both genetic and life cycle related. For practical reasons, research has been carried out almost exclusively on a few long-term-cultured strains of the erythrocytic stage of P. falciparum. This single model may be quite sufficient for studying basic mechanisms due to the apparent conservation of the epigenetic machinery across apicomplexa, but laboratory strains of P. falciparum are only poorly representative of the tremendous diversity of field strains, particularly in their variant gene families (10). Diversity of variant genes can certainly alter the effectiveness of the epigenetically regulated virulence processes in which these genes are expressed, but it has not been established whether and how genomic diversity might affect the underlying epigenetic mechanisms themselves. Most chromatin-modifying enzymes are well conserved among all sequenced genomes, but the duplication of at least one putative histone deacetylase has been observed (92). A recent study which used genomic microarrays to explore the extent of genomic and transcriptional variation in P. falciparum further suggested that genome diversity can have a significant impact on epigenetic processes. Recent field isolates were compared with a reference lab strain via both DNA and RNA microarrays, detecting very widespread amplifications and deletions in the field strains, together with altered patterns of gene expression during blood-stage development. Gene expression was often affected even outside the amplified or deleted regions, and this was proposed to represent an epigenetic “position effect,” although this remains to be proven mechanistically (69).
Disparate life cycle stages.
Furthermore, the well-studied erythrocytic stage is only one part of a complicated life cycle. It is, of course, the stage that causes human disease and undergoes antigenic variation and immune evasion, so its importance cannot be underestimated. Nevertheless, significant epigenetic pathways may operate in other life cycle stages which are important for malaria transmission, if not pathogenesis. It remains technically difficult to study the liver and insect stages of Plasmodium, because very limited amounts of parasite material can be obtained from hepatocyte cultures or dissected from mosquitoes. Nevertheless, as referenced above, transcriptional profiling has revealed distinct expression profiles in all life cycle stages, emphasizing the urgent need for functional studies of the underlying mechanisms. As yet, most of the transcription factors and all of the possible epigenetic factors which might control life cycle transitions remain to be characterized.
In vivo conditions.
A third issue that limits our present understanding of Plasmodium epigenetics is the nature of in vitro culture conditions. P. falciparum is grown in defined, energy-rich media representing only a small window of probable conditions in human bloodstreams (63). Furthermore, parasites are generally cultured in healthy O+ erythrocytes, another intentionally limited condition. Within these constraints, experimental manipulations can be carried out to mimic defined in vivo conditions, such as panning for the cytoadhesion of infected cells to particular receptors (97) or enzyme treating naïve cells to alter the availability of invasion ligands (21). Both these procedures lead to changes in the epigenetic environment of the parasites which survive them, thus yielding important data (58, 67). However, other forms of diversity which have yet to be replicated in vitro almost certainly exist in the human and insect hosts and these may affect gene expression in complicated and clinically important ways.
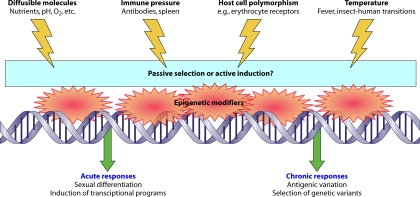
The in vivo stimuli which a parasite does not normally encounter in laboratory culture can be divided into physical effects (host erythrocyte polymorphisms that may affect merozoite invasion, immune pressure via antibody binding, and contact with the spleen), metabolic effects (fluctuations in diffusible molecules such nutrients, pH, and oxygen tension), and finally, temperature fluctuations within the human host and between insects and humans (Fig. 1). There is evidence that all these factors can alter parasites epigenetically, which might be expected because responding to adverse conditions such as fever or starvation could benefit the parasite. For example, in a febrile patient, parasite cytoadherence could be altered or sexual differentiation initiated to boost the chance of transmission.
Fig. 1.
Schematic showing in vivo stimuli which may affect the epigenetic control of gene transcription in Plasmodium falciparum.
Regarding the response to temperature, applying a heat shock to cultured P. falciparum alters the expression of many genes, including sexual-stage-specific rRNAs (38) and genes that are known to be subject to or involved in epigenetic regulation, such as var genes and the genes for histones, the histone deacetylase Sir2a, and a putative SWI2/SNF2 chromatin remodeler (82). The genes that are upregulated via heat shock are heavily biased toward subtelomeric regions, and epigenetic regulation is a common feature of these regions in Plasmodium as well as other organisms (68, 74). There is conflicting evidence as to whether heat shock actually alters the expression of PfEMP1 adhesins on the infected cell surface as a result of altered var transcription, but nevertheless an epigenetic response clearly occurs, at least at the transcriptional level (82, 107). Similarly, reducing the glucose concentration in culture media as a crude mimic of starvation affects the transcription of many genes, including sexual-stage rRNAs, var genes, and Sir2a genes (39).
With regard to immune pressure and host erythrocyte polymorphism, there is solid evidence that these can select for gene expression variants which are able to survive in the bloodstream: first, variants expressing PfEMP1s to which the host is not yet immune (57) and second, variants expressing invasion ligands suitable for the erythrocytes of a given host (71). What is not so clear is whether the parasite actually modulates the expression of var genes or invasion ligands epigenetically in response to immune pressure or whether expression simply varies stochastically, allowing immune selection to operate upon the results. In the P. knowlesi/rhesus macaque model, it has been reported that the immune status of the host and the presence or absence of a spleen actively influences the expression of the surface antigen “sicavar” (7–9), but no plausible mechanism has yet been proposed. The spleen of the squirrel monkey was likewise demonstrated to influence P. falciparum cytoadherence and antigenic variation (29), although this could be by passive selection. The squirrel monkey is a nonnatural host for P. falciparum, and the clinical relevance of this result is unclear, since splenectomized humans do not necessarily suffer from severe or abnormal presentations of falciparum malaria (66) despite anecdotal evidence of altered surface adhesin expression in such patients (5).
Conceptually, however, the parasite could certainly benefit from evolving a transcriptional response(s) to host immunity. For example, different PfEMP1 proteins adhere to different host endothelial ligands and are differently associated with severe malaria (60, 94), so modulating var gene expression to alter the strength or pattern of cytoadherence might help the parasite to avoid either killing or being killed. It is impossible to properly investigate epigenetic responses to the human immune system in vitro, but since var gene expression patterns in vitro do not represent those in vivo, host conditions probably do play a role in controlling var expression. Specifically, long-term-cultured strains of P. falciparum express chromosome-central var genes almost exclusively and very stably (43, 76), whereas patient isolates often express the telomere-proximal “UpsA” var genes (57) and also appear to switch between variants much more readily (50, 85). Again, the underlying mechanisms remain to be discovered, but genetic changes in laboratory strains do not seem to be entirely responsible, implicating epigenetic changes. One attractive candidate for a signaling molecule that might relay information from cell surface receptors into the parasite is the family of FIKK kinases. FIKK kinases have an unusual structure and no known function, but the family is greatly expanded in P. falciparum compared to other Plasmodium species and some members are exported to the erythrocyte cytoplasm, placing them appropriately to perform host-parasite signaling (81, 101).
Overall, the in vitro experiments described above offer some valuable leads on how Plasmodium might respond epigenetically to changing host conditions, although the real situation will, of course, be more multifactorial. Just how flexible the intraerythrocytic transcriptional “cascade” actually is remains controversial, since some studies have reported a very “hard-wired” transcriptional pattern: there is almost no transcriptional response when cultured parasites are treated with lethal antifolate drugs, for example (48). However, P. falciparum may well have evolved to modify its transcriptional program only in response to specific in vivo stimuli (not including the evolutionarily recent phenomenon of drug treatment). One genome-wide study of expression profiles in direct patient isolates certainly suggested that transcriptional flexibility does exist in a “real world” situation: three distinct groups of patient isolates were collected, each characterized by a widely different expression profile, two of which have not yet been replicated in vitro (27).
HOW CAN ANIMAL MODELS BE USED TO ADVANCE THE FIELD?
Epigenetic responses to host-parasite interactions remain underexplored and cannot be properly addressed using in vitro culture. It is technically and ethically difficult to study many of these questions in human patients because immune or metabolic manipulations of the host are generally not possible, infections cannot be followed over time without providing curative treatment, and in vitro conditions may interfere with the real patterns of in vivo gene expression once parasites are removed from the host. Epigenetic chromatin states are, by their nature, subject to rewriting, so even short-term culturing of patient isolates may yield results which are difficult to interpret. For example, a rare volunteer study of a laboratory strain of P. falciparum passed through a human showed that var gene expression profiles changed within days of the transfer from the bloodstream to in vitro culture (86). Thus, only snapshots of host conditions and parasite responses can usually be obtained from human subjects.
Experiments with mice and monkeys have gone some way toward filling this gap in our knowledge, although caution is always advisable when extrapolating from animal models to humans. Aotus and saimiri monkeys can both be infected, as nonnatural hosts, with P. falciparum (reviewed in references 18 and 52); these monkeys are used primarily in vaccine research, but studies of antigenic variation and cytoadherence have also been made. The rhesus macaque infected with P. knowlesi is a somewhat more accessible model and this system provided the first-ever data on clonal expression, silencing, and switching of variant antigens in vivo (reviewed in reference 47). The P. knowlesi sicavar antigen is not strictly analogous to PfEMP1, but it seems to undergo antigenic variation in a similar way (14), so it should be possible to study any epigenetic pathways regulating sicavar expression over the course of an infection and in response to altered host metabolic conditions. As an added advantage, P. knowlesi can be cultured in vitro and genetically modified, so antigenic switch rates could be compared in vitro versus in vivo and the roles of chromatin modifiers in the parasite could potentially be investigated. Whether simian models might also be helpful in the study of variant invasion pathways or other areas of epigenetic transcriptional control has yet to be explored, perhaps because very few researchers have access to such models.
Rodent species of Plasmodium grown in mice, i.e., P. berghei, P. chabaudi, and P. yoelli, have the great advantage of easy availability, and P. berghei, like P. knowlesi, is genetically tractable. The mouse offers a complete immune system as a context for infection, as well as the potential for both metabolic and genetic manipulation of the host. Furthermore, an entire insect-to-mammalian life cycle can be completed relatively easily with P. berghei, opening up the investigation of epigenetics in stages other than the erythrocyte stage. It is important to note, however, that none of the rodent parasites entirely resembles the human parasite P. falciparum. Although they have a comparable life cycle and intraerythrocytic developmental cycle, these species lack well-characterized surface-expressed adhesins and are not known to sequester in the microvasculature or to undergo antigenic variation or variant erythrocyte invasion. A large family of variantly expressed genes called “PIR” does exist in all Plasmodium species, but its function is unknown and its expression pattern in most species is not completely characterized (26).
Nevertheless, the mouse model does have great potential for use in studying epigenetic responses to host conditions. An elegant example of the use of a mouse model to demonstrate how a parasite can respond epigenetically to its host in vivo was recently published, with the parasite in this case being the fungus Candida glabrata (31). C. glabrata is an NAD+ auxotroph which senses external nicotinic acid levels and responds via the NAD+-dependent histone deacetylase Sir2. Surface adhesins, which are regulated epigenetically by Sir2, are expressed only when nicotinic acid, and therefore NAD+, is limited. Since nicotinic acid levels are high in the bloodstream but low in the urinary tract of the mouse, C. glabrata expresses adhesins only in the urinary tract, where they facilitate colonization. Feeding mice excess nicotinic acid which is excreted in the urine reverses this phenotype, thus confirming a functional relationship. Comparable experiments have not yet been carried out with Plasmodium, but feeding a histone deacetylase inhibitor to infected mice can cure them of P. berghei, an ambiguous result which nevertheless suggests that there is potential for functional epigenetic studies in the mouse model (1).
WHAT CAN BE GAINED FROM STUDYING P. FALCIPARUM IN HUMANS?
Finally, within ethical and practical limits, much work remains to be done in studying the epigenetic responses of P. falciparum to its human host. Although experimental manipulations may not be possible, associative studies comparing the transcriptional profiles and chromatin states of direct patient isolates with human factors such as metabolic and immune status, fever, and blood polymorphisms can be done. Such studies are vital to test in vitro findings for their in vivo relevance; disparities are likely to arise and should be rigorously examined to avoid making erroneous inferences from in vitro work. One such example, as previously discussed, is the apparent disparity between antigenic switch rates in vitro and rates calculated (albeit from limited evidence) in vivo.
A few important studies relating human factors to var gene expression have been published, this being one phenomenon that is well characterized as epigenetically regulated in vitro. The immune status of the host influences the type of var gene expressed, whether actively by host-parasite signaling or passively by immune selection, since naïve and semi-immune hosts favor parasites expressing telomere-proximal or “UpsA-type” var genes, while functionally immune adults favor chromosome-internal or “UpsC” types (57). var gene expression is presently one of the few examples which has been studied extensively in a patient context; however, numerous metabolic and immune factors have been correlated more generally with malaria infection and/or severity of disease, so additional patterns of host-responsive gene expression which may affect virulence probably remain undiscovered (reviewed in reference 63).
This last point highlights the importance of taking systems biology approaches to parasite gene expression both in vitro and in vivo. Only one study has thus far described the genome-wide transcriptional profiles of patient isolates of P. falciparum together with human metabolic factors, and this is certainly a costly and technically difficult approach (27). Three different transcriptional profiles were found, and these were interpreted as indicating a starvation response (a metabolic shift to oxidative respiration), a stress response, or growth in in vitro-like conditions. The stress response profile was correlated with markers of fever and inflammation, but considering the breadth of this study, it is perhaps surprising that more human factors did not emerge as correlates of parasite gene expression. Importantly, this study excluded the most highly polymorphic gene families such as var, since it is very difficult to study polymorphic gene families comprehensively in patient isolates using a single microarray. Furthermore, controversy has arisen regarding the interpretation of the results of this study (61). Replication in other cohorts and also with other experimental approaches may therefore be required to establish whether P. falciparum genuinely is refractory to changes in host conditions or whether the causative changes have been missed.
In conclusion, a combination of genome-wide and targeted transcriptional studies using both humans and animals will be necessary in the future to fully evaluate the in vivo relevance of our growing knowledge of Plasmodium epigenetics. Genome-wide studies should provide a picture of the overall transcriptional landscape under various host conditions, while targeted studies of particular genes or gene families should establish in detail how epigenetic mechanisms can influence parasite virulence. One of the greatest challenges may be integrating this picture with our expanding knowledge of parasite diversity from large-scale sequencing projects. This will, however, be vitally important if we are to understand Plasmodium epigenetics in the real world as well as in the laboratory.
ACKNOWLEDGMENTS
C.J.M. is a Charles H. Hood Foundation postdoctoral fellow. M.T.D. is a Burroughs Wellcome Fund New Investigator in the Pathogenesis of Infectious Diseases.
Footnotes
Published ahead of print on 18 June 2010.
REFERENCES
- 1.Agbor-Enoh S., Seudieu C., Davidson E., Dritschilo A., Jung M. 2009. Novel inhibitor of Plasmodium histone deacetylase that cures P. berghei-infected mice. Antimicrob. Agents Chemother. 53:1727–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews K. T., Tran T. N., Lucke A. J., Kahnberg P., Le G. T., Boyle G. M., Gardiner D. L., Skinner-Adams T. S., Fairlie D. P. 2008. Potent antimalarial activity of histone deacetylase inhibitor analogues. Antimicrob. Agents Chemother. 52:1454–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews K. T., Tran T. N., Wheatley N. C., Fairlie D. P. 2009. Targeting histone deacetylase inhibitors for anti-malarial therapy. Curr. Top. Med. Chem. 9:292–308 [DOI] [PubMed] [Google Scholar]
- 4.Andrews K. T., Walduck A., Kelso M. J., Fairlie D. P., Saul A., Parsons P. G. 2000. Anti-malarial effect of histone deacetylation inhibitors and mammalian tumour cytodifferentiating agents. Int. J. Parasitol. 30:761–768 [DOI] [PubMed] [Google Scholar]
- 5.Bachmann A., Esser C., Petter M., Predehl S., von Kalckreuth V., Schmiedel S., Bruchhaus I., Tannich E. 2009. Absence of erythrocyte sequestration and lack of multicopy gene family expression in Plasmodium falciparum from a splenectomized malaria patient. PLoS One 4:e7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balaji S., Babu M. M., Iyer L. M., Aravind L. 2005. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 33:3994–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnwell J. W., Howard R. J., Coon H. G., Miller L. H. 1983. Splenic requirement for antigenic variation and expression of the variant antigen on the erythrocyte membrane in cloned Plasmodium knowlesi malaria. Infect. Immun. 40:985–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnwell J. W., Howard R. J., Miller L. H. 1982. Altered expression of Plasmodium knowlesi variant antigen on the erythrocyte membrane in splenectomized rhesus monkeys. J. Immunol. 128:224–226 [PubMed] [Google Scholar]
- 9.Barnwell J. W., Howard R. J., Miller L. H. 1983. Influence of the spleen on the expression of surface antigens on parasitized erythrocytes. Ciba Found. Symp. 94:117–136 [DOI] [PubMed] [Google Scholar]
- 10.Barry A. E., Leliwa-Sytek A., Tavul L., Imrie H., Migot-Nabias F., Brown S. M., McVean G. A., Day K. P. 2007. Population genomics of the immune evasion (var) genes of Plasmodium falciparum. PLoS Pathog. 3:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baum J., Papenfuss A. T., Mair G. R., Janse C. J., Vlachou D., Waters A. P., Cowman A. F., Crabb B. S., de Koning-Ward T. F. 2009. Molecular genetics and comparative genomics reveal RNAi is not functional in malaria parasites. Nucleic Acids Res. 37:3788–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bougdour A., Maubon D., Baldacci P., Ortet P., Bastien O., Bouillon A., Barale J. C., Pelloux H., Menard R., Hakimi M. A. 2009. Drug inhibition of HDAC3 and epigenetic control of differentiation in Apicomplexa parasites. J. Exp. Med. 206:953–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bozdech Z., Llinas M., Pulliam B. L., Wong E. D., Zhu J., DeRisi J. L. 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1:E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown I. N., Brown K. N., Hills L. A. 1968. Immunity to malaria: the antibody response to antigenic variation by Plasmodium knowlesi. Immunology 14:127–138 [PMC free article] [PubMed] [Google Scholar]
- 15.Chaal B. K., Gupta A. P., Wastuwidyaningtyas B. D., Luah Y. H., Bozdech Z. Histone deacetylases play a major role in the transcriptional regulation of the Plasmodium falciparum life cycle. PLoS Pathog. 6:e1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Q., Fernandez V., Sundstrom A., Schlichtherle M., Datta S., Hagblom P., Wahlgren M. 1998. Developmental selection of var gene expression in Plasmodium falciparum. Nature 394:392–395 [DOI] [PubMed] [Google Scholar]
- 17.Chookajorn T., Dzikowski R., Frank M., Li F., Jiwani A. Z., Hartl D. L., Deitsch K. W. 2007. Epigenetic memory at malaria virulence genes. Proc. Natl. Acad. Sci. U. S. A. 104:899–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Contamin H., Behr C., Mercereau-Puijalon O., Michel J. 2000. Plasmodium falciparumin the squirrel monkey (Saimiri sciureus): infection of non-splenectomised animals as a model for exploring clinical manifestations of malaria. Microbes Infect. 2:945–954 [DOI] [PubMed] [Google Scholar]
- 19.Cortes A. 2008. Switching Plasmodium falciparum genes on and off for erythrocyte invasion. Trends Parasitol. 24:517–524 [DOI] [PubMed] [Google Scholar]
- 20.Cortes A., Carret C., Kaneko O., Yim Lim B. Y., Ivens A., Holder A. A. 2007. Epigenetic silencing of Plasmodium falciparum genes linked to erythrocyte invasion. PLoS Pathog. 3:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cowman A. F., Crabb B. S. 2006. Invasion of red blood cells by malaria parasites. Cell 124:755–766 [DOI] [PubMed] [Google Scholar]
- 22.Cui L., Fan Q., Cui L., Miao J. 2008. Histone lysine methyltransferases and demethylases in Plasmodium falciparum. Int. J. Parasitol. 38:1083–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui L., Miao J., Cui L. 2007. Cytotoxic effect of curcumin on malaria parasite Plasmodium falciparum: inhibition of histone acetylation and generation of reactive oxygen species. Antimicrob. Agents Chemother. 51:488–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui L., Miao J., Furuya T., Fan Q., Li X., Rathod P. K., Su X. Z., Cui L. 2008. Histone acetyltransferase inhibitor anacardic acid causes changes in global gene expression during in vitro Plasmodium falciparum development. Eukaryot. Cell 7:1200–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui L., Miao J., Furuya T., Li X., Su X. Z., Cui L. 2007. PfGCN5-mediated histone H3 acetylation plays a key role in gene expression in Plasmodium falciparum. Eukaryot. Cell 6:1219–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cunningham D., Lawton J., Jarra W., Preiser P., Langhorne J. The pir multigene family of Plasmodium: antigenic variation and beyond. Mol. Biochem. Parasitol. 170:65–73 [DOI] [PubMed] [Google Scholar]
- 27.Daily J. P., Scanfeld D., Pochet N., Le Roch K., Plouffe D., Kamal M., Sarr O., Mboup S., Ndir O., Wypij D., Levasseur K., Thomas E., Tamayo P., Dong C., Zhou Y., Lander E. S., Ndiaye D., Wirth D., Winzeler E. A., Mesirov J. P., Regev A. 2007. Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature 450:1091–1095 [DOI] [PubMed] [Google Scholar]
- 28.Darkin-Rattray S. J., Gurnett A. M., Myers R. W., Dulski P. M., Crumley T. M., Allocco J. J., Cannova C., Meinke P. T., Colletti S. L., Bednarek M. A., Singh S. B., Goetz M. A., Dombrowski A. W., Polishook J. D., Schmatz D. M. 1996. Apicidin: a novel antiprotozoal agent that inhibits parasite histone deacetylase. Proc. Natl. Acad. Sci. U. S. A. 93:13143–13147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.David P. H., Hommel M., Miller L. H., Udeinya I. J., Oligino L. D. 1983. Parasite sequestration in Plasmodium falciparum malaria: spleen and antibody modulation of cytoadherence of infected erythrocytes. Proc. Natl. Acad. Sci. U. S. A. 80:5075–5079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Silva E. K., Gehrke A. R., Olszewski K., Leon I., Chahal J. S., Bulyk M. L., Llinas M. 2008. Specific DNA-binding by apicomplexan AP2 transcription factors. Proc. Natl. Acad. Sci. U. S. A. 105:8393–8398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Domergue R., Castano I., De Las Penas A., Zupancic M., Lockatell V., Hebel J. R., Johnson D., Cormack B. P. 2005. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science 308:866–870 [DOI] [PubMed] [Google Scholar]
- 32.Duffy M. F., Brown G. V., Basuki W., Krejany E. O., Noviyanti R., Cowman A. F., Reeder J. C. 2002. Transcription of multiple var genes by individual, trophozoite-stage Plasmodium falciparum cells expressing a chondroitin sulphate A binding phenotype. Mol. Microbiol. 43:1285–1293 [DOI] [PubMed] [Google Scholar]
- 33.Duraisingh M. T., Voss T. S., Marty A. J., Duffy M. F., Good R. T., Thompson J. K., Freitas-Junior L. H., Scherf A., Crabb B. S., Cowman A. F. 2005. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell 121:13–24 [DOI] [PubMed] [Google Scholar]
- 34.Epp C., Li F., Howitt C. A., Chookajorn T., Deitsch K. W. 2009. Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum. RNA 15:116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan Q., An L., Cui L. 2004. Plasmodium falciparum histone acetyltransferase, a yeast GCN5 homologue involved in chromatin remodeling. Eukaryot. Cell 3:264–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan Q., Li J., Kariuki M., Cui L. 2004. Characterization of PfPuf2, member of the Puf family RNA-binding proteins from the malaria parasite Plasmodium falciparum. DNA Cell Biol. 23:753–760 [DOI] [PubMed] [Google Scholar]
- 37.Fan Q., Miao J., Cui L., Cui L. 2009. Characterization of PRMT1 from Plasmodium falciparum. Biochem. J. 421:107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang J., Sullivan M., McCutchan T. F. 2004. The effects of glucose concentration on the reciprocal regulation of rRNA promoters in Plasmodium falciparum. J. Biol. Chem. 279:720–725 [DOI] [PubMed] [Google Scholar]
- 39.Fang J., Zhou H., Rathore D., Sullivan M., Su X. Z., McCutchan T. F. 2004. Ambient glucose concentration and gene expression in Plasmodium falciparum. Mol. Biochem. Parasitol. 133:125–129 [DOI] [PubMed] [Google Scholar]
- 40.Fernandez V., Chen Q., Sundstrom A., Scherf A., Hagblom P., Wahlgren M. 2002. Mosaic-like transcription of var genes in single Plasmodium falciparum parasites. Mol. Biochem. Parasitol. 121:195–203 [DOI] [PubMed] [Google Scholar]
- 41.Flueck C., Bartfai R., Niederwieser I., Witmer K., Alako B. T., Moes S., Bozdech Z., Jenoe P., Stunnenberg H. G., Voss T. S. A major role for the Plasmodium falciparum ApiAP2 protein PfSIP2 in chromosome end biology. PLoS Pathog. 6:e1000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flueck C., Bartfai R., Volz J., Niederwieser I., Salcedo-Amaya A. M., Alako B. T., Ehlgen F., Ralph S. A., Cowman A. F., Bozdech Z., Stunnenberg H. G., Voss T. S. 2009. Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog. 5:e1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frank M., Dzikowski R., Amulic B., Deitsch K. 2007. Variable switching rates of malaria virulence genes are associated with chromosomal position. Mol. Microbiol. 64:1486–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freitas-Junior L. H., Bottius E., Pirrit L. A., Deitsch K. W., Scheidig C., Guinet F., Nehrbass U., Wellems T. E., Scherf A. 2000. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature 407:1018–1022 [DOI] [PubMed] [Google Scholar]
- 45.Freitas-Junior L. H., Hernandez-Rivas R., Ralph S. A., Montiel-Condado D., Ruvalcaba-Salazar O. K., Rojas-Meza A. P., Mancio-Silva L., Leal-Silvestre R. J., Gontijo A. M., Shorte S., Scherf A. 2005. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell 121:25–36 [DOI] [PubMed] [Google Scholar]
- 46.French J. B., Cen Y., Sauve A. A. 2008. Plasmodium falciparum Sir2 is an NAD+-dependent deacetylase and an acetyllysine-dependent and acetyllysine-independent NAD+ glycohydrolase. Biochemistry 47:10227–10239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galinski M. R., Corredor V. 2004. Variant antigen expression in malaria infections: posttranscriptional gene silencing, virulence and severe pathology. Mol. Biochem. Parasitol. 134:17–25 [DOI] [PubMed] [Google Scholar]
- 48.Ganesan K., Ponmee N., Jiang L., Fowble J. W., White J., Kamchonwongpaisan S., Yuthavong Y., Wilairat P., Rathod P. K. 2008. A genetically hard-wired metabolic transcriptome in Plasmodium falciparum fails to mount protective responses to lethal antifolates. PLoS Pathog. 4:e1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gardner M. J., Hall N., Fung E., White O., Berriman M., Hyman R. W., Carlton J. M., Pain A., Nelson K. E., Bowman S., Paulsen I. T., James K., Eisen J. A., Rutherford K., Salzberg S. L., Craig A., Kyes S., Chan M. S., Nene V., Shallom S. J., Suh B., Peterson J., Angiuoli S., Pertea M., Allen J., Selengut J., Haft D., Mather M. W., Vaidya A. B., Martin D. M., Fairlamb A. H., Fraunholz M. J., Roos D. S., Ralph S. A., McFadden G. I., Cummings L. M., Subramanian G. M., Mungall C., Venter J. C., Carucci D. J., Hoffman S. L., Newbold C., Davis R. W., Fraser C. M., Barrell B. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gatton M. L., Peters J. M., Fowler E. V., Cheng Q. 2003. Switching rates of Plasmodium falciparum var genes: faster than we thought? Trends Parasitol. 19:202–208 [DOI] [PubMed] [Google Scholar]
- 51.Gissot M., Kelly K. A., Ajioka J. W., Greally J. M., Kim K. 2007. Epigenomic modifications predict active promoters and gene structure in Toxoplasma gondii. PLoS Pathog. 3:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herrera S., Perlaza B. L., Bonelo A., Arevalo-Herrera M. 2002. Aotus monkeys: their great value for anti-malaria vaccines and drug testing. Int. J. Parasitol. 32:1625–1635 [DOI] [PubMed] [Google Scholar]
- 53.Horrocks P., Wong E., Russell K., Emes R. D. 2009. Control of gene expression in Plasmodium falciparum—ten years on. Mol. Biochem. Parasitol. 164:9–25 [DOI] [PubMed] [Google Scholar]
- 54.Issar N., Ralph S. A., Mancio-Silva L., Keeling C., Scherf A. 2009. Differential sub-nuclear localisation of repressive and activating histone methyl modifications in P. falciparum. Microbes Infect. 11:403–407 [DOI] [PubMed] [Google Scholar]
- 55.Issar N., Roux E., Mattei D., Scherf A. 2008. Identification of a novel post-translational modification in Plasmodium falciparum: protein sumoylation in different cellular compartments. Cell. Microbiol. 10:1999–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iyer L. M., Anantharaman V., Wolf M. Y., Aravind L. 2008. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int. J. Parasitol. 38:1–31 [DOI] [PubMed] [Google Scholar]
- 57.Jensen A. T., Magistrado P., Sharp S., Joergensen L., Lavstsen T., Chiucchiuini A., Salanti A., Vestergaard L. S., Lusingu J. P., Hermsen R., Sauerwein R., Christensen J., Nielsen M. A., Hviid L., Sutherland C., Staalsoe T., Theander T. G. 2004. Plasmodium falciparum associated with severe childhood malaria preferentially expresses PfEMP1 encoded by group A var genes. J. Exp. Med. 199:1179–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang L., Lopez-Barragan M. J., Jiang H., Mu J., Gaur D., Zhao K., Felsenfeld G., Miller L. H. Epigenetic control of the variable expression of a Plasmodium falciparum receptor protein for erythrocyte invasion. Proc. Natl. Acad. Sci. U. S. A 107:2224–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaiser K., Matuschewski K., Camargo N., Ross J., Kappe S. H. 2004. Differential transcriptome profiling identifies Plasmodium genes encoding pre-erythrocytic stage-specific proteins. Mol. Microbiol. 51:1221–1232 [DOI] [PubMed] [Google Scholar]
- 60.Kyriacou H. M., Stone G. N., Challis R. J., Raza A., Lyke K. E., Thera M. A., Kone A. K., Doumbo O. K., Plowe C. V., Rowe J. A. 2006. Differential var gene transcription in Plasmodium falciparum isolates from patients with cerebral malaria compared to hyperparasitaemia. Mol. Biochem. Parasitol. 150:211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lemieux J. E., Gomez-Escobar N., Feller A., Carret C., Amambua-Ngwa A., Pinches R., Day F., Kyes S. A., Conway D. J., Holmes C. C., Newbold C. I. 2009. Statistical estimation of cell-cycle progression and lineage commitment in Plasmodium falciparum reveals a homogeneous pattern of transcription in ex vivo culture. Proc. Natl. Acad. Sci. U. S. A. 106:7559–7564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Le Roch K. G., Zhou Y., Blair P. L., Grainger M., Moch J. K., Haynes J. D., De La Vega P., Holder A. A., Batalov S., Carucci D. J., Winzeler E. A. 2003. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301:1503–1508 [DOI] [PubMed] [Google Scholar]
- 63.LeRoux M., Lakshmanan V., Daily J. P. 2009. Plasmodium falciparum biology: analysis of in vitro versus in vivo growth conditions. Trends Parasitol. 25:474–481 [DOI] [PubMed] [Google Scholar]
- 64.Li F., Sonbuchner L., Kyes S. A., Epp C., Deitsch K. W. 2008. Nuclear non-coding RNAs are transcribed from the centromeres of Plasmodium falciparum and are associated with centromeric chromatin. J. Biol. Chem. 283:5692–5698 [DOI] [PubMed] [Google Scholar]
- 65.Llinas M., Bozdech Z., Wong E. D., Adai A. T., DeRisi J. L. 2006. Comparative whole genome transcriptome analysis of three Plasmodium falciparum strains. Nucleic Acids Res. 34:1166–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Looareesuwan S., Suntharasamai P., Webster H. K., Ho M. 1993. Malaria in splenectomized patients: report of four cases and review. Clin. Infect. Dis. 16:361–366 [DOI] [PubMed] [Google Scholar]
- 67.Lopez-Rubio J. J., Gontijo A. M., Nunes M. C., Issar N., Hernandez Rivas R., Scherf A. 2007. 5′ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol. Microbiol. 66:1296–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lopez-Rubio J. J., Mancio-Silva L., Scherf A. 2009. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe 5:179–190 [DOI] [PubMed] [Google Scholar]
- 69.Mackinnon M. J., Li J., Mok S., Kortok M. M., Marsh K., Preiser P. R., Bozdech Z. 2009. Comparative transcriptional and genomic analysis of Plasmodium falciparum field isolates. PLoS Pathog. 5:e1000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mai A., Cerbara I., Valente S., Massa S., Walker L. A., Tekwani B. L. 2004. Antimalarial and antileishmanial activities of aroyl-pyrrolyl-hydroxyamides, a new class of histone deacetylase inhibitors. Antimicrob. Agents Chemother. 48:1435–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maier A. G., Duraisingh M. T., Reeder J. C., Patel S. S., Kazura J. W., Zimmerman P. A., Cowman A. F. 2003. Plasmodium falciparum erythrocyte invasion through glycophorin C and selection for Gerbich negativity in human populations. Nat. Med. 9:87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mair G. R., Braks J. A., Garver L. S., Wiegant J. C., Hall N., Dirks R. W., Khan S. M., Dimopoulos G., Janse C. J., Waters A. P. 2006. Regulation of sexual development of Plasmodium by translational repression. Science 313:667–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matuschewski K., Ross J., Brown S. M., Kaiser K., Nussenzweig V., Kappe S. H. 2002. Infectivity-associated changes in the transcriptional repertoire of the malaria parasite sporozoite stage. J. Biol. Chem. 277:41948–41953 [DOI] [PubMed] [Google Scholar]
- 74.Merrick C. J., Duraisingh M. T. 2006. Heterochromatin-mediated control of virulence gene expression. Mol. Microbiol. 62:612–620 [DOI] [PubMed] [Google Scholar]
- 75.Merrick C. J., Duraisingh M. T. 2007. Plasmodium falciparum Sir2: an unusual sirtuin with dual histone deacetylase and ADP-ribosyltransferase activity. Eukaryot. Cell 6:2081–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Merrick C. J., Dzikowski R., Imamura H., Chuang J., Deitsch K., Duraisingh M. T. The effect of Plasmodium falciparum Sir2a histone deacetylase on clonal and longitudinal variation in expression of the var family of virulence genes. Int. J. Parasitol 40:35–43 [DOI] [PubMed] [Google Scholar]
- 77.Miao J., Fan Q., Cui L., Li J., Li J., Cui L. 2006. The malaria parasite Plasmodium falciparum histones: organization, expression, and acetylation. Gene 369:53–65 [DOI] [PubMed] [Google Scholar]
- 78.Miao J., Li J., Fan Q., Li X., Li X., Cui L. The Puf-family RNA-binding protein PfPuf2 regulates sexual development and sex differentiation in the malaria parasite Plasmodium falciparum. J. Cell Sci. 123:1039–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mok B. W., Ribacke U., Rasti N., Kironde F., Chen Q., Nilsson P., Wahlgren M. 2008. Default pathway of var2csa switching and translational repression in Plasmodium falciparum. PLoS One 3:e1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mok B. W., Ribacke U., Winter G., Yip B. H., Tan C. S., Fernandez V., Chen Q., Nilsson P., Wahlgren M. 2007. Comparative transcriptomal analysis of isogenic Plasmodium falciparum clones of distinct antigenic and adhesive phenotypes. Mol. Biochem. Parasitol. 151:184–192 [DOI] [PubMed] [Google Scholar]
- 81.Nunes M. C., Goldring J. P., Doerig C., Scherf A. 2007. A novel protein kinase family in Plasmodium falciparum is differentially transcribed and secreted to various cellular compartments of the host cell. Mol. Microbiol. 63:391–403 [DOI] [PubMed] [Google Scholar]
- 82.Oakley M. S., Kumar S., Anantharaman V., Zheng H., Mahajan B., Haynes J. D., Moch J. K., Fairhurst R., McCutchan T. F., Aravind L. 2007. Molecular factors and biochemical pathways induced by febrile temperature in intraerythrocytic Plasmodium falciparum parasites. Infect. Immun. 75:2012–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Patel V., Mazitschek R., Coleman B., Nguyen C., Urgaonkar S., Cortese J., Barker R. H., Greenberg E., Tang W., Bradner J. E., Schreiber S. L., Duraisingh M. T., Wirth D. F., Clardy J. 2009. Identification and characterization of small molecule inhibitors of a class I histone deacetylase from Plasmodium falciparum. J. Med. Chem. 52:2185–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Perez-Toledo K., Rojas-Meza A. P., Mancio-Silva L., Hernandez-Cuevas N. A., Delgadillo D. M., Vargas M., Martinez-Calvillo S., Scherf A., Hernandez-Rivas R. 2009. Plasmodium falciparum heterochromatin protein 1 binds to tri-methylated histone 3 lysine 9 and is linked to mutually exclusive expression of var genes. Nucleic Acids Res. 37:2596–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peters J., Fowler E., Gatton M., Chen N., Saul A., Cheng Q. 2002. High diversity and rapid changeover of expressed var genes during the acute phase of Plasmodium falciparum infections in human volunteers. Proc. Natl. Acad. Sci. U. S. A. 99:10689–10694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peters J. M., Fowler E. V., Krause D. R., Cheng Q., Gatton M. L. 2007. Differential changes in Plasmodium falciparum var transcription during adaptation to culture. J. Infect. Dis. 195:748–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ponts N., Harris E. Y., Prudhomme J., Wick I., Eckhardt-Ludka C., Hicks G. R., Hardiman G., Lonardi S., Le Roch K. G. Nucleosome landscape and control of transcription in the human malaria parasite. Genome Res. 20:228–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Prusty D., Mehra P., Srivastava S., Shivange A. V., Gupta A., Roy N., Dhar S. K. 2008. Nicotinamide inhibits Plasmodium falciparum Sir2 activity in vitro and parasite growth. FEMS Microbiol. Lett. 282:266–272 [DOI] [PubMed] [Google Scholar]
- 89.Raabe C. A., Sanchez C. P., Randau G., Robeck T., Skryabin B. V., Chinni S. V., Kube M., Reinhardt R., Ng G. H., Manickam R., Kuryshev V. Y., Lanzer M., Brosius J., Tang T. H., Rozhdestvensky T. S. A global view of the nonprotein-coding transcriptome in Plasmodium falciparum. Nucleic Acids Res. 38:608–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Raibaud A., Brahimi K., Roth C. W., Brey P. T., Faust D. M. 2006. Differential gene expression in the ookinete stage of the malaria parasite Plasmodium berghei. Mol. Biochem. Parasitol. 150:107–113 [DOI] [PubMed] [Google Scholar]
- 91.Ralph S. A., Bischoff E., Mattei D., Sismeiro O., Dillies M. A., Guigon G., Coppee J. Y., David P. H., Scherf A. 2005. Transcriptome analysis of antigenic variation in Plasmodium falciparum—var silencing is not dependent on antisense RNA. Genome Biol. 6:R93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ribacke U., Mok B. W., Wirta V., Normark J., Lundeberg J., Kironde F., Egwang T. G., Nilsson P., Wahlgren M. 2007. Genome wide gene amplifications and deletions in Plasmodium falciparum. Mol. Biochem. Parasitol. 155:33–44 [DOI] [PubMed] [Google Scholar]
- 93.Roberts D. J., Biggs B. A., Brown G., Newbold C. I. 1993. Protection, pathogenesis and phenotypic plasticity in Plasmodium falciparum malaria. Parasitol. Today 9:281–286 [DOI] [PubMed] [Google Scholar]
- 94.Rottmann M., Lavstsen T., Mugasa J. P., Kaestli M., Jensen A. T., Muller D., Theander T., Beck H. P. 2006. Differential expression of var gene groups is associated with morbidity caused by Plasmodium falciparum infection in Tanzanian children. Infect. Immun. 74:3904–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sacci J. B., Jr., Ribeiro J. M., Huang F., Alam U., Russell J. A., Blair P. L., Witney A., Carucci D. J., Azad A. F., Aguiar J. C. 2005. Transcriptional analysis of in vivo Plasmodium yoelii liver stage gene expression. Mol. Biochem. Parasitol. 142:177–183 [DOI] [PubMed] [Google Scholar]
- 96.Saksouk N., Bhatti M. M., Kieffer S., Smith A. T., Musset K., Garin J., Sullivan W. J., Jr., Cesbron-Delauw M. F., Hakimi M. A. 2005. Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Mol. Cell. Biol. 25:10301–10314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Salanti A., Staalsoe T., Lavstsen T., Jensen A. T., Sowa M. P., Arnot D. E., Hviid L., Theander T. G. 2003. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol. Microbiol. 49:179–191 [DOI] [PubMed] [Google Scholar]
- 98.Salcedo-Amaya A. M., Hoeijmakers W. A., Bartfai R., Stunnenberg H. G. Malaria: could its unusual epigenome be the weak spot? Int. J. Biochem. Cell Biol. 42:781–784 [DOI] [PubMed] [Google Scholar]
- 99.Salcedo-Amaya A. M., van Driel M. A., Alako B. T., Trelle M. B., van den Elzen A. M., Cohen A. M., Janssen-Megens E. M., van de Vegte-Bolmer M., Selzer R. R., Iniguez A. L., Green R. D., Sauerwein R. W., Jensen O. N., Stunnenberg H. G. 2009. Dynamic histone H3 epigenome marking during the intraerythrocytic cycle of Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 106:9655–9660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Scherf A., Hernandez-Rivas R., Buffet P., Bottius E., Benatar C., Pouvelle B., Gysin J., Lanzer M. 1998. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J. 17:5418–5426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schneider A. G., Mercereau-Puijalon O. 2005. A new Apicomplexa-specific protein kinase family: multiple members in Plasmodium falciparum, all with an export signature. BMC Genomics 6:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Siau A., Silvie O., Franetich J. F., Yalaoui S., Marinach C., Hannoun L., van Gemert G. J., Luty A. J., Bischoff E., David P. H., Snounou G., Vaquero C., Froissard P., Mazier D. 2008. Temperature shift and host cell contact up-regulate sporozoite expression of Plasmodium falciparum genes involved in hepatocyte infection. PLoS Pathog. 4:e1000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Silvestrini F., Bozdech Z., Lanfrancotti A., Di Giulio E., Bultrini E., Picci L., Derisi J. L., Pizzi E., Alano P. 2005. Genome-wide identification of genes upregulated at the onset of gametocytogenesis in Plasmodium falciparum. Mol. Biochem. Parasitol. 143:100–110 [DOI] [PubMed] [Google Scholar]
- 104.Sullivan W. J., Jr., Naguleswaran A., Angel S. O. 2006. Histones and histone modifications in protozoan parasites. Cell. Microbiol. 8:1850–1861 [DOI] [PubMed] [Google Scholar]
- 105.Tarun A. S., Peng X., Dumpit R. F., Ogata Y., Silva-Rivera H., Camargo N., Daly T. M., Bergman L. W., Kappe S. H. 2008. A combined transcriptome and proteome survey of malaria parasite liver stages. Proc. Natl. Acad. Sci. U. S. A. 105:305–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tonkin C. J., Carret C. K., Duraisingh M. T., Voss T. S., Ralph S. A., Hommel M., Duffy M. F., Silva L. M., Scherf A., Ivens A., Speed T. P., Beeson J. G., Cowman A. F. 2009. Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum. PLoS Biol. 7:e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Udomsangpetch R., Pipitaporn B., Silamut K., Pinches R., Kyes S., Looareesuwan S., Newbold C., White N. J. 2002. Febrile temperatures induce cytoadherence of ring-stage Plasmodium falciparum-infected erythrocytes. Proc. Natl. Acad. Sci. U. S. A. 99:11825–11829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Westenberger S. J., Cui L., Dharia N., Winzeler E., Cui L. 2009. Genome-wide nucleosome mapping of Plasmodium falciparum reveals histone-rich coding and histone-poor intergenic regions and chromatin remodeling of core and subtelomeric genes. BMC Genomics 10:610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yuda M., Iwanaga S., Shigenobu S., Kato T., Kaneko I. 2010. Transcription factor AP2-Sp and its target genes in malarial sporozoites. Mol. Microbiol. 75:854–863 [DOI] [PubMed] [Google Scholar]
- 110.Yuda M., Iwanaga S., Shigenobu S., Mair G. R., Janse C. J., Waters A. P., Kato T., Kaneko I. 2009. Identification of a transcription factor in the mosquito-invasive stage of malaria parasites. Mol. Microbiol. 71:1402–1414 [DOI] [PubMed] [Google Scholar]