Abstract
Candida albicans is an opportunistic pathogen that colonizes diverse mucosal niches with distinct environmental characteristics. To adapt to these different sites, C. albicans must activate and attenuate a variety of signal transduction pathways. A mechanism of signal attenuation is through receptor endocytosis and subsequent vacuolar degradation, which requires the endosomal sorting complex required for transport (ESCRT) pathway. This pathway comprises several polyprotein complexes (ESCRT-0, -I, -II, -III, and -DS) that are sequentially recruited to the endosomal membrane. The ESCRT pathway also activates the Rim101 transcription factor, which governs expression of genes required for virulence. Here, we tested the hypothesis that the ESCRT pathway plays a Rim101-independent role(s) in pathogenesis. We generated deletion mutants in each ESCRT complex and determined that ESCRT-I, -II, and -III are required for Rim101 activation but that ESCRT-0 and ESCRT-DS are not. We found that the ESCRT-0 member Vps27 and ESCRT-DS components are required to promote epithelial cell damage and, using a murine model of oral candidiasis, found that the vps27Δ/Δ mutant had a decreased fungal burden compared to that of the wild type. We found that a high-dose inoculum can compensate for fungal burden defects but that mice colonized with the vps27Δ/Δ strain exhibit less morbidity than do mice infected with the wild-type strain. These results demonstrate that the ESCRT pathway has Rim101-independent functions for C. albicans virulence.
Candida albicans is a ubiquitous human commensal that can cause infections in susceptible hosts (33, 41). In both disease and nondisease states, C. albicans colonizes diverse mucosal surfaces of its host, including the oral cavity, gastrointestinal tract, and urogenital tract (32). These niches can vary widely in pH, osmolarity, and available nutrients. C. albicans therefore survives in diverse environmental niches within its human host.
To adapt to these specific niches, C. albicans must be able to activate specific signal transduction pathways and concomitantly terminate other pathways to properly respond to environmental cues and regulate gene expression. One mechanism to terminate a signaling pathway is to degrade the cognate receptors in the vacuole. Thus, niche adaptation requires tight regulation of signaling pathway activation and termination, and receptor degradation is one mechanism to terminate signal propagation.
Prior to vacuolar degradation, the receptor protein is internalized through endocytosis, where it is subsequently incorporated into the endosomal lumen as an interluminal vesicle (ILV). Formation of the ILV requires the endosomal sorting pathway required for transport (ESCRT) complex pathway (3, 39, 42). ILVs increase in number, generating a mature multivesicular body (MVB), which then fuses with and releases its luminal content into the vacuole for degradation. MVB formation allows for transmembrane receptor downregulation and contributes to signal transduction arrest during adaptation to new environments.
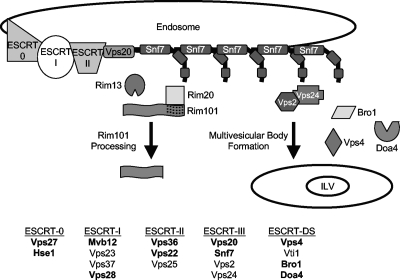
The ESCRT complex is composed of heterogeneous polyprotein complexes recruited to the endosomal membrane by the posttranslational modification pattern, frequently ubiquitination, of the receptor (or cargo) protein and the membrane lipid content (22). There are four core polyprotein complexes (Fig. 1): ESCRT-0 (Vps27 and Hse1), ESCRT-I (Mvb12, Vps23, Vps37, and Vps28), ESCRT-II (Vps36, Vps22, and Vps25), and ESCRT-III (Vps20, Snf7, Vps2, and Vps24) (2, 3, 7, 13, 21, 22). These complexes are recruited sequentially, and while ESCRT-0, -I, and -II are recruited as fully formed complexes, ESCRT-III arrives as two separate heterodimers (2). The first heterodimer, Vps20-Snf7, facilitates nucleation of additional Snf7 monomers around the ubiquitinated cargo protein (42), and Snf7 oligomerization is thought to mediate membrane involution necessary for ILV formation (16, 40, 42). The second ESCRT-III heterodimer, Vps2-Vps24, caps the Snf7 oligomer (2, 42) and recruits downstream proteins such as Bro1, Doa4, and Vps4 (4, 30). Bro1 recruits Doa4, which deubiquitinates the cargo protein before its incorporation into the ILV. Vps4, an AAA-ATPase, catalyzes ESCRT-III dissociation from the membrane and formation of the ILV (3, 4). The ESCRT complex function thereby results in ILV and subsequent MVB formation.
Fig. 1.
Model of ESCRT pathway and Rim101 pathway intersection. The Rim101 pathway intersects with the ESCRT pathway. ESCRT-0, -I, -II, and -III complexes are recruited to the endosomal membrane. ESCRT-III member Snf7 can interact either with Rim101 pathway members, resulting in proteolytic activation of Rim101, or with downstream ESCRT members, resulting in MVB formation. Yeast ESCRT complex components are listed below, and designations in bold are of those investigated in this study.
In addition to forming MVBs, the ESCRT-III member Snf7 plays a direct role in signal transduction in C. albicans and other fungi (24, 47, 49). Snf7 interacts with Rim20 (9, 20), a scaffold protein that binds the Rim101 transcription factor (48), and with Rim13 (8, 20), the putative protease responsible for Rim101 proteolytic activation (Fig. 1) (25). Activation of Rim101 is required for adaptation to neutral-alkaline environments (15), and this activation is mediated by Snf7 recruitment of Rim13 and Rim20. Snf7 is therefore required for both MVB formation and Rim101 processing in C. albicans.
Snf7 localization to the endocytic membrane is important for both MVB and Rim101 functions of Snf7 and is dependent on the upstream ESCRT complex members. In Saccharomyces cerevisiae, mutations in ESCRT-I or -II or the first heterodimer of ESCRT-III prevent Snf7 recruitment to the endosomal membrane and the ability of these strains to form MVBs. These mutants are also unable to process Rim101 (38, 49). ESCRT component mutations downstream of SNF7 prevent normal MVB formation but not Rim101 processing (38, 49). Likewise, mutation of the ESCRT-0 component VPS27 disrupts MVB formation but not Rim101 processing in S. cerevisiae. These data demonstrate that ESCRT-0 and ESCRT-DS are not required for Rim101 processing in S. cerevisiae but that ESCRT-I and -II and the first ESCRT-III heterodimer are (38, 49). This suggests that Snf7 must be recruited to the endosome for either MVB-specific or Rim101-specific function. Snf7 and Rim20 exhibit alkaline pH-dependent colocalization in a punctate pattern reminiscent of endosomal staining (10), suggesting that extracellular environment can influence Snf7 interactions. Snf7 therefore acts as a molecular hub at the endosome that can coordinate MVB-specific or Rim101-specific functions.
The role of ESCRT components in C. albicans Rim101 processing is relatively unexplored. Mutants in C. albicans ESCRT-I, -II, and -IIIa components displayed filamentation defects when tested for Rim101-dependent filamentation (49), suggesting that these genes are required for Rim101-dependent phenotypes. However, no ESCRT-0 C. albicans mutants have been tested for filamentation or Rim101 processing. Thus, the critical ESCRT complexes for C. albicans Rim101 processing are not fully understood.
We wished to investigate the relationship between the ESCRT and Rim101 pathways in C. albicans and to determine whether C. albicans pathogenesis requires ESCRT function independent of Rim101. Previous studies have suggested that ESCRT function plays an independent role in bloodstream candidiasis by demonstrating that mice infected with vps28Δ/Δ or vps32Δ/Δ C. albicans strains succumb to infection more slowly than do mice infected with a rim101Δ/Δ C. albicans strain (11). However, these mutant strains have not fully decoupled the Rim101 and ESCRT pathways, as these mutations affect both pathways. We hypothesized that, like S. cerevisiae, ESCRT-0 would not be required for Rim101 processing and would therefore be an ideal candidate in which to study MVB-specific contributions of the ESCRT pathway. Here, we generated a collection of ESCRT component mutants to characterize Rim101- and ESCRT-dependent phenotypes. We then used these mutants to investigate the role of ESCRT function in epithelial cell damage and C. albicans virulence. Our results demonstrate a role for ESCRT complex function in C. albicans pathogenesis independent of effects on Rim101 activation.
MATERIALS AND METHODS
Media and growth conditions.
All C. albicans strains were routinely passaged in YPD (1% yeast extract, 2% [wt/vol] Bacto peptone, 2% [wt/vol] dextrose) supplemented with 80 μg/ml uridine (Sigma). To select for Arg+, Ura+, or His+ transformants, synthetic medium (0.67% yeast nitrogen base plus ammonium sulfate and without amino acids; 2% dextrose; 80 μg of uridine per ml) was used, supplemented as required by the auxotrophic needs of the cells (1). To test for Rim101-dependent growth phenotypes, YPD was buffered with 150 mM HEPES to pH 9 with NaOH or contained 150 mM LiCl. For alkaline-induced filamentation assays, M199 medium (Gibco BRL) was buffered with 150 mM HEPES and pH adjusted to pH 4 or pH 8 and supplemented with 80 μg/ml uridine. For serum-induced filamentation assays, 4% bovine calf serum (BCS) was supplemented with 80 μg/ml uridine. For determining CFU from mouse tissue, YPD was supplemented with 1 μg/ml ampicillin (Invitrogen). Solid medium was prepared as described above with the addition of 2% Bacto agar.
Strain manipulation.
All strain manipulations were performed in the BWP17 genetic background and are listed in Table 1. All knockouts were engineered by subsequent rounds of allelic exchange with PCRs using a set of disrupt (DR) primers specific for the targeted gene (Table 2). For example, to generate the vps27Δ/Δ strain (DAY1177), the vps27::ARG4 cassette was amplified in a PCR using Vps27 5DR and Vps27 3DR primers and pRS-ARGΔSpe (46) template to generate the heterozygous mutant DAY1176. DAY1176 was transformed with the vps27::URA3-dpl200 cassette, which was amplified in a PCR using Vps27 5DR and Vps27 3DR primers and pDDB57 template (45), to generate the homozygous mutant DAY1177. To generate a prototrophic His+ strain, DAY1177 was transformed with NruI-digested pDDB78 plasmid to generate DAY1160. For the VPS27 complementation vector, genomic VPS27 plus promoter and terminator sequence was amplified in a PCR using Vps27 5comp and Vps27 3comp primers and DAY1 genomic DNA as a template. This sequence was transformed into S. cerevisiae along with NotI/EcoRI-digested pDDB78 to generate pDDB501 by in vivo recombination (28). NruI-digested pDDB501 was transformed into DAY1177 to generate DAY1264. All allelic exchange results were confirmed by diagnostic PCR (Fig. 2 and data not shown). To express RIM101-V5 in the vps27Δ/Δ strain, NruI-digested pDDB233 (25) was transformed into DAY1177 to generate DAY1178. All other ESCRT mutants were made similarly (25).
Table 1.
Strains used in these studies
| Name | Genotype | Reference |
|---|---|---|
| BWP17 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG his1::hisG/his1::hisG | 46 |
| DAY25 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG HIS1::DDB78::his1::hisG/his1::hisG rim101::ARG4/rim101::URA3 | 15 |
| DAY185 | ura3::λimm434/ura3::λimm434ARG4::URA3::arg4::hisG/arg4::hisG HIS1::DDB78::his1::hisG/his1::hisG | 14 |
| DAY534 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG his1::hisG/his1::hisG snf7::ARG4/snf7::URA3-dpl200 | 24 |
| DAY537 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG his1::hisG/his1::hisG vps4::ARG4/vps4::URA3-dpl200 | 24 |
| DAY568 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG RIM101-V5::HIS1::DDB233:::his1::hisG/his1::hisG vps4::ARG4/vps4::URA3-dpl200 | 24 |
| DAY576 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG RIM101-V5::HIS1::DDB233:::his1::hisG/his1::hisG vps4::ARG4 vps4::URA3-dpl200 | 24 |
| DAY653 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG his1::hisG/his1::hisG bro1::ARG4/bro1::URA3-dpl200 | 24 |
| DAY763 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG HIS1::DDB78::his1::hisG/his1::hisG snf7::ARG4/snf7::URA3-dpl200 | 24 |
| DAY1155 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG HIS1::DDB78::his1::hisG/his1::hisG vps4::ARG4/vps4::URA3-dpl200 | This study |
| DAY1156 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG HIS1::DDB78::his1::hisG/his1::hisG bro1::ARG4/bro1::URA3-dpl200 | This study |
| DAY1157 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG HIS1::DDB78::his1::hisG/his1::hisG vps20::ARG4/vps20::URA3-dpl200 | This study |
| DAY1158 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG HIS1::DDB78::his1::hisG/his1::hisG doa4::ARG4/doa4::URA3-dpl200 | This study |
| DAY1159 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG HIS1::DDB78::his1::hisG/his1::hisG bro1::ARG4/bro1::URA3-dpl200 | This study |
| DAY1160 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG HIS1::DDB78::his1::hisG/his1::hisG vps27::ARG4/vps27::URA3-dpl200 | This study |
| DAY1161 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG HIS1::DDB78::his1::hisG/his1::hisG vps28::ARG4/vps28::URA3-dpl200 | This study |
| DAY1162 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG HIS1::DDB78::his1::hisG/his1::hisG vps36::ARG4/vps36::URA3-dpl200 | This study |
| DAY1163 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG HIS1::DDB78::his1::hisG/his1::hisG mvb12::ARG4/mvb12::URA3-dpl200 | This study |
| DAY1176 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG his1::hisG/his1::hisG vps27::ARG4/VPS27 | This study |
| DAY1177 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG his1::hisG/his1::hisG vps27::ARG4/vps27::URA3-dpl200 | This study |
| DAY1178 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG RIM101-V5::DDB233::his1::hisG/his1::hisG vps27::ARG4/vps27::URA3-dpl200 | This study |
| DAY1181 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG his1::hisG/his1::hisG vps28::ARG4/VPS28 | This study |
| DAY1182 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG his1::hisG/his1::hisG vps28::ARG4/vps28::URA3-dpl200 | This study |
| DAY1183 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG RIM101-V5::DDB233::his1::hisG/his1::hisG vps28::ARG4/vps28::URA3-dpl200 | This study |
| DAY1186 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG his1::hisG/his1::hisG vps36::ARG4/VPS36 | This study |
| DAY1187 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG his1::hisG/his1::hisG vps36::ARG4/vps36::URA3-dpl200 | This study |
| DAY1188 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG RIM101-V5::DDB233::his1::hisG/his1::hisG vps36::ARG4/vps36::URA3-dpl200 | This study |
| DAY1191 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG his1::hisG/his1::hisG mvb12::ARG4/MVB12 | This study |
| DAY1192 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG his1::hisG/his1::hisG mvb12::ARG4/mvb12::URA3-dpl200 | This study |
| DAY1193 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG RIM101-V5::DDB233::his1::hisG/his1::hisG mvb12::ARG4/mvb12::URA3-dpl200 | This study |
| DAY1196 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG his1::hisG/his1::hisG vps22::ARG4/VPS22 | This study |
| DAY1197 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG his1::hisG/his1::hisG vps22::ARG4/vps22::URA3-dpl200 | This study |
| DAY1200 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG his1::hisG/his1::hisG hse1::ARG4/HSE1 | This study |
| DAY1201 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG his1::hisG/his1::hisG hse1::ARG4/hse1::URA3-dpl200 | This study |
| DAY1212 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG RIM101-V5::DDB233::his1::hisG/his1::hisG rim101::ARG4/rim101::URA3-dpl200 | This study |
| DAY1217 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG HIS1::DDB78::his1::hisG/his1::hisG vps22::ARG4/vps22::URA3-dpl200 | This study |
| DAY1218 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG HIS1::DDB78::his1::hisG/his1::hisG hse1::ARG4/hse1::URA3-dpl200 | This study |
| DAY1219 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG RIM101-V5::DDB233::his1::hisG/his1::hisG bro1::ARG4/bro1::URA3-dpl200 | This study |
| DAY1221 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG RIM101-V5::DDB233::his1::hisG/his1::hisG hse1::ARG4/hse1::URA3-dpl200 | This study |
| DAY1222 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG RIM101-V5::DDB233::his1::hisG/his1::hisG vps22::ARG4/vps22::URA3-dpl200 | This study |
| DAY1264 | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG VPS27::HIS1::DDB501::his1::hisG/his1::hisG vps27::ARG4/vps27::URA3-dpl200 | This study |
| DAY750 (CT128.1) | ura3::λimm434/ura3::λimm434arg4::hisG/arg4::hisG his1::hisG/his1::hisG ftr1::ARG4/ftr1::URA3-dpl200 | 23 |
Table 2.
Primers used in these studies
| Primer name | Primer sequence |
|---|---|
| Vps27 5DR | AATACTTATTAATTTCACATATAATATCATTTCATTAGTAGTAGGAAATACCTTTATATTTTTCCCAGTCACGACGTT |
| Vps27 3DR | AGAAGTAGAAGTTTGGCTAAATATTTCTAGATAAGCAAAAATTATTGCCAACTTTTATTAGTGGAATTGTGAGCGGATA |
| Hse1 5DR | TTTGCCTCCTCCCACTATTAATCAATTAGTAACCCCGATCATCAATTCCTTACCAACAACCTTTCCCAGTCACGACGTT |
| Hse1 3DR | AGTACTACAGCTTTATACTTCTTTCAATAGAAGAATATTAGTGAGACGTGTATTATATGACTGGAATTGTGAGCGGATA |
| Mvb12 5DR | GCAACCAACGAACGAACGAACGAACGAGCGAACCTATAAACACATACATTCACAACAACATTTCCCAGTCACGACGTT |
| Mvb12 3DR | TTATACTATTAATAATATGACCCATCTATTTACAAAAAAAAAACGTTTCATTATAATTCAGTGGAATTGTGAGCGGATA |
| Vps28 5DR | TTTCTTTAGTGTTTTGTTTATCCTTAGTCATAGAACAATCAACTTTGATTTATTGCCATTTTTCCCAGTCACGACGTT |
| Vps28 3DR | ATCGTATAAGCAAGAAACAGAGTATCCAACCAAACAGATAATTGTGTATTGTGTATATTAGTGGAATTGTGAGCGGATA |
| Vps36 5DR | TAGATGGGGAGTGGGAGCGAGTCAGATCTAATCTCTTATATATAGAAAAACCTCAATATTTTTCCCAGTCACGACGTT |
| Vps36 3DR | ATTAATACATAGTTCTATATATATGTCCATTTTTTTTTTTAAAAAAAACATATTACTTTAGTGGAATTGTGAGCGGATA |
| Vps22 5DR | ATTTTGATTAAAAGAGAAATCAAATCTTTATCTATCTTCCTATCACTTAACTATACTATTTTTCCCAGTCACGACGTT |
| Vps22 3DR | GGACAAGAAAAACTCAATAAAGAATAAGAAGTAAGTATGCCTGTATATATTTATTATTTAGTGGAATTGTGAGCGGATA |
| Vps20 5DR | TTATCAGGAACTGTAATTTGCTATCATAATCAATATAGATCTAATACATTCAAATTGACATTTCCCAGTCACGACGTT |
| Vps20 3DR | ATGTAAATGCAAAAATTTATTATTAAGTGTATTATGACTCACATAGAATTATGTTGTTGGGTGGAATTGTGAGCGGATA |
| Doa4 5DR | TTCGTACACTTTCTGATTCCAAACTAAATTACCACCAACTCTAACTTTTGTTCTTTGAAGTTTCCCAGTCACGACGTT |
| Doa4 3DR | GGGACACCACCCCAAGTTTCAATTTATTGACAAAAATAAATCTATGAGTTCTATTTAATAGTGGAATTGTGAGCGGATA |
| Vps27 5det | CCAGACGATATCAAACCTCC |
| Vps27 3det | CGAAAAAGATTAACACTACG |
| Hse1 5det | CCTCTGCTTATTCCAATTAGAACC |
| Hse1 3det | CGGTCAAGAAATCAAGGCTGACCC |
| Mvb12 5det | TCTCCAATCAAAGTGTTGTC |
| Mvb12 3det | GGTGTTCATCTTCCGATTAG |
| Vps28 5det | CGTCTGTGAATCTCATGGTG |
| Vps28 3det | CTCAAAGTTAGCATCGGACA |
| Vps36 5det | GACAAGAATAAGACCACGAG |
| Vps36 3det | CTCAACGTTATTTTTCTTC |
| Vps22 5det | GGCTCTGTATGGTCAATGAATAGC |
| Vps22 3det | CCCATCAATGGAATGGAAATCC |
| Vps20 5det | GAGGGGGATCAAGTTGCAAA |
| Vps20 3det | CCTTCCTGATTTTGCAAGAC |
| Doa4 5det | AAAATAAAGGAAATCCTGCA |
| Doa4 3det | ACGGCGAAATACACATAAAT |
| Vps27 5comp | AAGCTCGGAATTAACCCTCACTAAAGGGAACAAAAGCTGGCCTCTTCTTATCCAGTTTACTG |
| Vps27 3comp | ACGACGGCCAGTGAATTGTAATACGACTCACTATAGGGCGTTAATGAATTAATGATTTCTCC |
Fig. 2.
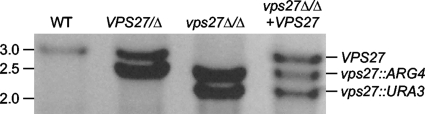
PCR genotyping of vps27Δ/Δ strains. VPS27 was amplified in a PCR using Vps27 5det and Vps27 3det primers (Table 2). Size markers in kilobases are noted on the left, and genotypes of each band are labeled on the right. Strains shown are VPS27/VPS27 (DAY185), VPS27/Δ (DAY1176), vps27Δ/Δ (DAY1160), and vps27Δ/Δ + VPS27 (DAY1264) strains. WT, wild type.
Growth assays.
Overnight YPD cultures were diluted 1:20 in sterile water and diluted subsequently 1:5 into sterile water using a 96-well sterile plate. Five microliters of each dilution was spotted onto YPD, YPD plus LiCl, and YPD pH 9 plates and incubated for 2 days at 37°C. For growth assays under iron-limiting conditions, strains were grown in YPD containing 50 μM iron chelator bathophenanthrolinedisulfonic acid (BPS) for 2 days before being spotted in 5-fold serial dilutions onto YPD plus 150 μM BPS plates. Plates were photographed with a Canon Powershot A560 camera and adjusted in Adobe Photoshop Elements 2.0.
Filamentation assays.
Strains were grown in 3 ml YPD overnight at 30°C. For alkaline-induced filamentation assays, 3-μl overnight cultures were spotted onto M199 pH 8 agar and incubated for 5 days at 37°C. For serum-induced filamentation, 3-μl overnight cultures were spread on 4% BCS plates and incubated for 24 h at 37°C. Strains were visualized and assessed for filamentation on a Zeiss Imager.M1 microscope.
Protein preparations.
Strains were grown to mid-log phase in 40 ml M199 pH 4 medium and either collected directly for protein preparation or collected and resuspended in 40 ml M199 pH 8 for 30 min before collection. Cells were collected by centrifugation and washed in 1 mM phenylmethylsulfonyl fluoride (PMSF), and cell pellets were resuspended in radioimmunoprecipitation assay buffer (50 mM Tris [pH 8], 150 mM NaCl, 1% NP-40, 3 mM EDTA, 0.5% deoxycholate, 0.1% sodium laurel sulfate) with protease inhibitors (1 mM PMSF and 1 μg/ml each of aprotinin, pepstatin, and leupeptin) and 10 mM dithiothreitol. Cells were lysed by glass bead disruption by being vortexed four times at 4°C. Lysates were then cleared by centrifugation at 15,000 × g for 15 min at 4°C to remove cell debris.
Western blot analysis.
A 50-μl protein preparation was separated by SDS-polyacrylamide gel electrophoresis (PAGE) in an 8% resolving gel. Gels were transferred to a nitrocellulose membrane and blocked for 1 h at room temperature with 5% dry milk dissolved in Tris-buffered saline containing 0.05% Tween 20 (TBS-T). Membranes were incubated with blocking solution containing 1:5,000 monoclonal anti-V5-horseradish peroxidase (HRP) conjugate antibody (Invitrogen). Membranes were washed three times in TBS-T, incubated with ECL reagent (Amersham), and exposed to film.
FaDu cell damage assay.
FaDu cells (ATCC) were plated in 24-well tissue culture dishes and incubated at 37°C and 5% CO2 in modified Eagle medium (MEM) with a 10% final concentration of fetal bovine serum (FBS) and 5 ml antibiotic/antimycotic cocktail (Invitrogen). At 90% monolayer confluence, cells were incubated in 0.5 ml medium containing 0.5 μCi Cr51 for 16 h. After the FaDu cells were washed with phosphate-buffered saline (PBS), 1 × 105 C. albicans cells were added in MEM with 10% FBS and 5 ml antibiotic cocktail (Invitrogen) and incubated for 10 h. Some FaDu cells were left uninfected to measure spontaneous Cr51 release. An 0.5-ml amount of supernatant was moved to a 13-ml glass test tube. An 0.5-ml amount of 6 M NaOH was added to the FaDu cells, the entire volume was moved to a separate test tube, and a final wash with 0.5 ml Liftaway (RPI Corp.) was performed. Specific release was calculated as [(2 × supernatant) − (2 × spontaneous release)]/[(2 × total) − (2 × spontaneous release)]. All samples were run in triplicate.
Oropharyngeal candidiasis (OPC) mouse model.
BALB/c mice 4 to 6 weeks old were obtained from Charles River (Wilmington, MA). On day −1, mice were immunosuppressed with 0.2 mg cortisone acetate/g of body weight. On day 0, 50 μl of 1 × 105-cell/ml or 1 × 106-cell/ml C. albicans cell suspensions was inoculated onto a sterilized cotton ball and incubated for 1 h in the oral cavity of mice anesthetized with 100 μg/g ketamine, 20 μg/g xylazine-HCl, and 2.8 μg/g acepromazine. On day 3, 10 mice per C. albicans strain were sacrificed, and the tongues were removed, weighed, and saved in 2 ml PBS. Tissues were homogenized and plated in 100, 10−1, and 10−2 dilutions on YPD plus ampicillin (AMP) plates. Plates were incubated at 37°C overnight. Colonies were counted and calculated to represent CFU/g tissue. On day 3, remaining mice were reimmunosuppressed with 0.2 mg/g cortisone acetate. On day 6, the remaining mice were sacrificed and tongues were homogenized and plated as described for day 3. All mice were weighed daily throughout the course of infection. All experiments were performed under the established University of Minnesota Institutional Animal Care and Use Committee guidelines.
RESULTS
ESCRT knockout mutations and ESCRT-dependent phenotypic defects.
We hypothesized that MVB formation plays a role in C. albicans pathogenesis independent of Rim101 processing. To test this hypothesis, we generated and characterized strains with mutations in individual members of the ESCRT-0 (Vps27 and Hse1), ESCRT-I (Mvb12 and Vps28), ESCRT-II (Vps36 and Vps22), or ESCRT-III (Vps20 and Snf7) complexes, as well as mutations in components that act downstream of Snf7 (Vps4, Bro1, and Doa4) (referred to as ESCRT-DS in this text) (Fig. 1 and Table 1). All mutant strains were made prototrophic and were then tested for ESCRT-dependent and Rim101-dependent phenotypes.
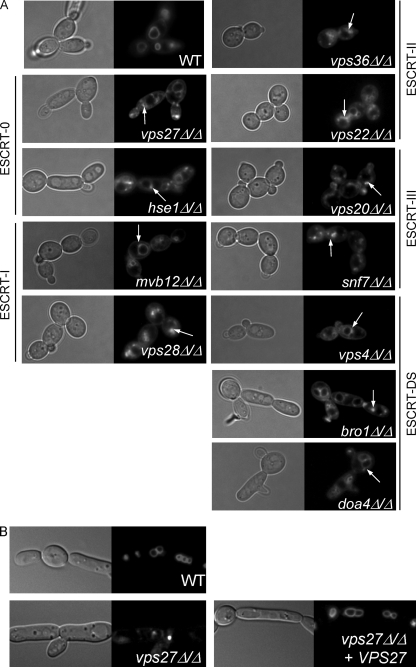
We first tested whether ESCRT homologs in C. albicans function in endosomal trafficking using the lipophilic dye FM 4-64 to visualize internalized vesicle trafficking (43). The plasma membrane was stained with FM 4-64, and the unbound dye was removed by washing. Following a 60-min chase, wild-type cells showed strong staining of the vacuole, with little to no cytoplasmic staining (Fig. 3 A). Although the mvb12Δ/Δ, vps4Δ/Δ, bro1Δ/Δ, and doa4Δ/Δ strains displayed weaker defects than did the other ESCRT mutants tested, all strains lacking ESCRT components displayed staining patterns containing accumulations of perivacuolar staining, indicating that each of the individual mutations in ESCRT complex genes leads to formation of class E-like exclusion bodies (Fig. 3A). In S. cerevisiae, most ESCRT components were initially identified as class E vps mutants, due to the formation of a “class E exclusion body” around the vacuole perimeter, which consists of accumulated immature and/or incomplete MVBs unable to fuse with the vacuole (36). This demonstrates that the homologs identified in C. albicans play a similar role in endosomal trafficking as they do in other organisms. We concluded that all ESCRT components identified play a role in MVB trafficking.
Fig. 3.
FM 4-64 trafficking defects of C. albicans ESCRT mutants. (A) Exponentially growing cultures were exposed to 16 mM FM 4-64 for 15 min on ice. Cells were washed with M199 pH 8, resuspended in fresh medium, and grown for 60 min at 30°C. Eighty microliters of culture was added to 10 μl each of 100 mM NaF and 100 mM NaN3 before microscopic examination. Strains analyzed include wild-type (WT) (DAY185), vps27Δ/Δ (DAY1160), hse1Δ/Δ (DAY1218), mvb12Δ/Δ (DAY1162), vps28Δ/Δ (DAY1161), vps36Δ/Δ (DAY1163), vps22Δ/Δ (DAY1217), vps20Δ/Δ (DAY1157), snf7Δ/Δ (DAY763), vps4Δ/Δ (DAY1155), bro1Δ/Δ (DAY1156), and doa4Δ/Δ (DAY1158) strains. Arrows denote class E-like exclusion bodies within the cell. (B) Complementation of the vps27Δ/Δ mutant. FM 4-64 staining was performed as described above using WT (DAY185), vps27Δ/Δ (DAY1160), and vps27Δ/Δ + VPS27 (DAY1264) strains.
Rim101-dependent phenotypic defects.
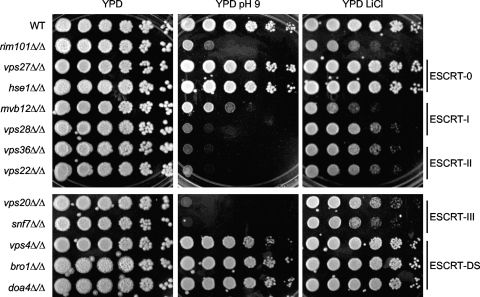
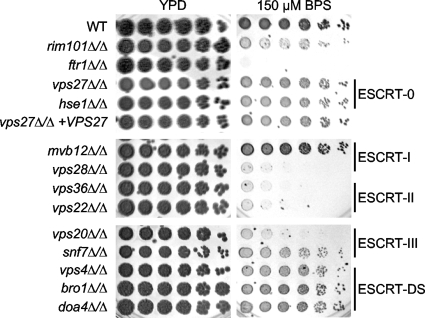
We next determined the effect of the ESCRT mutants on Rim101-dependent processes in C. albicans. Overnight YPD cultures were diluted 1:50 in PBS, and 5-fold dilutions were spotted onto YPD, YPD pH 9, or YPD plus LiCl agar medium. After 2 days at 37°C, all strains grew robustly on YPD, with similar colony formation numbers at higher dilutions, indicating that each dilution set represented roughly equivalent numbers of cells (Fig. 4). Thus, all mutants were able to grow well on rich medium and at a physiologically relevant temperature.
Fig. 4.
Effect of ESCRT mutants in Rim101-dependent growth assays. Fivefold dilutions of overnight YPD cultures were spotted onto YPD, YPD pH 9, or YPD plus LiCl agar medium. Plates were incubated at 37°C for 2 days before being photographed. Strains analyzed are the same as those in Fig. 3A.
As expected, we observed robust growth of the wild-type strain and poor growth of the rim101Δ/Δ strain on YPD pH 9 and YPD plus LiCl (Fig. 4). However, the vps27Δ/Δ and the hse1Δ/Δ strains, which lack individual ESCRT-0 components, grew similarly to wild type on both YPD pH 9 and YPD plus LiCl. Thus, ESCRT-0 is not required for Rim101-dependent growth.
As predicted, all strains lacking ESCRT-I (mvb12Δ/Δ or vps28Δ/Δ), ESCRT-II (vps36Δ/Δ or vps22Δ/Δ), or either component of the Vps20-Snf7 ESCRT-III heterodimer grew poorly on both YPD pH 9 and YPD plus LiCl (Fig. 4). This extends but is consistent with previously reported data from S. cerevisiae and C. albicans (11, 17, 47, 49). However, we noted that all of these mutants, excepting the mvb12Δ/Δ strain, grew more poorly than did the rim101Δ/Δ mutant on YPD pH 9, suggesting that these ESCRT pathway members have Rim101-dependent and -independent functions during alkaline growth. Although displaying an alkaline growth defect, the mvb12Δ/Δ mutant grew slightly better than did the rim101Δ/Δ strain on this medium, suggesting an intermediate role in Rim101-dependent alkaline growth. All ESCRT-I, -II, and -III mutants tested therefore displayed a growth defect on alkaline medium. Similarity in growth between these mutants and the rim101Δ/Δ mutant on YPD plus LiCl suggests that these components play only a Rim101-dependent role during LiCl growth (Fig. 4). We noted an exception with the mvb12Δ/Δ mutant, which grew slightly worse than did the rim101Δ/Δ mutant (Fig. 4). This suggests that Mvb12 contributes to LiCl growth in a Rim101-dependent and -independent manner and that this Rim101-independent function is also ESCRT independent.
We did not observe growth defects in any ESCRT-DS mutant strain (Fig. 4). The vps4Δ/Δ, bro1Δ/Δ, and doa4Δ/Δ strains grew well on YPD pH 9 and YPD plus LiCl. This is consistent with S. cerevisiae and our results, which demonstrate that strains lacking VPS4 constitutively process Rim101 (17; see also below). Further, neither the Δbro1 nor the Δdoa4 S. cerevisiae mutation prevents Rim101 processing, and a C. albicans bro1Δ/Δ mutant did not confer Rim101-dependent filamentation defects (49). These results support a model where ESCRT-DS members are not required for Rim101-dependent growth.
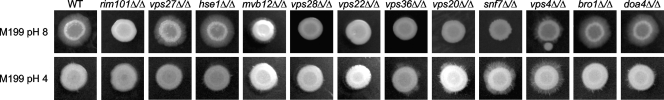
We then investigated the role of various ESCRT components in Rim101-dependent filamentation. We expected that ESCRT-I, -II, and -III would be required for Rim101 processing and thus have Rim101-dependent filamentation defects but that ESCRT-0 and ESCRT-DS components would not. To test this hypothesis, we spotted 3 μl of culture onto M199 pH 8 plates and incubated them at 37°C. After 6 days, a ring of filamentation was apparent around the periphery of the wild-type colony (Fig. 5; Table 3). No peripheral filamentation was observed around the rim101Δ/Δ mutant. We observed wild-type filamentation around the vps27Δ/Δ, hse1Δ/Δ, bro1Δ/Δ, and doa4Δ/Δ mutant colonies. However, we found intermediate filamentation, indicated by a shorter filament radius or an uneven filamentous periphery, around the mvb12Δ/Δ and vps4Δ/Δ colonies, respectively. No filamentation was observed around the vps28Δ/Δ, vps36Δ/Δ, vps22Δ/Δ, vps20Δ/Δ, or snf7Δ/Δ mutant colonies (Fig. 5). Thus, the mutant strains with the most severe filamentation defects were those with Rim101-dependent growth defects.
Fig. 5.
ESCRT mutant filamentation phenotypes. (Top) Three microliters of overnight YPD cultures was spotted on M199 pH 8 agar plates, and plates were incubated for 6 days at 37°C before being photographed. (Bottom) Three microliters of overnight YPD cultures was spotted onto M199 pH 4 agar plates, and plates were incubated for 6 days at 37°C before being photographed. Strains analyzed are the same as those in Fig. 3A.
Table 3.
Phenotypic analyses of ESCRT mutantsa
| Strain | Genotype | Phenotypeb |
% WT FaDu cell damage | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Growth on medium type: |
Filamentation on: |
BPS growth | Rim101 processing | ||||||
| Alkaline | LiCl | Alkaline agar | Acidic agar | BCS agar | |||||
| DAY185 | WT | + | + | + | − | + | + | + | NA |
| DAY25 | rim101Δ/Δ | − | − | − | − | − | − | − | 3 ± 5 |
| DAY1160 | vps27Δ/Δ | + | + | + | − | + | ± | + | 63 ± 17 |
| DAY1218 | hse1Δ/Δ | + | + | + | − | + | ± | + | 94 ± 16 |
| DAY1162 | mvb12Δ/Δ | ± | − | ± | − | − | + | − | 37 ± 15 |
| DAY1161 | vps28Δ/Δ | − | − | − | − | − | − | − | 12 ± 7 |
| DAY1163 | vps36Δ/Δ | − | − | − | ± | − | − | − | 6 ± 6 |
| DAY1217 | vps22Δ/Δ | − | − | − | − | − | − | − | 8 ± 8 |
| DAY1157 | vps20Δ/Δ | − | − | − | ± | − | − | − | 14 ± 3 |
| DAY763 | snf7Δ/Δ | − | − | − | ± | − | − | − | 4 ± 3 |
| DAY1155 | vps4Δ/Δ | + | − | ± | ± | + | − | + | 59 ± 7 |
| DAY1156 | bro1Δ/Δ | + | − | + | − | + | ± | + | 68 ± 12 |
| DAY1158 | doa4Δ/Δ | + | − | + | − | + | ± | + | 68 ± 11 |
WT, wild type; NA, not applicable.
+, positive; −, negative; ±, intermediate.
We next tested whether the ESCRT mutants affected filamentation at acidic pH. C. albicans does not normally filament in acidic conditions, but we have found that both snf7Δ/Δ and vps4Δ/Δ strains had peripheral filamentation on M199 pH 4 agar (47). We hypothesized that acidic filamentation may be suppressed by an ESCRT function and that other ESCRT knockout mutants would behave similarly. However, when spotted onto M199 pH 4 plates, only the snf7Δ/Δ and vps4Δ/Δ strains showed uniform peripheral rings after incubation at 37°C (Fig. 5; Table 3). We did note smaller areas of filamentation in uneven patches around the periphery of the vps36Δ/Δ, vps20Δ/Δ, bro1Δ/Δ, and doa4Δ/Δ colonies. Thus, Snf7 and Vps4 play the most critical roles in inhibiting acidic filamentation, and other ESCRT components play a small role or none.
Finally, we tested whether the ESCRT mutations affected serum-induced filamentation. A rim101Δ/Δ strain is unable to filament on 4% BCS agar after 24 h of incubation at 37°C. We streaked overnight cultures onto 4% BCS agar. After 24 h, the wild-type strain had produced a thick mesh of hyphae, while the rim101Δ/Δ strain was observed only in yeast form (Table 3). All ESCRT-0 and ESCRT-DS mutant strains were observed to form wild-type-like hyphal cells, while all ESCRT-I, -II, and -III mutant strains were observed only as yeast cells (Table 3). These results support the model that ESCRT-I, -II, and -III are required for Rim101 processing in C. albicans and that ESCRT-0 and ESCRT-DS are not.
Rim101 processing.
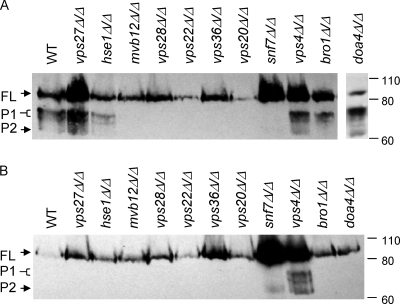
To confirm that the growth and filamentation defects observed in the ESCRT mutants were or were not associated with Rim101 processing, we introduced a RIM101-V5 allele, which encodes a functional Rim101 protein containing an internal V5 epitope tag, into each mutant background. Strains were grown to mid-log phase, and protein was collected for Western blot analysis. After growth in M199 pH 8 medium, we observed distinct Rim101-V5 bands in the wild-type strain: the full-length, unprocessed 84-kDa band (FL), the processed, active 74- to 78-kDa bands (P1), and the processed 65-kDa band of unknown function (P2) (Fig. 6 A) as reported previously (25). We noted similar banding patterns in the two ESCRT-0 knockout strains, the vps27Δ/Δ and hse1Δ/Δ strains (Fig. 6A). All ESCRT-I, -II, and -III knockout strains showed little to no processing to the P1 or P2 forms, indicating that Rim101 processing is defective in these strains (Fig. 6A). The vps4Δ/Δ, bro1Δ/Δ, and doa4Δ/Δ strains, whose products all act downstream of Snf7 recruitment to the endosome, showed a banding pattern similar to that of the wild-type strain (Fig. 6A). Since the Δvps4 mutant in S. cerevisiae processes Rim101 under noninducing conditions (17), we analyzed the RIM101-V5 processing in the ESCRT mutants after growth in M199 pH 4 medium. In wild-type cells, only full-length Rim101 was observed. Similar results were observed in all mutant strains except the vps4Δ/Δ mutant strain (Fig. 6B). The vps4Δ/Δ strain displayed both P1 and P2 bands at pH 4, indicating improper Rim101 activation. However, the other ESCRT-DS mutation did not affect Rim101 processing at pH 4. Regardless, the growth defects observed in the ESCRT mutants correlated strongly with the ability of the mutant strains to process Rim101 at pH 8.
Fig. 6.
Rim101 processing at alkaline and acidic pH. (A) Overnight YPD cultures were inoculated into 40 ml M199 pH 4 and grown for 5 h at 30°C. Cell pellets were collected and resuspended in 50 ml M199 pH 8 and grown for 30 min at 30°C, protein purified, and analyzed by Western blotting. Rim101-V5 was detected after 1 h of incubation with anti-V5-HRP antibody (Invitrogen) by chemiluminescence. Strains analyzed included wild-type (WT) (DAY1212), vps27Δ/Δ (DAY1178), hse1Δ/Δ (DAY1221), mvb12Δ/Δ (DAY1193), vps28Δ/Δ (DAY1183), vps36Δ/Δ (DAY1188), vps22Δ/Δ (DAY1222), vps20Δ/Δ (DAY1257), snf7Δ/Δ (DAY568), vps4Δ/Δ (DAY576), bro1Δ/Δ (DAY1219), and doa4Δ/Δ (DAY1260) strains. Molecular mass markers (in kilodaltons) are indicated to the right. (B) Overnight YPD culture was inoculated into 40 ml M199 pH 4 and grown for 5 h at 30°C. Protein preparations and Western blot analysis were done as described above.
Iron-dependent growth defects.
Rim101 promotes expression of genes involved in uptake of iron, which is insoluble in alkaline environments (5). Studies of several C. albicans ESCRT mutants demonstrated that members of ESCRT-I, -II, and -III are required for transporting hemoglobin to the vacuole, including the ESCRT-III member VPS2 (44). Since Rim101 is not required for vacuolar transport, we hypothesized that the ESCRT pathway may play a role in iron acquisition independent of Rim101 processing.
To test this hypothesis, we grew strains in YPD plus 50 μM iron chelator BPS and serially diluted them onto YPD or YPD plus 150 μM BPS agar medium. After 3 days of growth, all strains grew equally well on YPD medium. The wild-type strain grew well on YPD plus BPS plates, resulting in large thick colonies. The ftr1Δ/Δ mutant strain, which lacks a high-affinity iron permease (35), did not grow on YPD plus BPS medium, while the rim101Δ/Δ mutant strain showed an intermediate growth defect, able to form only thin colonies on the medium. Similar intermediate growth defects were seen in the vps27Δ/Δ, hse1Δ/Δ, snf7Δ/Δ, vps4Δ/Δ, bro1Δ/Δ, and doa4Δ/Δ mutants (Fig. 7), although a more severe growth defect was observed with the vps28Δ/Δ, vps36Δ/Δ, vps22Δ/Δ, and vps20Δ/Δ mutant strains (Fig. 7). The mvb12Δ/Δ mutant strain grew similarly to the wild-type strain, which likely reflects the limited phenotypes observed with this mutant (Fig. 3, 4, and 5). Since ESCRT-I, ESCRT-II, and ESCRT-III mutants have more severe growth defects than do ESCRT-0 and ESCRT-DS mutants, this suggests that the ESCRT pathway contributes to iron acquisition through Rim101-dependent and -independent mechanisms. Furthermore, since the snf7Δ/Δ mutant showed a growth defect similar to that of the rim101Δ/Δ, ESCRT-0, and ESCRT-DS mutants, this suggests that the snf7Δ/Δ mutation does not affect both Rim101-dependent and ESCRT-dependent iron acquisition.
Fig. 7.
ESCRT mutants have growth defects on iron-depleted medium. YPD plus BPS cultures were diluted 1:50 in PBS, and 5-fold serial dilutions were spotted onto YPD and YPD plus BPS agar plates. Plates were incubated at 37°C for 2 days before being photographed. Strains investigated included wild-type (WT) (DAY185), ftr1Δ/Δ (DAY750), rim101Δ/Δ (DAY25), vps27Δ/Δ (DAY1160), hse1Δ/Δ (DAY1218), mvb12Δ/Δ (DAY1162), vps28Δ/Δ (DAY1161), vps36Δ/Δ (DAY1163), vps22Δ/Δ (DAY1217), vps20Δ/Δ (DAY1157), snf7Δ/Δ (DAY763), vps4Δ/Δ (DAY1155), bro1Δ/Δ (DAY1156), and doa4Δ/Δ (DAY1158) strains.
Epithelial cell damage.
Epithelial cells line the skin and all mucosal surfaces and are the primary cell type on which C. albicans resides as a commensal, and Rim101 activation is required to damage epithelial cells (29). C. albicans causes damage to the epithelia by mediating its own endocytosis by the epithelial cells following germination and causing epithelial cell lysis (31, 34). With radiolabeling of a monolayer of FaDu oropharyngeal epithelial cell culture, damage can be monitored by measuring the radioactivity released to the culture supernatant. We used our ESCRT mutants to ask the question: does the ESCRT pathway play a Rim101-independent role in damaging epithelial cells?
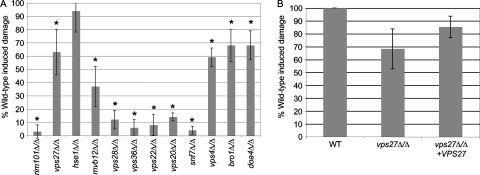
When incubated with wild-type C. albicans, we observed robust epithelial cell damage, with a typical assay resulting in 30 to 50% specific Cr51 release (14). To facilitate comparison between assays, we normalized the wild-type damage level in each assay and compared the amount of damage induced by the mutant strains with that induced by the wild type. As reported previously, the rim101Δ/Δ mutant showed a severe reduction in epithelial damage (Fig. 8 A), yielding ∼3% of wild-type damage. While we found that the ESCRT-0 hse1Δ/Δ mutant was not statistically different from the wild type, the vps27Δ/Δ mutant had an ∼35% decrease in wild-type damage (P < 0.001). We observed significant decreases in epithelial cell damage with all ESCRT-I, -II, and -III mutants, although the mvb12Δ/Δ mutant caused more damage than did the other ESCRT-I member. The vps4Δ/Δ, bro1Δ/Δ, and doa4Δ/Δ strains all showed an ∼35 to 40% decrease in damage compared to the wild type, like the vps27Δ/Δ strain. The vps27Δ/Δ, bro1Δ/Δ, and doa4Δ/Δ strains did not affect Rim101-dependent phenotypes or Rim101 processing but did affect FM 4-64 trafficking, demonstrating that ESCRT function plays a role in C. albicans-mediated epithelial cell damage independent of Rim101 activation.
Fig. 8.
(A) ESCRT mutants show defects in epithelial cell damage. C. albicans cells (105) were incubated for 10 h with 51Cr-labeled FaDu cells. Supernatant was collected and compared to that of uninfected FaDu cells. Samples were run in triplicate for each assay. Graph shows damage conferred by each mutant relative to a simultaneously tested wild-type (WT) strain. Mutants were tested in at least three separate assays. *, P < 0.05. Strains analyzed included wild-type (DAY185), ftr1Δ/Δ (DAY750), rim101Δ/Δ (DAY25), vps27Δ/Δ (DAY1160), hse1Δ/Δ (DAY1218), mvb12Δ/Δ (DAY1162), vps28Δ/Δ (DAY1161), vps36Δ/Δ (DAY1163), vps22Δ/Δ (DAY1217), vps20Δ/Δ (DAY1157), snf7Δ/Δ (DAY763), vps4Δ/Δ (DAY1155), bro1Δ/Δ (DAY1156), and doa4Δ/Δ (DAY1158) strains. (B) Complementation of the vps27Δ/Δ mutant using wild-type (DAY185), vps27Δ/Δ (DAY1160), and vps27Δ/Δ + VPS27 (DAY1264) strains.
OPC mouse model of disease.
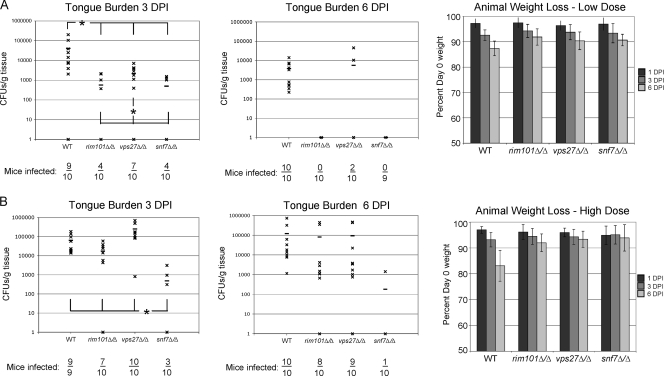
We next determined the contribution of ESCRT function to virulence using an oropharyngeal candidiasis (OPC) mouse model. Cortisone-immunosuppressed mice were infected orally with wild-type, rim101Δ/Δ, vps27Δ/Δ, or snf7Δ/Δ C. albicans. Ten mice per strain were sacrificed at 3 days postinfection (DPI) and at 6 DPI. Mouse tongues were harvested to assess oral fungal burden, and mice were weighed daily. This approach allowed us to assess fungal burden and mouse morbidity.
We first assessed the fungal burden of mice infected with 5 × 103 CFU. At 3 DPI, the oral cavity of mice infected with the wild-type strain showed an average fungal burden of 4 × 104 CFU/g tissue. A significant decrease in fungal burden was observed between the wild-type strain and the three C. albicans mutants (P < 0.04 for rim101Δ/Δ, vps27Δ/Δ, and snf7Δ/Δ strains) (Fig. 9 A). Mice infected with the vps27Δ/Δ strain carried 1 log less CFU/g tongue compared to mice infected with the wild type; the rim101Δ/Δ and snf7Δ/Δ strains had nearly a 2-log-lower burden than did wild-type C. albicans. The fact that all mutants colonized to lower burden levels suggests that all three genes play a role in C. albicans pathogenesis. Additionally, both the rim101Δ/Δ and snf7Δ/Δ strains colonized the oral cavity at a significantly lower burden than did the vps27Δ/Δ strain (P < 0.04 and < 0.03, respectively) (Fig. 9A). Thus, both Rim101 processing and ESCRT function are necessary for wild-type levels of in vivo tongue colonization, but defects in Rim101 processing result in more severe virulence defects than do defects in ESCRT function.
Fig. 9.
A vps27Δ/Δ mutant is attenuated in a mouse model of oral candidiasis. (A) Tongue fungal burden and animal weight loss after low-dose inoculation. Cortisone-immunosuppressed animals were infected with 50 μl of 105 CFU/ml of wild-type (WT) (DAY185), rim101Δ/Δ (DAY25), vps27Δ/Δ (DAY1160), and snf7Δ/Δ (DAY763) C. albicans. At 3 DPI half the animals were harvested and their tongues were collected for determination of fungal burden. The remaining animals were reimmunosuppressed with cortisone and harvested at 6 DPI for determination of fungal burden. *, P < 0.05. (B) Tongue fungal burden and animal weight loss after high-dose inoculation. The experiment was run identically but with an infectious dose of 50 μl of 106 CFU/ml C. albicans.
At 6 DPI, all mice infected with the wild-type strain were still colonized, although the fungal burden decreased by approximately a log (4 × 103 CFU/g tissue). All mice infected with the rim101Δ/Δ and snf7Δ/Δ mutants and all but two mice infected with the vps27Δ/Δ mutant cleared the infection to levels below detection (Fig. 9A). This indicates that both Rim101 processing and ESCRT function are necessary for maintaining infection and that Rim101 processing plays a more important role in maintenance than ESCRT function.
During the course of infection, we weighed the mice as a measure of mouse morbidity. Mice with oral candidiasis can lose weight quickly, and mice infected with the four C. albicans strains lost weight over the course of the experiment. However, by 6 DPI, mice infected with the wild-type strain had lost significantly more weight than did mice infected with any of the three mutant strains (P < 0.003, P < 0.03, and P < 0.01 for rim101Δ/Δ, vps27Δ/Δ, and snf7Δ/Δ strains, respectively) (Fig. 9A). There was no statistical difference in weight loss between mice infected with the rim101Δ/Δ, vps27Δ/Δ, and snf7Δ/Δ strains. This suggests that weight loss correlates with fungal burden and that mice more heavily colonized will manifest disease symptoms more quickly than those less heavily colonized.
Since the mutant strains showed a decreased burden at 3 DPI, we hypothesized that this might be due to poor initial colonization, which might be overcome with a higher initial inoculum. To address this possibility, we infected mice with a 10-fold-higher inoculum (5 × 104 CFU) and assessed fungal burden at 3 DPI and 6 DPI. At 3 DPI, the oral cavity of mice infected with the wild-type strain showed an average fungal burden of 6 × 104 CFU/g tissue, which was comparable to the colonization level for the lower dose (Fig. 9B). Mice infected with the rim101Δ/Δ strain showed a significant decrease in burden, 2 × 104 CFU/g tissue, compared to those infected with wild-type cells (P < 0.03). However, the rim101Δ/Δ strain-infected mice at the higher dose were colonized with a 2-log-higher burden than were the rim101Δ/Δ strain-infected mice at the lower dose. Mice infected with the vps27Δ/Δ strain also showed ∼2 logs more colonization than did mice infected with the lower dose of the vps27Δ/Δ strain. Unlike the rim101Δ/Δ mutant, the vps27Δ/Δ mutant had an ∼2-fold-higher average fungal burden, 2 × 105 CFU/g tissue, than did mice infected with wild-type C. albicans (Fig. 9B). Mice infected with the snf7Δ/Δ strain showed a significant decrease in fungal burden from that of wild-type-infected mice (P < 0.005), comparable to that of the snf7Δ/Δ strain-infected mice in the lower-dose experiment. Because an increase in the inoculum of wild-type C. albicans did not increase the fungal burden at 3 DPI, we deduce that there are a finite number of sites within the mouse oral cavity for C. albicans to reside in.
At 6 DPI, mice infected with the wild-type strain exhibited a fungal burden of 1 × 105 CFU/g tissue. Mice infected with the rim101Δ/Δ or vps27Δ/Δ strain showed no significant difference from wild-type-infected mice in oral fungal burden with an average of 8 × 104 and 9 × 104 CFU/g, respectively (P < 0.50 and P < 0.15, respectively) (Fig. 9B). This suggests that Rim101 processing and ESCRT function contribute independently to maintaining infection. All mice but one cleared the snf7Δ/Δ mutant from their oral cavity by 6 DPI, indicating that the effects of Rim101 processing and ESCRT function are additive. Alternatively, the snf7Δ/Δ mutant may have additional defects independent of either Rim101 or ESCRT function that decrease the viability of this strain. Since increasing the inoculum of the rim101Δ/Δ and vps27Δ/Δ mutants, but not the snf7Δ/Δ mutant, improved establishment and maintenance of infection, we conclude that the rim101Δ/Δ and vps27Δ/Δ strains have defects in initial oral colonization.
We also monitored weight loss during the course of the infection with the higher inocula. Again, mice lost weight over the course of the infection, except for snf7Δ/Δ strain-infected mice (Fig. 9B). While rim101Δ/Δ strain- and vps27Δ/Δ strain-infected mice lost weight, mice infected with wild-type C. albicans lost a significantly larger amount of weight than did mice infected with either mutant (P < 0.001, P < 0.0005, and P < 0.0005 for rim101Δ/Δ, vps27Δ/Δ, and snf7Δ/Δ mutants, respectively) (Fig. 9B). Although mice infected with the wild-type, rim101Δ/Δ, or vps27Δ/Δ strains are colonized to similar levels (Fig. 9B), mice infected with wild-type C. albicans show significantly more morbidity than do mice infected with either mutant. These results demonstrate that the ESCRT pathway plays a Rim101-independent role during C. albicans pathogenesis.
VPS27 complementation.
To confirm that the vps27Δ/Δ phenotypes observed were due to VPS27 specific mutation, we transformed the vps27Δ/Δ strain with a VPS27-containing complementation plasmid and compared it to the prototrophic vps27Δ/Δ mutant. Addition of VPS27 to the vps27Δ/Δ mutant completely restored the FM 4-64 trafficking phenotype to a wild-type staining pattern (Fig. 3B) and restored epithelial cell damage defect compared to the vps27Δ/Δ strain (P < 0.01) (Fig. 8B). However, the complemented strain did not restore epithelial cell damage to wild-type levels, nor did it restore growth on iron-limiting medium to wild-type levels (Fig. 7), suggesting that complementation is not complete. This partial rescue could be due to position effects, haploinsufficiency, or insufficient promoter sequence inclusion in the construct. However, the complementation of the epithelial cell damage and FM 4-64 trafficking phenotypes led us to conclude that the phenotypes associated with the vps27Δ/Δ mutant are indeed due to loss of VPS27.
DISCUSSION
We have used a series of ESCRT pathway mutants to investigate the role of individual ESCRT protein components in both Rim101 processing and MVB trafficking in C. albicans. We found that ESCRT-I, -II, and -III components, but not ESCRT-0 and ESCRT-DS components, are required for proteolytic processing of the transcription factor Rim101. Our in vitro epithelial cell damage studies demonstrate that ESCRT function plays a Rim101-independent role in epithelial cell damage, and our in vivo studies suggest that ESCRT function is required for wild-type C. albicans pathogenesis. We found that Vps27 is required for wild-type colonization levels when orally administered to mice at a relatively low dose but that this colonization defect can be overcome by administering a higher dose. However, even when mice are colonized with similar numbers of wild-type and vps27Δ/Δ C. albicans, wild-type-infected mice lose significantly more weight than do knockout-infected mice. Thus, ESCRT function is required for normal colonization and morbidity associated with C. albicans infection independent of Rim101.
Since the phenotypes of our ESCRT pathway mutants were internally consistent in general (see below) and based on the fact that our ESCRT mutants act similarly to ESCRT mutants in other systems, we conclude that the genes that we identified encode bona fide ESCRT pathway members. This idea is supported by the complementation test of the vps27Δ/Δ mutant, which restored the FM 4-64 trafficking and epithelial cell damage phenotypes. However, we did note that growth phenotypes differed between mutants in the ESCRT-I complex members Vps28 and Mvb12. While the vps28Δ/Δ mutant behaved similarly to the ESCRT-II mutants, the mvb12Δ/Δ mutant had intermediate phenotypes for alkaline growth and filamentation and more severe defects on LiCl-supplemented medium. This suggests that Mvb12 may play an accessory role in the ESCRT pathway and potentially have ESCRT-independent phenotypes, although further analyses are necessary to understand Mvb12 function. We also noted differences between mutants in the ESCRT-0 members Vps27 and Hse1. While the vps27Δ/Δ mutant conferred defects in epithelial cell damage, the hse1Δ/Δ mutant did not (Fig. 8). To the best of our knowledge, this is the first demonstration that Hse1 is not necessary for all Vps27-dependent phenotypes. Numerous analyses have been performed using a mutation within a single ESCRT complex subunit as representative of the behavior of the complex. The phenotypic differences between the mvb12Δ/Δ and vps28Δ/Δ mutant strains and vps27Δ/Δ and hse1Δ/Δ mutant strains stress the hazard in this assumption. Our data highlight the fact that molecular subunits of a complex do not necessarily have the same effect on function, and valuable information may be missed by using a single representative mutation.
Are all ESCRT complexes required for Rim101 processing?
ESCRT complex function plays a role in activating the transcription factor Rim101 by recruiting the processing machinery to the endosomal membrane in a number of fungi, including C. albicans, S. cerevisiae, and Aspergillus fumigatus (10, 37). In S. cerevisiae, the ESCRT-0 member Vps27 does not play a role in Rim101 activation (49), suggesting that the same may hold true in C. albicans. Our studies clearly establish that neither Vps27 nor Hse1, the other ESCRT-0 member, is required for Rim101-dependent growth or Rim101 processing (Fig. 2 and 4). While it is possible that C. albicans contains redundant VPS27 or HSE1 homologs, this is unlikely, as both vps27Δ/Δ and hse1Δ/Δ strains displayed FM 4-64 trafficking patterns associated with ESCRT defects. Additionally, neither VPS27 nor HSE1 paralogs were identified through BLAST search analyses. Thus, ESCRT-0 is not required for Rim101 processing in C. albicans.
Why does Rim101 processing bypass ESCRT-0 yet require ESCRT-I, -II, and -III? One possibility is that ESCRT-I, -II, and -III are recruited directly by the upstream Rim101 signaling molecules, Rim21 and Rim8. Rim21/PalH, a transmembrane protein found at the plasma membrane, interacts with ubiquitinated Rim8/PalF in Aspergillus nidulans (19), and Rim8 ubiquitination has recently been documented in S. cerevisiae (18). Further, ubiquitinated Rim8 can interact directly with ESCRT-I member Vps23 (18). The Rim21-Rim8 complex is thought to act as a surrogate “receptor” in alkaline conditions, initiating endocytosis and subsequent ESCRT complex assembly. This suggests that the Rim21-Rim8 complex bypasses ESCRT-0 by recruiting ESCRT-I directly. ESCRT-I recruitment appears to be mediated through Rim8 mimicry of ESCRT-0 component Vps27, as S. cerevisiae Rim8 interacts with Vps23 through an SXP motif similar to the Vps27 PSAP motif which interacts with Vps23 (18). However, Rim8 ubiquitination in S. cerevisiae is pH independent and thus does not explain how the cell can activate Rim101 only under neutral-alkaline conditions. Other systems are known to recruit ESCRT-I directly, bypassing ESCRT-0, to assemble the core ESCRT complexes at diverse cellular locations for distinct functions such as cytokinesis and viral budding (6, 26, 27). Direct recruitment of ESCRT-I by Rim101 signaling members is likely conserved across fungi containing Rim101.
ESCRT function in pathogenesis.
Previous studies from our lab analyzed the effects of a variety of snf7 alleles on C. albicans-stimulated epithelial cell damage (47). These studies suggested that the only contribution of Snf7 to epithelial cell damage is Rim101 dependent. However, these alleles are or are likely to be partially functional; thus, the damage assay may not be sensitive enough to detect differences with these partial disruptions. Thus, our previous experiments included several caveats and were not wholly conclusive.
The vps27Δ/Δ, hse1Δ/Δ, vps4Δ/Δ, bro1Δ/Δ, and doa4Δ/Δ mutant strains were all able to grow like the wild type on Rim101-dependent media, and all but the vps4Δ/Δ strain presented normal Rim101-dependent filamentation. Thus, we expected these strains to be the most representative of independent ESCRT function during C. albicans pathogenesis. The similar decreases in the vps27Δ/Δ, bro1Δ/Δ, and doa4Δ/Δ mutants in the FaDu epithelial cell damage assay suggest that the defect may be due to the common ESCRT function of these genes. The slightly larger decrease of the vps4Δ/Δ mutant may be due to the dysregulation of Rim101 processing in this strain (Fig. 6).
Previous studies have looked at the role of ESCRT complex proteins in C. albicans infection by using a bloodstream infection mouse model (12). The ESCRT complex members previously studied, Vps28 and Snf7/Vps32, function in both Rim101 processing and MVB trafficking. These mutant strains were found to be less pathogenic than the rim101Δ/Δ strain. These findings suggested that ESCRT function plays a role in C. albicans pathogenesis, but that study did not truly study ESCRT function independently from Rim101 function. The vps27Δ/Δ and hse1Δ/Δ strains used here study ESCRT function wholly uncoupled from Rim101 processing.
Data from the OPC mouse model demonstrate that the ESCRT pathway plays a Rim101-independent role in pathogenesis. At a lower inoculum, the vps27Δ/Δ strain was defective in colonization. This defect was overcome with an increased inoculum, but similar fungal burdens were not an indication of similar disease states, as mice infected with wild-type C. albicans lost significantly more weight than did mice infected with the vps27Δ/Δ strain. Thus, ESCRT plays a role both in establishing colonization during infection (at low doses) and in disease-associated morbidity (at high doses).
We compared the wild-type and vps27Δ/Δ strains in several assays for filamentation, adherence, and growth rate and observed no differences to account for morbidity differences (data not shown). Previous studies have shown that mutation of ESCRT complexes can increase sensitivity to the cell wall-targeting drug caspofungin more than can mutation of RIM101 alone (12), suggesting that the ESCRT pathway plays a role in cell wall function. The fungal cell wall initiates contact with host cells, resulting in damage (34), and may explain the damage defects of the ESCRT mutants. Additionally, we and others have shown that the ESCRT pathway is required for acquiring nutrients, such as iron (44). The role played by the ESCRT pathway during infection is therefore likely to be multifactorial, affecting both cell wall architecture and nutrient acquisition.
C. albicans niche size.
Are fungal colonization levels dependent on inoculum concentration? Lower- and higher-dose inocula lead to similar fungal burdens with the wild-type strain, while a higher-dose inoculum leads to a higher fungal burden with the mutant strains. However, mutant strain burden did not increase much beyond wild-type levels, if at all (Fig. 9). This potential upper limit of fungal colonization suggests that there exist a finite number of sites where C. albicans can grow within its host.
We were able to rescue colonization defects of both the vps27Δ/Δ and rim101Δ/Δ strains to wild-type levels by increasing the inoculum dose. However, we observed similar wild-type colonization numbers at 3 DPI in animals infected with low- or high-dose inocula. These facts indicate that C. albicans has a limited niche size in the oral cavity. We manipulated the niche size of our animals with cortisone and tetracycline treatments, and different manipulations could further alter this niche size. However, given a set of conditions in its host, there is an upper threshold to the C. albicans burden that it can bear.
Our studies have shown that C. albicans requires a fully functional ESCRT complex for wild-type pathogenic levels during in vitro and in vivo pathogenesis. Understanding the ESCRT complex function in C. albicans presents opportunities to molecularly characterize a nontraditional signal transduction cascade and to investigate the mechanism of ESCRT function during C. albicans interactions with the host.
ACKNOWLEDGMENTS
We thank the members of the Davis lab and Kirsten Nielson's lab for helpful discussions on this work. We thank Laura Okagaki for microscopy help.
The project described was supported by the NIH National Institute of Allergy and Infectious Disease award R01-AI064054-01, by NIH T32DE007288 from the National Institute of Dental and Craniofacial Research, and by the Investigators in Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Dental and Craniofacial Research or the National Institutes of Health.
Footnotes
Published ahead of print on 25 June 2010.
REFERENCES
- 1.Adams A., Gottschling D. E., Kaiser C. A., Stearns T. 1997. Methods in yeast genetics, 1997: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 2.Babst M., Katzmann D. J., Estepa-Sabal E. J., Meerloo T., Emr S. D. 2002. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3:271–282 [DOI] [PubMed] [Google Scholar]
- 3.Babst M., Katzmann D. J., Snyder W. B., Wendland B., Emr S. D. 2002. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3:283–289 [DOI] [PubMed] [Google Scholar]
- 4.Babst M., Wendland B., Estepa E. J., Emr S. D. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baek Y. U., Li M., Davis D. A. 2008. Candida albicans ferric reductases are differentially regulated in response to distinct forms of iron limitation by the Rim101 and CBF transcription factors. Eukaryot. Cell 7:1168–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barajas D., Jiang Y., Nagy P. D. 2009. A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus. PLoS Pathog. 5:e1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bilodeau P. S., Urbanowski J. L., Winistorfer S. C., Piper R. C. 2002. The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat. Cell Biol. 4:534–539 [DOI] [PubMed] [Google Scholar]
- 8.Blanchin-Roland S., Da Costa G., Gaillardin C. 2008. Ambient pH signalling in the yeast Yarrowia lipolytica involves YlRim23p/PalC, which interacts with Snf7p/Vps32p, but does not require the long C terminus of YlRim9p/PalI. Microbiology 154:1668–1676 [DOI] [PubMed] [Google Scholar]
- 9.Bowers K., Lottridge J., Helliwell S. B., Goldthwaite L. M., Luzio J. P., Stevens T. H. 2004. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic 5:194–210 [DOI] [PubMed] [Google Scholar]
- 10.Boysen J. H., Mitchell A. P. 2006. Control of Bro1-domain protein Rim20 localization by external pH, ESCRT machinery, and the Saccharomyces cerevisiae Rim101 pathway. Mol. Biol. Cell 17:1344–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornet M., Bidard F., Schwarz P., Da Costa G., Blanchin-Roland S., Dromer F., Gaillardin C. 2005. Deletions of endocytic components VPS28 and VPS32 affect growth at alkaline pH and virulence through both RIM101-dependent and RIM101-independent pathways in Candida albicans. Infect. Immun. 73:7977–7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornet M., Gaillardin C., Richard M. L. 2006. Deletions of the endocytic components VPS28 and VPS32 in Candida albicans lead to echinocandin and azole hypersensitivity. Antimicrob. Agents Chemother. 50:3492–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curtiss M., Jones C., Babst M. 2007. Efficient cargo sorting by ESCRT-I and the subsequent release of ESCRT-I from multivesicular bodies requires the subunit Mvb12. Mol. Biol. Cell 18:636–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis D., Edwards J. E., Jr., Mitchell A. P., Ibrahim A. S. 2000. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect. Immun. 68:5953–5959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis D., Wilson R. B., Mitchell A. P. 2000. RIM101-dependent and-independent pathways govern pH responses in Candida albicans. Mol. Cell. Biol. 20:971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghazi-Tabatabai S., Saksena S., Short J. M., Pobbati A. V., Veprintsev D. B., Crowther R. A., Emr S. D., Egelman E. H., Williams R. L. 2008. Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure 16:1345–1356 [DOI] [PubMed] [Google Scholar]
- 17.Hayashi M., Fukuzawa T., Sorimachi H., Maeda T. 2005. Constitutive activation of the pH-responsive Rim101 pathway in yeast mutants defective in late steps of the MVB/ESCRT pathway. Mol. Cell. Biol. 25:9478–9490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrador A., Herranz S., Lara D., Vincent O. 2010. Recruitment of the ESCRT machinery to a putative seven-transmembrane-domain receptor is mediated by an arrestin-related protein. Mol. Cell. Biol. 30:897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herranz S., Rodriguez J. M., Bussink H. J., Sanchez-Ferrero J. C., Arst H. N., Jr., Penalva M. A., Vincent O. 2005. Arrestin-related proteins mediate pH signaling in fungi. Proc. Natl. Acad. Sci. U. S. A. 102:12141–12146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. U. S. A. 98:4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katzmann D. J., Babst M., Emr S. D. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145–155 [DOI] [PubMed] [Google Scholar]
- 22.Katzmann D. J., Stefan C. J., Babst M., Emr S. D. 2003. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 162:413–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knight S. A., Vilaire G., Lesuisse E., Dancis A. 2005. Iron acquisition from transferrin by Candida albicans depends on the reductive pathway. Infect. Immun. 73:5482–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kullas A. L., Li M., Davis D. A. 2004. Snf7p, a component of the ESCRT-III protein complex, is an upstream member of the RIM101 pathway in Candida albicans. Eukaryot. Cell 3:1609–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M., Martin S. J., Bruno V. M., Mitchell A. P., Davis D. A. 2004. Candida albicans Rim13p, a protease required for Rim101p processing at acidic and alkaline pHs. Eukaryot. Cell 3:741–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin-Serrano J., Zang T., Bieniasz P. D. 2003. Role of ESCRT-I in retroviral budding. J. Virol. 77:4794–4804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morita E., Sandrin V., Chung H. Y., Morham S. G., Gygi S. P., Rodesch C. K., Sundquist W. I. 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26:4215–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muhlrad D., Hunter R., Parker R. 1992. A rapid method for localized mutagenesis of yeast genes. Yeast 8:79–82 [DOI] [PubMed] [Google Scholar]
- 29.Nobile C. J., Solis N., Myers C. L., Fay A. J., Deneault J. S., Nantel A., Mitchell A. P., Filler S. G. 2008. Candida albicans transcription factor Rim101 mediates pathogenic interactions through cell wall functions. Cell. Microbiol. 10:2180–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Odorizzi G., Katzmann D. J., Babst M., Audhya A., Emr S. D. 2003. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J. Cell Sci. 116:1893–1903 [DOI] [PubMed] [Google Scholar]
- 31.Park H., Myers C. L., Sheppard D. C., Phan Q. T., Sanchez A. A., J. E., Filler S. G. 2005. Role of the fungal Ras-protein kinase A pathway in governing epithelial cell interactions during oropharyngeal candidiasis. Cell. Microbiol. 7:499–510 [DOI] [PubMed] [Google Scholar]
- 32.Perlroth J., Choi B., Spellberg B. 2007. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med. Mycol. 45:321–346 [DOI] [PubMed] [Google Scholar]
- 33.Pfaller M. A., Diekema D. J. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phan Q. T., Myers C. L., Fu Y., Sheppard D. C., Yeaman M. R., Welch W. H., Ibrahim A. S., Edwards J. E., Jr., Filler S. G. 2007. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 5:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramanan N., Wang Y. 2000. A high-affinity iron permease essential for Candida albicans virulence. Science 288:1062–1064 [DOI] [PubMed] [Google Scholar]
- 36.Raymond C. K., Howald-Stevenson I., Vater C. A., Stevens T. H. 1992. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3:1389–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez-Galan O., Galindo A., Hervas-Aguilar A., Arst H. N., Penalva M. A. 2009. Physiological involvement in pH signaling of Vps24-mediated recruitment of Aspergillus PalB cysteine protease to ESCRT-III. J. Biol. Chem. 284:4404–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rothfels K., Tanny J. C., Molnar E., Friesen H., Commisso C., Segall J. 2005. Components of the ESCRT pathway, DFG16, and YGR122w are required for Rim101 to act as a corepressor with Nrg1 at the negative regulatory element of the DIT1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 25:6772–6788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saksena S., Sun J., Chu T., Emr S. D. 2007. ESCRTing proteins in the endocytic pathway. Trends Biochem. Sci. 32:561–573 [DOI] [PubMed] [Google Scholar]
- 40.Shim S., Merrill S. A., Hanson P. I. 2008. Novel interactions of ESCRT-III with LIP5 and VPS4 and their implications for ESCRT-III disassembly. Mol. Biol. Cell 19:2661–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Southern P., Horbul J., Maher D., Davis D. A. 2008. C. albicans colonization of human mucosal surfaces. PLoS One 3:e2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teis D., Saksena S., Emr S. D. 2008. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell 15:578–589 [DOI] [PubMed] [Google Scholar]
- 43.Vida T. A., Emr S. D. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128:779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weissman Z., Shemer R., Conibear E., Kornitzer D. 2008. An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans. Mol. Microbiol. 69:201–217 [DOI] [PubMed] [Google Scholar]
- 45.Wilson R. B., Davis D., Enloe B. M., Mitchell A. P. 2000. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16:65–70 [DOI] [PubMed] [Google Scholar]
- 46.Wilson R. B., Davis D., Mitchell A. P. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolf J. M., Davis D. A. 2010. Mutational analysis of Candida albicans SNF7 reveals genetically separable Rim101 and ESCRT functions and demonstrates divergence in bro1-domain protein interactions. Genetics 184:673–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu W., Mitchell A. P. 2001. Yeast PalA/AIP1/Alix homolog Rim20p associates with a PEST-like region and is required for its proteolytic cleavage. J. Bacteriol. 183:6917–6923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu W., Smith F. J., Jr., Subaran R., Mitchell A. P. 2004. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol. Biol. Cell 15:5528–5537 [DOI] [PMC free article] [PubMed] [Google Scholar]