Abstract
The increasing incidence of nosocomial infections caused by glycopeptide-resistant enterococci is a global concern. Enterococcal species are also difficult to eradicate with existing cleaning regimens; they can survive for long periods on surfaces, thus contributing to cases of reinfection and spread of antibiotic-resistant strains. We have investigated the potential use of copper alloys as bactericidal surfaces. Clinical isolates of vancomycin-resistant Enterococcus faecalis and Enterococcus faecium were inoculated onto copper alloy and stainless steel surfaces. Samples were assessed for the presence of viable cells by conventional culture, detection of actively respiring cells, and assessment of cell membrane integrity. Both species survived for up to several weeks on stainless steel. However, no viable cells were detected on any alloys following exposure for 1 h at an inoculum concentration of ≤104 CFU/cm2. Analysis of genomic and plasmid DNA from bacterial cells recovered from metal surfaces indicates substantial disintegration of the DNA following exposure to copper surfaces that is not evident in cells recovered from stainless steel. The DNA fragmentation is so extensive, and coupled with the rapid cell death which occurs on copper surfaces, that it suggests that mutation is less likely to occur. It is therefore highly unlikely that genetic information can be transferred to receptive organisms recontaminating the same area. A combination of effective cleaning regimens and contact surfaces containing copper could be useful not only to prevent the spread of viable pathogenic enterococci but also to mitigate against the occurrence of potential resistance to copper, biocides, or antibiotics and the spread of genetic determinants of resistance to other species.
Enterococci are an important cause of nosocomial infections worldwide (34), and in the United States, approximately one-third of enterococcal infections in intensive care units are caused by vancomycin-resistant enterococci (VRE) (35).
There is now a significant problem of enterococci acquiring resistance to clinically important antibiotics, particularly aminoglycosides; glycopeptides, including vancomycin; and quinolones (18, 23, 24, 45, 47, 50). Recent reports have identified enterococci resistant to the latest antimicrobial agents, including linezolid, daptomycin, tigecycline, and quinupristin-dalfopristin. This, coupled with the bacteriostatic properties of linezolid and tigecycline, is limiting the treatment options for severe infections with Gram-positive bacteria (1, 12, 29).
Enterococci have a propensity for genetic transfer via transposons and plasmids which has resulted in the dissemination of antibiotic resistance genes (28, 41, 54). In addition, a recent report has highlighted the gap between the development of new antibiotics and an increasing number of infections caused by multiantibiotic-resistant organisms (14).
Enterococci are intestinal commensals and can withstand the high salt concentrations and pH values found in the bowel and are known to be able to survive for long periods in the environment. They can also survive on soft surfaces, including hospital linens and plastics (36), upholstery, and floor and wall coverings (27), and can exhibit resistance to some routinely used cleaning agents, including sodium hypochlorite (10, 25). Any enterococci not removed by routine cleaning procedures can therefore persist in a viable state and pose a risk of further infection. Hayden et al. discovered that health care workers were almost as likely to contaminate their gloves or hands after touching the environment in a room occupied with VRE-colonized patients as after touching the patients themselves (9, 22, 40).
Contaminated surfaces are known to contribute to infection spread (4, 8, 45); therefore, the use of bactericidal surfaces along with rigorous disinfection protocols could potentially reduce the incidence of horizontal disease transmission.
The biocidal properties of copper have been known for centuries (3). More recently, the potential use of copper alloys as microbicidal surfaces has been described. Rapid killing of Escherichia coli O157 (38, 52), Listeria monocytogenes (53), methicillin-resistant Staphylococcus aureus (MRSA) (37), Clostridium difficile (48, 51), Mycobacterium tuberculosis (32), Candida albicans and other pathogenic fungi (49), and influenza A virus (39) has been observed on copper compared to the stainless steel surfaces which are prevalent throughout the health care environment. In the United States, Environmental Protection Agency approval has been granted for the use of alloys containing >65% copper as bactericidal surfaces (7) and has recently been extended down to alloys containing >60% copper. It is estimated that biofilm formation on surfaces is a contributing factor in 65% of cases of nosocomial infections (43). There is no evidence that pathogens can form biofilms on dry copper surfaces (37), and the effectiveness of copper alloys as bactericidal agents is not affected by temperature and humidity, unlike that of silver (33).
At present, trials of copper surfaces used along with conventional cleaning regimens in the clinical environment are under way. In the United Kingdom, a pilot study is investigating the effectiveness of corrosion-resistant copper alloys to reduce the microbial burden of “constant touch” bare metal surfaces in a busy hospital ward (tap handles, ward door push plates, grab rails, door handles, sink traps) (5, 6). In addition, plastic toilet seats were compared with those coated with a copper resin composite (approximately 70% copper). Preliminary results suggest a significant reduction in bacterial contamination on all copper alloy surfaces, and investigations are now proceeding to a larger-scale trial (5, 6). Other trials are in progress worldwide, including the United States, Germany, Chile, Japan, and South Africa (7).
We have investigated whether various copper-based alloys, compared with stainless steel, may kill pathogenic vancomycin-resistant and -sensitive enterococcal isolates. In particular, we have investigated the effect of exposure to copper on enterococcal DNA. This is significant because to effectively prevent the spread of nosocomial enterococcal infections and antibiotic resistance, not only do the cells have to be killed but the DNA must be compromised. If the DNA remains intact, there may still be the possibility of resistance mutations occurring with the potential transmission of genetic material to other species.
MATERIALS AND METHODS
Bacterial strains.
Vancomycin-resistant control strains Enterococcus faecalis ATCC 51299 (VanB phenotype) and Enterococcus faecium NCTC 12202 (VanA phenotype) were supplied by Oxoid.
Clinical isolates of vancomycin-resistant E. faecalis (n = 2), E. faecium (n = 5), and E. gallinarum (n = 1) were obtained from patients (aged 11 days to 70 years) between June 2004 and June 2006 at Southampton General Hospital, Southampton, United Kingdom (Table 1). The enterococcal genomes exhibited 54 to 93% similarity (Dice similarity coefficient) by pulsed-field gel electrophoresis of SmaI-digested DNA (data not shown).
TABLE 1.
Characteristics of the Enterococcus clinical isolates used in this study
| Isolate | Source | Sample type | Antimicrobial resistancea |
|---|---|---|---|
| E. faecalis 1 | Surgical ward | Wound swab | VAN, ERY, CHL, TET |
| E. faecalis 2 | Surgical ward | Feces | VAN, ERY, CHL, TET |
| E. faecium 1 | Intensive care unit | Ascitic fluid | VAN, PEN, ERY, CHL, TET, AMP |
| E. faecium 2 | Surgical ward | Intraabdominal drain swab | VAN, PEN, ERY, CHL, AMP |
| E. faecium 3 | Leukemia ward | Blood culture | VAN, PEN, ERY, CHL, TET, AMP |
| E. faecium 4 | Neonatal unit | Gastric aspirate | VAN, PEN, ERY, CHL, TET, AMP |
| E. faecium 5 | Intensive care unit | Central venous catheter swab | VAN, PEN, ERY, CHL, TET, AMP |
| E. gallinarum | Surgical ward | Feces | VAN, PEN, ERY, CHL, TET, AMP |
Antimicrobial abbreviations: VAN, vancomycin; PEN, penicillin; ERY, erythromycin; CHL, chloramphenicol, TET, tetracycline; AMP, ampicillin.
Vancomycin-sensitive E. faecalis NCTC 775 was supplied by a local water authority.
Culture preparation.
Bacteria were maintained on Glycerol Protect beads (Fisher Scientific) and also in 1-ml aliquots of VRE broth (VREB; Oxoid) containing 15% (wt/vol) glycerol at −80°C. For each experiment, one bead or vial of enterococci was inoculated into 15 ml sterile brain heart infusion broth (Oxoid) or VREB and incubated aerobically at 37°C for 18 ± 2 h.
Coupon preparation.
Various copper alloys were tested (Table 2). Before analysis, sheets (1 to 3 mm thick) of each metal alloy (supplied by the Copper Development Association Inc., New York, NY) were cut into coupons (10 by 10 mm). Coupons were degreased and cleaned by vortexing in approximately 10 ml acetone containing 20 to 30 glass beads (2-mm diameter) for 30 s and then immersed in absolute ethanol. Prior to use, coupons were flamed with a Bunsen burner and placed, using forceps, in sterile petri dishes for microbial inoculation.
TABLE 2.
Compositions of the alloys tested in this study
| UNSa no. | % of total composition that was: |
|||||
|---|---|---|---|---|---|---|
| Cu | Zn | Sn | Ni | Fe | Cr | |
| C11000 | 100 | |||||
| C26000 | 70 | 30 | ||||
| C28000 | 60 | 40 | ||||
| C51000 | 95 | 5 | ||||
| C70600 | 89 | 10 | 1 | |||
| C75200 | 65 | 17 | 18 | |||
| S30400 | 8 | 74 | 18 | |||
UNS, unified numbering system.
Inoculation.
For each exposure time, duplicate coupons were analyzed using either culture methods or staining methods; 20 μl of bacterial culture was spread evenly over the surface of each coupon, dried in a sterile airflow in a class II microbiological safety cabinet, and incubated on the bench at room temperature (21 ± 2°C) for various time periods. The use of bacteriological medium as an inoculation matrix was included as a “worst-case scenario” to mimic contamination of hospital surfaces with complex organic material.
Culture analysis.
Coupons were aseptically transferred to 5 ml phosphate-buffered saline (PBS) containing 2-mm-diameter glass beads and vortexed for 30 s. (Preliminary experiments using PBS with 20 mM EDTA to chelate free copper ions gave no significant difference in any of the viability testing methods used.) Serial dilutions were prepared, and 10 or 100 μl of each dilution was spread over 45- or 90-mm agar plates in triplicate. Slanetz and Bartley agar (Merck) and Columbia blood agar (CBA; bioMérieux) were used for the recovery of vancomycin-sensitive strains, and VRE agar containing 6 mg/liter vancomycin (Oxoid) and CBA were used for VRE strains (but with no meropenem added, as only pure cultures were investigated). Plates were allowed to dry before inversion and aerobic incubation at 37°C for 24 and 48 h. Colonies on plates were counted by eye, and the concentration per coupon was calculated and recorded as CFU per coupon (1 cm2).
Staining protocols used to detect actively respiring bacterial cells and membrane integrity in situ on metal surfaces. (i) SYTO 9 and CTC (5-cyano-2,3-ditolyl tetrazolium chloride).
A 30-μl volume of CTC (Sigma-Aldrich) at a final concentration of 5 mM was pipetted onto the surface of each inoculated coupon for the final 2 h of required contact time. Coupons were placed in a humid chamber and incubated in the dark at 37°C for 2 h. To stain all of the cells present on coupons, SYTO 9 (7 μM; Invitrogen) was pipetted onto the coupons and they were incubated at room temperature in the dark for the final 30 min.
(ii) BacLight (SYTO 9 and propidium iodide [PI]).
A staining solution containing 7 μM SYTO 9 and 40 μM PI (L7012; Invitrogen) was prepared in filter-sterilized deionized water (2 μl/ml each stain); 50 μl of this solution was pipetted onto the coupons, and they were incubated at room temperature for the final 30 min of required contact time in the dark.
After staining with SYTO 9/CTC or BacLight, coupons were tipped to remove the stain and 1 drop of sterile deionized water was gently pipetted onto the coupon from a disposable Pastette and the coupon was tipped to remove the remaining excess stain.
Episcopic differential interference contrast (EDIC) microscopy combined with epifluorescence microscopy was used to scan the coupons directly (Nikon Eclipse ME600; Best Scientific, Swindon, United Kingdom) (26). A minimum of 10 fields of view were photographed using a digital camera (Coolsnap CF; Roper Industries) connected to a computer with digital image analysis software (Image-Pro Plus, version 4.5.1.22; Media Cybernetics). Total cells (SYTO 9 stained), respiring cells (CTC stained), and membrane-damaged/intact cells (PI/SYTO 9 stained) were enumerated.
(iii) Genomic DNA assay.
The protocol used is described in detail elsewhere (17) and allows analysis of whole bacterial genomes. Briefly, approximately 107 bacterial cells left untreated, heat/alcohol killed, or exposed to copper or stainless steel surfaces were trapped in low-melting-point agarose on a slide previously coated with standard agarose. The bacteria had been pretreated with lysozyme (4 mg/ml [approximately 30,000 U]) for 15 min at 37°C. The cell membrane was permeabilized with a lysing solution (2% sodium dodecyl sulfate, 0.05 M EDTA, 0.1 M dithiothreitol, pH 11.5) for 5 min at 37°C before drying in ethanol baths and overnight baking in a 65°C oven. Dried slides were heated in a microwave oven (4 min, 750 W) before staining with the sensitive nucleic acid stain SYBR Gold (Invitrogen), which detects single- and double-stranded DNA, for 5 min at room temperature in the dark. Epifluorescence microscopy was used to analyze the DNA fragments produced.
(iv) Purification of bacterial DNA and separation of fragments by gel electrophoresis.
Enterococcal DNA (50-kb fragments) were purified using the Qiagen DNeasy Blood and Tissue kit (following pretreatment with lysozyme), and preparations were separated on a 1, 2, or 3% (wt/vol) agarose gel containing the DNA stain SYBRsafe (Invitrogen) exposed to a current of 300 mA for 90 min. Plasmid DNA was extracted using the QIAprep Spin Miniprep kit (Qiagen), and preparations were separated on 0.9% agarose gels. Gels were observed in a UV light box and photographed using GeneScan software.
Statistical analysis.
Data are expressed as means ± the standard errors of the means. Differences between duplicate samples were assessed using the Mann-Whitney rank t test. Group comparisons were analyzed using the Mann-Whitney U test, where statistical significance was considered P < 0.05. Statistical analyses were performed using Sigma Stat version 3.5, and graphical representations were performed using SigmaPlot version 10.
RESULTS
Culture analysis.
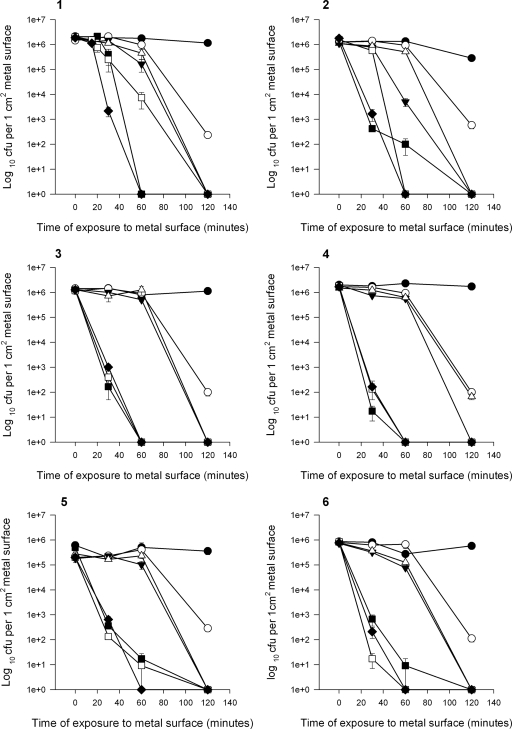
The survival time of all of the strains of enterococcal species tested was significantly less on copper and copper alloys containing 60 to 95% copper than on stainless steel for all of the inoculum concentrations studied (Fig. 1 to 4).
FIG. 1.
Survival of vancomycin-resistant E. faecium NCTC 12202 (graph 1) and clinical isolates 1 (graph 2), 2 (graph 3), 3 (graph 4), 4 (graph 5), and 5 (graph 6) on stainless steel, pure copper, and copper alloys (S30400 [•], C28000 [○], C75200 [▾], C26000 [▵], C70600 [▪], C51000 [□], and copper C11000 [⧫]) at 22°C.
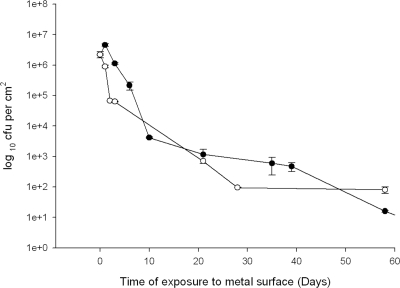
On stainless steel using the highest inoculum concentration of 106 CFU/cm2 of metal surface, there was only a 1-log reduction in viable cells after 2 and 6 days for vancomycin-resistant E. faecium and E. faecalis, respectively, and viable cells of both species were still present at 60 days (Fig. 3).
FIG. 3.
Survival of vancomycin-resistant E. faecalis (ATCC 51299) (•) and E. faecium (NCTC 12202) (○) on stainless steel at 22°C.
In contrast, at the same inoculum concentration, no viable cells were detected on pure copper at a contact time of 1 h and on alloys after 1 to 3 h for both of the species tested (Fig. 1 and 2).
FIG. 2.
Survival of vancomycin-resistant E. faecalis ATCC 51299 (graph 1), E. faecalis clinical isolates 1 (graph 2) and 2 (graph 3), E. gallinarum (graph 4), and vancomycin-sensitive E. faecalis NCTC775 (graph 5) on stainless steel, pure copper, and copper alloys (S30400 [•], C28000 [○], C75200 [▾], C26000 [▵], C70600 [▪], C51000 [□], and copper C11000 [⧫]) at 22°C.
Although all of the alloys are very effective at killing control and clinical strains of E. faecium, compared to stainless steel, survival appears to be related to the percentage of copper in the alloys (Fig. 1). The most rapid killing occurs on pure copper and alloys containing >90% copper (C51000 and C70600), with increased survival times on the remaining alloys containing 60 to 70% copper (C28000, C75200, and C26000) (Fig. 1). Alloys containing >95% copper (C51000, C70600) were as effective as pure copper for isolates 2 and 3, resulting in cell death at 1 h, but small numbers of cells of isolates 1and 4 remained viable (86% similarity) at up to 2 h after contact. Viable cells of all of the isolates were detected following 2 h of contact with alloy C28000, which has the lowest copper content tested here (60%). There are, however, exceptions: viable cells of clinical isolate 3 were detected following 2 h of contact on alloy C26000 (cartridge brass, 70% copper), but C75200 (nickel silver), which contains 5% less copper, was a more effective bactericidal surface.
The results are similar for control and clinical strains of E. faecalis (Fig. 2), with rapid killing achieved with all of the copper alloys compared to stainless steel. However, unlike E. faecium, no viable cells were detected on alloy C28000 at 2 h of contact. It is interesting that alloy C75200 is once again a more effective bactericidal surface than alloy C26000 for the E. gallinarum isolate tested (Fig. 2, graph 4). This species, although not as prevalent as E. faecium and E. faecalis, was reported to be isolated in 2.6% of the enterococcal bacteremia infections in the United Kingdom in 2007 (23). More-rapid killing was observed with the vancomycin-sensitive E. faecalis strain (Fig. 2, graph 5), all of the cells of which were killed on pure copper and copper alloys at 30 and 80 min, respectively (C28000 not tested), but it is difficult to assess the significance of this unless further sensitive strains are tested, and the results may not be clinically relevant.
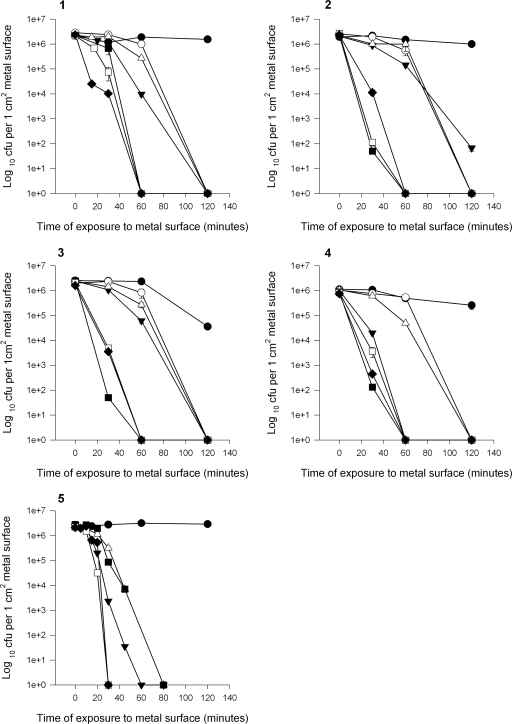
Survival time is also dependent on the inoculum concentration. If the inoculum was increased to 5 × 107 CFU/cm2, low numbers of viable E. faecalis cells could be recovered after 2.5 h of contact with alloys C26000 and C70600, but not C75200, after 2.5 h of contact (data not shown). Reducing the inoculum concentration of the E. faecium control strain to 105 CFU/cm2 and below minimized the discrepancy between pure copper and the copper alloys, as all of the alloy surfaces were even more effective at killing enterococcal cells (Fig. 4). All of the cells were killed in 60 min, except for those on alloy C70600, where a few cells remained viable from an inoculum concentration of 105 CFU/cm2. However, contact for 20 min on pure copper killed all of the cells at an inoculum concentration of 1,000 cells/cm2.
FIG. 4.
Effect of inoculum concentration on survival of vancomycin-resistant E. faecium (NCTC 12202) on stainless steel, pure copper, and copper alloys (S30400 [•], C75200 [○], C26000 [▾], C70600 [▵], C5100 [▪], and C11000 [□]) at 22°C. Inoculum concentrations tested: 105 CFU/cm2 (graph 1), 104 CFU/cm2 (graph 2), and 103 CFU/cm2 (graph 3).
Analysis of respiring cells (CTC) and membrane integrity (SYTO 9/PI) following contact with copper and stainless steel.
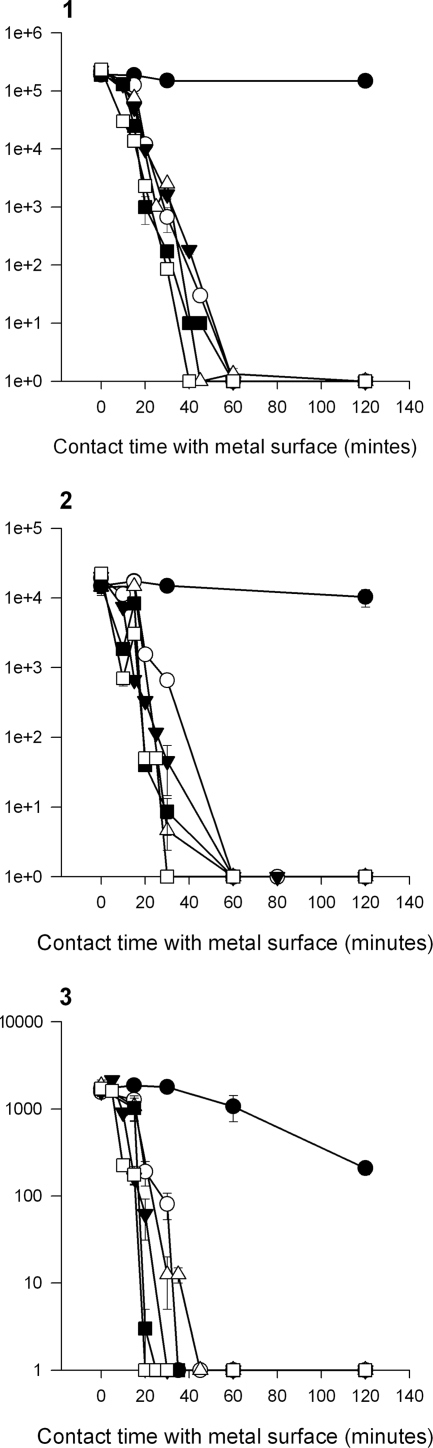
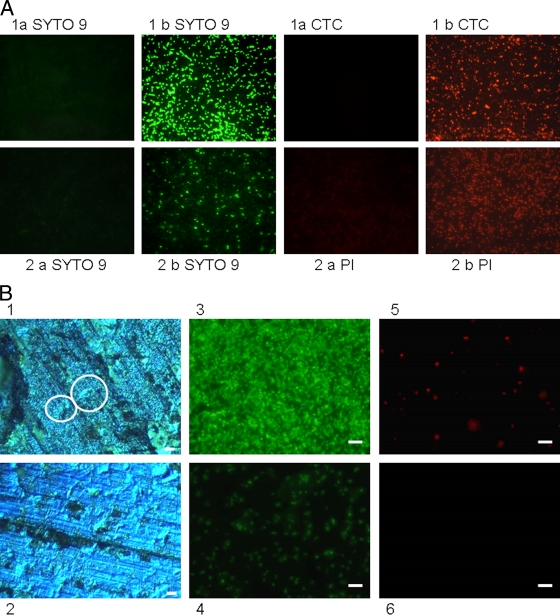
To determine if noncultivable cells were indeed dead and to investigate the potential mechanism of copper's antibacterial action, copper and steel coupons were inoculated with vancomycin-resistant E. faecalis control strain ATCC 51299 and directly stained with the redox dye CTC at various time points. Actively respiring cells accumulate the insoluble red fluorescent product, CTC-formazan, which can be visualized in situ using epifluorescence microscopy and a long-working-distance objective lens. Figure 5 A demonstrates CTC staining of the cells after 4 h of contact with copper and steel at room temperature. Actively respiring bacterial cells can be seen on stainless steel coupons (Fig. 5A, part 1b CTC) but not on copper (Fig. 5A, part 1a CTC). The membrane-permeating DNA stain SYTO 9 was used to determine the total bacterial count, but the staining intensity on copper (Fig. 5A, part 1a SYTO 9) appeared greatly diminished relative to that on stainless steel (Fig. 5A, part 1b SYTO 9). Similar results were obtained for the control strain of E. faecium (not shown). Actively respiring cells were also present on alloys C26000 (Fig. 5B) and C70600, as well as stainless steel, when a higher inoculum concentration of 5 × 107 CFU/cm2 was used for 2 h of contact time but not on copper alloys C51000 and C75200 (data not shown; alloy C28000 not tested). At this higher concentration, the cells are piled on top of each other and are not necessarily in direct contact with the alloy surface (Fig. 5B). This appears not to be a problem for pure copper and alloys C75200 (65% Cu) and C51000 (95% Cu) but may be important if a particular alloy has a reduced copper release rate.
FIG. 5.
(A) Assessment of viability of E. faecalis (ATCC 51299) on copper (a) and stainless steel (b) surfaces using the redox dye CTC (positive for respiring cells) and SYTO 9 (total cell count, regardless of viability) (row 1) or BacLight (row 2) to detect bacterial membrane integrity. (B) Effects of inoculum concentration and cell stacking on the susceptibility of alloy C26000 to inhibit respiration. The alloy was inoculated with VRE strain E. faecium NCTC 12202 at inoculum concentrations of 5 ×107 CFU/cm2 (images 1, 3, and 5) and 5 × 106 CFU/cm2 (images 2, 4, and 6) for 2 h. Images 1 and 2 were captured using EDIC microscopy. Circled in image 1 are areas where bacterial cells at the higher inoculum concentration may not be in direct contact with the metal surface, as they are stacked on top of each other. At the lower inoculum concentrations (image 2), the cells are spread in small clumps and are all exposed to the alloy directly (this spreading of individual cells is also clearly visible in epifluorescence image 4). Epifluorescence images 3 and 4 represent SYTO 9 total cell staining. CTC-positive staining of respiring cells is present at the higher (image 5) but not at the lower (image 6) inoculum concentration. Bars, 10 μm.
Enumeration of CTC-positive cells on stainless steel gave consistently higher counts than results obtained from culture, but the difference was not statistically significant.
BacLight comprises two nucleic acid stains, SYTO 9 and the higher-affinity stain PI. SYTO 9 can permeate the membrane and will stain all of the cells regardless of viability. If the cell membrane is damaged, PI can enter the cell and displace the lower-affinity stain SYTO 9. As the cells die, the ratio of cells staining green (membranes intact) changes to predominantly red (dead cells with compromised membranes). The vancomycin-resistant E. faecalis control strain was inoculated onto all of the metal samples. At the earliest time point of 30 min, green staining in situ revealed a large number of viable cells and few red-staining cells with damaged membranes in all of the samples. After 4 h of contact with stainless steel, the number of bright SYTO 9-stained cells was reduced, while red PI staining had increased, suggesting that cells die slowly on stainless steel and their cytoplasmic membranes are compromised (Fig. 5A, part 2b). It was not possible to enumerate viable cells in situ beyond 30 min on copper and copper alloys because preparations were negative for both stains. (Fig. 5A, part 2a). If the cells were removed from the coupons prior to staining, there was still no uptake of PI, suggesting that membranes were not damaged. A similar finding was demonstrated for MRSA (L. Weaver et al., unpublished data), where contact with copper resulted in respiratory failure but not damage to the bacterial cell membrane.
However, at a higher inoculum concentration of 5 × 107 CFU/cm2, viable cells were detectable not only on stainless steel after 2 h of contact but again also on alloys C26000 and C70600, with significantly higher counts compared to culture for C70600 (P < 0.001).
Therefore, enterococci at an inoculum concentration of ≤106 CFU/cm2 on copper and copper alloy surfaces for at least 2 h showed no detectable viable cells by culture and also demonstrated inhibition of respiration but membrane integrity did not appear to be compromised.
Analysis of bacterial DNA following exposure to copper and stainless steel surfaces. (i) DNA size separation by agarose gel electrophoresis.
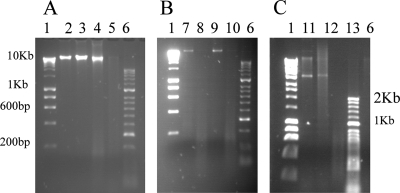
The reduced SYTO 9 staining data suggested that exposure to copper could be affecting bacterial DNA. This was investigated further using DNA purified from bacteria exposed to copper surfaces. Genomic DNA was purified using the Qiagen DNeasy Blood and Tissue kit, which cuts the bacterial genome into approximately 50-kb fragments before separation by agarose gel electrophoresis. The results obtained with DNA purified from the vancomycin-resistant E. faecium control strain are shown in Fig. 6 A, and those obtained with DNA purified from E. faecium clinical isolate 5 (lanes 7 and 8) and E. faecalis clinical isolate 2 (lanes 9 and 10) are shown in Fig. 6B. DNA purified from cells that had not been exposed to metal surfaces (live cells, lane A2) and dead cells (not shown) exhibited fragments that are too large to migrate through the gel (>10 kb), remaining at the site of loading. The same pattern is seen with DNA purified from cells exposed to stainless steel for 2 h (Fig. 6A, lane 3, and B, lanes 7 and 9). However, the DNA from all of the strains tested, when exposed to copper for 2 h, exhibit a pronounced smearing effect characteristic of nucleic acid degradation (Fig. 6A, lane 4, and B, lanes 8 and 10). This suggests that the DNA is being broken down but not at specific points because there is no accumulation of particular-sized fragments. If cells are exposed to copper for a longer time, 4 h, the denaturation has continued and no DNA can be detected at all (lane A5). This may be because the fragments are too small to be visualized and may have run straight through the gel. Increasing the agarose concentration did not allow further visualization of the small DNA fragments generated on contact with copper surfaces, suggesting that extensive denaturation of the DNA has occurred (Fig. 6A and B show 2% agarose gels).
FIG. 6.
Agarose gel electrophoresis of purified enterococcal DNA. (A) Purified genomic DNA of E. faecium NCTC 12202. Lane 2, cells not exposed to metal surfaces; lane 3, cells exposed to stainless steel for 2 h; lane 4, cells exposed to copper for 2 h; lane 5, cells exposed to copper for 4 h. (B) Purified genomic DNA of clinical isolates E. faecium 5 (lanes 7 and 8) and E. faecalis 2 (lanes 9 and 10) exposed to stainless steel (lanes 7 and 9) or copper (lanes 8 and 10) for 2 h at 22°C. (C) Purified plasmid DNA of E. faecium NCTC 12202 not exposed to metal (lane 11) or exposed to stainless steel (lane 12) or copper (lane 13) for 2 h at 22°C. Control lanes are Bioline Hyperladder I (lanes 1) and Hyperladder II (lanes 6). Genomic DNA was purified using the Qiagen DNeasy Blood and Tissue kit (2% agarose) and the Qiaprep Spin Miniprep kit for plasmid DNA (0.9% agarose).
No viable cells were present at either time point on copper surfaces when they were assessed by any of the viability testing methods described previously. This suggests that disintegration of the DNA into very small fragments is continuing after the death of the bacteria.
Similar results were obtained for plasmid DNA extracted from the E. faecium control strain exposed to copper and stainless steel (Fig. 6C). Lanes 11 and 12 demonstrate plasmid DNA present in untreated cells and those exposed to stainless steel for 2 h. No plasmid DNA bands are detected in those cells exposed to copper surfaces (lane 13). It must be noted that this is a crude, undigested plasmid preparation and therefore it includes supercoiled and linear fractions of the same plasmids. However, the results suggest that both genomic and plasmid DNAs are denatured upon exposure to copper surfaces.
(ii) Genomic DNA fragmentation assay.
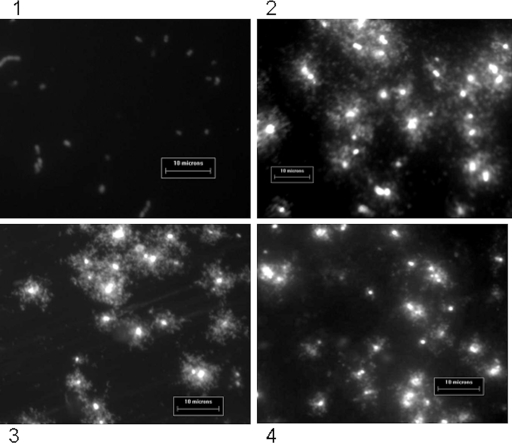
To determine if DNA disintegration into very small fragments was due to copper alone and not a preparation artifact, the DNA fragmentation assay was used to gently visualize the entire genome of individual cells in situ (Fig. 7). Analysis of the untreated bacterial cells prior to inoculation of coupons revealed a pattern of DNA loops protruding through and close to the lysed cell membrane in live and heat-killed cells (Fig. 7, panels 3 and 4, respectively), characteristic of an intact genome. Following 2 h of contact with stainless steel, the pattern of DNA staining was similar but there were more discrete fragments that had diffused away from the bacterial cells (Fig. 7, image 2).
FIG. 7.
Analysis of genomic DNA of E. faecalis ATCC 51299 in situ by DNA fragmentation assay following 2 h of exposure to copper (image 1) or steel (image 2). Images 3 and 4 show bacterial cells not exposed to metal surfaces and are of live and dead (heat-killed) cells, respectively, which were then used in the DNA fragmentation assay. Loops of DNA are visible in all of the samples except that in image 1, suggesting that exposure to copper has resulted in disintegration of the bacterial DNA into fragments too small to be visualized even by the sensitive nucleic acid stain SYBR Gold used for this assay.
The pattern of DNA staining after 2 h of contact with copper surfaces was very different (Fig. 7, image 1). The absence of visible DNA strands suggests that extensive damage to the nucleic acid has resulted in fragments that are too small to be visualized and have dispersed throughout the slide.
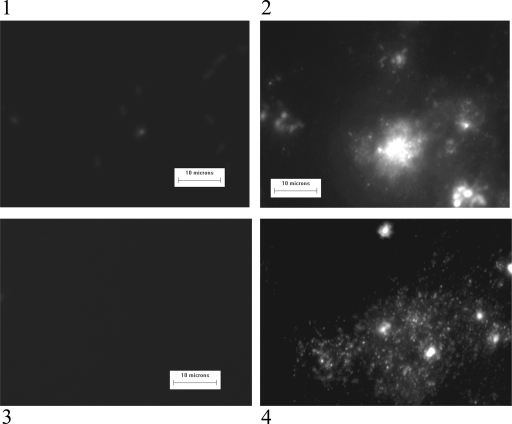
A similar result was observed with clinical isolates of E. faecalis and E. faecium exposed to copper surfaces for 1 h at room temperature (Fig. 8). Intact DNA was visible following contact with stainless steel (images 2 and 4) but not visible on cells isolated from copper surfaces (images 1 and 3). Preliminary results suggest that DNA damage may begin much earlier (data not shown).
FIG. 8.
Analysis of genomic DNA of clinical isolates E. faecalis 2 (images 1 and 2) and E. faecium 5 (images 3 and 4) in situ by DNA fragmentation assay following a 1-h exposure to copper (images 1 and 3) or steel (images 2 and 4). No DNA loops are visible on cells exposed to copper, whereas DNA fragments are visible emanating from cells isolated from stainless steel surfaces.
DISCUSSION
Contamination of hospital surfaces with pathogenic microorganisms contributes to reinfection and spread of disease. Not only are enterococci hardy and able to survive on many types of “touch” surfaces (27, 36) for several weeks, but the ability of these organisms to easily transfer antibiotic resistance means that it is essential that any contamination of the environment from infected individuals with viable cells be effectively destroyed (41). A combination of bactericidal surfaces providing a constant “killing surface” and regular effective disinfection could greatly reduce the spread of disease. Stainless steel is a commonly used hospital surface for many reasons, including resistance to corrosion and the ability to withstand regular disinfection. However, our research has shown that vancomycin-resistant isolates of the two main pathogenic enterococcal species are able to survive for several months on stainless steel surfaces, which could potentially contribute to the reinfection of personnel and especially of vulnerable patients.
In our studies on alloy surfaces containing at least 65% copper, enterococci at a high contamination concentration of 106 CFU/cm2 were rapidly killed over a few hours of contact, compared to survival for several months on stainless steel. Pure copper was the most effective surface at killing bacterial cells.
In general, alloys containing >90% copper were as effective as pure copper, with all of the isolates of E. faecalis and two of E. faecium achieving complete cell death by 1 h (with a few cells of the remaining E. faecium isolates remaining viable for another hour but still with a 3- to 4-log reduction in number). For both species, no viable cells were detected after 2 h of contact with alloys containing 60 to 70% copper. Occasionally, alloy C75200 (nickel-silver) outperformed alloys with a higher copper content, e.g., C26000 (cartridge brass). The other metal constituents and physical properties of each alloy, particularly the rate of release of copper, may have a role in the bactericidal activity reported.
However, at bacterial contamination levels of ≤105 CFU/cm2, all of the copper alloys tested were virtually indistinguishable from pure copper and were very effective at killing pathogenic enterococcal cells within 1 h and in 20 min at <103 CFU/cm2. This reinforces the potential for the use of these alloys in the clinical environment, because a recent study in the United States determined VRE contamination on surfaces to be, at most, a few hundred cells/100 cm2 at three large hospitals (M. Schmidt, personal communication; 42).
Our experiments were done under worst-case scenario conditions: inoculation of surfaces with aqueous samples in a nutritious and isotonic medium. It has been reported that the presence of biological fluids or meat juices can delay the mechanism of Staphylococcus aureus and E. coli O157 killing by copper, respectively (38, 46). Recent work in our laboratories has also determined the importance of chelating substances on bacterial survival on copper surfaces (unpublished data). Experiments with a rapidly drying swabbed inoculum in PBS or water suggest that killing is even more rapid (data not shown), but these conditions may be not entirely relevant to actual contamination with organic specimens in a clinical environment.
Assessment of the number of viable cells recovered from metal surfaces by respiratory staining was not significantly different from that obtained by culture, providing more evidence that enterococci survive for long periods on stainless steel but also suggesting the absence of a viable-but-nonculturable state on copper surfaces under these conditions.
Assessment of the viability of cells recovered from stainless steel using BacLight SYTO 9/PI staining also demonstrated no significant difference from results obtained by culture but suggested that some damage to the cell membrane does occur on this surface. However, staining with BacLight does not always produce distinct “live” and “dead” populations, particularly because cells with an intact membrane are not necessarily alive (2). The BacLight staining method was difficult to interpret for enterococci exposed to copper alloys because of the diminished and frequently absent staining with SYTO 9 and PI, respectively, except at the time of inoculation. This suggested that the bacterial DNA has been affected to the extent that intercalating DNA stains cannot now bind. There does not appear to be any uptake of PI by enterococci exposed to copper alloys, suggesting that damage to the cell membrane is not occurring, but these results may be misleading if the DNA is too damaged to bind PI.
Targets of copper toxicity are thought to include nucleic acid, structural and functional proteins, lipids, and inhibition of metabolic processes such as respiration and osmotic stress resulting in cell lysis. In mammalian cells, soluble copper(II) ions are known to bind to DNA bases, resulting in unwinding of the double helix, and under aerobic conditions, the Fenton reaction with bound and free ions and hydrogen peroxide results in the production of reactive oxygen species (ROS) that cause double- and single-strand breaks and intrastrand cross-linking (30). Macomber et al. (31). reported that exposure of E. coli mutants lacking in copper export systems to copper solutions resulted in copper-overloaded cells and no detectable oxidative damage to the DNA using a gene-specific PCR assay. The reasons suggested were compartmentalization of hydroxyl radicals generated in the periplasm of the cell. The effect of hydroxyl radical damage is short reaching, and therefore, damage to the DNA could not occur if the sites of hydroxyl radical generation are spatially separated from the nucleic acid. They also described the existence of ligands, perhaps glutathione, complexing with copper ions and suggested that copper toxicity may primarily be due to damage of metalloenzymes by the ROS, i.e., dihydrogen peroxide, hydroxyl radicals, and superoxides.
Exposure to relatively low soluble copper concentrations, described by Macomber et al. (31), is very different from continual contact with copper and copper alloy surfaces. In our system, we have reported extensive disintegration of the genomic and plasmid DNAs of Gram-positive enterococci exposed to copper and copper alloy surfaces because the DNA from cells exposed to copper appears to be (i) denatured over time in agarose gel electrophoresis, (ii) does not bind intercalating stains SYTO 9 and PI, and (iii) genomic DNA cannot be detected in the DNA fragmentation assay. These effects were not observed on stainless steel. The DNA fragmentation assay is a useful tool because it allows observation of the entire genome of individual cells without a purification step that could result in damage to DNA from stressed bacterial cells and lead to spurious results. This technology has been successfully used for the analysis of eukaryotic nucleic acid, for example, in the analysis of sperm DNA and the efficacy of specific anticancer agents on patient DNA. Fernández et al. (17). successfully adapted the method to investigate damage to bacterial DNA following treatment with quinolone antibiotics. We have also shown how exposure to copper resulted in the inhibition of respiration with minimal damage to the integrity of the bacterial cell membrane. We suggest that the absence of an outer membrane in Gram-positive cells and lack of a periplasmic space may facilitate the access of copper(I)/(II) and generated ROS to the DNA directly and rapidly inflict damage.
The ligands, described by Macomber et al. (31), responsible for removing copper(II) in copper-overloaded cells may still be present, but the effect is insignificant when bacteria are constantly in contact with the copper surface and binding sites are saturated with copper ions.
Espírito Santo et al. (13) determined that ROS are generated when E. coli is exposed to copper surfaces. They identified hydroxyl radicals generated under aerobic conditions, presumably by Haber-Weiss and Fenton reactions of reduced copper ions [supplied by redox cycling of copper(I) and copper(II)].
Preliminary experiments in our laboratory with E. coli O157 exposed to copper surfaces using the DNA fragmentation assay have indicated that genomic DNA is also destroyed in this species but more slowly (manuscript in preparation). The DNA stability reported by Macomber et al. (31) is probably due to exposure to soluble Cu(II) rather than the Cu(I)/Cu(II) redox cycling proposed in our studies. This DNA degradation effect on E. coli and other species will be investigated in future studies.
Concerns have been expressed about the possibility of the development of copper resistance if alloy surfaces are constantly in use. Mutations in bacterial copper homeostasis mechanisms do affect survival times on copper alloys (11, 13), but because survival times on stainless steel and other commonly in-use surfaces are so much greater, the significance may not be relevant in a real-life situation. Further studies are required to elucidate the mechanism of copper killing and investigate this possibility.
There has been much concern recently that the frequency of antimicrobial resistance in bacteria has increased in concert with increasing usage of antimicrobial compounds. A recent European Commission report (16) has summarized the scientific evidence from bacteriological, biochemical, and genetic data indicating that the use of active molecules in biocidal products may contribute to the increased occurrence of antibiotic-resistant bacteria. The selective stress exerted by biocides may favor bacteria expressing resistance mechanisms and their dissemination. Some biocides have the capacity to maintain the presence of mobile genetic elements that carry genes involved in cross-resistance between biocides and antibiotics. In enterococci, up to 25% of the genome has been found to contain mobile elements (41). The dissemination of these mobile elements, their genetic organization, and the formation of biofilms provide conditions that could create a potential risk of development of cross-resistance between antibiotics and biocides. The case for the use of copper in antimicrobial products was considered, but there was no evidence that this might lead to antibiotic resistance in the way that the widespread use of Triclosan has been associated with the emergence of triclosan and mupirocin resistance in MRSA, although evidence for this is limited (15, 16, 44). The plasmid-localized copper resistance tcrB gene has been identified in E. faecium and E. faecalis thought to originate from pigs fed with copper sulfate-supplemented food (19). The Tn1546 element and erm genes conferring glycopeptide and macrolide resistance are located on the same plasmid, but there is no significant evidence that use of copper in animal feeds coselected for antibiotic resistance (20) except under experimental conditions in piglets fed a high concentration of copper sulfate (21). However, continued use of copper sulfate was not able to maintain high levels of antimicrobial resistance (21).
The current study indicates that DNA is rapidly destroyed in enterococci exposed to copper surfaces, meaning that there is little chance of high-level copper or antibiotic resistance developing. Consequently, this disintegration of bacterial nucleic acid supports the use of copper alloys as contact surfaces in clinical environments to actively kill bacterial cells without the occurrence of DNA mutation and transfer of genetic material carrying antibiotic resistance genes.
Acknowledgments
This research was supported by the Copper Development Association, New York, NY, and the International Copper Association, Ltd., New York, NY.
Footnotes
Published ahead of print on 25 June 2010.
REFERENCES
- 1.Amyes, S. G. B. 2007. Enterococci and streptococci. Int. J. Antimicrob. Agents 29:S43-S52. [DOI] [PubMed] [Google Scholar]
- 2.Berney, M., F. Hammes, F. Bosshard, H.-U. Weilenmann, and T. Egli. 2007. Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight kit in combination with flow cytometry. Appl. Environ. Microbiol. 73:3283-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borkow, G., and J. Gabbay. 2004. Putting copper into action: copper impregnated products with potent biocidal activities. FASEB J. 18:1728-1730. [DOI] [PubMed] [Google Scholar]
- 4.Boyce, J. M. 2007. Environmental contamination makes an important contribution to hospital infection. J. Hosp. Infect. 65(Suppl. 2):50-54. [DOI] [PubMed] [Google Scholar]
- 5.Casey, A. L., P. A. Lambert, L. Miruszenko, and T. S. J. Elliot. 2008. Copper for preventing microbial contamination, poster K-4121, p. 202. 48th ICAAC. American Society for Microbiology, Washington, DC.
- 6.Casey, A. L., D. Adams, T. J. Karpanen, P. A. Lambert, B. D. Cookson, P. Nightingale, L. Miruszenko, R. Shillam, P. Christian, and T. S. J. Elliot. 2010. The role of copper in the reduction of contamination of the hospital environment. J. Hosp. Infect. 74:72-77. [DOI] [PubMed] [Google Scholar]
- 7.Copper Development Association. 2010. Reducing the risk of healthcare associated infections: the role of copper touch surfaces. CDA publication 196. Copper Development Association, New York, NY. http://www.copperinfo.co.uk/antimicrobial/downloads/pub-196-reducing-risk-hcais-report.pdf.
- 8.Drees, M., D. R. Syndman, C. H. Schmid, L. Barefoot, K. Hansjosten, P. M. Vue, M. Cronin, A. Nasraway, and Y. Golan. 2008. Prior environmental contamination increases the risk of acquisition of vancomycin-resistant enterococci. Clin. Infect. Dis. 46:678-685. [DOI] [PubMed] [Google Scholar]
- 9.Duckro, A. N., D. W. Blom, E. A. Lyle, R. A. Weinstein, and M. K. Hayden. 2005. Transfer of vancomycin-resistant enterococci via health care worker hands. Arch. Intern. Med. 165:302-307. [DOI] [PubMed] [Google Scholar]
- 10.Eckstein, B. C., D. A. Adams, E. C. Eckstein, A. Rao, A. K. Sethi, G. K. Yadavalli, and C. J. Donskey. 2007. Reduction of Clostridium difficile and vancomycin-resistant Enterococcus contamination of environmental surfaces after an intervention to improve cleaning methods. BMC Infect. Dis. 7:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elguindi, J., J. Wagner, and C. Rensing. 2009. Genes involved in copper resistance influence survival of Pseudomonas aeruginosa on copper surfaces. J. Appl. Microbiol. 106:1448-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eliopoulos, G. M. 2009. Microbiology of drugs for treating multiply drug-resistant Gram-positive bacteria. J. Infect. 59:S17-S24. [DOI] [PubMed] [Google Scholar]
- 13.Espírito Santo, C. E., N. Taudte, D. H. Nies, and G. Grass. 2008. Contribution of copper ion resistance to survival of Escherichia coli on metallic copper surfaces. Appl. Environ. Microbiol. 74:977-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.European Centre for Disease Prevention and Control and European Medicines Agency. 2009. ECDC/EMEA joint technical report. The bacterial challenge: time to react. A call to narrow the gap between multidrug-resistant bacteria in the EU and the development of new antibacterial agents. http://www.ema.europa.eu/pdfs/human/antimicrobial_resistance/EMEA-576176-2009.pdf.
- 15.European Commission. 2002. Opinion on triclosan resistance. European Commission Health & Consumer Protection Directorate-General, Brussels, Belgium. http://ec.europa.eu/food/fs/sc/ssc/out269_en.pdf.
- 16.European Commission. 2009. Assessment of the antibiotic resistance effects of biocides. Scientific Committee on Emerging and Newly Identified Health Risks, Brussels, Belgium. http://ec.europa.eu/health/ph_risk/committees/04_scenihr/docs/scenihr_0_021.pdf.
- 17.Fernández, J. L., M. Cartelle, L. Muriel, R. Santiso, M. Tamayo, V. Goyanes, J. Gosálvez, and G. Bou. 2008. DNA fragmentation in microorganisms assessed in situ. Appl. Environ. Microbiol. 74:5925-5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gould, I. M. 2008. The epidemiology of antibiotic resistance. Int. J. Antimicrob. Agents 32(Suppl. 1):S2-S9. [DOI] [PubMed] [Google Scholar]
- 19.Hasman, H., and F. M. Aaresstrup. 2002. tcrB, a gene conferring transferable copper resistance in Enterococcus faecium: occurrence, transferability and linkage to macrolide and glycopeptide resistance. Antimicrob. Agents Chemother. 46:1410-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hasman, H., and F. M. Aaresstrup. 2005. Relationship between copper, glycopeptide, and macrolide resistance among Enterococcus faecium strains isolated from pigs in Denmark between 1997 and 2003. Antimicrob. Agents Chemother. 49:454-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasman, H., I. Kempf, B. Chidaine, R. Cariolet, A. J. Ersboll, H. Houe, H. C. B. Hansen, and F. M. Aaresstrup. 2006. Copper resistance in Enterococcus faecium, mediated by the tcrB gene, is selected by supplementation of pig feed with copper sulfate. Appl. Environ. Microbiol. 72:5784-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayden, M. K., D. W. Blom, E. A. Lyle, C. G. Moore, and R. A. Weinstein. 2008. Risk of hand or glove contamination after contact with patients colonized with vancomycin-resistant enterococcus or the colonized patients environment. Infect. Control Hosp. Epidemiol. 29:149-154. [DOI] [PubMed] [Google Scholar]
- 23.Health Protection Agency. 2008. Antimicrobial resistance and prescribing in England, Wales and Northern Ireland, 2008. Health Protection Agency, London, United Kingdom.
- 24.Health Protection Agency. 2008. Health protection report 2, no. 42. Health Protection Agency, London, United Kingdom.
- 25.Kearns, A. M., R. Freeman, and N. F. Lightfoot. 1995. Nosocomial enterococci: resistance to heat and sodium hypochlorite. J. Hosp. Infect. 30:193-199. [DOI] [PubMed] [Google Scholar]
- 26.Keevil, C. W. 2003. Rapid detection of biofilms and adherent pathogens using scanning confocal laser microscopy and episcopic differential interference contrast microscopy. Water Sci. Technol. 47:105-116. [PubMed] [Google Scholar]
- 27.Lankford, M. G., S. Collins, L. Youngberg, D. M. Rooney, J. R. Warren, and G. A. Noskin. 2006. Assessment of materials commonly utilized in health care: implications for bacterial survival and transmission. Am. J. Infect. Control 34:258-263. [DOI] [PubMed] [Google Scholar]
- 28.Launay, A., S. A. Ballard, P. D. R. Johnson, M. L. Grayson, and T. Lambert. 2006. Transfer of vancomycin resistance transposon Tn1549 from Clostridium symbiosum to Enterococcus spp. in the gut of gnotobiotic mice. Antimicrob. Agents Chemother. 50:1054-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livermore, D. M. 2009. Has the era of untreatable infections arrived? J. Antimicrob. Chemother. 64(Suppl. 1):i29-i36. [DOI] [PubMed] [Google Scholar]
- 30.Lloyd, D. R., and D. H. Phillips. 1999. Oxidative DNA damage mediated by copper(II), iron(II) and nickel(II) Fenton reactions: evidence for site-specific mechanisms in the formation of double-strand breaks, 8-hydroxydeoxyguanosine and putative intrastrand cross-links. Mutat. Res. 424:23-36. [DOI] [PubMed] [Google Scholar]
- 31.Macomber, L., C. Rensing, and J. A. Imlay. 2007. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J. Bacteriol. 189:1616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehtar, S., I. Wiid, and S. D. Todorov. 2008. The antimicrobial activity of copper and copper alloys against nosocomial pathogens and Mycobacterium tuberculosis isolated from healthcare studies in the Western Cape: an in vitro study. J. Hosp. Infect. 68:45-51. [DOI] [PubMed] [Google Scholar]
- 33.Michels, H. T., J. O. Noyce, and C. W. Keevil. 2009. Effects of temperature and humidity on the efficacy of methicillin-resistant Staphylococcus aureus challenged antimicrobial materials containing silver and copper. Lett. Appl. Microbiol. 49:191-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray, B. E. 1998. Diversity of multidrug-resistant enterococci. Emerg. Infect. Dis. 4:37-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.National Nosocomial Infections Surveillance System. 2004. National Nosocomial Infections Surveillance (NNIS) system report, data summary from January 1992 through June 2004, issued October 2004. Am. J. Infect. Control 32:470-485. [DOI] [PubMed] [Google Scholar]
- 36.Neely, A., and M. P. Maley. 2000. Survival of enterococci and staphylococci on hospital fabrics and plastic. J. Clin. Microbiol. 38:724-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noyce, J. O., H. Michels, and C. W. Keevil. 2006. Potential use of copper surfaces to reduce survival of epidemic methicillin-resistant Staphylococcus aureus in the healthcare environment. J. Hosp. Infect. 63:289-297. [DOI] [PubMed] [Google Scholar]
- 38.Noyce, J. O., H. Michels, and C. W. Keevil. 2006. Use of copper cast alloys to control Escherichia coli O157 cross-contamination during food processing. Appl. Environ. Microbiol. 72:4239-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noyce, J. O., H. Michels, and C. W. Keevil. 2007. Inactivation of influenza A virus on copper versus stainless steel surfaces. Appl. Environ. Microbiol. 73:2748-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ott, M., and H. Wirick. 2008. Vancomycin-resistant enterococci (VRE) and the role of the healthcare worker. Can. Oper. Room Nurs. J. 26:21-24, 26-29, 32. [PubMed] [Google Scholar]
- 41.Paulsen, I. T., L. Banerjei, G. S. A. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Varmathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 42.Salgado, C. D., K. A. Sepkowitz, T. Plaskett, J. F. John, J. R. Cantey, H. H. Attaway, L. L. Steed, H. T. Michels, and M. G. Schmidt. 2008. Microbial burden of objects in ICU rooms, poster K-4117, p. 202. 48th ICAAC. American Society for Microbiology, Washington, DC.
- 43.Smith, K., and I. S. Hunter. 2008. Efficacy of common hospital biocides with biofilms of multi-drug resistant clinical isolates. J. Med. Microbiol. 57:966-973. [DOI] [PubMed] [Google Scholar]
- 44.Suller, M. T., and A. D. Russell. 2000. Triclosan and antibiotic resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 46:11-18. [DOI] [PubMed] [Google Scholar]
- 45.Tacconelli, E., and M. A. Cataldo. 2008. Vancomycin-resistant enterococci (VRE): transmission and control. Int. J. Antimicrob. Agents 31:99-106. [DOI] [PubMed] [Google Scholar]
- 46.Tolba, O., A. Loughrey, C. E. Goldsmith, B. C. Millar, P. J. Rooney, and J. E. Moore. 2007. Survival of epidemic strains of nosocomial- and community-acquired methicillin-resistant Staphylococcus aureus on coins. Am. J. Infect. Control 35:342-346. [DOI] [PubMed] [Google Scholar]
- 47.Top, J., R. Willems, and M. Bonten. 2008. Emergence of CC17 Enterococcus faecium: from commensal to hospital adapted pathogen. FEMS Immunol. Med. Microbiol. 52:297-308. [DOI] [PubMed] [Google Scholar]
- 48.Weaver, L., H. T. Michels, and C. W. Keevil. 2008. Survival of Clostridium difficile on copper and steel. J. Hosp. Infect. 68:145-151. [DOI] [PubMed] [Google Scholar]
- 49.Weaver, L., H. T. Michels, and C. W. Keevil. 2010. Potential for preventing spread of fungi in air conditioning systems constructed using copper instead of aluminium. Lett. Appl. Microbiol. 50:18-23. [DOI] [PubMed] [Google Scholar]
- 50.Werner, G., T. M. Coque, A. M. Hammerum, R. Hope, W. Hryniewicz, A. Johnson, I. Klare, K. G. Kristinsson, R. Leclerq, C. H. Lester, M. Lillie, C. Novais, B. Olsson-Liljuquist, L. V. Peixe, E. Sadowy, G. S. Simonsen, J. Top, J. Vuopio-Varkila, R. J. Willems, W. Witte, and N. Woodford. 2008. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill. 13:1-11. [PubMed] [Google Scholar]
- 51.Wheeldon, L. J., T. Worthington, P. A. Lambert, A. C. Hilton, C. J. Lowden, and T. S. Elliot. 2008. Antimicrobial efficacy of copper surfaces against spores and vegetative cells of Clostridium difficile: the germination theory. J. Antimicrob. Chemother. 62:522-525. [DOI] [PubMed] [Google Scholar]
- 52.Wilks, S. A., H. Michels, and C. W. Keevil. 2005. The survival of Escherichia coli O157 on a range of metal surfaces. Int. J. Food Microbiol. 105:445-454. [DOI] [PubMed] [Google Scholar]
- 53.Wilks, S. A., H. T. Michels, and C. W. Keevil. 2006. Survival of Listeria monocytogenes Scott A on metal surfaces: implications for cross-contamination. Int. J. Food Microbiol. 111:93-98. [DOI] [PubMed] [Google Scholar]
- 54.Witte, W. 2004. Glycopeptide resistant Staphylococcus. J. Vet. Med. B Infect. Dis. Vet. Public Health 51:370-373. [DOI] [PubMed] [Google Scholar]