Abstract
The cloning and expression of a native gene encoding a Bacillus subtilis phytase using Pichia pastoris as the host is described. In addition, the influence of N-glycosylation on the biochemical properties of the B. subtilis phytase, the influence of pH on the thermostability of the recombinant and native B. subtilis phytases, and the resistance of both phytases to shrimp digestive enzymes and porcine trypsin are also described. After 48 h of methanol induction in shake flasks, a selected recombinant strain produced and secreted 0.82 U/ml (71 mg/liter) recombinant phytase. This phytase was N-glycosylated, had a molecular mass of 39 kDa after N-deglycosylation, exhibited activity within a pH range of 2.5 to 9 and at temperatures of 25 to 70°C, had high residual activity (85% ± 2%) after 10 min of heat treatment at 80°C and pH 5.5 in the presence of 5 mM CaCl2, and was resistant to shrimp digestive enzymes and porcine trypsin. Although the recombinant Bacillus phytase had pH and temperature activity profiles that were similar to those of the corresponding nonglycosylated native phytase, the thermal stabilities of the recombinant and native phytases were different, although both were calcium concentration and pH dependent.
Phytases catalyze the release of phosphate from phytate (myo-inositol-1,2,3,4,5,6-hexakisphosphate), thereby generating less-phosphorylated myo-inositol derivatives (22). Phytate, the major storage form of phosphorus in plant-derived feedstuffs, is regarded as an antinutrient factor since it forms insoluble complexes with proteins and a variety of nutritionally important metal ions such as calcium, zinc, magnesium, and iron and decreases phosphorus bioavailability (9). During the last 2 decades, phytases have been used as feed additives for monogastric animals to enhance the utilization of plant-derived feedstuffs, improving the utilization of phytate-phosphorus in diets and reducing their manure phosphorus excretion to the environment (22). In addition, phytase supplementation leads to improved availability of other minerals and trace elements and enhances protein digestibility through phytate hydrolysis during digestion in the stomach or during food processing (22). Recently, research on phytases has been focused on the processing of human food (9, 12, 22) and the synthesis of lower inositol phosphates (12).
Phytases have been isolated from a number of sources, including plants, animals, and microorganisms (22). Based on the presence of a specific consensus motif and their tridimensional structure, phytases are classified into four major classes: histidine acid, beta-propeller, cysteine, and purple acid phytases (22, 26). Alternatively, based on the carbon ring position where removal of phosphate groups from phytate is initiated, the ENZYME database (available through the ExPASy Proteomics Server; http://www.expasy.ch/enzyme/) classifies phytases into three different groups: 3-phytase (alternative name, 1-phytase; EC 3.1.3.8), 4-phytase (alternative name, 6-phytase; EC 3.1.3.26), and 5-phytase (EC 3.1.3.72).
Phytases with a beta-propeller structure are mainly isolated from Bacillus species; they are nonglycosylated proteins with high thermal stability, an optimal pH in the neutral range, and an optimal temperature of 55 to 70°C and exhibit unique Ca2+-dependent catalytic properties (11, 17, 19, 20, 27). Inositol trisphosphate was proposed as the final product of the action of Bacillus phytases on the phytate (16). Nevertheless, from a crystal structure analysis of a Bacillus amyloliquefaciens phytase, Shin et al. (34) suggested that under test tube conditions, where the phosphate produced is not removed, the further degradation of inositol trisphosphate would be very slow. In a physiological situation, the less-phosphorylated myo-inositols could be further degraded by the enzyme, allowing the utilization of the phosphate ions produced. This is a highly desirable property for the application of a phytase as a feed additive.
Although commercial production of phytases is currently focused on fungal histidine acid phytases from Aspergillus species (1, 11), bacterial phytases from Bacillus species are an alternative to fungal enzymes because of their high thermal stability, calcium-phytate complex substrate specificity, pH profile, and proteolysis resistance (15, 19, 27). In contrast to the Aspergillus phytases, Bacillus phytases are specific for phytate. Therefore, important phosphate compounds other than phytate are not hydrolyzed by the Bacillus phytases and remain available for animal uptake.
While the addition of phytase is widely used to improve the utilization of plant phosphorus in poultry and swine, phytase utilization in feed for aquatic species is still at an early stage (1). Aquaculture species may be either agastric or monogastric, with digestive system pHs that are either near 7 or much lower, respectively. Since phytase activity is highly dependent on the gut's pH, the effects of phytase on growth and nutrient uptake would vary from species to species (23). Therefore, no single phytase can meet all of the needs of a feed for all aquaculture species. The phytases from Bacillus are suitable as feed additives for animals with neutral digestive tracts, such as some aquatic species, since these enzymes have an optimal pH near neutrality. In addition, these phytases are quite stable at the high temperatures that are encountered in the feed-pelleting process (2, 19).
The methylotrophic yeast Pichia pastoris has been successfully used as a host for heterologous gene expression, producing high levels of recombinant proteins, including phytases (13, 14, 31, 32, 40). A coding sequence cloned under the control of the AOX1 promoter is highly expressed when methanol is used as the sole carbon source and is repressed by most other carbon sources (14). P. pastoris can grow in simple defined media, reach a very high cell density, and accumulate extremely high concentrations of intra- or extracellular protein under the control of the AOX1 promoter (14). In addition, P. pastoris, as a eukaryotic expression system, can carry out protein processing, folding, and posttranslational modifications (14, 30).
In this paper, we report the molecular cloning of the phyC gene from Bacillus subtilis VTT E-68013 and its expression in P. pastoris GS115 strains. In addition, the influence of N-glycosylation on the biochemical properties of the B. subtilis phytase, the influence of pH on the thermostability of the recombinant and native B. subtilis phytases, and the resistance of both phytases to shrimp digestive enzymes and porcine trypsin are described.
MATERIALS AND METHODS
Strains, plasmids, medium composition, and chemicals.
B. subtilis VTT E-68013 was obtained from the culture collection of the Technical Research Centre of Finland. P. pastoris GS115 (his4) and Escherichia coli TOP10F′ and the plasmids TOPO pCR 2.1 and pPIC9 were purchased from Invitrogen (San Diego, CA). Regeneration dextrose base (RDB), buffered minimal glycerol (BMG), and buffered minimal methanol (BMM) media were prepared according to the manual of the Pichia Expression kit (Invitrogen, San Diego, CA). BMM medium was also supplemented with 0.1% (wt/vol) CaCl2 (BMM-CaCl2). Methanol was used at a final concentration of 0.75% (vol/vol). Isis proofreading DNA polymerase was from Qbiogene (Carlsbad, CA), and the XhoI restriction endonuclease, T4 DNA ligase, the PCR Clean-Up System (Wizard SV Gel), and the DNA Purification System (Wizard Plus SV Minipreps) were purchased from Promega (Madison, WI). The AvrII restriction endonuclease and endoglycosidase (Endo Hf) were purchased from New England Biolabs (Beverly, MA). Oligonucleotides were purchased from MWG (Laredo, TX). All chemicals were analytical grade and purchased from Sigma-Aldrich Co. (St. Louis, MO) and Productos Químicos Monterrey (Monterrey, N.L., México).
Construction of P. pastoris phytase C strains.
Genomic DNA of B. subtilis VTT E-68013, extracted by standard protocols, was the source of the phyC gene. The complete sequence encoding mature phytase C (GenBank accession no. AF029053) was amplified by PCR using a forward primer (5′-CAGCTCGAGAAAAGACTGTCCGATCCTTATCAT-3′) containing an XhoI site and a reverse primer (5′-TTTCCTAGGTTATTTTCCGCTTCTGTC-3′) containing an AvrII restriction site. The PCR was performed with Isis proofreading DNA polymerase and a 30-cycle amplification program under the following conditions: 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min, with a first denaturation step of 94°C for 2 min. The 1,089-bp amplified product was adenylated and cloned into the plasmid TOPO pCR 2.1 using standard protocols. The new plasmid, TOPOphyC, was cut with XhoI and AvrII. The released 1,077-bp fragment was ligated to vector pPIC9, which had been previously digested with the same restriction enzymes. The resulting new expression vector, pPIC9phyC, harbors the phyC gene sequence. The correct construction of this vector was confirmed by PCR and sequence analysis. All DNA manipulations were performed according to standard methods (33).
P. pastoris host strain GS115 (his4) was transformed with SalI-digested pPIC9phyC DNA by the spheroplasting protocol according to the manual of the Pichia Expression kit (Invitrogen, San Diego, CA). Transformants were selected for the ability to grow on histidine-deficient medium (RDB-agar plates) at 30°C until colonies appeared (His+ selection). Twenty His+ colonies were randomly selected (called GS115-PhyC). The integration of the expression cassette into the genome of GS115-PhyC strains was verified by PCR using the 5′ and 3′ AOX1 primers as previously described (5).
Expression and preparation of recombinant and native phytase C.
A selected Mut+ transformant strain was grown in 600 ml of BMG medium at 30°C and 250 rpm for 12 h until cultures reached an optical density at 600 nm (OD600) of 9 to 10. Cells were subsequently harvested by centrifugation (1,500 × g, 5 min) and used to inoculate 120 ml of BMM-CaCl2 medium at an initial OD600 of 50 to 52. Further incubation was carried out at 30°C for 48 h with continuous shaking at 250 rpm. In the induction step, methanol was added to a final concentration of 0.75% (vol/vol) every 24 h.
Recombinant B. subtilis phytase C (PhyC-R) was obtained from cell-free culture medium that was concentrated 20-fold and diafiltered by ultrafiltration using 10-kDa Amicon Centricon Plus-70 centrifugal filter devices (Millipore, MA) and a 100 mM Tris-HCl-50 mM NaCl-5 mM CaCl2-2% glycerol buffer (pH 8.5). This enzyme preparation was analyzed by Bradford protein and enzymatic activity assays, Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and Western blotting and used for biochemical assays. A preparation of native B. subtilis phytase C (PhyC-N) was obtained from B. subtilis VTT E-68013 cell-free culture medium by precipitation with ethanol as previously reported (28).
Enzymatic activity assays.
Phytase activity was measured in a reaction mixture that contained the enzyme preparation, 1.6 mM sodium phytate, 100 mM Tris-HCl buffer (pH 7.5), and 1 mM CaCl2 and was incubated at 37°C for 30 min. The reaction was stopped by adding an equal volume of 15% trichloroacetic acid, and the inorganic phosphate released was measured using the ascorbic acid method (7). One unit of phytase activity was defined as the amount of enzyme required to liberate 1 μmol of phosphate/min from sodium phytate under the assay conditions used.
N-deglycosylation, SDS-PAGE, Western blotting, and N-terminal analyses.
Endoglycosylase Hf (Endo Hf) was used to N-deglycosylate PhyC-R and PhyC-N by incubating both phytase preparations according to the manufacturer's instructions. The N-deglycosylated phytases were analyzed by SDS-PAGE and Western blotting. A rabbit-specific Bacillus phytase C polyclonal antibody, purchased from GenScript Corporation (Scotch Plains, NJ), was used as the primary antibody for Western blot analysis. Proteins separated by SDS-PAGE were transferred to Immobilon-NC transfer membranes (Millipore, Bedford, MA) using a Trans-Blot SD semidry electrophoretic transfer cell (Bio-Rad Laboratories, Inc., Hercules, CA), blocked, and incubated overnight with the primary antibody (1:5,000). Afterwards, the membrane was washed with phosphate-buffered saline, incubated with the polyclonal goat anti-rabbit IgG-horseradish peroxidase conjugate used as the secondary antibody (Sigma-Aldrich Co., St. Louis, MO), and stained with a 3,3′,5,5′-tetramethylbenzidine liquid substrate system for membranes (Sigma-Aldrich Co., St. Louis, MO).
For N-terminal analysis, N-deglycosylated PhyC-R was resolved by 12% SDS-PAGE and blotted onto a polyvinylidene difluoride membrane (Millipore, Bedford, MA). The N-deglycosylated PhyC-R band was cut out and subjected to sequencing of the 10 amino-terminal residues by automated Edman degradation in a protein sequencer (Applied Biosystems, Foster City, CA).
Biochemical characterization of recombinant and native phytase C.
The effect of pH on enzymatic activity at 37°C was determined using 250 mM glycine-HCl (pH 2.5), 360 mM sodium acetate (pH 5.5), or 100 mM Tris-HCl (pH 7.5 and 9.0). The effect of temperature on enzymatic activity was determined at various temperatures ranging from 25 to 80°C. A buffer of 100 mM Tris-HCl (pH 7.5) with sodium phytate was preincubated at the relevant temperature for 5 min, and enzyme reactions were started by adding the enzyme preparation (0.02 U/ml, CaCl2 at a final concentration of 1 mM). Relative activity was expressed as a percentage with respect to the highest value of phytase activity reached at the pH or temperature evaluated.
Enzyme kinetics were determined for PhyC-R. Phytase activity was tested using sodium phytate (ranging from 0.03 to 1.60 mM) in 100 mM Tris-HCl buffer (pH 7.5) with 1 mM CaCl2.
Thermal stability was evaluated by measuring phytase activity after incubation at 37, 60, or 80°C for 10 min in 100 mM Tris-HCl (pH 7.5) or in 360 mM sodium acetate (pH 5.5) in the presence of 1 or 5 mM CaCl2. Residual activity was calculated as phytase activity measured immediately after heat treatment and expressed as a percentage of the phytase activity before heat treatment (0.10 U/ml).
Susceptibility to shrimp digestive enzymes was tested by incubating a mixture containing the phytase preparations at 0.08 U/ml and 1× (0.045 U Nα-benzoyl-dl-arginine-p-nitroanilide [BAPNA]/ml of trypsin), 10×, or 40× shrimp digestive enzyme extract in the presence of 1 mM CaCl2-100 mM Tris-HCl (pH 7.5) at 37°C for 30 min, followed by phytase activity measurement to test for residual phytase activity. Similar assays were performed with 0.12 (1×) and 1.20 (10×) U/ml porcine trypsin (Sigma-Aldrich, St. Louis, MO) instead of the shrimp digestive enzymes. Trypsin activity determinations were carried out by evaluating amidase activity in a reaction mixture containing porcine pancreas trypsin or shrimp (Litopenaeus vannamei) digestive enzymes, 1 mM BAPNA as the substrate, 20 mM Tris-HCl (pH 7.6), and 20 mM CaCl2 at 37°C. The amount of p-nitroaniline released was monitored for 3 min by measuring the increase in absorbance at 405 nm (ɛ = 8,270 M−1 cm−1).
All results were compared between groups using analysis of variance and Tukey's multiple comparisons with a significance level of P < 0.05.
RESULTS
Construction of the expression vector pPIC9phyC.
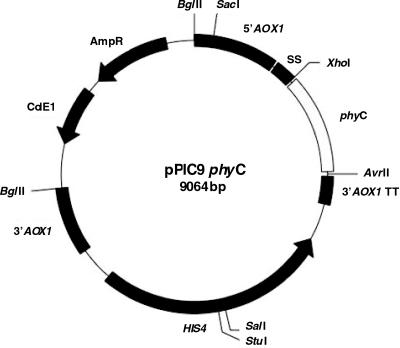
The constructed expression vector pPIC9phyC (Fig. 1) harbors the 1,062-bp phyC gene sequence in frame with the Saccharomyces cerevisiae alpha-factor pre-pro secretion signal and between the promoter and transcriptional terminator of the AOX1 gene. The forward primer, containing the XhoI site, was designed to restore the proteolytic processing site, Leu-Glu-Lys-Arg, placed prior to the first amino acid of the mature protein and to remove the sequence of Glu-Ala repeats. DNA sequencing analysis of the pPIC9phyC vector showed the integrity of the phyC gene sequence coding for the 354 amino acids of mature phytase C, in accordance with the phyC sequence from B. subtilis VTT E-68013 described previously (17).
FIG. 1.
pPIC9phyC expression vector. 5′AOX1, AOX1 promoter; 3′AOX1 TT, AOX1 transcriptional terminator; 3′AOX1, AOX1 downstream region; AmpR, ampicillin resistance gene; HIS4, P. pastoris wild-type gene coding for histidinol dehydrogenase; ColE1, E. coli origin of replication.
Expression and preparation of recombinant and native phytase C.
Transformation of P. pastoris GS115 with SalI-digested pPIC9phyC gave about 500 His+ transformants. After screening for the highest phytase activity, one colony was selected for shake flask expression. After 48 h of methanol induction, the GS115-PhyC culture showed an extracellular phytase activity of 0.82 U/ml (71 mg of PhyC-R/liter), with a specific activity of 8.9 U/mg protein and an extracellular protein concentration of 0.1 g/liter. The culture of B. subtilis VTT E-68013 showed an extracellular phytase activity of 0.40 U/ml (35 mg of PhyC-N/liter) with a specific activity of 1.2 U/mg protein. Although intracellular phytase activity was not detected in the P. pastoris strains, a dot blot analysis with the specific phytase antibody was positive (data not shown), indicating that under the culture conditions used, the secretion of phytase was not complete. However, the percentage of recombinant protein retained intracellularly was very low (less than 10%).
The enzyme preparations obtained gave 14 U/ml, with a specific activity of 11.0 U/mg protein for PhyC-R and 46.0 U/g with a specific activity of 1.4 U/mg protein for PhyC-N.
Biochemical properties of recombinant and native phytase C.
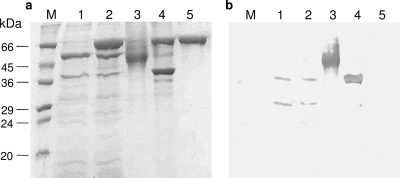
The SDS-PAGE analysis of the PhyC-N preparation showed two bands with apparent molecular sizes of nearly 39 and 58 kDa (Fig. 2a, lane 1), but neither was N-deglycosylated by Endo Hf (Fig. 2a, lane 2), since no size differences between Endo Hf-treated and untreated PhyC-N were seen. Nevertheless, PhyC-R showed a broad band with a molecular size between 45 and 66 kDa (Fig. 2a, lane 3) before N-deglycosylation treatment, which shifted to a molecular size of 39 kDa in a defined band (Fig. 2a, lane 4) after N-deglycosylation by Endo Hf. Thus, the percentage of N-glycosylation was approximately between 14 and 34%. Two bands of the PhyC-N preparation reacted positively with the specific phytase C antibody in the Western blot analysis: a band of 39 kDa and a band of 32 to 34 kDa (Fig. 2b, lanes 1 and 2). For the PhyC-R preparation, both the N-glycosylated (Fig. 2b, lane 3) and N-deglycosylated (Fig. 2b, lane 4) forms showed strong positive signals in the Western blot analysis.
FIG. 2.
SDS-PAGE (a) and Western blot analysis (b) of PhyC-R and PhyC-N preparations before and after N-deglycosylation by Endo Hf: Lanes M, protein molecular mass markers; lanes 1, PhyC-N before N-deglycosylation; lanes 2, PhyC-N after N-deglycosylation; lanes 3, PhyC-R before N-deglycosylation; lanes 4, PhyC-R after N-deglycosylation; lanes 5, Endo Hf.
The N-terminal sequencing of secreted and N-deglycosylated PhyC-R confirmed the identity of the 10 N-terminal residues corresponding to mature phytase C (LSDPYHFTVN) with leucine as the first amino acid at the N-terminal end. Therefore, efficient signal sequence processing was attained.
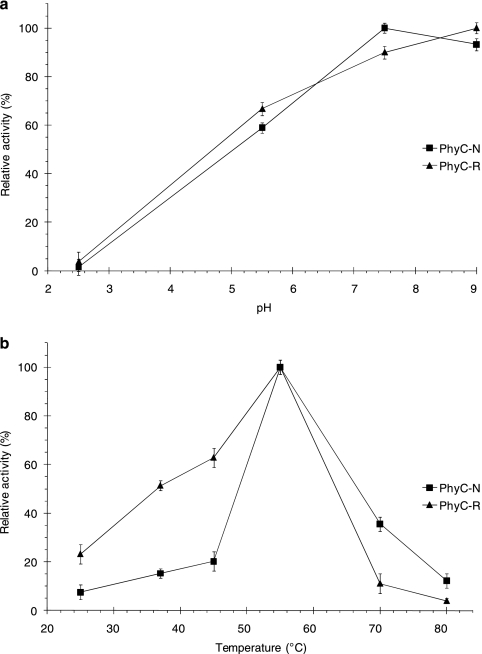
The native and recombinant phytases had similar pH-dependent activity profiles, although the pH values for maximum activity were different (7.5 for PhyC-N and 9.0 for PhyC-R). The activities of PhyC-N and PhyC-R at pH 5.5 decreased 41% and 33% from the maximum values, respectively (Fig. 3a). Although both phytases had an optimal temperature of 55°C (Fig. 3b), these enzymes had different temperature profiles. While PhyC-N displayed a sharp decrease in relative activity at temperatures other than 50°C, PhyC-R showed a relative activity decrease that was less acute and had higher relative activities at temperatures below 55°C, in comparison to those shown by PhyC-N.
FIG. 3.
Effects of pH (a) and temperature (b) on the phytase activity of PhyC-R and PhyC-N preparations. Phytase activities are expressed as relative values. The data are the means ± the standard deviations of at least three independent assays.
PhyC-R followed typical Michaelis-Menten kinetics. Using the Lineweaver-Burk plot, a Km value of 0.45 mM was estimated for phytate hydrolysis.
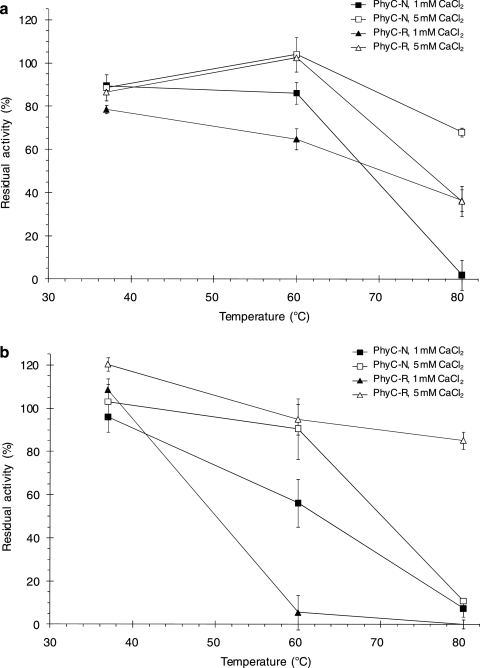
Thermostability (expressed as residual activity) after a 10-min heat treatment at 37, 60, or 80°C in 1 or 5 mM CaCl2 at pH 7.5 or 5.5 (Fig. 4 a and b) was pH dependent and Ca2+ concentration dependent and was different for the two phytases. Although PhyC-N had residual activity at pH 7.5, 80°C, and 5 mM CaCl2 that was higher than that of PhyC-R (68 versus 36%), PhyC-R was far more stable at pH 5.5, 80°C, and 5 mM CaCl2 than PhyC-N (85 versus 11%). PhyC-R showed less thermal stability than did PhyC-N at 60°C in 1 mM CaCl2 at both pH values; however, both phytases were stable at 60°C when the assays were carried out at pH 5.5 or 7.5 in 5 mM CaCl2. Although PhyC-N showed extensive denaturation at 80°C and pH 5.5, even in the presence of 5 mM CaCl2, PhyC-R exhibited high residual activity at this temperature and pH, but only in the presence of 5 mM CaCl2 (Fig. 4b). PhyC-R was partially denatured at 80°C and pH 7.5, even in the presence 5 mM CaCl2, exhibiting the same low residual activity in both calcium concentrations tested (Fig. 4a). Under the same conditions, PhyC-N was completely denatured in 1 mM CaCl2, while at 5 mM CaCl2 the thermal stability of PhyC-N was improved (Fig. 4a).
FIG. 4.
Residual phytase activities of PhyC-R and PhyC-N preparations after heat treatment at pH 7.5 (a) and pH 5.5 (b). The data are the means ± the standard deviations of at least three independent assays.
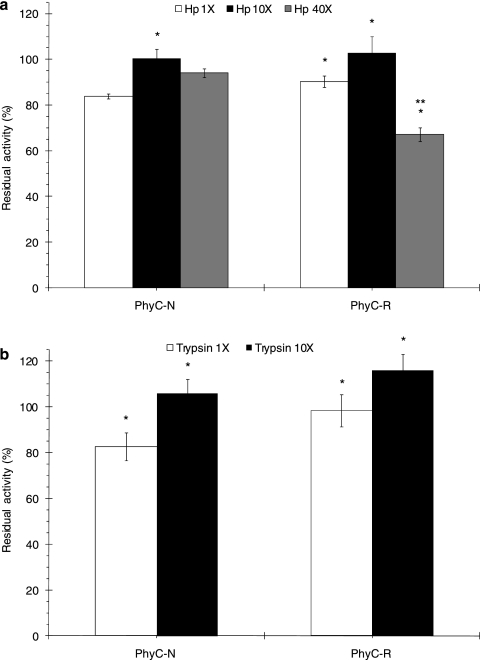
Both phytases were resistant to shrimp digestive enzymes and to porcine trypsin. The residual activities of PhyC-N and PhyC-R after shrimp digestive enzyme treatment were higher than 84% and higher than 67%, respectively (Fig. 5a). After 30 min of porcine trypsin treatment, PhyC-N and PhyC-R showed residual activities that were higher than 83% and 98%, respectively (Fig. 5b). Residual activity increases were observed when the protease concentration was increased from 1× to 10× (Fig. 5a and b): 16% for PhyC-N and 13% for PhyC-R after shrimp digestive enzyme incubation and 23% for PhyC-N and 18% for PhyC-R after porcine trypsin treatment. However, a 33% depletion of residual activity was observed after 40× shrimp digestive enzyme treatment of PhyC-R (Fig. 5a).
FIG. 5.
Residual phytase activities of PhyC-R and PhyC-N preparations after shrimp digestive enzyme (Hp) treatment (a) and after porcine trypsin treatment (b). The data are the means ± the standard deviations of at least three independent assays. *, P < 0.05 (differences for the effect of the protease treatment concentration on each phytase). **, P < 0.05 (differences between PhyC-R and PhyC-N at the same protease treatment concentration).
The results show that Ca2+ has a stabilizing effect that protects the recombinant phytase against thermal denaturation, as described elsewhere for native Bacillus phytases (15, 19). Although some slight differences between the temperature and pH activity profiles and stabilities of PhyC-N and PhyC-R were observed, the overall properties of the two phytases were similar under the conditions tested. The thermal stability profiles of the two enzymes were significantly different, maybe due to the N-glycosylation present in PhyC-R, as has been described for other proteins (3, 13).
DISCUSSION
The phytases from Bacillus species are beta-propeller phytases requiring calcium for activity and stability and exhibiting an optimum pH between 6.0 and 9.0 and are suitable for animals with neutral digestive tracts. The stability of Bacillus phytases in a high temperature range, between 60 and 95°C, is another important and useful characteristic for their application as animal feed additives because the process of pelleting uses steam and high temperatures (8, 27).
In this paper, we report the cloning and expression of the phyC gene from B. subtilis VTT E-68013 using P. pastoris as the host. After 48 h of methanol induction in shake flasks, the selected P. pastoris GS115-PhyC strain produced and secreted an N-glycosylated form of phytase C with an extracellular phytase activity of 0.82 U/ml (71 mg of PhyC-R/liter) and a specific activity of 8.9 U/mg protein, representing 68% of the total extracellular protein (0.1 g/liter). At present, while the bioprocess has not yet been fully optimized, we are producing PhyC-R using a 5-liter bioreactor yielding up to 12.5 U/ml phytase activity (1 g of PhyC-R/liter) in the cell-free culture medium (unpublished results).
Under our assay conditions, the culture of B. subtilis VTT E-68013 showed 50% less extracellular phytase activity (0.4 U/ml) than the GS115-PhyC strain. The low specific activity (1.2 U/mg protein) indicates that a high concentration of proteins other than phytase were present in the culture medium from the B. subtilis culture. The extracellular phytase activity from the B. subtilis VTT E-68013 culture was similar to or higher than the activity reported by other authors for other Bacillus strains cultured on the shake flask scale (6, 17, 28).
The PhyC-N preparation (Fig. 2a, lanes 1 and 2) had another main protein of around 58 kDa and other proteins in smaller proportions. The 58-kDa band of the PhyC-N preparation was not a phytase, as indicated by the Western blot analysis (Fig. 2b, lanes 1 and 2). Nevertheless, this analysis also showed that the PhyC-N preparation had a 32- to 34-kDa band, probably from the degradation of PhyC-N by B. subtilis proteases. Although proteins other than phytase were present in the PhyC-N preparation, the biochemical properties of PhyC-N were found to be very similar to those described in the literature for native PhyC (15, 17). Furthermore, all of the biochemical properties described in this paper are relative to the maximum value (Fig. 3) or to the initial value (Fig. 4 and 5); therefore, any effect of impurities in the phytase preparation would not be expected to affect the relative values of the biochemical properties.
Heterologous expression of Bacillus phytases has been used as an alternative enzyme source because of the low production levels and high protease activity exhibited by wild strains, in addition to high levels of secreted proteins other than phytases. The phyC gene from B. subtilis VTT E-68013 has been expressed in Lactobacillus plantarum 755 (18), although the recombinant phytase was produced at very low levels. The expression of Bacillus phytases in E. coli resulted in the formation of inclusion bodies (17, 29), which required downstream processing to recover the active enzyme (29). Recombinant Bacillus phytase production with the B. subtilis expression system has been studied by several authors. Bacillus sp. strain DS11 phytase was produced with an activity of 2 U/ml (21), B. subtilis VTT E-68013 phytase was overexpressed at an activity of 47.7 U/ml (37), and the phytase gene (phyL) from B. licheniformis and a phytase gene (168phyA) identified from B. subtilis 168 were overexpressed at activity levels of up to 35 U/ml (36).
Recently, the extracellular expression of a phyC gene from B. subtilis WHNB02 in P. pastoris was reported as yielding 2.4 U/ml phytase (38). This phytase differs from the phytase from B. subtilis VTT E-68013 at 24 amino acid residues. The same research group designed and synthesized the phyC gene from B. subtilis WHNB02 according to the P. pastoris codon usage bias without altering the protein sequence (40). The yield of total extracellular phytase activity after 120 h of methanol induction was 18.5 U/ml in wheat bran extract induction medium in a high-cell-density culture (OD600, ∼80) on the shake flask scale. Several factors, like differences in amino acid sequence, secretion efficiency, and a better overproducer strain, with improved culture conditions and codon usage improvement, could explain the different production levels, in comparison to our findings. In any case, our culture conditions rendered a high percentage of extracellular phytase (68% of the total protein), allowing phytase preparations to be recovered with a high degree of purity using only a downstream ultrafiltration process. The SDS-PAGE analysis of N-glycosylated PhyC-R (Fig. 2a, lane 3) showed a broad band that could not be used to observe nonphytase proteins. Nevertheless, in the N-deglycosylated PhyC-R analysis (Fig. 2a, lane 4), two bands were mainly observed: a 70-kDa band from Endo Hf (compare lanes 4 and 5) and another band of 39 kDa from N-deglycosylated PhyC-R, indicating that the PhyC-R preparation had a high degree of purity. In addition, since the PhyC-R preparation obtained by ultrafiltration had a specific activity of 11.0 U/mg protein and PhyC-R purified by anion-exchange chromatography has a specific activity of 11.5 ± 1.1 U/mg protein (unpublished results), the PhyC-R preparation appeared to have a high degree of purity.
In this paper, we also describe the influence of N-glycosylation on the biochemical properties of the Bacillus phytase by comparing the properties of the N-glycosylated recombinant phytase to those of the corresponding nonglycosylated native phytase, showing the influence of pH on the thermostability of the recombinant and native Bacillus phytases and showing the resistance of both phytases to shrimp digestive enzymes and porcine trypsin. These findings are valuable for decisions on how to best handle a recombinant Bacillus phytase for applications as feed additives or for industrial food processes.
The NetNGlyC 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/) predicted four potential N-glycosylation sites for PhyC-R (residues N121, N281, N138, and N243). In any case, experimental data are needed to verify whether or not a recombinant protein is N-glycosylated by the host, since the presence of N-glycosylation sites is not sufficient to conclude that an asparagine residue would be N-glycosylated. Our results from Endo Hf treatment clearly demonstrate that PhyC-N is non-N-glycosylated, while PhyC-R is N-glycosylated. The N-glycosylation pattern of PhyC-R should be similar to that described for other recombinant proteins produced in P. pastoris (4, 24, 25). Two of the four N-glycosylation sites are more likely to be N-glycosylated, since N138 and N243 are at the protein surface, based on the three-dimensional structure of B. amyloliquefaciens phytase (Protein Data Bank codes 1POO, 2POO, 1QLG, 1CVM, and 1H6L) (11, 34). After N-deglycosylation by Endo Hf, PhyC-R showed a molecular mass of 39 kDa, the same as that of the native form (Fig. 2a, lane 4). The correspondence between the 10 N-terminal residues of PhyC-R and those of mature phytase C supports the protein's identification and the efficiency of the signal sequence processing.
PhyC-N and PhyC-R have exactly the same amino acid sequence. Therefore, any differences in biochemical proprieties between the two phytases (maximum-activity pH values, temperature-dependent activity profiles, and thermostability) must be due to their differing degrees of N-glycosylation. Although both phytases displayed an optimal temperature of 55°C, PhyC-R had a higher relative activity than PhyC-N over a wider temperature range, which would be advantageous for possibly diverse applications. N-glycosylation affects biochemical properties such as molecular mass, isoelectric point, and pH optima (10) and changes the charge distribution on the glycoprotein's surface, which has been proposed to explain this effect (10, 35).
Native and recombinant phytases showed high residual activity at 37°C for at least 2 h, even after storage at −20°C for at least a year, in both cases, in the presence of 5 mM CaCl2. This information is important for the efficient handling of recombinant Bacillus phytases. In addition, the thermostability of the phytases was calcium concentration dependent, as reported by others (15). At 60 and 80°C, the two phytases had different stabilities, which were pH dependent. The influence of pH and the effect of buffer composition on heat treatment have been reported by some authors to affect protein thermal denaturation (3, 30). Likely, the instability of PhyC-N at pH 5.5 and 80°C is caused by disruption of the interactions between Ca2+ ions and amino acid residues that confer structural stability on the protein. Three calcium ions interact with 11 amino acid residues to confer the structural stability of calcium-dependent phytases, and of these 11 residues, 2 are glutamates and 4 are aspartates (11, 34). Because the pI of PhyC-N is 6.6 (17), the negative net charge of the protein would change to a positive net charge when the pH changes from 7.5 to 5.5. Protonation of amino acid residues in PhyC-N at pH 5.5 would lead to partial disruption of the salt bridges and interactions with the Ca2+ ions. In addition, the influence of pH on the unfolding process would be enhanced by the disruption of hydrogen bonds and nonpolar hydrophobic interactions caused by heat treatment. Under the same denaturation conditions, PhyC-R showed high thermal stability, with 85% residual activity, in comparison to only 11% for PhyC-N. This indicates that the N-glycosylation in PhyC-R enhanced the protein's thermal stability at pH 5.5, possibly by stabilizing the interactions with Ca2+ ions. The probable protonation of some amino acid residues on PhyC-R at pH 5.5 that would be absent at pH 7.5 could produce a different surface distribution charge at pH 7.5, compared to that at pH 5.5, which could lead to differences in the kind and number of interactions between carbohydrate chains and the protein surface, rendering PhyC-R more thermostable at pH 5.5.
PhyC-R was more stable at pH 5.5, 80°C, and 5 mM CaCl2 than PhyC-N was (85% versus 11%), which is an important property since the enzyme is incorporated into many feeds prior to pelleting and after the feed reaches a temperature near 80°C for 2 min during the feed-pelleting process (19, 39).
Phytase C from B. subtilis has been shown to be resistant to papain, pancreatin, and trypsin, even under conditions of calcium depletion. Nevertheless, this phytase has been found to be susceptible to pepsin (15), an effect that was explained by the denaturation of the enzyme at low pH, making it more susceptible to pepsin. In the present work, we tested the susceptibility of Bacillus phytases to shrimp (L. vannamei) digestive enzymes. Both phytases, PhyC-N and PhyC-R, were resistant to shrimp digestive enzymes (residual activities of 67 to 100%) and to exposure to porcine trypsin (activity above 83%). In addition, both phytases showed increased residual activities when the concentrations of the protease preparations were increased from 1× to 10×. This result may be due to conformational changes occurring in the phytases because of the hydrolysis of some peptide bonds. An increase in residual activity (30%) due to exposure to proteolytic enzymes (pepsin) has been previously described for the phytase r-AppA from E. coli (32). The authors concluded that stable polypeptides of r-AppA with phytase activity were generated after pepsin treatment.
In conclusion, the recombinant form of the neutral phytase C from B. subtilis VTT E-68013 produced in P. pastoris retained biochemical properties that were similar to those of the native form, except for thermal stability. The different characteristics, in terms of the maximum-activity pH and the temperature-dependent activity profile, give advantages to PhyC-R for the diversification of phytase applications. Moreover, PhyC-R is resistant to thermal denaturation and to shrimp digestive enzyme or porcine trypsin treatment. Our findings indicate that PhyC-R is more suitable than the commercially available Aspergillus and E. coli phytases as a feed additive for species with basic or neutral digestive tracts, such as agastric aquaculture species (shrimp and several fishes), since PhyC-R has an optimum pH near neutrality, while the Aspergillus and E. coli phytases have optimum pHs in the acidic range and the pHs of the digestive systems from agastric aquaculture species are near 7. Furthermore, the Aspergillus and E. coli phytases are less thermostable than PhyC-R. Therefore, PhyC-R would enhance the utilization of plant-derived feedstuffs for aquaculture species, improving the utilization of phytate-phosphorus in diets and reducing their manure phosphorus excretion into the environment. In addition, PhyC-R can be used in industrial food processes under gentle conditions (neutral pH and low temperatures) for phytate hydrolysis.
Acknowledgments
This study was supported by grants CN1114-05 (PAICYT) from the Universidad Autónoma de Nuevo León and 2003-02-141 (CONACYT-SAGARPA). J.G.C.-T. and J.A.G.-L. thank the Consejo Nacional de Ciencia y Tecnología (CONACYT) for their fellowship, and L.R.-B. thanks PROMEP-UJAT for her fellowship.
We thank Programa Maricultura, Facultad de Ciencias Biológicas, Universidad Autónoma de Nuevo León, for providing L. vannamei specimens used for the preparation of the shrimp digestive gland extract and José Antonio Fuentes-Garibay and Mauricio Castillo-Galván for technical support. We are grateful to Glen D. Wheeler for his stylistic suggestions in the preparation of the manuscript.
Footnotes
Published ahead of print on 2 July 2010.
REFERENCES
- 1.Cao, L., W. Wang, C. Yang, Y. Yang, J. Diana, A. Yakupitiyage, Z. Luo, and D. Li. 2007. Application of microbial phytase in fish feed. Enzyme Microb. Technol. 40:497-507. [Google Scholar]
- 2.Choi, Y. M., H. J. Suh, and J. M. Kim. 2001. Purification and properties of extracellular phytase from Bacillus sp. KHU-10. J. Protein Chem. 20:287-292. [DOI] [PubMed] [Google Scholar]
- 3.Clark, S. E., E. H. Muslin, and C. H. Henson. 2004. Effect of adding and removing N-glycosylation recognition sites on the thermostability of barley α-glucosidase. Protein Eng. Des. Sel. 17:245-249. [DOI] [PubMed] [Google Scholar]
- 4.Daly, R., and M. T. Hearn. 2005. Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production. J. Mol. Recognit. 18:119-138. [DOI] [PubMed] [Google Scholar]
- 5.Ecamilla-Treviño, L. L., J. M. Viader-Salvadó, H. A. Barrera-Saldaña, and M. Guerrero-Olazarán. 2000. Biosynthesis and secretion of recombinant human growth hormone in Pichia pastoris. Biotechnol. Lett. 22:109-114. [Google Scholar]
- 6.Farhat, A., H. Chouayekh, M. B. Farhat, K. Bouchaala, and S. Bejar. 2008. Gene cloning and characterization of a thermostable phytase from Bacillus subtilis US417 and assessment of its potential as a feed additive in comparison with a commercial enzyme. Mol. Biotechnol. 40:127-135. [DOI] [PubMed] [Google Scholar]
- 7.Fiske, C. H., and Y. Subarrow. 1925. The colorimetric determination of phosphorus. J. Biol. Chem. 66:375-400. [Google Scholar]
- 8.Fu, S., J. Sun, L. Qian, and Z. Li. 2008. Bacillus phytases: present scenario and future perspectives. Appl. Biochem. Biotechnol. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 9.Greiner, R., and U. Konietzny. 2006. Phytase for food application. Food Technol. Biotechnol. 44:125-140. [Google Scholar]
- 10.Guo, M., H. Hang, T. Zhu, Y. Zhuang, J. Chu, and S. Zhang. 2008. Effect of glycosylation on biochemical characterization of recombinant phytase expressed in Pichia pastoris. Enzyme Microb. Technol. 42:340-345. [Google Scholar]
- 11.Ha, N. C., B. C. Oh, S. Shin, H. J. Kim, T. K. Oh, Y. O. Kim, K. Y. Choi, and B. H. Oh. 2000. Crystal structures of a novel thermostable phytase in partially and fully calcium-loaded states. Nat. Struct. Biol. 7:147-153. [DOI] [PubMed] [Google Scholar]
- 12.Haefner, S., A. Knietsch, E. Scholten, J. Braun, M. Lohscheidt, and O. Zelder. 2005. Biotechnological production and applications of phytases. Appl. Microbiol. Biotechnol. 68:588-597. [DOI] [PubMed] [Google Scholar]
- 13.Han, Y. M., and X. G. Lei. 1999. Role of glycosylation in the functional expression of an Aspergillus niger phytase (phyA) in Pichia pastoris. Arch. Biochem. Biophys. 364:83-90. [DOI] [PubMed] [Google Scholar]
- 14.Ilgen, C., J. Lin-Cereghino, and J. M. Cregg. 2004. Pichia pastoris, p. 143-162. In G. Gellissen (ed.), Production of recombinant proteins—novel microbial and eukaryotic expression systems. Wiley-VCH, Weinheim, Germany.
- 15.Kerovuo, J., I. Lappalainen, and T. Reinikainen. 2000. The metal dependence of Bacillus subtilis phytase. Biochem. Biophys. Res. Commun. 268:365-369. [DOI] [PubMed] [Google Scholar]
- 16.Kerovuo, J., J. Rouvinen, and F. Hatzack. 2000. Analysis of myo-inositol hexakisphosphate hydrolysis by Bacillus phytase: indication of a novel reaction mechanism. Biochem. J. 352:623-628. [PMC free article] [PubMed] [Google Scholar]
- 17.Kerovuo, J., M. Lauraeus, P. Nurminen, N. Kalkkinen, and J. Apajalahti. 1998. Isolation, characterization, molecular gene cloning, and sequencing of a novel phytase from Bacillus subtilis. Appl. Environ. Microbiol. 64:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerovuo, J., and S. Tynkkynen. 2000. Expression of Bacillus subtilis phytase in Lactobacillus plantarum 755. Lett. Appl. Microbiol. 30:325-329. [DOI] [PubMed] [Google Scholar]
- 19.Kim, D. H., B. C. Oh, W. C. Choi, J. K. Lee, and T. K. Oh. 1999. Enzymatic evaluation of Bacillus amyloliquefaciens phytase as a feed additive. Biotechnol. Lett. 20:925-927. [Google Scholar]
- 20.Kim, Y. O., H. K. Kim, K. S. Bae, J. H. Yu, and T. K. Oh. 1998. Purification and properties of a thermostable phytase from Bacillus sp. DS11. Enzyme Microb. Technol. 22:2-7. [Google Scholar]
- 21.Kim, Y. O., J. K. Lee, B. C. Oh, and T. K. Oh. 1999. High-level expression of a recombinant thermostable phytase in Bacillus subtilis. Biosci. Biotechnol. Biochem. 63:2205-2207. [DOI] [PubMed] [Google Scholar]
- 22.Lei, X. G., J. M. Porres, E. J. Mullaney, and H. Brinch-Pedersen. 2007. Phytase: source, structure and application, p. 505-529. In J. Polaina and A. P. MacCabe (ed.), Industrial enzymes: structure, function and applications. Springer, Dordrecht, Netherlands.
- 23.Liebert, F., and L. Portz. 2005. Nutrient utilization of Nile tilapia Oreochromis niloticus fed plant based low phosphorus diets supplemented with graded levels of different sources of microbial phytase. Aquaculture 248:111-119. [Google Scholar]
- 24.Macauley-Patrick, S., M. L. Fazenda, B. McNeil, and L. M. Harvey. 2005. Heterologous protein production using the Pichia pastoris expression system. Yeast 22:249-270. [DOI] [PubMed] [Google Scholar]
- 25.Montesino, R., R. García, O. Quintero, and J. A. Cremata. 1998. Variation in N-linked oligosaccharide structures on heterologous proteins secreted by the methylotrophic yeast Pichia pastoris. Protein Expr. Purif. 14:197-207. [DOI] [PubMed] [Google Scholar]
- 26.Mullaney, E. J., and A. H. Ullah. 2007. Phytases: attributes, catalytic mechanisms and applications, p. 97-110. In L. Turner, A. E. Richardson, and E. J. Mullaney (ed.), Inositol phosphates: linking agriculture and the environment. CAB International, Oxfordshire, United Kingdom.
- 27.Oh, B. C., W. C. Choi, S. Park, Y. O. Kim, and T. K. Oh. 2004. Biochemical properties and substrate specificities of alkaline and histidine acid phytases. Appl. Microbiol. Biotechnol. 63:362-372. [DOI] [PubMed] [Google Scholar]
- 28.Powar, V. K., and V. Jagannathan. 1982. Purification and properties of phytase-specific phosphatase from Bacillus subtilis. J. Bacteriol. 151:102-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao, D. E., K. V. Rao, and V. D. Reddy. 2008. Cloning and expression of Bacillus phytase gene (phy) in Escherichia coli and recovery of active enzyme from the inclusion bodies. J. Appl. Microbiol. 105:1128-1137. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez, E., E. J. Mullaney, and X. G. Lei. 2000. Expression of the Aspergillus fumigatus phytase gene in Pichia pastoris and characterization of the recombinant enzyme. Biochem. Biophys. Res. Commun. 268:373-378. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez, E., J. M. Porres, Y. Han, and X. G. Lei. 1999. Different sensitivity of recombinant Aspergillus niger phytase (r-PhyA) and Escherichia coli pH 2.5 acid phosphatase (r-AppA) to trypsin and pepsin in vitro. Arch. Biochem. Biophys. 365:262-267. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez, E., Z. A. Wood, P. A. Karplus, and X. G. Lei. 2000. Site-directed mutagenesis improves catalytic efficiency and thermostability of Escherichia coli pH 2.5 acid phosphatase/phytase expressed in Pichia pastoris. Arch. Biochem. Biophys. 382:105-112. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Shin, S., N. C. Ha, B. C. Oh, T. K. Oh, and B. H. Oh. 2001. Enzyme mechanism and catalytic property of beta propeller phytase. Structure 9:851-858. [DOI] [PubMed] [Google Scholar]
- 35.Spiro, R. G. 2002. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12:43R-56R. [DOI] [PubMed] [Google Scholar]
- 36.Tye, A. J., F. K. Siu, T. Y. Leung, and B. L. Lim. 2002. Molecular cloning and the biochemical characterization of two novel phytases from B. subtilis 168 and B. licheniformis. Appl. Microbiol. Biotechnol. 59:190-197. [DOI] [PubMed] [Google Scholar]
- 37.Vuolanto, A., N. V. Weymarn, J. Kerovuo, H. Ojamo, and M. Leisola. 2001. Phytase production by high cell density culture of recombinant Bacillus subtilis. Biotechnol. Lett. 23:761-766. [Google Scholar]
- 38.Wang, H. N., Q. Wu, H. X. Zhao, H. Chen, and P. Liu. 2005. Secretory expression of Bacillus subtilis phytase phyC in Pichia pastoris. J. Zhejiang Univ. (Agric. Life Sci.) 31:621-627. [Google Scholar]
- 39.Wyss, M., L. Pasamontes, R. Rémy, J. Kohler, E. Kusznir, M. Gadient, F. Müller, and A. P. G. M. van Loon. 1998. Comparison of the thermostability properties of three acid phosphatases from molds: Aspergillus fumigatus phytase, A. niger phytase, and A. niger pH 2.5 acid phosphatase. Appl. Environ. Microbiol. 64:4446-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zou, L. K., H. N. Wang, X. Pan, T. Xie, Q. Wu, Z. W. Xie, and W. R. Zhou. 2006. Design and expression of synthetic phyC gene encoding the neutral phytase in Pichia pastoris. Acta Biochim. Biophys. Sin. (Shanghai) 38:803-811. [DOI] [PubMed] [Google Scholar]