Abstract
The physiology of filamentous fungi at growth rates approaching zero has been subject to limited study and exploitation. With the aim of uncoupling product formation from growth, we have revisited and improved the retentostat cultivation method for Aspergillus niger. A new retention device was designed allowing reliable and nearly complete cell retention even at high flow rates. Transcriptomic analysis was used to explore the potential for product formation at very low specific growth rates. The carbon- and energy-limited retentostat cultures were highly reproducible. While the specific growth rate approached zero (<0.005 h−1), the growth yield stabilized at a minimum (0.20 g of dry weight per g of maltose). The severe limitation led to asexual differentiation, and the supplied substrate was used for spore formation and secondary metabolism. Three physiologically distinct phases of the retentostat cultures were subjected to genome-wide transcriptomic analysis. The severe substrate limitation and sporulation were clearly reflected in the transcriptome. The transition from vegetative to reproductive growth was characterized by downregulation of genes encoding secreted substrate hydrolases and cell cycle genes and upregulation of many genes encoding secreted small cysteine-rich proteins and secondary metabolism genes. Transcription of known secretory pathway genes suggests that A. niger becomes adapted to secretion of small cysteine-rich proteins. The perspective is that A. niger cultures as they approach a zero growth rate can be used as a cell factory for production of secondary metabolites and cysteine-rich proteins. We propose that the improved retentostat method can be used in fundamental studies of differentiation and is applicable to filamentous fungi in general.
The filamentous fungus Aspergillus niger is one of the most important microorganisms in industrial biotechnology because of its ability to produce high levels of enzymes and organic acids (6, 50, 57). Apart from their biotechnological importance, filamentous fungi in the Aspergillus section Nigri (the black aspergilli) represent some of the most widespread storage molds which contaminate food and feedstocks with mycotoxins (26, 37, 48).
Both metabolic engineering approaches and the search for optimal cultivation conditions have long been used to improve A. niger as a production host (e.g., 14, 22, 41). With the availability of the A. niger genome sequence (50), systems biology tools are being developed (4, 5, 33) which, together with new efficient methods for constructing gene knockout mutants (43), open new possibilities for further improvement of A. niger as a cell factory.
A major ongoing challenge for microbial production processes is to minimize the amount of biomass formed while maintaining high productivity. Solutions to uncouple product formation from biomass accumulation or growth are therefore highly desirable. However, production at zero growth is difficult to achieve when nutrients are supplied to allow formation of a desired product. Carbon- and energy-limited retentostat cultivation is a method that makes it possible to approach a condition of zero growth with a stable supply of nutrients (11, 68). A retentostat culture is basically a chemostat culture to which cell retention is applied. This leads to transient biomass accumulation in the culture while at the same time allowing metabolites and other soluble molecules to leave with the dilute effluent. The zero-growth condition is based on the concept of maintenance energy (52), which suggests that a cell uses a certain minimum amount of energy to sustain basal household processes and viability. Thus, theory predicts that carbon- and energy-limited retentostat cultures will approach a zero growth rate as the energy source consumed by the individual cell nears its maintenance ration (68). Previous studies of product formation in carbon- and energy-limited retentostat cultures of A. niger (60, 61, 69) have focused on products associated with vegetative growth, such as the major secreted glycoprotein glucoamylase and organic acids. It was also noted that A. niger was subject to differentiation as it approached a growth rate of zero (69).
The physiology of A. niger and other filamentous fungi at specific growth rates approaching zero has been subject to limited study and exploitation. During subaerial growth, A. niger produces chains of black spores from biseriate conidiophores (55), but like other molds (15, 16, 56, 62) it also conidiates in submerged culture in response to severe nitrogen or carbon limitation (15, 27, 46). More recent molecular studies have shown that the transcription factor BrlA plays a central role as a positive regulator of conidiation in liquid as well as aerial environments (1, 2, 25, 40, 67, 74) and that nutrient starvation is associated with induction of brlA transcription (62).
This study revisits carbon- and energy-limited retentostat cultivation of A. niger with the aim of increasing our knowledge of its physiology during differentiation and to explore the perspectives for product formation at growth rates near zero. We present a new device which allowed efficient and reliable retention of a filamentous microorganism at high flow rates. Maltose-limited retentostat cultivation of A. niger induced hyphal compartmentalization and submerged conidiation, and the continuous supply of substrate fuelled the differentiation processes. Genome-wide transcriptional analysis was applied to three physiologically distinct phases during retentostat cultivation. The transcriptomes were used to identify leads for new products which can be efficiently formed at low growth rates during submerged asexual development. The transcriptomic analysis revealed high-level transcription of genes encoding small cysteine-rich proteins and suggested adaptation of the secretory pathway to facilitate their processing. In addition, several gene clusters with apparent but currently uncharacterized roles in isoprene, polyketide, and nonribosomal peptide synthesis became transcriptionally activated. Our results indicate that retentostat cultivation is a promising method to study asexual development and to produce conidiospores and sporulation related molecules.
MATERIALS AND METHODS
Strain and inoculum.
A. niger N402 (cspA), a standard laboratory strain with short conidiophores (13), was propagated on solidified complete medium (CM) to produce conidial inoculum for submerged cultivation. CM agar plates contained the following (per liter):10.0 g of glucose, 6.0 g of NaNO3, 1.5 g of KH2PO4, 0.5 g of KCl, 0.5 g of Mg SO4·7H2O, 1.0 g of Casamino Acids, 5.0 g of yeast extract, 20 g of agar, and 1 ml of trace metal solution. The trace metal solution contained the following per liter: 10 g of EDTA, 4.4 g of ZnSO4·7H2O, 1.01 g of MnCl2·4H2O, 0.32 g of CoCl2·6H2O, 0.315 g of CuSO4·5H2O, 0.22 g of (NH4)6Mo7O24·4H2O, 1.47 g of CaCl2·2H2O, and 1 g of FeSO4·7H2O (modified from composition given by Vishniac and Santer [71]). The pH of the medium was adjusted to 5.8 with NaOH prior to autoclaving. The solid medium cultures were incubated for at least 6 days at 30°C to allow adequate conidium formation. Spore plates were stored for no more than 3 months at 4°C. For preparation of inoculum, conidia were harvested and washed with a sterile detergent solution containing 0.05% (wt/vol) Tween 80 and 0.9% (wt/vol) NaCl.
Batch cultures.
Bioreactors holding 5 liters of minimal medium (MM) were inoculated with conidial suspension to give 109 conidia liter−1. The MM contained the following (per liter): 4.5 g of NH4Cl, 1.5 g of KH2PO4, 0.5 g of KCl, 0.5 g of MgSO4·7H2O, and 1 ml of trace metal solution (described above). The pH was adjusted to 3. The final cell density-limiting (growth-limiting) substrate was 4 g of maltose (monohydrate) per liter. Concentrated solutions (20×) of carbon source were heat sterilized separately and added aseptically to sterile MM. During cultivation, the temperature was 30°C at pH 3, kept constant by computer-controlled addition of 2 M NaOH. Sterile air was supplied at 1 liter min−1 through a ring sparger. Dissolved oxygen tension was above 40% of air saturation at any time, ensuring oxygen-sufficient growth. Germination of the dormant conidia was induced by addition of 0.003% (wt/wt) yeast extract before inoculation. During the first 6 h of cultivation, the culture was aerated through the headspace of the reactor, and stirrer speed was kept low at 250 rpm. These precautions minimized the gas-liquid interface, thereby reducing loss of hydrophobic conidia to the exhaust gas. After 6 h and germination of most conidia (now hydrophilic), air was sparged into the culture broth, and mixing was intensified (750 rpm) for more efficient mass transfer. At this stage, 0.01% (vol/vol) polypropylene glycol (PPG) P2000 was added as an antifoaming agent. Submerged cultivation was performed in 6.6-liter BioFlo3000 bioreactors (New Brunswick Scientific, NJ). Culture mass was monitored with a Mettler Toledo KCC150s scale, which has a maximum load capacity of 150 kg and an accuracy of 10 g. The glass surface of the headspace was cooled to minimize wall growth.
Cell retention.
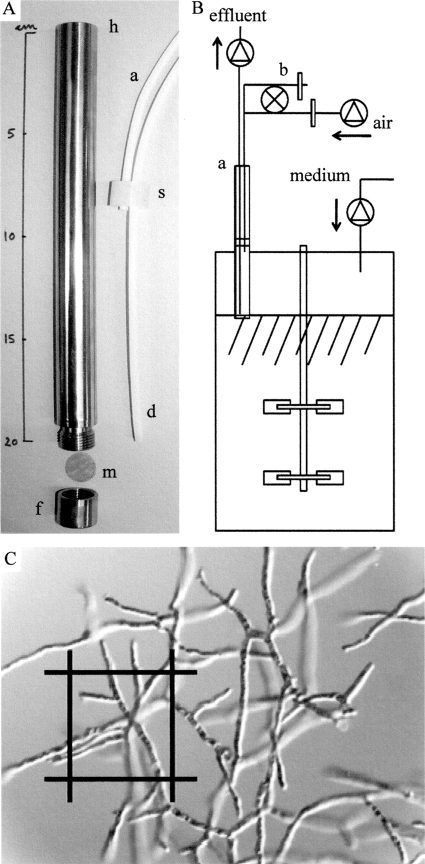
A special cell retention device (Fig. 1 A) was developed to enable efficient separation of filamentous biomass and allow reliable operation of retentostat cultures (this study). The retention principle is simple (Fig. 1B and C). Dilute culture broth rises, by unforced filtration over a coarse filter, into a cylinder inserted from the top plate (through existing 19-mm injection port). The dilute is constantly drained from the cylinder, thereby maintaining constant culture volume. The vertical position of the filter can be adjusted aseptically to facilitate filtration as biomass density increases. An antifouling mechanism prevents blockage of the filter by intermittent sparging (2 s every 12 s) sterile air (2 liter min−1) through the cylinder and filter. Biomass retention was nearly complete (95 to 99%), and filtrate could be drained at a rate of at least 0.25 liters h−1 (0.19 liters h−1 per cm2 of filter area) at biomass densities of at least 10 g of dry weight (gDW) per liter of culture broth (see Fig. SA1 in the supplemental material). The device was constructed from durable materials with high temperature resistance; this allowed repeated use and heat sterilization. The filter housing (frame and cylinder, 19 by 200 mm) was made of stainless steel, the septum was made of silicone, and the drain tubing was made of Teflon. The filter itself was cut from sheets of Spectra Mesh (Spectrum Laboratories, CA). Stainless steel mesh with a pore size of 51 μm and 42% open area was used. The cell retention device was crafted at the Department of Biochemistry and Molecular Biology (BMB) workshop at the University of Southern Denmark according to instructions from T. R. Jørgensen.
FIG. 1.
Cell retention device for in situ filtration of mycelial culture broth. (A) Scaled picture of filter components: housing (h), mesh (m), frame nut (f), septum (s), air inlet (a), and drain (d) for continuous removal of filtrate (effluent). (B) Schematic illustration of separation principle: a, adjustable cell retention device; b, antifouling mechanism regulated by a computer controlled solenoid valve. (C) Comparison of pore size (51 μm) and organism.
Retentostat cultures.
Continuous cultivation was commenced in the late exponential growth phase of a batch culture, when 90% of the maltose had been consumed, at a biomass concentration of about 2.1 gDW kg−1 of culture. The exact time for starting the flow was based on alkali addition (32, 34). Carbon-limited MM, containing 8 g of maltose (monohydrate) per liter and 0.01% (vol/vol) PPG, was fed to the culture from two interconnected 20-liter reservoirs. The flow rate was 0.125 liter h−1, which corresponded to a dilution rate (D) of 0.025 h−1 and a substrate feed rate of 0.2 g of maltose h−1 kg−1. The cell retention device was inserted aseptically just before the medium flow was started (to avoid growth from spores in the cylinder), and the level was adjusted to maintain a culture mass of 5 kg. The corrected airflow rate during retentostat cultivation was 1.3 liters min−1. Samples were withdrawn regularly to ensure good resolution of transient variables. A total of 75 to 100 g of culture broth was harvested during a 24-h period, which is <3% of the medium in-flow. Samples were also taken of the dilute effluent from the retention device.
Online measurements.
The content of CO2 and O2 in the exhaust gas was analyzed using a Xentra 4100C Gas Purity Analyser (Servomex BV, Netherlands). The pH was measured with an autoclavable glass electrode (Mettler Toledo), and the dissolved oxygen tension was measured with an lnPro-6000 series O2 sensor (Mettler Toledo).
Sample preparation and biochemical analyses of dry cells and supernatant.
Dry weight biomass concentration was determined by weighing lyophilized mycelium separated from a known mass of culture broth. Culture broth was filtered through GF/C glass microfiber filters (Whatman). The filtrate was collected and frozen for use in solute analyses. The mycelium was washed with demineralized water, rapidly frozen in liquid nitrogen, and stored at −80°C until lyophilization. Mycelium intended for gene expression analyses was separated from culture medium and frozen in liquid nitrogen within 10 to 15 s from sampling. Samples were analyzed as follows.
(i) Analysis of dry-cell composition.
Total RNA was extracted with perchloric acid according to the method of Herbert et al. (31) and determined with the orcinol method (63) for pentoses using Saccharomyces cerevisiae RNA as a standard. Biomass protein was extracted with hot alkali (16) and determined using the alkali-compatible DC Protein Assay (Bio-Rad) with bovine serum albumin (BSA) as a standard. Lipids were quantified from chloroform and methanol extractions. Carbohydrate content was determined with the phenol-sulfuric acid method (24), and glucose was used as a standard. Melanin, which in this study refers to an alkali-soluble brown pigment which precipitates at low pH, was also quantified. Melanin was extracted with 0.5 M NaOH while incubating for at least 8 min in a bath of boiling water. Melanin from a large quantity of biomass harvested at the end of retentostat cultivation was extracted with hot alkali and purified according to the methods of de Cássia et al. (21). This was used to establish a relation between the amount of pure melanin and absorbance at 425 nm of alkaline extracts of melanin from biomass. This relation was used to determine melanin content of smaller biomass samples harvested during cultivation and extracted with hot alkali for spectrometric analysis.
(ii) Analysis of supernatant.
Total solute carbon in the supernatant was measured with a total organic carbon (TOC) analyzer (TOC-Vcsn; Shimadzu, Japan), using glucose as a standard. Extracellular protein was determined with a Quick Start Bradford protein assay (Bio-Rad) with BSA as a standard.
Microscopy.
Broth samples rapidly frozen in liquid nitrogen and stored at −80°C, were thawed on ice for microscopic analyses, as follows.
(i) Light microscopy.
Diluted samples were fixed, stained, and mounted in lactophenol blue solution (Fluka) according to the manufacturer's protocol. Micrographs of mycelium morphology were taken with a DC200 camera (Leica) using a DMRBE microscope (Leica).
(ii) SEM.
Material was fixed in 4% (wt/vol) glutaraldehyde in 0.1 M sodium cacodylate buffer for 3 h at room temperature and overnight at 4°C, rinsed in buffer, treated for 1 h with 1% (wt/vol) OsO4 in buffer, and rinsed in buffer, followed by dehydration in a graded series of ethanol from 70 to 100%. Critical-point drying was performed using a BAL-TEC CPD-030; after drying, sputtering with gold was done with a Polaron scanning electron microscope (SEM) coating unit E5100. Examination of the material was done with a JSM-6400 scanning electron microscope.
(iii) Transmission electron microscopy (TEM).
Material was fixed with 4% (wt/vol) glutaraldehyde in 0.1 M sodium cacodylate buffer for 3 h at room temperature and overnight at 4°C, rinsed in buffer, and spun down in low-melting-point agar; after blocks were cut, they were fixed in 1% (wt/vol) OsO4 for 1 h and then rinsed in buffer, followed by dehydration in a graded series of ethanol (70 to 100%), before being embedded in resin (Agar 100). Ultrathin sections were examined with a JEM-1010 transmission electron microscope and photographed.
RNA isolation and quality control.
Total RNA for Northern and microarray analyses was isolated from frozen ground mycelium by Trizol extraction according to the manufacturer's instructions. Following extraction, RNA was purified on NucleoSpin RNA II columns (Machery-Nagel), including a DNase I digestion step. RNA was eluted in 60 μl of MilliQ water. RNA quantity and quality were determined on a Nanodrop spectrophotometer, and integrity was tested on an Agilent 2100 Bioanalyser. The spectrum generated by the Agilent Bioanalyser was visually inspected for possible RNA degradation and contamination with genomic DNA to ensure good sample quality.
Microarray analysis.
Probe synthesis and fragmentation were performed at ServiceXS (Leiden, Netherlands) according to the GeneChip Expression Analysis Technical Manual (3). DSM (Delft, Netherlands) proprietary A. niger gene chips were hybridized, washed, stained, and scanned as described in the GeneChip Expression Analysis Technical Manual (3). The Affymetrix program Microarray Suite, version 5 (MAS5), was used for condensation. The 3′ to 5′ signal ratios of probe sets of internal control genes, like gpdA (glyceraldehyde-3-phosphate dehydrogenase), pkiA (pyruvate kinase), hxkA (hexokinase), and γ-actin, were below 3 on all arrays (nine arrays). The percentages of probe sets (of 14,554 probe sets) evaluated as present were 37% at day 0 (three arrays), 44% at day 2 (three arrays), and 43% at day 8 (three arrays). Day 0 array samples were from the exponential growth phase of batch cultures of the same strain under the same culture conditions as used in batch cultures prior to retentostat cultivation; the only difference was a starting substrate concentration of 8 g of maltose (monohydrate) per liter. Array data were analyzed as follows.
(i) Normalization, filtering, statistical significance, and comparisons.
Handling of microarray results and statistical comparisons were performed with GeneData Expressionist software (GeneData AG). Genes with detection calls of present (P < 0.04) or marginal (0.04 ≤ P < 0.06) in at least one of three replicate measurements were accepted as expressed and used in further analyses. Raw signal values were subjected to scaling normalization to the arithmetic mean of a reference group (day 0 sample) prior to comparison. RNA samples representing three different time points (0, 2, and 8 days of retentostat cultivation) were compared to each other; each time point was represented by three independent biological replicates. Differential gene expression was evaluated by a t test of expressed genes, multiple testing correction was applied, and a Benjamini Hochberg false discovery rate (FDR) (8) of <0.05 was used to define differentially expressed genes. The ratio of transcript levels of a gene was calculated from normalized values.
(ii) GO and enrichment analysis.
Gene Ontology (GO) annotation for the A. niger CBS513.88 genome (49) was obtained with Blast2GO (20), version 1.2.7 (available at www.Blast2GO.de). This application was also used for enrichment analysis of GO terms in uploaded lists of differentially expressed genes. Fisher's exact test with an FDR of <0.05 was used to identify overrepresented GO terms.
Northern blot analysis.
RNA (5 μg) from each sample was used for Northern blot analysis of the expression of three selected genes, An01g10540 (homolog of the central regulator of sporulation, brlA), An03g06550 (glucoamylase; glaA), and An08g09880 (probable spore-specific hydrophobin). The protocol described by Meyer et al. (44) was used. 18S rRNA was used as loading control. Hybridization probes were generated by PCR using the primers brlAP1 (5′-CCTCCATGGCTTCCAGCTT-3′) and brlAP2 (5′-CAATTGAGTCTGGCTGCGG-3′) for An01g10540 and coh1P1 (5′-CATATCCTTGCCGTTCTCGC-3′) and coh1P2 (5′-ACCTGGACGGTAAGGCAGTTC-3′) for An08g09880. For 18S rRNA hybridization, a 0.7-kb EcoRI fragment from plasmid pMN1 (12) was used as a probe, and for An03g06550, a 0.9-kb NcoI-PstI fragment from pAN56-1H (GenBank accession number Z32700.1) was used.
Microarray data accession number.
The microarray data have been deposited in the Gene Expression Omnibus (GEO [http://ncbi.nlm.nih/gov/geo/]) database under accession number GSE21752.
RESULTS
Retentostat cultivation induced asexual differentiation.
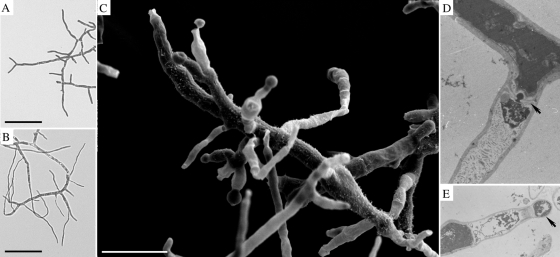
During exponential growth in the batch cultures (day 0), the vegetative mycelia consisted of thick hyphae with uniformly stained cytoplasm (Fig. 2 A). The severe carbon and energy limitation of retentostat cultivation resulted in clear morphological changes, and within the first day the newly formed hyphae were thin and branched infrequently (Fig. 2B). The first conidiogenic structures were observed on day 2. After day 4, differentiation accelerated, with conidia budding from flask-shaped cells formed directly at the apex of lateral branches (Fig. 2C) or from reduced conidiophores. By day 8 most apices had differentiated to produce conidia, and the spore concentration in the effluent was about 1010 conidia liter−1. Germination of newly formed conidia was never observed although they were viable as these conidia were capable of forming colonies on CM plates. Mycelium compartmentalization after the batch phase was apparent, especially with the onset of conidiation. Plugged septa (Fig. 2D) separated different processes in adjacent compartments. The appearance of empty hyphal compartments in lateral hyphae coincided with conidiogenesis. Special, thick septations delineated the newly formed conidium (Fig. 2E). As conidiation increased, the culture turned dark brown, and the odor changed to musty. After 8 to 10 days of retentostat cultivation, the cultures suddenly foamed. The experiments were stopped at this time since the foaming caused significant amounts of biomass to deposit in the headspace of the reactor vessel. Late in the cultures (7 to 8 days) vegetative mycelia reappeared in low proportions. Plating out dilute culture broth did not reveal asporogenic mutants, and the reappearance of vegetative mycelia was interpreted as a physiological phenomenon. The cultures consisted of free filamentous and, in the end, sporulating mycelia throughout cultivation (a time series of micrographs showing submerged development is given in Fig. SA2 in the supplemental material).
FIG. 2.
Morphological differentiation of A. niger N402 in maltose-limited retentostat cultures. (A) Mycelium from exponentially growing culture. Scale bar, 50 μm. (B) Mycelium after 1 day of substrate-limited growth. Scale bar, 50 μm. (C) SEM of conidiospore-forming mycelium after 8 days of substrate-limited growth. Scale bar, 10 μm. (D) TEM showing compartmentalization within a mycelium after 8 days. Arrow, septum plugged with Woronin body. (E) TEM of conidiogenous cell and newly formed conidium (arrow).
Physiology as the specific growth rate approaches zero.
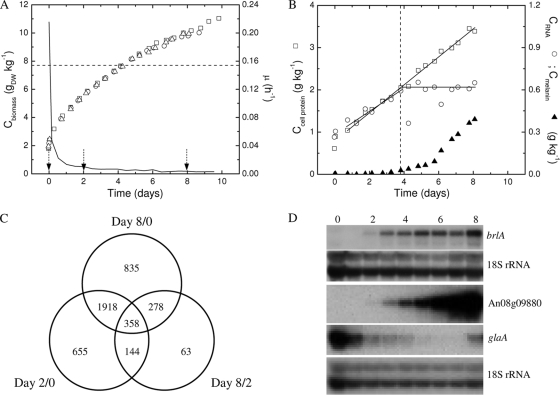
The accumulation of biomass in the three replicate retentostat cultures was highly reproducible (Fig. 3 A). The cultures were performed at two different geographical locations, one in Odense (Denmark) and two in Leiden (Netherlands). The retention efficiency dropped to 95% during sporulation as the small spores were washed out with the effluent. Physiological variables and coefficients were corrected for harvested biomass (sampling and effluent). The specific growth rate (μ) dropped from μmax (0.24 h−1) in the batch culture to <0.02 h−1 within the first 24 h of retentostat cultivation (Fig. 3A; also Table 1). Although μ approached zero, the growth yield (Y) stabilized at a minimum value (Table 1) after 4 to 6 days with the onset of prolific sporulation. A major part of the substrate was converted into biomass in the exponentially growing cultures (day 0) while substrate utilization as the specific growth rate approached zero shifted to respiration as the primary sink (Table 1).
FIG. 3.
Physiological and molecular differentiation of A. niger N402 in maltose-limited retentostat cultures. (A) Biomass accumulation in three replicate cultures (□, Odense, Denmark; ○ and ▵, Leiden, Netherlands) and the specific growth rate (μ; solid line). The dashed line indicates maximum theoretical maintained cell density calculated from a maintenance requirement of 0.026 g gDW−1 h−1; the three arrows mark time points used in the transcriptomic analysis. (B) Accumulation of cell protein (□), total RNA (○), and melanin (▴) in a retentostat culture (per kg of culture); the dashed line marks a transition in biomass composition. (C) Venn diagram showing the number of genes differentially expressed (4,251 genes in total) in transcriptomic comparisons of 0, 2, and 8 days of retentostat cultivation. (D) Northern blot analyses of the central regulator of conidiation, brlA (An01g10540), and the most highly transcribed reproductive and vegetative genes, respectively, a hydrophobin gene (An08g09880) and the glucoamylase gene (glaA [An03g06550]). Exposure time was 5 h for all three genes, 1 h for the first 18S rRNA panel (brlA comparison), and overnight for the second 18S rRNA panel.
TABLE 1.
Growth on the limiting carbon and energy source, maltosea
| Period and/or days | μ (h−1) | Y (gDW g−1 maltose) | C respired (%) |
|---|---|---|---|
| Exponential growth | 0.24 ± 0.02 | 0.58 ± 0.03 | 32 ± 3 |
| Starvation | |||
| Day 1 to day 4 | 0.014 ± 0.001 | 0.27 ± 0.01 | 50 ± 2 |
| Day 4 to day 10 | 0.005 | 0.20 ± 0.01 | 64 ± 1 |
Data for μ and Y are mean values (± standard deviation) for a given period, corrected for cell loss due to sampling or washout. Values were determined from triplicate cultures. The Y value and substrate utilization for maltose are based on glucose equivalents.
Changes in biomass composition reflect the shift to reproductive growth.
Analyses of the dry-cell composition revealed that the content of cytoplasmic components (protein and total RNA) decreased rapidly within the first 24 h of substrate limitation. The biomass composition was stable from day 1 to day 4, and growth appeared balanced, with cell protein and RNA accumulating in proportion to each other (Fig. 3B). Melanin was first observed to increase in the effluent from day 2, concomitant with the first observations of sporulation. The biomass composition changed markedly as sporulation accelerated around day 4, with cessation in the accumulation of total cell RNA and increased accumulation of particle-bound melanin (Fig. 3B). Net growth after day 4 to day 5 was accounted for by small melanized particles, as shown by the equal relations of melanin and biomass accumulation in the culture and in the spore-enriched effluent (particles of <51 μm), which were 0.21 ± 0.03 g gDW−1 of biomass (n = 3) and 0.19 ± 0.02 g gDW−1 (n = 2), respectively (the relation of melanin and biomass is demonstrated in Fig. SA3 in the supplemental material).
Transcriptomic analysis of the retentostat cultures.
Transcriptomic analysis was applied to explore processes involved in the observed submerged differentiation near a specific growth rate of zero. Global transcriptional profiles were established for samples representing three different phenotypic phases in the cultures (see the time points marked in Fig. S3A in the supplemental material). The three triplicate transcriptomes (see Table SA1 in the supplemental material) represented exponential vegetative growth (day 0), slow and predominantly vegetative growth (day 2), and slow reproductive growth (day 8). The biological reproducibility of gene transcription was high (34, 51), characterized by mean coefficient of variation values of 0.14, 0.15, and 0.18 for days 0, 2, and 8, respectively. A total of 4,251 unique genes were identified as differentially expressed (defined in Materials and Methods) between the three phenotypic phases (Fig. 3C). This corresponds to 30% of all genes (about 14,000) or 55% of all genes evaluated as expressed (7,722 genes). The complete list of differentially expressed genes, including statistical significance and transcript ratios, is given in the supplemental material (see Table SA1). Nearly half of the differentially expressed genes displayed the same regulation when expression levels of days 2 and 8 were compared to those of day 0 (day 2/day 0 and day 8/day 0, respectively). A total of 1,371 genes were commonly upregulated, whereas 874 genes were commonly downregulated during severe carbon and energy limitation. Strikingly, 83% of the commonly downregulated genes had at least one associated Gene Ontology (GO) term. For comparison, this proportion was 54% for the upregulated genes and 42% for the genomic average.
Overview of cellular processes and functions regulated at the level of transcription.
To obtain an overview of processes affected on the transcriptional level, overrepresented GO terms were identified in regulation groups (up-, down-, and up-/downregulated) of differentially expressed genes. All overrepresented GO terms among genes differentially expressed between the three phases of the cultures are given in the supplemental material (see Table SA2 in the supplemental material). Common for the slow-growing cultures at days 2 and 8 was downregulation of conserved structural, biosynthetic, and energy-generating functions residing in organelles and cytoplasm (347 GO terms), whereas genes involved in nutrient mobilization (e.g., autophagy, N-acetylglucosamine metabolism, and carbohydrate transport) and reproduction were upregulated. Transcription regulator activity was also enriched among commonly upregulated genes, stressing the ramifying consequences of severe carbon and energy limitation. During prolific sporulation at day 8, additional upregulation (compared to day 2) of genes involved in reproductive and structural differentiation was a particularly significant feature, together with further downregulation of some biosynthetic processes. Glycosyl hydrolase activity was enriched among upregulated genes at day 2 (day 2/day 0), while it was among the downregulated genes at day 8 (day 8/day 2). Cellular differentiation during starvation is evidently highly complex, and it is not the aim of this investigation to give a full interpretation of all processes involved. However, from the overrepresented GO terms (see Table SA2 in the supplemental material), we have listed selected genes to represent some of the most characteristic processes affected by severe carbon and energy limitation (Table 2). These include genes involved in endogenous nutrient mobilization, structural rearrangement, conidiation, and regulation of cell proliferation and development. The supplemental material includes detailed lists of differentially expressed genes with roles in autophagy or microbody processes (36) (see Table SA3 in the supplemental material) and cell wall biosynthesis (50) (see Table SA4).
TABLE 2.
A selection of differentially expressed genes with characterized or putative role in cellular differentiation of A. niger
| ORF and functional groupa | Description (homologue and organism)b | Fold changec |
||
|---|---|---|---|---|
| Day 2/day 0 | Day 8/day 0 | Day 8/day 2 | ||
| Endogenous nutrient mobilization and structural rearrangement | ||||
| An04g03950 | atg1, autophagy-related serine/threonine kinase | 4.0 | 5.3 | |
| An07g10020 | atg8, autophagy-related ubiquitin modifier | 3.9 | 3.5 | |
| An16g08980 | Regulator of fatty acid catabolism (farB, A. nidulans) | 2.7 | 2.0 | 0.76 |
| An01g01830 | Catalase-peroxidase (katG, Aspergillus terreus) | 12 | 14 | |
| An07g03880 | pepC vacuolar serine proteinase | 2.4 | 2.6 | |
| An09g04010 | chsB, chitin synthase, class III | 2.3 | 2.4 | |
| An02g07020 | cfcA, endochitinase, class V | 7.2 | 25 | 3.5 |
| An01g09270 | Isocitrate lyase (acuD, A. nidulans) | 9.9 | 8.5 | |
| An04g05720 | Ketothiolase, β-oxidation (pot1, Yarrowia lipolytica) | 2.0 | 2.5 | |
| An18g04100 | exgA, exo-β-1,3-glucanase | 52 | 16 | |
| An07g04570 | Woronin body protein (hex1, A. nidulans) | 0.81 | 0.45 | 0.56 |
| An02g07770 | Trehalose phosphorylase (ccg9, Neurospora crassa) | 3.7 | 8.2 | 2.2 |
| Spore-related genes | ||||
| An09g05730 | Polyketide synthase (alb1, A. fumigatus) | 21 | 12 | |
| An14g05350 | Hydrolase (ayg1, A. fumigatus) | 32 | 374 | 12 |
| An14g05370 | Multicopper oxidase (abr1, A. fumigatus) | 28 | 345 | 12 |
| An01g13660 | Laccase (abr2, A. fumigatus) | 237 | 330 | |
| An03g02360 | Hydrophobin (dewA, A. nidulans) | 173 | 3,096 | |
| Regulation of conidiation | ||||
| An11g01000 | Histidine kinase, two-component system (tscA, A. nidulans) | 3.6 | 7.4 | 2.1 |
| An02g03160 | Regulator of G-protein signaling (flbA, A. nidulans) | 1.7 | ||
| An02g05420 | C2H2 transcription factor (flbC, A. nidulans) | 2.8 | 3.0 | |
| An01g04830 | Myb-like DNA-binding protein (flbD, A. nidulans) | 12 | 17 | |
| An02g02150 | Transcription factor (medA, A. nidulans) | 1.5 | ||
| An05g00480 | Transcription factor (stuA, A. nidulans) | 3.1 | 5.0 | |
| An01g10540 | Transcription factor (brlA, A. nidulans) | 12 | 178 | 14 |
| An01g03750 | Transcription factor (abaA, A. nidulans) | 9.3 | 7.2 | |
| An01g08900 | Transcription regulator (wetA, A. nidulans) | 8.9 | 3.2 | |
| Cell cycle and macromolecule metabolism | ||||
| An01g03330 | Pcl-like cyclin (pclA, A. nidulans) | 4.5 | 9.7 | 2.2 |
| An01g04720 | DNA primase, large SU (pri2, S. cerevisiae) | 0.38 | 0.46 | |
| An02g03010 | Condensin-complex SU (smc2, S. cerevisiae) | 0.57 | 0.54 | |
| An11g01890 | Phosphotyrosine phosphatase (nimT, A. nidulans) | 0.73 | ||
| An07g10400 | Histone chaperone FACT, SU (spt16 or CDC68, S. cerevisiae) | 0.36 | 0.39 | |
| An01g00040 | TFIIF, α-SU (tfg1, S. cerevisiae) | 0.26 | 0.27 | |
Open reading frame identifier (50).
Description of gene or closest homologue (BLASTP).
Fold change of differentially expressed (FDR < 0.05) genes based on a comparison of transcription levels at day 2 or day 8 to day 0 (day 2/day 0 and day 8/day 0, respectively) and at day 8 to day 2 (day 8/day 2).
Retentostat cultivation induced transcription of up- and downstream elements in brlA regulatory pathway and structural sporulation genes.
Temporal and spatial control of asexual reproduction is complex, and in Aspergillus nidulans involves a number of DNA-binding regulators (reviewed in references 2 and 25). Homologs of many of these regulators were upregulated during submerged sporulation (Table 2) including homologs of early fluffy genes, flbA, flbC, and flbD, and modulators, medA and stuA. Homologs of positive regulators of development of conidiogenic structures and conidia peaked in expression late in the cultures (day 8); expression of brlA, the central regulator of conidiation, was upregulated 12-fold at day 2 compared to day 0 and had increased a further 14-fold in expression at day 8. Downstream (of brlA) regulators of phialide and conidium maturation, abaA and wetA, were upregulated at day 8. Several other homologs of genes encoding elements involved in regulation of sporulation, via signaling (e.g., An11g01000) (70) or cell cycle control (e.g., An01g03330) (58), were upregulated as retentostat cultivation progressed. Hydrophobins (e.g., An03g02360), which are important for escaping moist substrate during formation of aerial mycelium for attachment and aerial dispersion of conidia (reviewed in reference 73), appeared as a major group of upregulated cell wall-associated genes by day 8 (Table SA4 in the supplemental material). The basal genetic elements involved in synthesis of conidial melanins have been described for A. nidulans and Aspergillus fumigatus (38, 66). A. niger contains homologous genes (e.g., An09g05730, An14g05350, An14g05370, and An01g13360) (Table 2), which were strongly upregulated late in the retentostat cultures, in correlation with sporulation, melanin accumulation, and expression profiles of regulators of conidium maturation (abaA and wetA homologs). Genes with roles in regulation of the balance between sexual and asexual reproduction were also induced during severe carbon limitation and conidiation and are included with other genes involved in reproduction (lists from reference 50) in Table SA5 in the supplemental material.
Downregulation of essential mitosis and general transcription facilitator genes during conidiation.
Macromolecular metabolic process was a major enriched GO category among genes downregulated at day 8 compared to day 2 (see Table SA2 in the supplemental material). Transcription of affected genes showed little response to the nearly 20-fold reduction in the specific growth rate from day 0 to day 2 but displayed a consistent and a significant reduction during prolific conidiation at day 8. Especially apparent was the abundance of genes showing strong identity with genes involved in DNA processing and cell division. Most of these genes are essential and represent functions associated with initiation of DNA replication (pri2 [An01g04720]) (54) transition to M phase (nimT [An11g01890]) (9), and chromosome condensation and segregation (smc2 [An02g03010]) (39) (Table 2). Table SA6 in the supplemental material lists additional differentially expressed cell cycle genes. Other genes in this list are, for example, homologs of the replication licensing factor (nimQ [An09g04640]), cyclin B (nimE [An01g07420]), cell cycle kinases (nimX [An11g06960] and nimA [An12g08100]), tubulin (mipA [An08g06460]), kinesin motor protein (bimC [An12g01500]), and many genes involved DNA repair and replication, e.g., e.g., kusA (An15g02700) (43), nimO (An11g06950), and pol3 (An15g07150) (18). Characterized cell cycle genes in Aspergillus are reviewed by Doonan (23) and Osmani and Mirabito (49). Other cell cycle genes were expressed at higher levels during nutrient-limited growth and conidiation, for instance, pclA (An01g03330), a NimX-interacting cyclin specifically involved in conidiospore formation (58, 59) (Table 2). Homologs of other genes essential for growth and viability in Saccharomyces cerevisiae, were uniquely downregulated during conidiation (Table 2); among these the FACT (facilitates chromatin transcription) complex histone chaperone, spt16 (or CDC68 [An07g10400]), which facilitates transcription by RNA polymerase I (Pol I), Pol II, and Pol III from chromatin (10); another general transcription factor of RNA Pol II, tfg1 (An01g00040), encoding a subunit of TFIIF, which is required for transcription initiation and elongation (30), was severely downregulated at day 8. The lists of genes specifically downregulated during conidiation is longer, and the above mentioned genes represent some of the most apparent essential functions. Although a number of ribosomal protein genes are further downregulated from day 2 to day 8, the expression levels of these genes were already affected by the early reduction in specific growth rate.
Transiently expressed life-form-specific genes draw an outline of differentiation.
Based on the transcriptomic results, three genes (An01g10540, or brlA; An08g09880, or hydrophobin-like; and An03g06550, or glaA) were selected for a more detailed analysis of temporal changes in expression over the 8-day period of retentostat cultivation by Northern blot analysis (Fig. 3D). Transcription of brlA (An01g10540) was first detected on day 2, concomitant with the first observations of conidiophores, and it increased drastically over the next 2 days before reaching a high stable level during prolific sporulation (days 5 to 8). The glucoamylase gene (glaA, or An03g06550) and an uncharacterized hydrophobin-encoding gene (An08g09880) were also included because they were the two most highly expressed genes during vegetative (day 0) and reproductive (day 8) growth, respectively, and because the function they encode each represents life-form-characteristic functions. Expression of the hydrophobin gene followed brlA expression, and both were inversely related to glaA expression (Fig. 3D). The Northern analysis of these genes showed potential saturation of, at least, the glaA array signal at day 0, since there was a clear reduction in expression compared to day 2 in the Northern blot analysis, which was not apparent from the microarray data (Table 3). The temporal profile of glaA expression in a selected retentostat culture also revealed very low expression during prolific sporulation while in the sample of day 8, also used for transcriptomic analysis, the level had increased again. glaA expression in the corresponding samples of the two other cultures had not increased to the same level at day 8, which contributed significantly to the variation around the mean transcript level of the triplicate (see replicate transcript levels in Table SA1 in the supplemental material).
TABLE 3.
Expression of genes encoding major secreted proteins and key proteins involved in protein secretion
| ORF and functional groupa | Description (homologue and organism)b | mRNA abundance atc: |
Fold changed |
||||
|---|---|---|---|---|---|---|---|
| Day 0 | Day 2 | Day 8 | Day 2/day 0 | Day 8/day 0 | Day 8/day 2 | ||
| Hydrolases | |||||||
| An03g06550 | glaA, glucoamylase | 91 | 85 | 57 | |||
| An04g06920 | agdA, extracellular alpha-glucosidase | 65 | 76 | 55 | |||
| An11g03340 | aamA, acid alpha-amylase | 14 | 15 | 4 | |||
| An12g08280 | inu1, exo-inulinase | 1 | 10 | 1 | 14 | 0.11 | |
| An14g04710 | pepA, aspartic proteinase | 1 | 6 | 2 | 10 | 3.0 | 0.29 |
| Small cysteine-rich proteinse | |||||||
| An08g09880 | Hypothetical protein (hydrophobin motif); 9/91 | 0 | 9 | 100 | 91 | 1,500 | |
| An07g01320 | Antifungal protein; 6/92 | 0 | 88 | 93 | 239 | 251 | |
| An03g02400 | Hydrophobin (dewA, A. nidulans); 8/131 | 0 | 7 | 87 | 16 | 265 | |
| An15g02250 | Hypothetical protein, 6/83 | 0 | 5 | 69 | 11 | 211 | |
| An03g02360 | Hydrophobin (dewA, A. nidulans); 8/122 | 0 | 4 | 52 | 173 | 3,096 | |
| An04g04460 | Hypothetical protein; 6/97 | 0 | 11 | 39 | 88 | 317 | |
| An16g01850 | Hypothetical protein (BYS1 domain); 6/159 | 0 | 10 | 36 | 58 | 203 | |
| An16g06570 | Hypothetical protein; 4/125 | 0 | 2 | 34 | 18 | 249 | 14 |
| An18g06360 | Hypothetical protein (CFEM domain); 10/198 | 1 | 4 | 33 | 8 | 63 | 8 |
| Secretory pathway proteins | |||||||
| An11g04180 | bipA, ER chaperone | 10 | 5 | 6 | 0.53 | 0.59 | |
| An01g08420 | clxA, calnexin | 24 | 7 | 9 | 0.29 | 0.39 | |
| An02g14800 | pdiA, protein disulfide isomerase | 26 | 17 | 27 | 0.67 | 1.6 | |
| An01g00160 | hacA, UPR transcription factor | 8 | 9 | 19 | 2.4 | 2.1 | |
| An04g05100 | Glycosylphosphatidylinositol biosynthetic protein (gpi13, S. cerevisiae) | 0 | 0 | 1 | 1.6 | 31 | 20 |
| An01g12550 | α-1,2-Mannosidase (msdS, Aspergillus saitoi) | 0 | 11 | 7 | 102 | 60 | |
Open reading frame identifier (50).
Description of gene or closest homologue (BLASTP).
Relative to mean of most abundant transcript (An08g09880 at day 8) in the transcriptome listed for days 0, 2 and 8.
Fold change of differentially expressed (FDR < 0.05) genes, based on a comparison of transcription levels at day 2 or day 8 to day 0 and at day 8 to day 2.
Genes selected from Table SA8 in the supplemental material as described in the text. Numbers indicate number of cysteine residues/total amino acid length of the predicted protein.
Sporulation is accompanied by a shift in expression of genes encoding secreted proteins.
We decided to take closer look at genes encoding probable secreted proteins since the two most highly expressed genes (glucoamylase and a hydrophobin) both fall into this class and had inversely related transcription patterns during retentostat cultivation. Furthermore, secreted proteins are continuously eluted from the retentostat cultures and thus represent an obvious product group which may be targeted for exploitation. Transcription of genes encoding major secreted and glycosylated hydrolases was generally negatively affected by the prolific sporulation at day 8 (Table 3). The severe substrate limitation also led to reduced transcription of a multitude of genes involved in synthesis and secretion of glycoproteins (see Table SA7 in the supplemental material). Key elements in glycoprotein folding and modification, such as bipA and clxA (Table 3), were downregulated together with other elements residing in the endoplasmic reticulum during retentostat cultivation (see Table SA7).
Sporulation and the structural genes involved are poorly characterized, and it is important to realize that these hypothetical or uncharacterized genes are not subject to analysis and sorting based on functional annotation. To learn more about highly expressed secreted proteins in the sporulating retentostat cultures, we generated a list of genes (see Table SA8 in the supplemental material), which were upregulated by >10-fold at day 8 relative to day 0 and contain secretion signals in the form of a signal sequence or a primary structure consistent with nonclassical protein export (7). The genes in the list were ranked according to transcript abundance relative to the gene with the highest level of expression (An08g09880). The sorted list reveals that the most highly expressed genes upregulated during conidiation encode small cysteine-rich proteins. Fourteen of the 20 most abundant transcripts encode secreted proteins; 12 of these consist of fewer than 200 amino acid residues, and nine have a high cysteine content (>2%). These nine genes are identified as three putative hydrophobins (conserved C-CC-C-C-CC-C motif), a gene highly homologous to the Aspergillus giganteus antifungal protein (72), and five hypothetical genes (Table 3). Many additional genes encoding secreted small or Cys-rich proteins are expressed at higher levels during conidiation (see Table SA8). Interestingly, protein disulfide isomerase (pdiA) and hacA, the master regulator of the unfolded protein response (UPR) (45, 47), were both upregulated with high significance during sporulation at day 8 compared to day 2, quite opposite to other secretory pathway and endoplasmic reticulum (ER)-associated genes (see Table SA7 in the supplemental material). The regulation of pdiA appeared stringent as it was the second most significant observation (P = 0.000004) among upregulated genes.
When transcription in the biological triplicates was compared, it was characteristic that transcript levels of hydrolase genes (e.g., glaA and agdA) varied at day 8 while transcription levels of hydrophobin genes were in an ascending trend and consequently varied at day 2. Thus, these genes were not called as differentially expressed from day 2 to day 8 due to the high relative variation of these transcripts at one time and the stringency of the statistical test.
Transcription of secondary metabolism genes is induced by retentostat cultivation.
Secondary metabolites constitute another interesting and, in A. niger, unexploited group of excreted molecules which appear relevant products for a near-zero growth rate condition. Conidial melanin is, for example, derived from polyketide metabolites, and it appears to be a significant product in the sporulating retentostat cultures. For this reason, we searched the transcriptomes for genes potentially involved in biosynthesis of secondary metabolites. Secondary metabolism genes are generally poorly functionally annotated, but the associated products are often derived from polyketides or nonribosomal peptides and are products of metabolic pathways encoded by clustered and coregulated genes (i.e., aflatoxins, fumonisins, and dihydroxynaphthalene [DHN]-melanin in A. nidulans and A. fumigatus). In the retentostat cultures, apparently coexpressed and clustered genes, encode potential polyketide synthase (PKS), nonribosomal peptide synthetase NRPS, transporters, transcription factors, or oxidoreductases. In this way at least 14 probable secondary metabolite gene clusters were identified from their expression profiles (see Table SA9 in the supplemental material). Most of these gene clusters were expressed at very low levels or not expressed during vegetative growth (except a hyphal fusarine-siderophore cluster) but were then upregulated at day 2 or during sporulation at day 8. Notable examples are the A. niger fumonisin (a polyketide-derived sphingosine analog) gene cluster (26, 34, 50) and clusters encoding homologs of the recently described asperfuranone (19) and asperthecin (64) biosynthesis genes in A. nidulans. Most of the clusters probably encode biosynthetic routes of unknown metabolites or are not linked to known products of A. niger (48). Some secondary metabolites are produced from nonclustered pathway genes. This is the case with the polyketide-derived, conidial melanin in A. niger (Table 2). The polyketide synthase genes involved in melanin (An09g05730) and fumonisin (An01g06930) biosynthesis were by far the two most highly transcribed polyketide synthases. Formation of polyketide metabolites consumes acetyl-coenzyme A (CoA) and its derivative, malonyl-CoA. The gene encoding acetyl-CoA carboxylase (accA, or An12g04020), which catalyzes formation of malonyl-CoA, was significantly upregulated during sporulation (day 8). We identified additional gene clusters encoding functions not clearly related to secondary metabolism. One of these is a sporulation-specific gene cluster (An07g00010-An07g00130), which contains highly expressed hypothetical genes (e.g., An07g00070) and homologs of bacterial genes.
DISCUSSION
In revisiting retentostat cultivation of A. niger, we used bioreactors suited for 5-liter cultures and hence needed a retention device which could support a high flow of effluent. Continuous cell retention can be a major technical challenge when standard microbiological filter materials and techniques are applied, and this is likely a contributing reason for the small culture volumes (1 liter) and low effluent flow rates used in other retentostat studies (11, 60). The newly designed retention device presented here exploits the size and structure of filamentous fungi. The relatively large pore size (51 μm) and unforced filtration resulted in reliable and efficient cell retention at high flow rates, which allowed high reproducibility of continuous cultivation. The retention method also allowed small particles to exit with the effluent, a property which can be used to assess sporulation and to perform continuous harvesting of conidia. Due to the simple construction and principle, we propose that the retention method can be applied successfully to smaller- or larger-scale applications by adapting the dimensions of the device. In addition, the method may also be applied to other filamentous microorganisms, and retention efficiency can be altered by using other filter materials.
The theory of maintenance energy predicts a transient decrease to zero growth in carbon- and energy-limited retentostat cultures (52, 68). However, this model was not applicable to the maltose-limited retentostat cultures of A. niger, and although the specific growth rate (μ) did approach zero, the growth yield (Y) stabilized at a minimum value in the sporulating cultures. The maintenance concept assumes that metabolically active and growing cells conform to a single metabolic regime of fully viable cultures. This was clearly not the case in the retentostat cultures of A. niger; the mycelia differentiated in response to the limited substrate availability. Support for a basal maintenance energy requirement is often derived from chemostat studies where the specific substrate consumption rate shows a linear dependency on μ, and the maintenance ration is defined by extrapolation to zero μ (17, 46, 56). However, some studies have documented deviations from the maintenance concept at low μ values in sporulating cultures (42, 61). The specific growth rate observed in the maltose-limited retentostat cultures (0.005 h−1 < μ < 0.05 h−1) was below the range investigated in most chemostat-based studies. Pirt (53) proposed the existence of a critical dilution rate below which the original concept of maintenance energy requires correction for the proportion of the population which has differentiated into dormant cells. However, it is quite clear from the present study that submerged differentiation of A. niger near zero μ was not a process passively undertaken by a minor proportion of the population but, rather, a commitment of the entire culture to grow by producing reproductive cells in a metabolic context, which was very different from the preceding vegetative state. It seems reasonable to argue that a basal maintenance energy determined for growing vegetative cells does not practically or conceptually apply to A. niger during reproductive growth.
Many factors are likely to contribute to the minimum growth yield observed in the retentostat cultures. During prolific sporulation (after day 4) the actively growing proportion of the culture changed, as indicated by the cessation in total RNA accumulation and increased formation of melanin. That these changes reflect formation of dormant conidiospores is further supported by the transcriptional downregulation of many homologs of essential genes involved in cell division (e.g., nim genes) and in regulation of transcription of the different RNA species (FACT and TFIIF complex subunits), which is consistent with the fact that conidia are arrested in G1 phase of the cell cycle (9). The dormant spores, melanin, and empty hyphal compartments all contributed to the accumulating biomass but consumed little or none of the supplied substrate, which apparently allowed regrowth of vegetative hyphae after day 7.
Sporulation of A. niger was induced by retentostat cultivation and was at the same time fuelled by the continuous external supply of maltose. This is opposite to truly starved cultures, which would rely purely on mobilized endogenous nutrients. Other investigations (15, 56, 65) have shown that the maximal extent of submerged sporulation in carbon-limited cultures of Aspergillus and Penicillium is achieved with a low external supply of the carbon source. In this study we have shown that net growth in the retentostat cultures was accounted for by small melanized particles from days 4 to 7, which suggests that the cultures were fully committed to conidiospore formation in this period. This highlights the potential value of the cultivation system for studies of submerged conidiation and production of conidiation-associated products.
Carbon starvation or severe limitation induces submerged conidiation from reduced conidiogenous structures (15, 16, 27, 46, 62), as observed in this study. Under these conditions conidium-producing phialides are formed directly from the mycelium while conidiophore stalk and vesicles appear absent. The abbreviated sporulation program is consistent with phenotypes associated with high-level expression of BrlA, the central regulator of conidiation (74). In the maltose-limited retentostat cultures, the brlA transcript level followed the extent of sporulation, with both reaching a maximum after day 4. Regulators upstream of BrlA (e.g., flbC and flbD) and associated with conidiophore development were also induced by day 2, and this is different from the current model, which suggests that carbon starvation bypasses the fluffy genes and acts directly at brlA (2). The early response to starvation or severe carbon limitation and factors antecedent to BrlA may have more prominent roles in submerged development. Although this needs further investigation, our results reveal the complexity of early transcription events (day 2/day 0) as μ had decreased 25-fold. In this period more genes were upregulated than downregulated, and many of these genes are poorly characterized with respect to function.
Downstream of BrlA, transcription factors AbaA and WetA activate expression of phialide and conidium-specific genes sometimes referred to as class A and B genes (2). A. niger orthologues of class A and B genes and their activators were upregulated during retentostat cultivation in a similar way (at day 8). In addition to what has previously been described, the transcriptomes revealed other genes encoding hypothetical cysteine-rich proteins and secondary metabolite genes which were also expressed in a conidiation-related manner. Cysteine-rich proteins and polyketide metabolites (e.g., melanin pigment) produced by a conidium may have a synergistic function. It has been suggested by Griffith (28) that formation of the protective outer layer of a conidium involves oxidative cross-linking of protein sulfhydryl groups and conidial pigment.
From an application point of view, sporulation during severe carbon limitation appears to be a generally unproductive physiological state since nearly two-thirds of the substrate was respired. However, the biomass yield was significantly reduced, and unique physiological adaptations appeared to take place inside the organism to accommodate synthesis of special proteins and metabolites associated with conidiospore formation. Examples are the transcription profiles of genes encoding secreted proteins and elements involved in the secretion process. During vegetative growth, genes encoding glycosylated substrate hydrolases were the most highly expressed while this role was taken over by genes encoding small cysteine-rich proteins in the conidiating cultures. Concomitantly with this shift we observed an unusual uncoupling of transcriptional regulation of the protein disulfide isomerase (pdiA) from the normally coregulated endoplasmic reticulum-associated secretory pathway genes (29, 34). It is known that correct disulfide bond formation is essential for efficient secretion of conidial hydrophobins in Magnaporthe grisea (35), and it is likely that enhanced activity of protein disulfide isomerase may ensure efficient secretion of heavily disulfide-bonded proteins, such as hydrophobins, during conidiation. Another example is the upregulation of acetyl-CoA carboxylase (accA) in the sporulating retentostat cultures. An increase in the activity of this enzyme could increase the flux of malonyl-CoA to secondary metabolic pathways, like the one involved in formation of a major carbon sink, conidial melanin. Melanin biosynthesis is, from a metabolic engineering point of view, interesting because the organism as part of asexual development is forced to use the scarce substrate to produce significant amounts a costly metabolite. The perspectives for applying near-zero growth conditions is greatly increased if developmental decisions such as this can be exploited to direct metabolic fluxes into desired pathways and products or to allow efficient synthesis of special molecules of homologous or heterologous origin during development.
The present investigation has shown that retentostat cultivation both induces and fuels submerged asexual differentiation of A. niger and that genes associated with conidiation become highly expressed as the specific growth rate approaches zero. This introduces new perspectives for a classical microbial production host, where retentostat cultivation is a promising method to produce conidia, certain secondary metabolites, and cysteine-rich proteins. We further propose that the improved retentostat method can be used in fundamental studies of differentiation and is applicable to filamentous fungi in general.
Supplementary Material
Acknowledgments
We thank Thomas Rubæk and the BMB metal workshop (University of Southern Denmark) for technical assistance and Jaap Visser for his comments to the manuscript. T.R.J. thanks Jens J. L. Iversen and the Zero Growth project group (Kluyver Centre, Netherlands) for support and valuable discussion.
This work was carried out within the research program of the Kluyver Centre for Genomics of Industrial Fermentation, which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research.
Footnotes
Published ahead of print on 18 June 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adams, T. H., and W. E. Timberlake. 1990. Developmental repression of growth and gene expression in Aspergillus. Proc. Natl. Acad. Sci. U. S. A. 87:5405-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, T. H., J. K. Wieser, and J. H. Yu. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62:35-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Affymetrix Inc. 2002. GeneChip Expression Analysis Technical Manual. Affymetrix, Santa Clara, CA.
- 4.Andersen, M. R., W. Vongsangnak, G. Panagiotou, M. P. Salazar, L. Lehmann, and J. Nielsen. 2008. A trispecies Aspergillus microarray: comparative transcriptomics of three Aspergillus species. Proc. Natl. Acad. Sci. U. S. A. 105:4387-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen, M. R., and J. Nielsen. 2009. Current status of systems biology in aspergilli. Fungal Genet. Biol. 46:S180-190. [DOI] [PubMed] [Google Scholar]
- 6.Archer, D. B., and G. Turner. 2006. Genomics of protein secretion and hyphal growth in Aspergillus, p. 75-96. In A. J. P. Brown (ed.), The Mycota XIII. Springer-Verlag, Berlin, Germany.
- 7.Bendtsen, J. D., L. J. Jensen, N. Blom, G. von Heijne, and S. Brunak. 2004. Feature based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 17:349-356. [DOI] [PubMed] [Google Scholar]
- 8.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. Series B Stat. Methodol. 57:289-300. [Google Scholar]
- 9.Bergen, L. G., and N. R. Morris. 1983. Kinetics of the nuclear division cycle of Aspergillus nidulans. J. Bacteriol. 156:155-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birch, J. L., B. C. M. Tan, K. I. Panov, T. B. Panova, J. S. Andersen, T. A. Owen-Hughes, J. Russell, S. C. Lee, and J. C. B. M. Zomerdijk. 2009. FACT facilitates chromatin transcription by RNA polymerases I and III. EMBO J. 28:854-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boender, L. G. M., E. A. F. Hulster, A. J. A. van Maris, A. S. P. Daran-Lapujade, and J. T. Pronk. 2009. Quantitative physiology of Saccharomyces cerevisiae at near-zero specific growth rates. Appl. Environ. Microbiol. 75:5607-5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borsuk, P., M. Nagiéc, P. Stephień, and E. Bartnik. 1982. Organization of the ribosomal RNA gene cluster in Aspergillus nidulans. Gene 17:147-152. [DOI] [PubMed] [Google Scholar]
- 13.Bos, C. J., A. J. M. Debets, K. Swart, A. Huybers, G. Kobus, and S. M. Slakhorst. 1988. Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr. Genet. 14:437-443. [DOI] [PubMed] [Google Scholar]
- 14.Braaksma, M., A. K. Smilde, M. J. van der Werf, and P. J. Punt. 2009. The effect of environmental conditions on extracellular protease activity in controlled fermentations of Aspergillus niger. Microbiology 155:3430-3439. [DOI] [PubMed] [Google Scholar]
- 15.Broderick, A. J., and R. N. Greenshields. 1981. Sporulation of Aspergillus niger and Aspergillus ochraceus in continuous submerged liquid culture. J. Gen. Microbiol. 126:193-202. [Google Scholar]
- 16.Bushell, M. E., and A. T. Bull. 1999. Sporulation at minimum specific growth rate in Aspergillus nidulans chemostat culture predicted using protein synthesis efficiency estimations. J. Basic. Microbiol. 39:293-298. [DOI] [PubMed] [Google Scholar]
- 17.Carlsen, M., J. Nielsen, and J. Villadsen. 1996. Growth and α-amylase production by Aspergillus oryzae during continuous cultivations. J. Biotechnol. 45:81-93. [Google Scholar]
- 18.Chan, C. Y., A. Galli, and R. H. Schiestl. 2008. Pol3 is involved in nonhomologous end-joining in Saccharomyces cerevisiae. DNA Repair 7:1531-1541. [DOI] [PubMed] [Google Scholar]
- 19.Chiang, Y. M., E. Szewczyk, A. D. Davidson, N. Keller, B. R. Oakley, and C. C. Wang. 2009. A gene cluster containing two fungal polyketide synthases encodes the biosynthetic pathway for a polyketide, asperfuranone, in Aspergillus nidulans. J. Am. Chem. Soc. 131:2965-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conesa, A., S. Götz, J. M. García-Gómez, J. Terol, M. Talón, and M. Robles. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674-3676. [DOI] [PubMed] [Google Scholar]
- 21.de Cássia, R., R. Goncalves, and S. R. Pombeiro-Sponchiado. 2005. Antioxidant activity of the melanin-pigment extracted from Aspergillus nidulans. Biol. Pharm. Bull. 28:1129-1131. [DOI] [PubMed] [Google Scholar]
- 22.de Jongh, W. A., and J. Nielsen. 2008. Enhanced citric acid production through gene insertion in Aspergillus niger. Metab. Eng. 10:87-96. [DOI] [PubMed] [Google Scholar]
- 23.Doonan, J. H. 1992. Cell division in Aspergillus. J. Cell Sci. 103:599-611. [DOI] [PubMed] [Google Scholar]
- 24.Dubois, M., K. A. Gilles, J. K. Hamilton, P. A. Rebers, and F. Smith. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 26:350-356. [Google Scholar]
- 25.Fischer, R. 2002. Conidiation in Aspergillus nidulans, p. 59-88. In H. Osiewacz (ed.), Molecular development of fungal biology. Marcel Dekker, New York, NY.
- 26.Frisvad, J. C., J. Smedsgaard, R. A. Samson, T. O. Larsen, and U. Thrane. 2007. Fumonisin B2 production by Aspergillus niger. J. Agric. Food Chem. 55:9727-9732. [DOI] [PubMed] [Google Scholar]
- 27.Galbraith, J. C., and J. E. Smith. 1969. Sporulation of Aspergillus niger in submerged liquid culture. J. Gen. Microbiol. 59:31-45. [DOI] [PubMed] [Google Scholar]
- 28.Griffith, G. W. 1994. Phenoloxidases, p. 763-788. In S. D. Martinelli and J. R. Kinghorn (ed.), Aspergillus nidulans: 50 years on. Progress in industrial microbiology, vol. 29. Elsevier Science Publishers, Amsterdam, Netherlands. [Google Scholar]
- 29.Guillemette, T., N. N. van der Peij, T. Goosen, K. Lanthaler, G. D. Robson, C. A. van den Hondel, H. Stam, and D. B. Archer. 2007. Genomic analysis of secretion stress response in the enzyme-producing cell factory Aspergillus niger. BMC Genomics 8:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henry, N. L., A. M. Campbell, W. J. Feaver, D. Poon, P. Anthony, and R. D. Kornberg. 1994. TFIIF-TAF-RNA polymerase II connection. Genes Dev. 8:2868-2878. [DOI] [PubMed] [Google Scholar]
- 31.Herbert, D., P. J. Phipps, and R. E. Strange. 1969. Chemical analysis of microbial cells, p. 209-344. In J. R. Norris and D. W. Ribbons (ed.), Methods in microbiology 5B. Academic Press, London, United Kingdom.
- 32.Iversen, J. J. L., J. K. Thomsen, and R. P. Cox. 1994. On-line growth measurements in bioreactors by titrating metabolic proton exchange. Appl. Microbiol. Biotechnol. 42:256-262. [Google Scholar]
- 33.Jacobs, D. I., M. M. Olsthoorn, I. Maillet, M. Akeroyd, S. Breestraat, S. Donkers, R. A. van der Hoeven, C. A. van den Hondel, R. Kooistra, T. Lapointe, H. Menke, R. Meulenberg, M. Misset, W. H. Müller, N. N. van Peij, A. Ram, S. Rodriguez, M. S. Roelofs, J. A. Roubos, M. W. van Tilborg, A. J. Verkleij, H. J. Pel, H. Stam, and C. M. Sagt. 2009. Effective lead selection for improved protein production in Aspergillus niger based on integrated genomics. Fungal Genet. Biol. 46:S141-152. [DOI] [PubMed] [Google Scholar]
- 34.Jørgensen, T. R., T. Goosen, C. A. van den Hondel, A. F. J. Ram, and J. J. L. Iversen. 2009. Transcriptomic comparison of Aspergillus niger growing on two different sugars reveals coordinated regulation of the secretory pathway. BMC Genomics 10:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kershaw, M. J., C. R. Thornton, G. E. Wakley, and N. J. Talbot. 2005. Four conserved intramolecular disulphide linkages are required for secretion and cell wall localization of a hydrophobin during fungal morphogenesis. Mol. Microbiol. 56:117-125. [DOI] [PubMed] [Google Scholar]
- 36.Kiel, J. A. K. W., and I. J. van der Klei. 2009. Proteins involved in microbody biogenesis and degradation in Aspergillus nidulans. Fungal Genet. Biol. 46:S62-S71. [DOI] [PubMed] [Google Scholar]
- 37.Kozakiewicz, Z. 1989. Aspergillus species on stored products. Mycol. Papers 161:1-188. [Google Scholar]
- 38.Langfelder, K., M. Streibel, B. Jahn, G. Haase, and A. A. Brakhage. 2003. Biosynthesis of fungal melanins and their importance for human pathogenic fungi. Fungal Genet. Biol. 38:143-158. [DOI] [PubMed] [Google Scholar]
- 39.Losada, A., and T. Hirano. 2001. Shaping the metaphase chromosome: coordination of cohesion and condensation. BioEssays 23:924-935. [DOI] [PubMed] [Google Scholar]
- 40.Mah, J. H., and J. H. Yu. 2006. Upstream and downstream regulation of asexual development in Aspergillus fumigatus. Eukaryot. Cell 5:1585-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McIntyre, M., C. Müller, J. Dynesen, and J. Nielsen. 2001. Metabolic engineering of the morphology of Aspergillus. Adv. Biochem. Eng. Biotechnol. 73:103-128. [DOI] [PubMed] [Google Scholar]
- 42.Metwally, M., M. El Sayed, H. Osman, P. P. F. Hanegraaf, A. H. Stouthamer, and H. W. van Verseveld. 1991. Bioenergetic consequences of glucoamylase production in carbon-limited chemostat cultures of Aspergillus niger. Antonie van Leeuwenhoek 59:35-43. [DOI] [PubMed] [Google Scholar]
- 43.Meyer, V., M. Arentshorst, A. El-Ghezal, A. C. Drews, R. Kooistra, C. A. van den Hondel, and A. F. J. Ram. 2007. Highly efficient gene targeting in the Aspergillus niger kusA mutant. J. Biotechnol. 128:770-775. [DOI] [PubMed] [Google Scholar]
- 44.Meyer, V., R. A. Damveld, M. Arentshorst, U. Stahl, C. A. van den Hondel, and A. F. Ram. 2007. Survival in the presence of antifungals: genome-wide expression profiling of Aspergillus niger in response to sublethal concentrations of caspofungin and fenpropimorph. J. Biol. Chem. 282:32935-32948. [DOI] [PubMed] [Google Scholar]
- 45.Mulder, H. J., M. Saloheimo, M. Penttilä, and S. M. Madrid. 2004. The transcription factor HACA mediates the unfolded protein response in Aspergillus niger, and up-regulates its own transcription. Mol. Genet. Genomics 271:130-140. [DOI] [PubMed] [Google Scholar]
- 46.Ng, A. M. L., J. E. Smith, and A. F. McIntosh. 1973. Conidiation of Aspergillus niger in continuous culture. Arch. Mikrobiol. 88:119-126. [DOI] [PubMed] [Google Scholar]
- 47.Ngiam, C., D. J. Jeenes, P. J. Punt, C. A. van den Hondel, and D. B. Archer. 2000. Characterization of a foldase, protein disulfide isomerase A, in the protein secretory pathway of Aspergillus niger. Appl. Environ. Microbiol. 66:775-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nielsen, K. F. N., J. M. Mogensen, M. Johansen, T. O. Larsen, and J. C. Frisvad. 2009. Review of secondary metabolites and mycotoxins from the Aspergillus niger group. Anal. Bioanal. Chem. 395:1225-1242. [DOI] [PubMed] [Google Scholar]
- 49.Osmani, S. A., and P. M. Mirabito. 2004. The early impact of genetics on our understanding of cell cycle regulation in Aspergillus nidulans. Fungal Genet. Biol. 41:401-410. [DOI] [PubMed] [Google Scholar]
- 50.Pel, H. J., J. H. de Winde, D. B. Archer, P. S. Dyer, G. Hofmann, P. J. Schaap, G. Turner, R. P. de Vries, R. Albang, K. Albermann, M. R. Andersen, J. D. Bendtsen, J. A. Benen, M. van den Berg, S. Breestraat, M. X. Caddick, R. Contreras, M. Cornell, P. M. Coutinho, E. G. Danchin, A. J. Debets, P. Dekker, P. W. van Dijck, A. van Dijk, L. Dijkhuizen, A. J. Driessen, C. d'Enfert, S. Geysens, C. Goosen, G. S. Groot, P. W. de Groot, T. Guillemette, B. Henrissat, M. Herweijer, J. P. van den Hombergh, C. A. van den Hondel, R. T. van der Heijden, R. M. van der Kaaij, F. M. Klis, H. J. Kools, C. P. Kubicek, P. A. van Kuyk, J. Lauber, X. Lu, M. J. van der Maarel, R. Meulenberg, H. Menke, M. A. Mortimer, J. Nielsen, S. G. Oliver, M. Olsthoorn, K. Pal, N. N. van Peij, A. F. Ram, U. Rinas, J. A. Roubos, C. M. Sagt, M. Schmoll, J. Sun, D. Ussery, J. Varga, W. Vervecken, P. J. van de Vondervoort, H. Wedler, H. A. Wösten, A. P. Zeng, A. J. van Ooyen, J. Visser, and H. Stam. 2007. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25:221-231. [DOI] [PubMed] [Google Scholar]
- 51.Piper, M. D. W., P. Daran-Lapujade, C. Bro, B. Regenberg, S. Knudsen, J. Nielsen, and J. T. Pronk. 2002. Reproducibility of oligonucleotide microarray transcriptome analyses—an interlaboratory comparison using chemostat cultures of Saccharomyces cerevisiae. J. Biol. Chem. 277:37001-37008. [DOI] [PubMed] [Google Scholar]
- 52.Pirt, S. J. 1965. The maintenance energy of bacteria in growing cultures. Proc. R. Soc. Lond. B Biol. Sci. 163:224-231. [DOI] [PubMed] [Google Scholar]
- 53.Pirt, S. J. 1987. The energetics of microbes at slow growth rates: maintenance energies and dormant organisms. J. Ferment. Technol. 65:173-177. [Google Scholar]
- 54.Pollok, S., J. Stoepel, C. Bauerschmidt, E. Kremmer, and H. P. Nasheuer. 2003. Regulation of eukaryotic DNA replication at the initiation step. Biochem. Soc. Trans. 31:266-269. [DOI] [PubMed] [Google Scholar]
- 55.Raper, K. B., and D. I. Fennell. 1965. The genus Aspergillus. Williams and Wilkins, Baltimore, MD.
- 56.Righelato, R. C., A. P. J. Trinci, and S. J. Pirt. 1968. The influence of maintenance on energy and growth rate on the metabolic activity, morphology and conidiation of Penicillium chrysogenum. J. Gen. Microbiol. 50:399-412. [DOI] [PubMed] [Google Scholar]
- 57.Ruijter, G. J. G., C. P. Kubicek, and J. Visser. 2002. Production of organic acids by fungi, p. 213-230. In H. D. Osiewacz (ed.), The Mycota X. Springer-Verlag, Berlin, Germany.
- 58.Schier, N., R. Liese, and R. Fischer. 2001. A Pcl-like cyclin of Aspergillus nidulans is transcriptionally activated by developmental regulators and is involved in sporulation. Mol. Cell. Biol. 21:4075-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schier, N., and R. Fischer. 2002. The Aspergillus nidulans cyclin PclA accumulates in the nucleus and interacts with the central cell cycle regulator NimXCdc2. FEBS Lett. 523:143-146. [DOI] [PubMed] [Google Scholar]
- 60.Schrickx, J. M., A. S. Krave, J. C. Verdoes, C. A. M. J. J. van den Hondel, A. H. Stouthamer, and H. W. Verseveld. 1993. Growth and product formation in chemostat and recycling cultures by Aspergillus niger N402 and a glucoamylase overproducing transformant provided with multiple copies of the glaA gene. J. Gen. Microbiol. 139:2801-2810. [DOI] [PubMed] [Google Scholar]
- 61.Schrickx, J. M., M. J. Raedts, A. H. Stouthamer, and H. W. van Verseveld. 1995. Organic acid production by Aspergillus niger in recycling culture analyzed by capillary electrophoresis. Anal. Biochem. 231:175-181. [DOI] [PubMed] [Google Scholar]
- 62.Skromne, I., O. Sánchez, and J. Aquirre. 1995. Starvation stress modulates the expression of the Aspergillus nidulans brlA regulatory gene. Microbiology 141:21-28. [DOI] [PubMed] [Google Scholar]
- 63.Standing, C. N., A. G. Fredrickson, and H. M. Tsuchiya. 1972. Batch- and continuous-culture transients for two substrate systems. Appl. Microbiol. 23:354-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Szewczyk, E., Y. M. Chiang, C. E. Oakley, A. D. Davidson, C. C. Wang, and B. R. Oakley. 2008. Identification and characterization of the asperthecin gene cluster of Aspergillus nidulans. Appl. Environ. Microbiol. 74:7607-7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trinci, A. P. J., and R. C. Righelato. 1970. Changes in constituents and ultrastructure of hyphal compartments during autolysis of glucose-starved Penicillium chrysogenum. J. Gen. Microbiol. 60:239-249. [DOI] [PubMed] [Google Scholar]
- 66.Tsai, H. F., M. H. Wheeler, Y. C. Chang, and K. J. Kwong-Chung. 1999. A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J. Bacteriol. 181:6469-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Twumasi-Boateng, K., Y. Yu, D. Chen, F. N. Gravelat, W. C. Nierman, and D. C. Sheppard. 2009. Transcriptional profiling identifies a role for BrlA in the response to nitrogen depletion and for StuA in the regulation of secondary metabolite clusters in Aspergillus fumigatus. Eukaryot. Cell 8:104-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Verseveld, H. W., J. A. de Hollander, J. Frankena, M. Braster, F. J. Leeuwerik, and A. H. Stouthamer. 1986. Modelling of microbial substrate conversion, growth and product formation in a recycling fermentor. Antonie van Leeuwenhoek 52:325-342. [DOI] [PubMed] [Google Scholar]
- 69.van Verseveld, H. W., M. Metwally, M. el Sayed, M. Osman, J. M. Schrickx, and A. H. Stouthamer. 1991. Determination of the maximal product yield from glucoamylase-producing Aspergillus niger grown in recycling fermentor. Antonie van Leeuwenhoek 60:313-323. [DOI] [PubMed] [Google Scholar]
- 70.Virginia, M., C. L. Appleyard, W. L. McPheat, and M. J. Stark. 2000. A novel two-component protein containing histidine kinase and response regulator domains required for sporulation in Aspergillus nidulans. Curr. Genet. 37:364-372. [DOI] [PubMed] [Google Scholar]
- 71.Vishniac, W., and M. Santer. 1957. The thiobacilli. Bacteriol. Rev. 21:195-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wnendt, S., N. Ulbrich, and U. Stahl. 1994. Molecular cloning, sequence analysis and expression of the gene encoding an anti-fungal protein from Aspergillus giganteus. Curr. Genet. 25:519-523. [DOI] [PubMed] [Google Scholar]
- 73.Wösten, H. A. 2001. Hydrophobins: multipurpose proteins. Annu. Rev. Microbiol. 55:625-646. [DOI] [PubMed] [Google Scholar]
- 74.Yamada, O., B. R. Lee, K. Gomi, and Y. Iimura. 1999. Cloning and functional analysis of the Aspergillus oryzae conidiation regulator gene brlA by its disruption and misscheduled expression. J. Biosci. Bioeng. 87:424-429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.