Abstract
Pyruvate carboxylase is the sole anaplerotic enzyme in glucose-grown cultures of wild-type Saccharomyces cerevisiae. Pyruvate carboxylase-negative (Pyc−) S. cerevisiae strains cannot grow on glucose unless media are supplemented with C4 compounds, such as aspartic acid. In several succinate-producing prokaryotes, phosphoenolpyruvate carboxykinase (PEPCK) fulfills this anaplerotic role. However, the S. cerevisiae PEPCK encoded by PCK1 is repressed by glucose and is considered to have a purely decarboxylating and gluconeogenic function. This study investigates whether and under which conditions PEPCK can replace the anaplerotic function of pyruvate carboxylase in S. cerevisiae. Pyc− S. cerevisiae strains constitutively overexpressing the PEPCK either from S. cerevisiae or from Actinobacillus succinogenes did not grow on glucose as the sole carbon source. However, evolutionary engineering yielded mutants able to grow on glucose as the sole carbon source at a maximum specific growth rate of ca. 0.14 h−1, one-half that of the (pyruvate carboxylase-positive) reference strain grown under the same conditions. Growth was dependent on high carbon dioxide concentrations, indicating that the reaction catalyzed by PEPCK operates near thermodynamic equilibrium. Analysis and reverse engineering of two independently evolved strains showed that single point mutations in pyruvate kinase, which competes with PEPCK for phosphoenolpyruvate, were sufficient to enable the use of PEPCK as the sole anaplerotic enzyme. The PEPCK reaction produces one ATP per carboxylation event, whereas the original route through pyruvate kinase and pyruvate carboxylase is ATP neutral. This increased ATP yield may prove crucial for engineering of efficient and low-cost anaerobic production of C4 dicarboxylic acids in S. cerevisiae.
Interest in biotechnological production of the four-carbon dicarboxylic acids fumarate, succinate, and malate from sugars has strongly increased in recent years (19), as these sugar-derived acids are seen as potential replacements for oil-derived chemical intermediates such as maleic anhydride (41). Metabolic engineering of Escherichia coli has resulted in strains capable of producing 73 g liter−1 succinate at pH 7.0 with a yield that, at 1.61 mol per mol glucose (21), is at 94% of the theoretical maximum. The crucial role of carboxylation reactions in this biotechnological process is illustrated by a carbon yield of 1.07 C mol succinate per C mol of glucose. Despite the high product yields obtained with these prokaryotes, other microorganisms are also under investigation in view of possible gains in process economy and robustness. The yeast Saccharomyces cerevisiae might offer such gains due to its high tolerance to organic acids and to low pH and its insensitivity to bacteriophages.
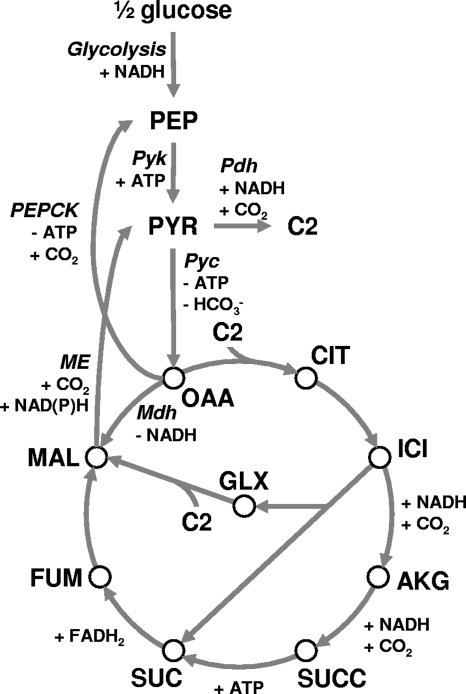
Metabolic engineering of S. cerevisiae has recently resulted in a strain able to produce malate and succinate at yields of 0.48 mol and 0.29 mol per mol glucose, respectively (43). Whereas phosphoenolpyruvate carboxykinase (PEPCK) is the main enzyme catalyzing the carboxylating reaction in the metabolically engineered E. coli strains referred to above (45) and in natural succinate producers such as Actinobacillus succinogenes (25, 35), dicarboxylic acid production in this S. cerevisiae strain depended on (over)expression of native pyruvate carboxylase. Although PEPCK and pyruvate carboxylase can both produce oxaloacetate, the choice of enzyme strongly affects the overall ATP balance of dicarboxylic acid formation. PEPCK directly converts phosphoenolpyruvate (PEP) into oxaloacetate while generating one ATP per carboxylation event (Fig. 1). In contrast, no net ATP is recovered in the route through pyruvate kinase and pyruvate carboxylase, where the ATP produced by the first enzyme is consumed by the second (Fig. 1). Introducing carboxylating PEPCK activity in S. cerevisiae would thus have significant benefits for dicarboxylic acid production by improving the ATP stoichiometry of the anaplerotic reaction.
FIG. 1.
Metabolic routes between PEP and malate in S. cerevisiae. Signs indicate consumption (−) or production (+) in the direction of the arrow. Abbreviations of metabolites: PEP, phosphoenolpyruvate; PYR, pyruvate, OAA, oxaloacetate; MAL, malate; CIT, citrate; ICI, isocitrate; AKG, alpha-ketoglutarate; SUCC, succinyl-CoA; SUC, succinate; FUM, fumarate; C2, acetyl-CoA; GLX, glyoxylate. Abbreviations of enzymes: Pyk, pyruvate kinase; Pyc, pyruvate carboxylase; Mdh, malate dehydrogenase; PEPCK, phosphoenolpyruvate carboxykinase; ME, malic enzyme; Pdh, pyruvate dehydrogenase.
In S. cerevisiae, PEPCK is generally considered to be a decarboxylating enzyme with a function in gluconeogenesis (14). Expression of PCK1, which encodes PEPCK, is repressed by glucose (26, 42) and induced by gluconeogenic substrates (11), while the Pck1 protein is inactivated in the presence of glucose (31). This multilayered regulation has presumably evolved to avoid futile cycling due to simultaneous decarboxylating PEPCK and carboxylating pyruvate carboxylase activities. In line with this, the central enzymes in the glyoxylate cycle, another anaplerotic alternative to pyruvate carboxylase, are also repressed by glucose (16, 17, 20, 32). As a consequence, S. cerevisiae strains lacking the PYC1 and PYC2 genes, encoding the pyruvate carboxylase isoenzymes, are unable to fulfill their anaplerotic requirements during growth on glucose and are auxotrophic for 4-carbon molecules, such as aspartate (34). In a mutagenized pyc1Δ pyc2Δ S. cerevisiae strain able to grow on glucose, derepression of the glyoxylate cycle enzymes was shown to have suppressed the 4-carbon auxotrophy (5). However, using the glyoxylate cycle for dicarboxylic acid production results in suboptimal product yields (44) and is therefore not interesting from an industrial viewpoint.
The aim of the present study is to investigate whether and under which conditions PEPCK can replace pyruvate carboxylase as the sole anaplerotic enzyme in S. cerevisiae. To this end, PEPCKs from S. cerevisiae and A. succinogenes (ScPEPCK and AsPEPCK, respectively) were constitutively (over)expressed in pyc1Δ pyc2Δ S. cerevisiae strains, thereby avoiding the carbon source-dependent transcription of the native gene and, in the case of AsPEPCK, also glucose catabolite inactivation of the protein. Subsequently, growth conditions and second-site mutations required for an in vivo contribution to oxaloacetate formation by PEPCK were investigated.
MATERIALS AND METHODS
Strains and maintenance.
All strains constructed in this study (Table 1) were derived from CEN.JB27, an S. cerevisiae strain from the CEN.PK family (36), containing targeted deletions of URA3 and of the two pyruvate carboxylase genes, PYC1 and PYC2, and carrying a plasmid expressing Escherichia coli PEP carboxylase and the URA3 marker (3).
TABLE 1.
S. cerevisiae strains used in this study
| Strain(s) | Description |
|---|---|
| CEN.PK 113-7D | MATa; reference strain |
| CEN.JB27 | MATapyc1::kanMx pyc2::ILV2smr ura3-52 (pAN-10ppc) |
| IMK157 ura3Δ | MATapyc1::kanMx pyc2::ILV2smr ura3-52 |
| IMY002 | MATapyc1::kanMx pyc2::ILV2smr ura3-52 (MB4917) |
| IMY007 empty vector | MATapyc1::kanMx pyc2::ILV2smr ura3-52 (pUDC1) |
| IMY050 and IMY051 | MATapyc1::kanMx pyc2::ILV2smr ura3-52 (MB4917); evolved |
| IMW001 and IMW002 | MATapyc1::kanMx pyc2::ILV2smr ura3-52 (cured); derived from IMY050 and IMY051, respectively |
| IMY011 | MATapyc1::kanMx pyc2::ILV2smr ura3-52 (pUDC5) |
| IMY012 and IMY013 | MATapyc1::kanMx pyc2::ILV2smr ura3-52 (pUDC5); derived from IMW001 and IMW002, respectively |
| IMY014 and IMY015 | MATapyc1::kanMx pyc2::ILV2smr ura3-52; with G436A or G1006T point mutations in PYK1, respectively |
Strains were maintained on YPD (demineralized water, 10 g liter−1 yeast extract [BD Difco], 20 g liter−1 peptone [BD Difco], and 20 g liter−1 glucose) or, for strains carrying plasmids, on synthetic medium consisting of demineralized water, 3 g liter−1 KH2PO4, 0.5 g liter−1 MgSO4, 6.6 g liter−1 l-aspartic acid (from a filter-sterilized 7.4-g liter−1 solution set to pH 6 with KOH), 6.6 g liter−1 K2SO4, 20 g liter−1 glucose, trace elements, and filter-sterilized vitamins (39). Culture stocks, prepared from shake flask cultures by the addition of glycerol (20%, vol/vol), were stored at −80°C in 1-ml aliquots. Incubations were performed at 30°C, and shake flasks were kept in orbital shakers at 200 rpm.
Plasmid construction and transformation.
Plasmid MB4917 (Table 2) was constructed by cloning PEPCK from Actinobacillus succinogenes (codon optimized for expression in S. cerevisiae by Blue Heron Biotechnology, Bothell, WA), preceded by an AACAAA Kozak sequence, into pRS416-GPD (27) via the XbaI and XhoI sites. MB4917 was subsequently used as the template for a PCR with primers 5′-pUDC1 and 3′-pUDC1-XbaI (Table 3). The resulting PCR product was inserted into MB4917 via the EcoRI and XhoI sites, which introduced a second XbaI site directly upstream of the XhoI site. Then the AsPEPCK gene was excised via digestion with XbaI and self ligation, resulting in the empty vector plasmid pUDC1. Construction of plasmid pUDC5 was started by isolating PCK1 from genomic DNA of CEN.PK 113-7D using primers 5′-pUDC5-XbaI and 3′-pUDC5-XhoI (Table 3). After ligation of the blunt PCR fragment into the pCR-Blunt II-TOPO vector (Invitrogen), the PCK1 open reading frame (ORF) was inserted into MB4917 via the XbaI and XhoI sites, replacing the AsPEPCK gene and thereby creating pUDC5.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pRS406 | Integrating yeast vector, URA3 | 33 |
| pRS416-GPD | Centromeric yeast vector, URA3, PTDH3-TCYC1 | 27 |
| MB4917 | Centromeric yeast vector, URA3, PTDH3-A. succinogenes PEPCK-TCYC1 | This work |
| pUDC1 | Centromeric yeast vector, URA3, PTDH3-TCYC1 | This work |
| pUDC5 | Centromeric yeast vector, URA3, PTDH3- S. cerevisiae PCK1-TCYC1 | This work |
| MB5633 | Integrating yeast vector, PYK1 allele replacement (G436A), URA3 | This work |
| MB5635 | Integrating yeast vector, PYK1 allele replacement (G1006T), URA3 | This work |
TABLE 3.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| 5′-pUDC1 | GCCGACTTTACAGTCCTAAACG |
| 3′-pUDC1-XbaI | ATTCATCTCGAGTCTAGATTAGGCCTTGGGTCCAGCTCCAAC |
| 5′-pUDC5-XbaI | ATCATCTAGACATGTCCCCTTCTAAAATGAATGC |
| 3′-pUDC5-XhoI | GTTACTCGAGTTACTCGAATTGAGGACCAGCGGCTAATAC |
| 5′-PYK1-HindIII | CACACAAGCTTGACATCGGGCTTCCACAATT |
| 3′-PYK1-NotI | CACACGCGGCCGCTGCAACACCTCATCGTTATG |
The mutated PYK1 alleles were isolated from strains IMW001 and IMW002 using primers 5′-PYK1-HindIII and 3′-PYK1-NotI. Each PCR fragment was ligated into the pRS406 vector via the HindIII and NotI restriction sites, resulting in plasmids MB5633 and MB5635.
Yeast transformations were performed as described previously (8). Transformants were selected on synthetic medium plates without uracil and with aspartate as the nitrogen source. Transformations were verified by PCR.
Plasmid curing.
Strain IMK157 (Table 1) was obtained by curing CEN.JB27 of its PEP carboxylase expression vector. Loss of the URA3-based plasmid was induced by growth on agar plates containing 5-fluoroorotic acid (5-FOA). These plates were prepared by combining an 8% agar solution (autoclaved for 20 min at 121°C) with a filter-sterilized solution containing the other medium components (synthetic medium with aspartate as the nitrogen source, 1 g liter−1 5-FOA, and 0.03 g liter−1 uracil). No pH correction was made to the medium components, as it was found that 5-FOA exerts its toxic effect only at low pH (consistent with diffusional entry of the undissociated acid into the cell). Plasmid loss was confirmed by testing for uracil and aspartate auxotrophies.
Strains IMW001 and IMW002 were obtained by curing strains IMY050 and IMY051, respectively. Here, the plasmid-carrying strains were grown under nonselective conditions on liquid YPD. After several serial transfers, the culture was plated on YPD agar. Single-colony isolates were tested for uracil auxotrophy, and plasmid loss was subsequently confirmed by PCR.
Isolation and characterization of evolved strains.
Samples of the continuous culture selection experiments were incubated under a CO2 atmosphere on synthetic medium-agar with 2 g liter−1 (NH4)2SO4 as the nitrogen source. Single-colony isolates, one per selection cultivation, were designated IMY050 and IMY051. Plasmids from IMY050 and IMY051 were isolated using the Zymoprep II yeast plasmid miniprep kit. The PYK1 ORF (extending 700 bp upstream and 300 bp downstream) of the cured IMW001 and IMW002 strains was sequenced by Beckman Coulter Genomics.
Introduction of mutated pyruvate kinase alleles.
Mutated PYK1 alleles were introduced into IMK157 by transformation with XbaI digests of the integration plasmids MB5633 and MB5635 (Table 2). Transformants were selected on synthetic medium plates without uracil and with aspartate as the nitrogen source. Single-colony “pop-in” isolates were incubated on 5-FOA-agar plates (see above for medium composition) to select for the “pop out” of the URA3 marker gene and one of the two PYK1 alleles. Several single-colony “pop-out” isolates were transformed with MB4917 and screened for growth on synthetic medium plates with (NH4)2SO4 as the nitrogen source under a CO2 atmosphere. For a number of growing transformants, sequencing of PYK1 was performed by BaseClear, Netherlands.
Plate cultivation.
Solid media were prepared by addition of 20 g liter−1 Bacto agar. Apart from the nitrogen source [either 4 g liter−1 aspartate or 2 g liter−1 (NH4)2SO4)] and the addition of Tween 80 (0.42 g liter−1) and ethanol-dissolved ergosterol (10 mg liter−1), added to allow for growth under anaerobic conditions, solid synthetic media were identical to the synthetic stock culture medium. Gas-tight jars with 20 kPa of overpressure were used for incubation of agar plates under a CO2 atmosphere.
Continuous cultivation.
Nitrogen-limited chemostat cultivation was carried out as described previously (38) at a dilution rate of 0.1 h−1 and a culture pH of 5 (set with 2 M KOH). Cultures were continuously sparged with 200 ml min−1 CO2. The medium was identical to the synthetic stock culture medium, with the following modifications: medium contained either 2 g liter−1 l-aspartic acid−1 and 6.6 g liter−1 K2SO4 or 1 g liter−1 (NH4)2SO4 and 5.3 g liter−1 K2SO4, as well as 30 g liter−1 glucose, 0.15 ml liter−1 silicon antifoam (BDH, Poole, England) to control foaming, Tween 80 (0.42 g liter−1), and ethanol-dissolved ergosterol (10 mg liter−1) to allow for growth under anaerobic conditions. Chemostat cultures were inoculated with 100 ml preculture, obtained from shake flasks inoculated with 1-ml aliquots of frozen stock culture.
Batch cultivation.
Batch cultivation was performed at 30°C in 2-liter bioreactors (Applikon, Schiedam, Netherlands) at a working volume of 1 liter. The pH was controlled by automatic addition of 2 M KOH. Bioreactors were sparged with 250 ml CO2 per minute and stirred at 800 rpm. The medium was identical to the synthetic stock culture medium, except for the replacement of the aspartic acid and K2SO4 by 5 g liter−1 (NH4)2SO4 and the addition of 0.15 ml liter−1 silicon antifoam (BDH, Poole, England) to control foaming. Tween 80 (0.42 g liter−1) and ethanol-dissolved ergosterol (10 mg liter−1) were added to the medium to allow for growth under anaerobic conditions. Maximum growth rates were determined from optical density (OD; 660 nm) measurements. Preculture shake flasks were inoculated with 1-ml aliquots of frozen stock culture. Cells from exponentially growing shake flask precultures were washed twice with demineralized water and used to inoculate batch cultures at an initial OD of 0.1 or less.
Metabolite analysis.
Extracellular concentrations of glucose were determined by high-performance liquid chromatography (HPLC) using a Bio-Rad Aminex HPX-87H column, eluted with 5 mM H2SO4 at a flow rate of 0.6 ml min−1 and kept at 60°C, coupled to a Waters 2410 refractive index detector.
In vitro enzyme activity assays.
Cell extracts for the in vitro measurement of enzyme activities were prepared from culture samples (each containing approximately 60 mg biomass dry weight) as described earlier (44). Activities of pyruvate carboxylase, phosphoenolpyruvate carboxykinase, and pyruvate kinase were determined as previously described (13), except that 2 mM instead of 10 mM PEP was used to start the pyruvate kinase assay. Protein concentrations in cell extracts were determined with the Lowry method (24), using bovine serum albumin as the standard.
RESULTS
AsPEPCK expression alone is insufficient to rescue C4 auxotrophy of Pyc− S. cerevisiae.
Pyruvate carboxylase-negative (Pyc−) strains of S. cerevisiae are unable to grow on glucose as the sole carbon source. Consistent with an essential anaplerotic role of pyruvate carboxylase, growth on glucose can be restored by the addition of the four-carbon compound aspartate (34). To test whether overexpression of PEPCK can restore C4 prototrophy in Pyc− S. cerevisiae, a codon-optimized PEPCK gene from the natural succinate producer Actinobacillus succinogenes on a centromeric plasmid was introduced into the Pyc− S. cerevisiae strain IMK157, resulting in strain IMY002 (Table 1). This heterologous gene was chosen because it has been shown to function as an anaplerotic enzyme in A. succinogenes (25, 35) and because it was expected to circumvent glucose-induced repression and glucose catabolite inactivation, which affect expression of the native S. cerevisiae PEPCK (26, 31, 42).
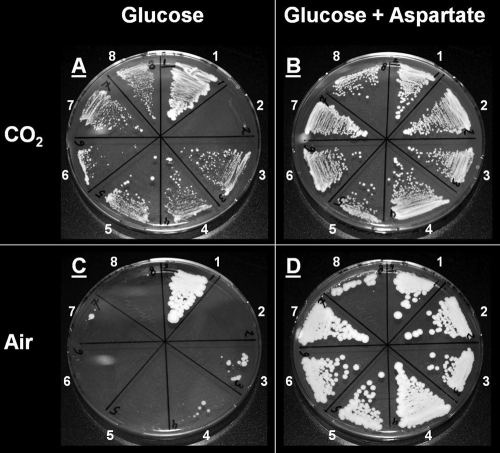
Enzyme activity measurements on glucose-aspartate-grown shake flask cultures of strain IMY002 showed a 20-fold-increased PEPCK activity (0.38 ± 0.11 mmol·min−1·g protein−1) compared to that of the laboratory reference strain CEN.PK 113-7D and the empty vector strain IMY007 (both 0.02 ± 0.00 mmol·min−1·g protein−1) (Table 4). However, despite successful overexpression of PEPCK, strain IMY002 was not able to grow in the absence of aspartate. This growth defect was shown on synthetic medium-agar plates with glucose as the sole carbon source, even when incubated under an atmosphere of 120 kPa CO2 (Fig. 2 A and C), which increases the thermodynamic potential of carboxylation reactions.
TABLE 4.
In vitro enzyme activitiesa
| Straina | Enzyme activityb (mmol·min−1·g protein−1) of: |
||
|---|---|---|---|
| PEPCK | PK | PYC | |
| CEN.PK 113-7D (reference) | 0.02 ± 0.00 | 9.8 ± 0.3 | 0.02 ± 0.00 |
| CEN.PK 113-7D (grown on ethanol) | 0.53 ± 0.04 | 0.9 ± 0.3 | 0.14 ± 0.00 |
| IMY007 (empty vector) | 0.02 ± 0.00 | 9.2 ± 0.3 | 0.01 ± 0.00 |
| IMY002 (AsPEPCK) | 0.38 ± 0.11 | 8.4 ± 0.6 | 0.01 ± 0.00 |
| IMY011 (ScPEPCK) | 0.55 ± 0.04 | 9.4 ± 0.8 | 0.01 ± 0.00 |
| IMY050 (AsPEPCK, evolved) | 0.87 ± 0.23 | 4.1 ± 0.4 | 0.01 ± 0.01 |
| IMY051 (AsPEPCK, evolved) | 0.72 ± 0.28 | 5.1 ± 0.4 | 0.01 ± 0.01 |
| IMY012 (ScPEPCK, evolved) | 0.40 ± 0.01 | 3.5 ± 0.3 | 0.01 ± 0.00 |
| IMY013 (ScPEPCK, evolved) | 0.29 ± 0.00 | 4.2 ± 0.3 | 0.01 ± 0.00 |
Yeast strains were grown in shake flasks on glucose and aspartate synthetic medium unless indicated otherwise.
PEPCK, phosphoenolpyruvate carboxykinase; PK, pyruvate kinase; PYC, pyruvate carboxylase. Errors are deviations from means (for each condition, two shake flask cultures were run).
FIG. 2.
Synthetic medium-agar plates, incubated for 7 days under an atmosphere of either 120 kPa CO2 (A and B) or air (C and D). Plates contained glucose, Tween 80, ergosterol, and, as the nitrogen source, either ammonium sulfate (A and C) or aspartate (B and D). Strains: 1, CEN.PK 113-7D; 2, IMY002 (Pyc−; AsPEPCK); 3, IMY050 (Pyc− evolved; AsPEPCK; 4, IMY051 (Pyc− evolved; AsPEPCK; 5, IMY012 (Pyc− evolved; ScPEPCK; 6, IMY013 (Pyc− evolved; ScPEPCK; 7, IMY014 (Pyc−; AsPEPCK, G436A), 8, IMY015 (Pyc−; AsPEPCK, G1006T).
Evolutionary engineering for increased anaplerotic flux.
The experiments described above demonstrated that PEPCK overexpression alone did not provide a sufficiently high anaplerotic flux to allow for observable growth after 7 days. Since an increase in the anaplerotic flux would allow for an increased growth rate, evolutionary engineering provides an opportunity to increase the flux through PEPCK. A nitrogen-limited chemostat culture of strain IMY002 with aspartate as the sole nitrogen source, maintained at pH 5 and flushed with CO2, provided a reproducible starting point for the evolutionary engineering and allowed for a quick switch of the nitrogen source without nitrogen starvation.
After a steady state was achieved, the selection procedure was started by switching the nitrogen source in the medium from aspartate to ammonium sulfate. Directly after the switch, growth of the culture ceased. To prevent complete washout of the culture, the medium inflow was stopped after 24 h and the experiment was continued as a batch culture. Three days into this batch cultivation, the rate of base addition started to increase exponentially, indicating ammonium consumption and thereby growth. When base addition stopped, glucose was depleted and dry weight measurements confirmed that growth had occurred. The base addition profile suggested a maximum specific growth rate of circa 0.11 h−1, and continuous cultivation was resumed at a dilution rate of 0.1 h−1. The evolved culture could indeed be maintained at this dilution rate, and a steady state was obtained. When, subsequently, the gas used to sparge the bioreactor was switched from CO2 to N2, complete washout of the culture occurred, indicating that a high CO2 concentration was required for growth. A second selection experiment yielded essentially the same results.
Samples from both cultivations (taken during steady state from cultures on ammonium sulfate with CO2 as the sparging gas) were plated on synthetic medium-agar plates with glucose as the sole carbon source and incubated under a CO2 atmosphere. In contrast to results for the unevolved IMY002 strain, growth was now observed, and single-colony isolates (one per experiment) were designated IMY050 and IMY051 (Fig. 2A and C, strains 3 and 4). With each of these isolates, duplicate bioreactor batch cultures were performed on synthetic medium at pH 5 with glucose as the sole carbon source. Cultures were continuously sparged with pure CO2. Optical density (660 nm) measurements showed a specific growth rate of 0.15 ± 0.01 h−1 for the evolved AsPEPCK-expressing strains IMY050 and IMY051. This corresponded to 50% of the specific growth rate observed for the pyruvate carboxylase-positive reference strain CEN.PK 113-7D (0.30 ± 0.01 h−1) under identical conditions.
Reverse engineering: the role of pyruvate kinase.
Even though growth of the Pyc− IMY002 S. cerevisiae strain was already observed only 4 days into the selection experiment, this period is long enough for a single mutated yeast cell, present at the start of the experiment, to go through 20 generations and produce close to a million daughter cells. Since, in addition, IMY050 and IMY051 retained their phenotype after sequential transfers on nonselective synthetic medium-agar plates (containing glucose and aspartate), it is likely that mutations were responsible for the observed physiological changes. As these mutations could be located in the genome, the AsPEPCK-expressing plasmid, or both, the plasmid was isolated from both strains. In addition, IMY050 and IMY051 were cured of their plasmids, resulting in strains IMW001 and IMW002, respectively.
To test whether the mutations responsible for the observed phenotype were located on the plasmid, the nonevolved Pyc− IMK157 S. cerevisiae strain was transformed with the plasmids isolated from the evolved strains. However, no growth of these strains was observed on synthetic medium-agar plates with glucose as the sole carbon source in the presence of a CO2 atmosphere (data not shown). To check whether changes in the genomes of the evolved strains are responsible for the observed phenotype, the cured strains (IMW001 and IMW002) were retransformed with either the original plasmid (MB4917), the empty vector plasmid (pUDC1), or the plasmids isolated from the evolved strains. After incubation on synthetic medium-agar plates with glucose as the sole carbon source in the presence of a CO2 atmosphere, only the cured strains retransformed with either the original or the isolated AsPEPCK plasmids showed growth. The cured strains transformed with the empty vector did not grow, indicating that the phenotype of the evolved strains not only requires the genomic mutation(s) but also depended on AsPEPCK overexpression.
In the selection experiments it was found that growth depended on the presence of high CO2 concentrations, suggesting that substrate and product concentrations strongly affect the anaplerotic flux through PEPCK. In Pyc− S. cerevisiae the PEP concentration depends not only on the activity of PEPCK but also on the flux through pyruvate kinase (Fig. 1). A lower activity or lower affinity of pyruvate kinase would result in increased PEP levels, which would benefit carboxylation of PEP by PEPCK. Indeed, in vitro enzyme activity measurements of samples taken during the selection of IMY051 showed that the specific activity of pyruvate kinase had dropped significantly (from 10 to 2 mmol·min−1·g protein−1) after a new steady state had been achieved on ammonium sulfate and that the Km had increased from 0.12 to 0.52 mM. Pyruvate kinase activity was also measured for strains IMY002, IMY050, and IMY051 grown in shake flasks on glucose and aspartate medium (Table 4). Here, however, the specific activity in the evolved strains was reduced less markedly (by 45%), while Km actually decreased from 0.13 to 0.08 mM. In contrast, PEPCK activity increased to 0.79 mmol·min−1·g protein−1 under these conditions. If it is assumed that IMY051 is representative of the steady-state culture from which it was derived, it might be concluded that culture conditions strongly affect the properties of pyruvate kinase, possibly by changing the isozyme distribution (6) or by posttranslational modifications.
Subsequent sequencing of PYK1, which encodes the main pyruvate kinase isozyme, revealed single point mutations in the coding region of the gene in both evolved strains (IMY050 and IMY051). Both single-base substitutions resulted in an amino acid change: G436A (in IMY050) giving Asp146Asn and G1006T (in IMY051) giving Ala336Ser. To test whether these mutations accounted for the observed change in phenotype, single point mutation allele replacements (G436A or G1006T) were carried out in the original Pyc− IMK157 strain via pop-in, pop-out recombination (29). With this technique, some of the recombined pop-out transformants will carry the mutated allele. The AsPEPCK-expressing plasmid was introduced into a number of the transformants (successful transformation of the plasmid was verified by PCR), and a phenotypic screening was performed on synthetic medium-agar plates for growth on glucose under a CO2 atmosphere. Sequencing of the PYK1 gene showed that 7 pop-out transformants that grew had incorporated a mutated allele (these included IMY014 [G436A] and IMY015 [G1006T]) (Fig. 2), whereas 2 pop-out transformants that showed no growth both contained the original allele. Together, these findings indicated that the combination of a mutated PYK1 allele and the expression of AsPEPCK enabled growth on glucose as the sole carbon source by IMY050 and IMY051.
ScPEPCK can support anaplerosis in evolved Pyc− S. cerevisiae.
For the previous experiments, (codon-optimized) PEPCK from A. succinogenes was used to circumvent the glucose-induced repression and inactivation that have been described for ScPEPCK (26, 42). Based on the results discussed in the preceding paragraph, we tested whether also the endogenous ScPEPCK (encoded by PCK1) could function in an anaplerotic role in Pyc− S. cerevisiae strains. To this end, plasmid pUDC5 (overexpressing PCK1 isolated from the reference strain CEN.PK 113-7D) was introduced into the cured Pyc− strains IMK157 (not evolved) and IMW001 and IMW002 (both evolved), resulting in strains IMY011, IMY012, and IMY013, respectively. No growth was observed when ScPEPCK was overexpressed in the nonevolved IMY011 strain. In contrast, expression of ScPEPCK in the evolved IMY012 and IMY013 strains did result in growth on synthetic medium-agar plates with glucose under a CO2 atmosphere (Fig. 2). In duplicate glucose-grown and CO2-sparged batch bioreactor cultivations, IMY012 and IMY013 grew at a specific rate of 0.13 ± 0.01 h−1, which is comparable to the rates obtained with the AsPEPCK-expressing S. cerevisiae strains IMY050 and IMY051.
DISCUSSION
PEPCK as an anaplerotic enzyme in S. cerevisiae.
Pyruvate carboxylase-negative strains of S. cerevisiae cannot grow on glucose as the sole carbon source (7, 34). When a Pyc− S. cerevisiae strain was grown in chemostat culture on a mixture of glucose and ethanol, washout occurred when ethanol made up less than 30% of the substrate carbon (12), indicating that the glyoxylate cycle, which synthesizes C4 building blocks from acetyl-coenzyme A (CoA), cannot easily assume the anaplerotic role in place of pyruvate carboxylase. However, suppressor mutants of Pyc− strains could be obtained by UV mutagenesis, and a derepressed glyoxylate cycle was shown to functionally perform the anaplerotic function in place of pyruvate carboxylase (5). In addition, overexpression of the Escherichia coli ppc gene, encoding PEP carboxylase, in a Pyc− mutant restored growth on glucose (18). However, neither alternative for anaplerosis is of particular interest for metabolic engineering of C4 dicarboxylic acid production. The glyoxylate cycle does not involve a net carboxylation (44), which reduces maximum theoretical product yields, while conversion of PEP to oxaloacetate via PEP carboxylase is ATP neutral. In contrast, PEPCK, which in this study was shown to be a third alternative for anaplerosis in Pyc− S. cerevisiae, both fully benefits from carboxylation and generates one ATP per carboxylation event.
The additional ATP that is produced compared to the original anaplerotic route via pyruvate carboxylase is both PEPCK's main advantage and disadvantage. While the additional ATP increases metabolic flexibility and might allow for anaerobic dicarboxylic acid production, it also increases the free energy change (ΔG′) of the reaction by at least 32 kJ mol−1. In addition, PEPCK uses CO2 as the source of inorganic carbon, whereas pyruvate carboxylase uses bicarbonate (10). In S. cerevisiae, inorganic carbon is transported over the cell membrane as dissolved CO2 by (facilitated) diffusion (2). Intracellularly, Nce103p carbonic anhydrase catalyzes the conversion between CO2 and bicarbonate (1). Assuming a cytosolic pH higher than the pKa1 of carbonic acid (pH 6.4) (9), dissolved CO2 will be less abundant than bicarbonate when this reaction is in equilibrium. The less-favorable thermodynamics of carboxylation by PEPCK most likely explain why wild-type S. cerevisiae strains use pyruvate carboxylase instead and are also consistent with the decreased pyruvate kinase activity in the evolved Pyc− PEPCK-overexpressing strains. Although this might come at the cost of a reduced maximum specific growth rate, a lower pyruvate kinase activity will increase the concentration of the shared substrate PEP, which benefits PEPCK. The thermodynamics of the PEPCK reaction also explain why the evolved strains require high CO2 concentrations for growth on glucose and why several natural succinate producers, which are known to use PEPCK for anaplerosis (23, 25, 30, 35), are found in the cow rumen, an anaerobic and CO2-rich environment. For example, at atmospheric pressure and equilibrium between the gas and liquid phases, an increase in the concentration of CO2 in the gas phase from 1% to 100% would decrease the ΔG by 12 kJ mol−1.
Whereas in previous selection experiments with (mutagenized) Pyc− S. cerevisiae the glyoxylate cycle became derepressed (5), this cycle did not seem to play a role in our evolution experiments. The multiple dependencies for growth on glucose exhibited by the evolved Pyc− PEPCK-expressing S. cerevisiae strains (requirements for CO2 enrichment, for PEPCK overexpression, and for a lowered pyruvate kinase activity) can be explained only if anaplerosis occurred via PEPCK. The absence of glyoxylate cycle derepression mutants during selection might be explained by inhibition of succinate dehydrogenase by CO2 (4, 15, 40). As this enzyme is required for a functional glyoxylate cycle, its inhibition by CO2 would represent a second hurdle in addition to the glucose repression of the glyoxylate cycle enzymes isocitrate lyase and malate synthase (16, 17, 20, 32).
PEPCK for C4 dicarboxylic acid production by S. cerevisiae.
The main attraction of using PEPCK to perform the carboxylating role in place of pyruvate carboxylase in S. cerevisiae is the improved ATP yield of C4 dicarboxylic acid production. If acid production has a positive ATP yield, it may be possible to completely eliminate additional pathways for sugar dissimilation, such as ethanolic fermentation or respiration. This should strongly improve product yields and is a prerequisite for “carbon-negative” processes, in which a net fixation of CO2 takes place. However, although this study shows that PEPCK can indeed sustain anaplerosis in S. cerevisiae, a higher carboxylating flux is required for successful application in metabolic engineering strategies for production of C4 dicarboxylic acids.
The Pyc− AsPEPCK-expressing strains constructed in this study grew slower on glucose under a CO2 atmosphere than an isogenic reference strain (0.15 ± 0.01 h−1 versus 0.30 ± 0.01 h−1). If this is taken as evidence that the anaplerotic PEPCK activity in these strains is growth limiting, the maximum anaplerotic capacity can then be estimated to be circa 0.3 mmol h−1 g dry biomass−1 (assuming a protein mass fraction in dry biomass of 0.4 [22], an average amino acid molecular mass of 109 g mol−1 [22], and a C4 requirement of 0.5 mol mol amino acid−1 [11, 22, 28]). In comparison, in a previous study, an engineered strain of S. cerevisiae overexpressing pyruvate carboxylase was found to be able to produce malate at a 7-fold-higher rate of ca. 2 mmol h−1 g dry biomass−1 (43). The anaplerotic flux might be increased by selecting for mutants with a higher growth rate (e.g., via sequential batch reactor cultivation). However, even if a growth rate of 0.30 h−1 could be reached with these Pyc− AsPEPCK strains, the increase in anaplerotic capacity would only be 2-fold.
An interesting approach to further increase the carboxylating PEPCK flux would be natural selection. If PEPCK-catalyzed carboxylation can be made solely responsible for fulfilling the ATP requirements of the cell, there would be a strong selective pressure to increase the rate of C4 dicarboxylic acid production. The first demand, that no other pathway should supply the cell with ATP, can be met by applying anaerobic conditions and by deleting the pyruvate decarboxylase genes, which, respectively, eliminate respiration and ethanolic fermentation. However, the second demand is more challenging. For the production of intracellular malate from glucose, the use of PEPCK increases the ATP yield from 0 to 1 mol mol malate−1. Although such a yield might be sufficient for growth, it is unclear whether malate can be exported efficiently from the cell without a free energy investment. Any export mechanism that depends on ATP hydrolysis or proton translocation (37) will consume all the ATP generated by PEPCK. Therefore, the success of this approach depends on the availability of energetically neutral acid export or, alternatively, on an increase in free energy conservation in the production of intracellular acid.
Acknowledgments
The Ph.D. research of R.M.Z. is financed by Tate & Lyle Ingredients Americas. This project was carried out within the research program of the Kluyver Centre for Genomics of Industrial Fermentation, which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research.
We acknowledge Eveline Vreeburg for her contributions to the experimental work and Peter Niederberger for granting permission to use strain CEN.JB27.
Footnotes
Published ahead of print on 25 June 2010.
REFERENCES
- 1.Aguilera, J., J. P. van Dijken, J. H. de Winde, and J. T. Pronk. 2005. Carbonic anhydrase (Nce103p): an essential biosynthetic enzyme for growth of Saccharomyces cerevisiae at atmospheric carbon dioxide pressure. Biochem. J. 391:311-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahn, Y., and F. A. Mühlschlegel. 2006. CO2 sensing in fungi and beyond. Curr. Opin. Microbiol. 9:572-578. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, J., M. A. H. Luttik, C. Flores, J. P. van Dijken, J. T. Pronk, and P. Niederberger. 1999. By-product formation during exposure of respiring Saccharomyces cerevisiae cultures to excess glucose is not caused by a limited capacity of pyruvate carboxylase. FEMS Microbiol. Lett. 179:107-113. [DOI] [PubMed] [Google Scholar]
- 4.Bendall, D. S., S. L. Ranson, and D. A. Walker. 1960. Effects of carbon dioxide on the oxidation of succinate and reduced diphosphopyridine nucleotide by Ricinus mitochondria. Biochem. J. 76:221-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blázquez, M. A., F. Gamo, and C. Gancedo. 1995. A mutation affecting carbon catabolite repression suppresses growth defects in pyruvate carboxylase mutants from Saccharomyces cerevisiae. FEBS Lett. 377:197-200. [DOI] [PubMed] [Google Scholar]
- 6.Boles, E., F. Schulte, T. Miosga, K. Freidel, E. Schluter, F. Zimmermann, C. Hollenberg, and J. Heinisch. 1997. Characterization of a glucose-repressed pyruvate kinase (Pyk2p) in Saccharomyces cerevisiae that is catalytically insensitive to fructose-1,6-bisphosphate. J. Bacteriol. 179:2987-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brewster, N. K., D. L. Val, M. E. Walker, and J. C. Wallace. 1994. Regulation of pyruvate carboxylase isozyme (PYC1, PYC2) gene expression in Saccharomyces cerevisiae during fermentative and nonfermentative growth. Arch. Biochem. Biophys. 311:62-71. [DOI] [PubMed] [Google Scholar]
- 8.Burke, D., D. Dawson, and T. Stearns (ed.). 2000. Methods in yeast genetics, p. 103-105. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 9.Canelas, A. B., W. M. van Gulik, and J. J. Heijnen. 2008. Determination of the cytosolic free NAD/NADH ratio in Saccharomyces cerevisiae under steady-state and highly dynamic conditions. Biotechnol. Bioeng. 100:734-743. [DOI] [PubMed] [Google Scholar]
- 10.Cooper, T. G., T. T. Tchen, H. G. Wood, and C. R. Benedict. 1968. The carboxylation of phosphoenolpyruvate and pyruvate. I. The active species of “CO2” utilized by phosphoenolpyruvate carboxykinase, carboxytransphosphorylase, and pyruvate carboxylase. J. Biol. Chem. 243:3857-3863. [PubMed] [Google Scholar]
- 11.Daran-Lapujade, P., M. L. A. Jansen, J. Daran, W. van Gulik, J. H. de Winde, and J. T. Pronk. 2004. Role of transcriptional regulation in controlling fluxes in central carbon metabolism of Saccharomyces cerevisiae: a chemostat culture study. J. Biol. Chem. 279:9125-9138. [DOI] [PubMed] [Google Scholar]
- 12.de Jong-Gubbels, P., J. Bauer, P. Niederberger, I. Stückrath, P. Kötter, J. P. van Dijken, and J. T. Pronk. 1998. Physiological characterisation of a pyruvate-carboxylase-negative Saccharomyces cerevisiae mutant in batch and chemostat cultures. Antonie Van Leeuwenhoek 74:253-263. [DOI] [PubMed] [Google Scholar]
- 13.de Jong-Gubbels, P., P. Vanrolleghem, S. Heijnen, J. P. Van Dijken, and J. T. Pronk. 1995. Regulation of carbon metabolism in chemostat cultures of Saccharomyces cerevisiae grown on mixtures of glucose and ethanol. Yeast 11:407-418. [DOI] [PubMed] [Google Scholar]
- 14.de Torrontegui, G., E. Palacián, and M. Losada. 1966. Phosphoenolpyruvate carboxykinase in gluconeogenesis and its repression by hexoses in yeasts. Biochem. Biophys. Res. Commun. 22:227-231. [DOI] [PubMed] [Google Scholar]
- 15.Drake, B. G., J. Azcon-Bieto, J. Berry, J. Bunce, P. Dijkstra, J. Farrar, R. M. Gifford, M. A. Gonzalez-Meler, G. Koch, H. Lambers, J. Siedow, and S. Wullschleger. 1999. Does elevated atmospheric CO2 concentration inhibit mitochondrial respiration in green plants? Plant Cell Environ. 22:649-657. [Google Scholar]
- 16.Duntze, W., D. Neumann, J. M. Gancedo, W. Atzpodien, and H. Holzer. 1969. Studies on the regulation and localization of the glyoxylate cycle enzymes in Saccharomyces cerevisiae. Eur. J. Biochem. 10:83-89. [DOI] [PubMed] [Google Scholar]
- 17.Fernández, E., F. Moreno, and R. Rodicio. 1992. The ICL1 gene from Saccharomyces cerevisiae. Eur. J. Biochem. 204:983-990. [DOI] [PubMed] [Google Scholar]
- 18.Flores, C., and C. Gancedo. 1997. Expression of PEP carboxylase from Escherichia coli complements the phenotypic effects of pyruvate carboxylase mutations in Saccharomyces cerevisiae. FEBS Lett. 412:531-534. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg, I., J. S. Rokem, and O. Pines. 2006. Organic acids: old metabolites, new themes. J. Chem. Technol. Biotechnol. 81:1601-1611. [Google Scholar]
- 20.Hartig, A., M. M. Simon, T. Schuster, J. R. Daugherty, H. S. Yo, and T. G. Cooper. 1992. Differentially regulated malate synthase genes participate in carbon and nitrogen metabolism of S. cerevisiae. Nucleic Acids Res. 20:5677-5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jantama, K., M. J. Haupt, S. A. Svoronos, X. Zhang, J. C. Moore, K. T. Shanmugam, and L. O. Ingram. 2008. Combining metabolic engineering and metabolic evolution to develop nonrecombinant strains of Escherichia coli C that produce succinate and malate. Biotechnol. Bioeng. 99:1140-1153. [DOI] [PubMed] [Google Scholar]
- 22.Lange, H. C., and J. J. Heijnen. 2001. Statistical reconciliation of the elemental and molecular biomass composition of Saccharomyces cerevisiae. Biotechnol. Bioeng. 75:334-344. [DOI] [PubMed] [Google Scholar]
- 23.Lee, S. J., H. Song, and S. Y. Lee. 2006. Genome-based metabolic engineering of Mannheimia succiniciproducens for succinic acid production. Appl. Environ. Microbiol. 72:1939-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 25.McKinlay, J. B., Y. Shachar-Hill, J. G. Zeikus, and C. Vieille. 2007. Determining Actinobacillus succinogenes metabolic pathways and fluxes by NMR and GC-MS analyses of 13C-labeled metabolic product isotopomers. Metab. Eng. 9:177-192. [DOI] [PubMed] [Google Scholar]
- 26.Mercado, J. J., R. Smith, F. A. Sagliocco, A. J. P. Brown, and J. M. Gancedo. 1994. The levels of yeast gluconeogenic mRNAs respond to environmental factors. Eur. J. Biochem. 224:473-481. [DOI] [PubMed] [Google Scholar]
- 27.Mumberg, D., R. Müller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 28.Oura, E. 1972. Reactions leading to the formation of yeast cell material from glucose and ethanol. Alkon Keskuslaboratorio, Helsinki, Finland.
- 29.Rothstein, R. 1991. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 194:281-301. [DOI] [PubMed] [Google Scholar]
- 30.Samuelov, N. S., R. Lamed, S. Lowe, and J. G. Zeikus. 1991. Influence of CO2-HCO3− levels and pH on growth, succinate production, and enzyme activities of Anaerobiospirillum succiniciproducens. Appl. Environ. Microbiol. 57:3013-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santt, O., T. Pfirrmann, B. Braun, J. Juretschke, P. Kimmig, H. Scheel, K. Hofmann, M. Thumm, and D. H. Wolf. 2008. The yeast Gid complex, a novel ubiquitin ligase (E3) involved in the regulation of carbohydrate metabolism. Mol. Biol. Cell 19:3323-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schöler, A., and H. Schüller. 1993. Structure and regulation of the isocitrate lyase gene ICL1 from the yeast Saccharomyces cerevisiae. Curr. Genet. 23:375-381. [DOI] [PubMed] [Google Scholar]
- 33.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stucka, R., S. Dequin, J. Salmon, and C. Gancedo. 1991. DNA sequences in chromosomes II and VII code for pyruvate carboxylase isoenzymes in Saccharomyces cerevisiae: analysis of pyruvate carboxylase-deficient strains. Mol. Gen. Genet. 229:307-315. [DOI] [PubMed] [Google Scholar]
- 35.van der Werf, M. J., M. V. Guettler, M. K. Jain, and J. G. Zeikus. 1997. Environmental and physiological factors affecting the succinate product ratio during carbohydrate fermentation by Actinobacillus sp. 130Z. Arch. Microbiol. 167:332-342. [DOI] [PubMed] [Google Scholar]
- 36.van Dijken, J. P., J. Bauer, L. Brambilla, P. Duboc, J. M. Francois, C. Gancedo, M. L. F. Giuseppin, J. J. Heijnen, M. Hoare, H. C. Lange, E. A. Madden, P. Niederberger, J. Nielsen, J. L. Parrou, T. Petit, D. Porro, M. Reuss, N. van Riel, M. Rizzi, H. Y. Steensma, C. T. Verrips, J. Vindeløv, and J. T. Pronk. 2000. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb. Technol. 26:706-714. [DOI] [PubMed] [Google Scholar]
- 37.van Maris, A. J. A., W. N. Konings, J. P. van Dijken, and J. T. Pronk. 2004. Microbial export of lactic and 3-hydroxypropanoic acid: implications for industrial fermentation processes. Metab. Eng. 6:245-255. [DOI] [PubMed] [Google Scholar]
- 38.van Maris, A. J., B. M. Bakker, M. Brandt, A. Boorsma, M. Teixeira de Mattos, L. A. Grivell, J. T. Pronk, and J. Blom. 2001. Modulating the distribution of fluxes among respiration and fermentation by overexpression of HAP4 in Saccharomyces cerevisiae. FEMS Yeast Res. 1:139-149. [DOI] [PubMed] [Google Scholar]
- 39.Verduyn, C., E. Postma, W. A. Scheffers, and J. P. van Dijken. 1992. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501-517. [DOI] [PubMed] [Google Scholar]
- 40.Wanders, R. J. A., A. J. Meijer, A. K. Groen, and J. M. Tager. 1983. Bicarbonate and the pathway of glutamate oxidation in isolated rat-liver mitochondria. Eur. J. Biochem. 133:245-254. [DOI] [PubMed] [Google Scholar]
- 41.Werpy, T., and G. Petersen. 2004. Top value added chemicals from biomass: I. Results of screening for potential candidates from sugars and synthesis gas. U.S. Department of Energy, Washington, DC.
- 42.Yin, Z., R. J. Smith, and A. J. P. Brown. 1996. Multiple signalling pathways trigger the exquisite sensitivity of yeast gluconeogenic mRNAs to glucose. Mol. Microbiol. 20:751-764. [DOI] [PubMed] [Google Scholar]
- 43.Zelle, R. M., E. de Hulster, W. Kloezen, J. T. Pronk, and A. J. A. van Maris. 2010. Key process conditions for production of C4 dicarboxylic acids in bioreactor batch cultures of an engineered Saccharomyces cerevisiae strain. Appl. Environ. Microbiol. 76:744-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zelle, R. M., E. de Hulster, W. A. van Winden, P. de Waard, C. Dijkema, A. A. Winkler, J. A. Geertman, J. P. van Dijken, J. T. Pronk, and A. J. A. van Maris. 2008. Malic acid production by Saccharomyces cerevisiae: engineering of pyruvate carboxylation, oxaloacetate reduction, and malate export. Appl. Environ. Microbiol. 74:2766-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, X., K. Jantama, J. C. Moore, L. R. Jarboe, K. T. Shanmugam, and L. O. Ingram. 2009. Metabolic evolution of energy-conserving pathways for succinate production in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 106:20180-20185. [DOI] [PMC free article] [PubMed] [Google Scholar]