Abstract
The human gastrointestinal (GI) tract provides home to a complex microbial community, collectively termed microbiota. Although major efforts have been made to describe the diversity and stability of the microbiota, functional studies have been largely restricted to intestinal isolates and include few community studies. The aim of this study was to explore the in situ gene expression of the fecal microbiota and to evaluate the RNA fingerprinting method cDNA-AFLP (cDNA amplified fragment length polymorphism) for this purpose. To this end, cDNA-AFLP analysis of enriched mRNA revealed that two healthy subjects showed highly divergent expression profiles with considerable fluctuations in time. Subsequent excision and sequence determination of bands from the mRNA-enriched profiles resulted in 122 identifiable sequences (transcripts and rRNAs). The classification of retrieved transcripts into functional clusters based on COG (cluster of orthologous genes) annotation showed that most assigned transcripts belonged to the metabolism cluster (26% of all sequences), underlining that even at the very end of the intestinal tract the microbiota is still very active. This study furthermore revealed that cDNA-AFLP is a useful tool to compare gene expression profiles in time in complex microbial communities.
The human gastrointestinal (GI) tract provides a niche for a complex community of microorganisms, commonly referred to as microbiota. Until recently, research focusing on the human GI tract microbiota was mostly aimed at describing its diversity and, to some extent, its functionality (49). Studies that investigated the functionality of this microbiota focused mainly on predominating species found in the large intestine and feces, including members of the genera Clostridium, Bacteroides, and Bifidobacterium (13, 19, 31, 40). The few studies that investigated the functionality of a complete microbial ecosystem used metaproteomics (14, 41), the construction and analysis of a cDNA library (24), or microarrays (17, 44), whereas recent studies applied next-generation sequencing technologies (NGT) (10, 37). In the metaproteome study described by Klaassens et al., temporal changes in production of a protein with high similarity to bifidobacterial aldolase were found in the feces of an unweaned infant (14). A shotgun metaproteomics approach of extracted proteins from two fecal samples revealed that proteins with functions related to translation, carbohydrate metabolism, and energy production were among the most abundantly present proteins (41). Analysis of a cDNA library is an approach at the transcriptome level that can be used to investigate the gene expression of mixtures of microorganisms. The application of this approach to bacterioplankton communities already showed that several functional genes of environmentally important processes could be detected (24). Although that study provided useful information about the activity of the ecosystem under study, application to the far more complex bacterial ecosystems such as in the GI tracts of human adults with a large number of different bacterial genomes present (>1,100 species) (48) will be far more complicated.
Over the last decade, the microarray platform has seen a rapid development toward applicability for environmental microorganisms (43). So-called functional gene arrays have been developed to investigate genes encoding key enzymes representative for certain metabolic pathways (17, 35, 44), including metal resistance, biodegradation (28), and dechlorinating activity (7). Nevertheless, since only differences in gene expression of already known sequences (at the time of platform construction) can be detected, application is still limited to communities for which the microbial diversity is largely known (43). Furthermore, the application of such a mixed-genome platform to a complex ecosystem such as the GI tract microbiota would have the drawback that sequence variation in closely related genes causes artifacts in the quantitative interpretation of the gene specific signals obtained by array analysis, as observed before in other microbial ecosystems (35, 44).
An increasing number of studies applied NGT to analyze the metatranscriptome of environmental samples (2, 9, 10, 39), including the murine distal gut ecosystem (37). This revolutionizing approach enabled access to both known and previously unknown transcripts in microbial communities (10). In addition, community structure and function can be efficiently linked in a single experiment without biases inherent to both methods (39). However, NGT sequencing still faces big challenges, including the pressing need for bioinformatics algorithms and the very high sequence coverage crucial for the detection of differences in gene expression levels (2).
The RNA fingerprinting method cDNA amplified fragment length polymorphism (cDNA-AFLP) has been shown to be a very powerful tool for investigating (temporal) gene expression in several biological processes of a single specimen (3). In terms of sensitivity, reproducibility, and specificity, cDNA-AFLP results match very well with GeneChip results (27). Relative to microarray-based approaches, cDNA-AFLP has the advantage that any unknown genome or set of genomes can be studied without prior sequence knowledge (3). Therefore, this technique should in principle be applicable to any microbial ecosystem. Moreover, the cDNA-AFLP approach allows the detection of lowly expressed genes and may allow the discrimination between homologous genes. Thus far, this method has been applied most extensively (more than 100 studies) to analyze transcription profiles of single species, mostly plants (6). In addition, cDNA-AFLP was applied to a lesser extent to study the gene expression in mammalian cells (8, 21, 29), yeasts (27), nematodes (22), and some bacteria, mainly plant pathogens (32). The aim of the present study was to gain insight into the in situ gene expression of the human GI tract microbiota and to evaluate cDNA-AFLP as a potentially suitable technology for this purpose. To this end, total DNA and RNA extracts, as well as mRNA enriched fractions from fecal samples, were used for cDNA-AFLP profiling. The technical reproducibility of the nucleotide isolation method (DNA or RNA), the mRNA enrichment, and the AFLP profiling were quantified. In addition, profiles of two fecal samples from two individuals were compared to get insight into the temporal stability of the gene expression. Excision of bands from the mRNA-enriched profiles and subsequent sequence analysis was performed to investigate the functional distribution of genes expressed by the GI tract microbiota. Overall, the present study explores the applicability of cDNA-AFLP to analyze the composition and stability of the human fecal microbiota metatranscriptome.
MATERIALS AND METHODS
Sample collection.
Fresh fecal samples were collected from two healthy volunteers (26-year-old female; 33-year-old male) with a time interval of 3 days. Informed consent was obtained from each volunteer before the sampling. The subjects had not been subjected to any antibiotic treatment, dietary intervention, or specific diet for at least 1 year prior to sampling and considered themselves healthy. Samples were stored at −20°C for DNA isolation or quenched with methanol-HEPES buffer at −80°C (±1:4 [vol/vol]) after thorough homogenization for RNA isolation (23) and processed within 24 h after collection.
DNA and RNA isolation, quantity and quality check.
DNA was isolated from fecal samples as described by Zoetendal et al. (47) by using a stool DNA isolation kit (Qiagen, Venlo, Netherlands) and quantified spectrophotometrically (Nanodrop; Isogen Life Science B.V., IJsselstein, Netherlands). Total RNA was isolated from fresh fecal samples quenched in methanol-HEPES by using a Macaloid-based RNA isolation procedure (46). Total RNA was treated with RNase-free DNase I (Roche, Almere, Netherlands; 10 U of DNase per 20 μg of RNA) for 20 min at room temperature. The RNA was heat denatured at 65°C for 10 min. DNA absence was verified by using 100 ng of total RNA in a PCR with 16S rRNA gene-specific primers. The quality of RNA was determined by electrophoresis (2100 Bioanalyzer; Agilent Technologies, Amstelveen, Netherlands).
Adaptation of the cDNA-AFLP procedure for profiling of prokaryotic transcripts.
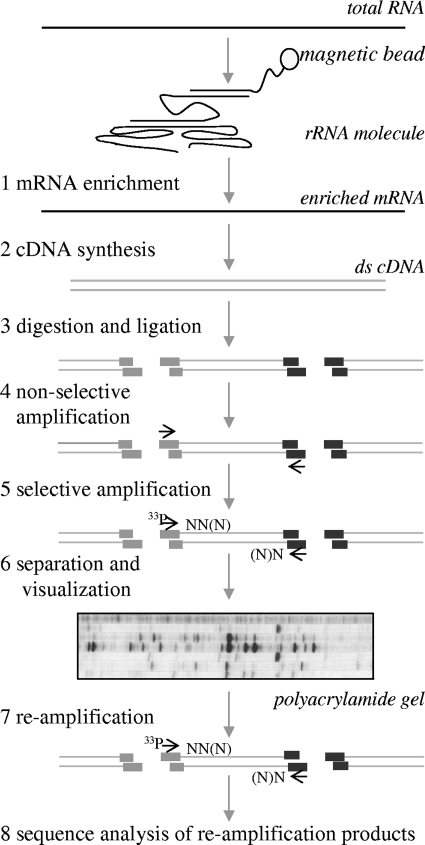
The cDNA-AFLP method was originally developed to study gene expression in eukaryotic species. Since the selective cDNA synthesis of mRNAs based on reverse transcription of the 3′-poly(A) tail is not applicable to prokaryotic messengers, several methods to enrich for prokaryotic mRNAs were investigated. Subtractive hybridization by rRNA probe-based fishing (MicrobExpress bacterial mRNA enrichment kit; Ambion/Applied Biosystems, Nieuwerkerk a/d IJssel, Netherlands) was found to be most efficient in enriching for mRNA from total RNA and was therefore added to the original cDNA-AFLP procedure (3) prior to cDNA-synthesis (Fig. 1). An aliquot of 10 μg of DNase-treated RNA was used for mRNA enrichment by MicrobExpress (Ambion); the enriched mRNA was quantified, and its quality was checked based on the presence of the tRNA and 5S rRNA peaks, since these molecules should remain unaffected during the mRNA enrichment procedure.
FIG. 1.
Schematic overview of the cDNA-AFLP procedure applied to RNA isolated from GI tract samples, adapted from Bachem et al. (4). (Step 1) Total RNA isolated from GI tract samples (feces) is enriched for mRNA by subtractive hybridization as indicated by the manufacturer (MicrobExpress; Ambion, Huntington, United Kingdom). (Step 2) cDNA is synthesized by reverse transcription using a mixture of random decamers, followed by second-strand synthesis. (Step 3) The double-stranded cDNA is digested by two different 4-bp cutting enzymes, followed by ligation of adaptors. The adaptors used are designed in such a way that once ligated to the sticky ends of the fragments, the sequence is changed and no longer recognized by the restriction enzymes. Therefore, restriction and ligation can be performed together. (Step 4) Nonselective primers (indicated by arrows), which anneal to the ligated adaptors, are used to nonselectively amplify the cDNA molecules. (Step 5) In the selective amplification, a small aliquot of the nonselectively amplified fragments is used in a second PCR. These selective primers extend one, two, or three bases inward. This reduces the number of fragments by a factor 64, 256, or 1,024, respectively. One of the primers contains a radioactive dye (33P). (Step 6) The fragments are size separated and visualized on a polyacrylamide gel. (Step 7) The fragments of the selected profiles are reamplified with the corresponding selective primer pairs used in step 5. (Step 8) Sequence analysis of reamplified fragments is performed.
Single-stranded cDNA was synthesized by using avian myeloblastosis virus reverse transcriptase (Invitrogen Life Technologies B.V., Breda, Netherlands) and random hexamers (200 ng μl−1). Double-stranded cDNA was generated by using protocols described by Sambrook et al. (30). The double-stranded cDNA was purified and concentrated in 10 μl by using a MinElute PCR purification kit (Qiagen).
cDNA-AFLP analysis.
Double-stranded cDNA (50 ng) was digested with TaqI and MseI or with TaqI and BfaI, respectively (New England Biolabs, Westburg, Leusden, Netherlands) (4, 42). After digestion, ligation of the adaptors (TaqI [3] and MseI [4, 42] or BfaI [identical to the MseI adaptor]), and nonselective amplification, the amplification mixtures were diluted to get equal concentrations for all samples, and 5 μl was used as a template for the final amplifications using a 33P-labeled TaqI primer (with one/two selective nucleotides) and BfaI or MseI (with one or two/three selective nucleotides, respectively). In all, 15 different sets of selective primer combinations were used for cDNA-AFLP analysis (Table 1).
TABLE 1.
Restriction enzymes and selective nucleic acids used for profiling the nucleotides (DNA, total RNA, and enriched mRNA) extracted from the fecal microbiota of two healthy subjects
| Primer codea | Primer1 | Primer2 |
|---|---|---|
| TR14/B01 | TaqI+AT | BfaI+A |
| TR14/B03* | TaqI+AT | BfaI+G |
| TR14/B04* | TaqI+AT | BfaI+T |
| TR01/M20* | TaqI+A | MseI+GC |
| TR01/M23* | TaqI+A | MseI+TA |
| TR01/M24* | TaqI+A | MseI+TC |
| TR03/M17 | TaqI+A | MseI+CG |
| TR12/M24 | TaqI+AC | MseI+TC |
| TR18/M17 | TaqI+CT | MseI+CG |
| TR14/M20 | TaqI+AT | MseI+GC |
| TR14/M23 | TaqI+AT | MseI+TA |
| TR14/M24 | TaqI+AT | MseI+TC |
| TR14/M82* | TaqI+AT | MseI+TAT |
| TR14/M85* | TaqI+AT | MseI+TCG |
| TR20/M55 | TaqI+GC | MseI+CGA |
Primer combinations that were used for sequence analysis of the profiled RNA molecules are indicated with an asterisk (*).
Visualization on the gel, fragment isolation, and sequence analysis.
Polyacrylamide gels (4.5%; 7 M urea-1.0× Tris-borate-EDTA) were prepared according to the manufacturer's instructions (SequaGel; AGCT Bioproducts, Hessle, United Kingdom). Amplification products (1.4 μl) were loaded onto the prewarmed gel (45°C), and Sequamark 10-bp ladder (Invitrogen Life Technologies) was loaded every 10th lane for normalization purposes. The gel was run for 2 h at 45°C, and all well-separated bands within the lanes of enriched mRNA profiles were excised and reamplified with the selective primers corresponding to the profile, as described previously (27). Reamplification products were purified prior to sequencing to remove excess primers and deoxynucleoside triphosphates. Sequencing reactions were performed by using a Dyenamic ET dye terminator cycle sequencing kit and were analyzed on a MegaBace automated sequencer (GE Healthcare, Diegum, Belgium). The sizes of the retrieved sequences were compared to the sizes of the corresponding bands on the cDNA-AFLP gel, and only those that corresponded in size were taken into account for further analysis.
Phylogenetic analysis.
For analysis of the microbial composition of the fecal samples, the phylogenetic microarray human intestinal tract chip (HITChip) was used that covers more than 1,100 intestinal microbial phylotypes (26). The procedure for hybridization and analysis was performed as described previously (26).
Statistical analysis and cDNA-AFLP reliability analysis.
Normalization of the AFLP profiles was performed based on the 10-bp ladder profiles with the software package BioNumerics (version 4.0; Applied Maths N.V., Sint-Martens-Latem, Belgium). Subsequent cluster analysis of the technical duplicates was conducted by using the pairwise Pearson correlation coefficient based on densitometric curves of the cDNA-AFLP profiles. Dendrograms were generated by UPGMA (unweighted pair group method with arithmetic mean) clustering, which describes the relationship among the fecal transcription profiles. The similarity between samples was deduced from the pairwise comparison of cDNA-AFLP profiles.
To analyze the fingerprint variation caused by the (non)selective amplification and subsequent profiling step of the cDNA-AFLP procedure, Pearson-based correlation coefficients were calculated between mRNA samples collected in time and profiled in duplicate for two of the +2/+2 selective primer pairs used. In addition, the fingerprint variation caused by the choice of primers (either +1/+2, +2/+2, or +2/+3) and random profile comparison were analyzed as follows. The ranges and standard deviations of the Pearson-based correlation coefficients were calculated between both samples collected in time for all of the 15 primer pairs included for further sequence analysis.
Bioinformatics.
The taxonomic assignment of the cDNA-AFLP sequences was performed by searching for homologous sequences in the GenBank database (including bacterial and human ST sequences, STS, GSS, environmental samples, and phase 0, 1, or 2 HTGS sequences) using BLAST (1) with an e-value threshold of 1e−10 or >40% identity with protein sequences. Putative proteins encoded by the cDNA sequences were assigned to orthologous groups of the COG database (36) as described previously (33).
Quantification of transcripts by quantitative PCR.
To quantify the expression level of the transcripts detected by cDNA-AFLP analysis relative to total 16S rRNA gene copies, quantitative PCR was used for six transcripts detected at both time points (t0 and t3). These transcripts had been detected in profiles generated with the primer TaqI+A/MseI+GC during cDNA-AFLP analysis. Quantitative PCRs (25 μl) were performed by using 1× SYBR green PCR Master Mix (Applied Biosystems, Foster City, CA) and 100 nM concentrations of each of the primers (MWG, Ebersberg, Germany). In each reaction, 5 μl of 10-fold-diluted cDNA was used as a template, and plasmids containing the six chosen transcript fragments were used as a standard. Total 16S rRNA was determined with the primers Bact-1369f and Prok-1492r (34), with a plasmid containing the complete 16S rRNA gene of L. gasseri as a standard. Quantitative PCR amplification and detection of cDNA were performed with the iQ5 (Bio-Rad Laboratories B.V., Veenendaal, Netherlands). The data analysis was conducted with iQ5 Optical System Software version 1.1.
Nucleotide sequence accession numbers.
The mRNA sequences determined in the present study were deposited in the GenBank database under accession numbers GW391682 to GW391721.
RESULTS AND DISCUSSION
mRNA enrichment from total RNA.
The mRNA enrichment procedure based on selective removal of 16S and 23S rRNA molecules with magnetic beads (MicrobExpress) was used on the total RNA extracted from the fecal samples. The RNA samples were analyzed both by conventional agarose gel electrophoresis and by capillary electrophoresis (BioAnalyzer) before and after the enrichment procedure to determine the efficiency of the procedure and the quality of the RNA. The quality of the isolated RNA with a 16S/23S rRNA ratio of 1.7 or higher was judged as good (see Fig. SA1 in the supplemental material). After enrichment the peaks corresponding to 16S and 23S rRNA disappeared almost completely, whereas the peak corresponding to tRNAs and 5S rRNA remained quantitatively equal compared to its original level in the total RNA sample. This result indicates that the enrichment procedure selectively removed 16S and 23S rRNA molecules, supporting that mRNA remained unaffected.
For identification of the genes expressed by the fecal microbiota, a subset of the dominant and well-separated bands (93 in total) from the total RNA and mRNA enriched profiles was excised and subjected to comparative sequence analysis using BLAST searches against DNA, protein, and expressed sequence tag (EST) databases. This revealed that 42% of the sequences obtained from the mRNA enriched profiles showed the highest homology to sequences from the mRNA origin (EST or encoded protein) versus 20% of the sequences obtained from the total RNA profiles, underlining the importance of the mRNA enrichment procedure. Subsequently, bands were excised only from the mRNA-enriched profiles to maximize the recovery of protein-encoding gene fragments of the fecal metatranscriptome.
Selection of restriction enzyme-primer combination.
The optimal number of bands per lane within a cDNA-AFLP profile for subsequent profile comparison, as well as excision of bands and sequence analysis, is around 50 to 70 (4). To investigate which combination of restriction enzymes was needed to obtain this number of bands for fecal samples, four combinations of restriction enzymes—MboI-TaqI, TaqI-Csp6I, TaqI-BfaI, and TaqI-MseI—were used for initial transcript profiling of fecal samples. The latter two showed ∼50 more or less equally distributed and distinctive bands and were selected as enzyme combinations of choice for further transcript profiling, whereas the other two primer combinations in general gave fewer than 50 bands that were unevenly distributed over the gel. The selectivity of the primer pairs was varied between one to three additional nucleotides on either one of the primers.
Technical reproducibility and reliability of cDNA-AFLP profiles.
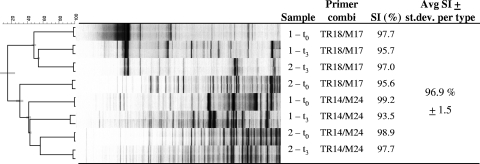
To analyze the fingerprint variation caused by the cDNA-AFLP profiling, the average Pearson-based correlation coefficient was calculated for 8 pairwise comparisons of mRNA profiles. The technical reproducibility of the cDNA-AFLP profiling after mRNA enrichment showed an average reproducibility of 97% (±1.5) based on Pearson-based correlation coefficients (Fig. 2). This indicates that the reproducibility of the profiling procedure is very high, which is essential for the comparison of gene expression profiles over time.
FIG. 2.
Cluster analysis based on UPGMA of technical profiling duplicates of fecal cDNA-AFLP profiles from subjects 1 and 2. Enriched mRNA of two time samples (t0 and t3) were used as the starting material per subject. For cDNA-AFLP profiling of these four samples, two different primer pairs were used: TR18/M17, Taq+CT/Mse+CG; and TR14/M24, Taq+AT/Mse+TC. Similarity index (SI) values based on Pearson correlation for the technical profiling replicates are given with the overall average for +2/+2 primer selectivity. The tree on the left indicates the similarity; the cophenetic correlation coefficients indicated as error bars at the root of each cluster indicate the relatedness of the tree.
In addition, the fingerprint variation caused by the choice of primers was analyzed by calculating the range of the Pearson-based correlation coefficients between both samples collected in time for all 15 primer pairs included for further sequence analysis. This revealed that for the DNA profiles, similarity coefficients between the DNA fingerprints of fecal samples ranged from 64 to 86% for all included primer combinations (indicated by gray shading in the distance tree of Fig. 3). This indicates that the choice of primers introduced a variation of 22% for the DNA profiles. Correlation analysis of the mRNA enriched fingerprints profiled with the same set of primer combinations indicated that the similarity coefficients ranged between 41 and 79% (indicated by gray shading in the distance tree of Fig. 3), introducing a variation of 38%. Nevertheless, this is significantly higher than values observed for random profile comparisons (6.3 to 19.5%). These results stress the importance of the inclusion of a wide range of selective primers for the (cDNA-)AFLP profiling, since each primer represents only a snapshot of the total metagenome or metatranscriptome.
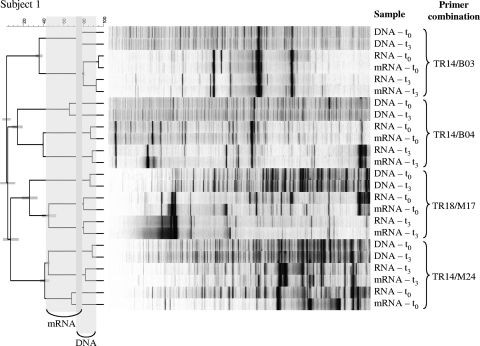
FIG. 3.
Cluster analysis based on UPGMA of the temporal fecal cDNA-AFLP profiles of subject 1. DNA, total RNA, and enriched mRNA were used as starting material. On the left, similarity coefficients based on Pearson correlation are indicated, whereas the gray bars at the root of each cluster indicate the cophenetic correlation coefficients. Four different primer combinations are displayed: TR14/B03, Taq+AT/Bfa+G; TR14/B04, Taq+AT/Bfa+T; TR18/M17, Taq+CT/Mse+CG; TR14/M24, Taq+AT/Mse+TC, respectively. The different time points are indicated by t0 and t3. The gray shading in the distance tree indicates the variation introduced by DNA or mRNA profiling, respectively.
Temporal stability of the metatranscriptome.
To gain insight into the temporal stability of the fecal metatranscriptome, two samples per subject taken with an interval of 3 days were compared. This revealed that for almost all restriction enzymes and primer combinations used, the DNA profiles clustered together as expected, whereas the enriched mRNA profiles were more similar to the total RNA profiles of the same time point, as shown for subject 1 in Fig. 3. Cluster analysis of the profiles of subject 2 revealed the same clustering based on sample type although with slightly lower similarity coefficients. This profile-type-based clustering is most likely due to the residual rRNA present in the enriched mRNA extracts. This is supported by the relatively low efficiency of rRNA removal, which was ca. 50% (the retrieval of mRNA sequences increased from 20 to 42% upon mRNA enrichment). Thus, the residual rRNA will still contribute significantly to the enriched mRNA profiles, resulting in clustering more closely to the RNA profiles than the corresponding time points.
Other studies that used subtractive hybridization with the same method prior to metatranscriptome analysis of environmental samples found mRNA enrichment efficiencies of 80% (24) and 99.9% (11). Since the efficiency of rRNA removal is dependent on the species compatibility of the Microbe Express kit (Ambion), differences in microbial diversity of the microbial ecosystem under study can explain this difference in efficiency.
Subject-specific profiles.
Profiling of both DNA and RNA by (cDNA-)AFLP revealed individual specific differences, which is in line with previous observations based on 16S rRNA gene diversity (45). Remarkably, the temporal variation with respect to DNA- and mRNA-based profiles was different between the two individuals. The DNA profiles were more stable in time than the mRNA profiles in subject 1 (the average similarities between temporal profiles based on Pearson correlation of 15 different selective primer pairs were 65.0% [±18.9] for DNA and 42.8% [±20.7] for mRNA-enriched profiles), which might indicate that the microbiota composition is more stable in time compared to its activity. In contrast, subject 2 showed far lower stability of the microbiota, but a less fluctuating transcriptome pattern in time (the similarities for DNA and enriched mRNA profiles were 36.5% [±21.0] and 47.9% [±19.0], respectively), which might indicate that the microbiota composition varies more over time compared to subject 1. For the DNA-based profiles, this finding was confirmed by the phylogenetic HITChip profiles, although the difference between individuals was not as large (see Fig. SA2 in the supplemental material). Although only a fraction of the total diversity in DNA or RNA sequences was profiled and compared by (cDNA-)AFLP profiling due to the selective amplification step (1/64 of the total metatranscriptome is profiled per each of the seven +1/+2 selective primer combinations used), this snapshot already demonstrates the usefulness of cDNA-AFLP profiling to visualize dynamic trends within the fecal metatranscriptomes.
Retrieval of sequences from the mRNA profiles.
Distinct and well-separated bands of seven enriched mRNA profiles of both subjects profiled at both time points were excised from the gel and used for sequence analysis. Of the 273 excised bands, 58% (156 sequences) resulted in high-quality sequences. For the +2/+3 primer combinations, the success rate was on average higher than for the +2/+2 primer combinations, most likely due to less interference of weak transcript signals in the background of the +2/+3 profiles. BLAST analysis of the retrieved sequences revealed that 11% (n = 11) and 50% (n = 29) of the sequences obtained from the mRNA-enriched profiles of subjects 1 and 2, respectively, showed the highest number of hits with prokaryotic protein-encoding gene sequences, whereas 21% (n = 21) and 24% (n = 13) had no significant hits in the database. The residual 68% (n = 67) and 26% (n = 15) of the sequences obtained from the mRNA-enriched profiles of subjects 1 and 2, respectively, displayed the highest homology to rRNA sequences (mostly with 16S or 23S rRNA gene sequences of the genera Bacteroides and Clostridium).
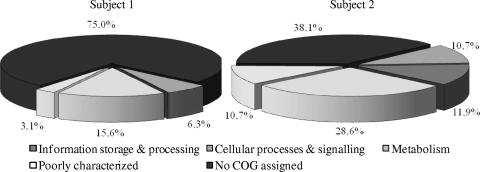
The 74 bacterial protein-encoding and no-hit sequences that were retrieved were investigated to determine their distribution among the functional clusters as defined by COG classification. This allowed the functional annotation of 51% of all expressed genes. The functional distribution of the protein-encoding transcripts into COGs is depicted in Fig. 4. This relatively high percentage of COG-assigned genes is comparable to functional assignments of sequences retrieved from fecal metagenomes: on average, 48.1% assigned COGs to sequences obtained from 13 Japanese subjects (16), and an average of 54.5% assigned COGs to the sequences retrieved from two American subjects (12).
FIG. 4.
Classification of the sequences retrieved from the mRNA enriched cDNA-AFLP profiles of fecal samples of subject 1 (32 sequences) and subject 2 (43 sequences), respectively, based on the COG as defined by Tatusov et al. (36).
High recovery of transcripts involved in metabolism.
Based on COG distribution, most genes found to be expressed by the fecal microbiota were involved in metabolism, in particular of carbohydrate (15 and 11%, respectively, for subjects 1 and 2), making up 26% on average of all sequenced and annotated transcripts (Fig. 4 and see Table SA1 in the supplemental material). This observation is in line with metaproteomics studies on the fecal microbiota of adults (41) and infants (15) and suggests that carbohydrate metabolism is an important function exerted by the microbiota in the colon. In addition, metagenome studies revealed that genes involved in carbohydrate metabolism were relatively enriched in fecal samples of adults (16) and twins (38). Qin et al. recently demonstrated that the metabolism of complex carbohydrates is one of the important functions in host-bacterial interaction, together with the synthesis of short-chain fatty acids, indispensable amino acids, and vitamins (25).
Within the carbohydrate metabolism cluster, sequences encoding for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and pyruvate flavodoxin/ferredoxin oxidoreductase were detected most frequently. Furthermore, response regulators, hydrogenases, SSS sodium solute transporters, and hypothetical proteins were among the retrieved sequences from these fecal samples. Remarkably, many of the transcripts from subject 1 encoded enzymes involved in the glycolysis and pyruvate synthesis (3× GAPDH and 3× pyruvate ferredoxin-oxidoreductase), and almost all of these originated from species of the order Clostridiales. The colon has a lower transit rate compared to the preceding part of the GI tract (20) and, as a result, the simple sugars are completely absorbed at the end of the colon, leaving fecal microbes such as, for example, Clostridium cluster XIVa, with the more complex molecules for fermentation (18). Species belonging to this cluster were also found to be dominantly present in the living fractions of fecal samples (5).
In contrast to subject 1, transcripts encoding enzymes involved in amino acid conversion were observed in the microbiota of subject 2 (see Table SA1 in the supplemental material). The amino acid metabolism was indicated as one of importance for the microbiota by other researchers (38). This might indicate that the GI tract microbiota in subject 2 is involved in the fermentation of proteins. However, this remains speculative, since only a fraction of the total fecal metatranscriptome of two individuals was studied. Furthermore, environmental factors, such as diet, could also contribute to the differences observed between both individuals as noted before (38).
Transcript quantification by quantitative reverse transcription-PCR.
Profile comparison and sequence analysis revealed that six pairs of bands observed at the same height in the temporal profiles of subjects 1 and 2 on the cDNA-AFLP gels resulted in sequences that were identical, with minor differences in sequence length. The six identified genes detected at both time points (three for each person) were analyzed by quantitative PCR. Two investigated transcripts (2F02 with a significant hit to phosphonate ABC transporter and 2G02 with a significant hit to an unknown protein within the phylum Proteobacteria) had transcription levels too low to be analyzed by quantitative PCR. Two other transcripts (1E07a with no significant hits and 2G01 with a significant hit to aspartate kinase) were expressed just above the detection level. The latter two transcripts (1E04a and 1E09a; both with no significant hits compared to GenBank) were detected in numbers as high as 2.3 × 107 and 1.9 × 107 cDNA copies for transcripts 1E04a and 1E07a, respectively, on t0, whereas these levels were 1.9 × 107 and 2.0 × 107 on t3, respectively.
Conclusion.
In the present study we evaluated cDNA-AFLP as a fingerprinting method for the characterization of the GI tract microbiota metatranscriptome. We have shown here that the average similarity of duplicate profiles was 97%, indicating a high level of reproducibility. Furthermore, individual-specific differences with respect to population composition, gene expression, and their variation over time were found. The majority of annotated transcripts of both individuals were found within the metabolism functional cluster, in particular the carbohydrate metabolism. However, the retrieval of sequences was limited due to the interference of background transcripts in the profiles. In addition, only a snapshot of the total metatranscriptome can be analyzed when a subset of the available selective primers is used. Therefore, cDNA-AFLP analysis seems to be most suitable for determining whether environmental variables (such as space and time) have an influence on the gene expression profile and composition, since this technology has the advantages that gene expression profiles can be compared relatively easily for both highly abundant and rare transcripts. Subsequent in-depth metatranscriptome analysis to determine the mRNA identity should be performed with the more high-throughput next-generation sequencing tools (2) as soon as bioinformatic challenges are overcome. Metatranscriptomic studies can eventually be used in a complementary manner to achieve a better understanding of the ecology and functionality of the human GI tract microbiota in health and disease.
Supplementary Material
Acknowledgments
We thank Christian Bachem, Asun Fernandez del Carmen, and Louisa Trindade for useful suggestions for modifications of the cDNA-AFLP procedure for whole community analysis. We are very grateful to Michiel Wels for performing part of the bioinformatics analysis.
Footnotes
Published ahead of print on 18 June 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Asmann, Y. W., M. B. Wallace, and E. A. Thompson. 2008. Transcriptome profiling using next-generation sequencing. Gastroenterology 135:1466-1468. [DOI] [PubMed] [Google Scholar]
- 3.Bachem, C. W., R. S. van der Hoeven, S. M. de Bruijn, D. Vreugdenhil, M. Zabeau, and R. G. Visser. 1996. Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: analysis of gene expression during potato tuber development. Plant J. 9:745-753. [DOI] [PubMed] [Google Scholar]
- 4.Bachem, C. W. B., R. J. F. J. Oomen, and R. G. F. Visser. 1998. Transcript imaging with cDNA-AFLP: a step-by-step protocol. Plant Mol. Biol. Rep. 16:157-173. [Google Scholar]
- 5.Ben-Amor, K., H. Heilig, H. Smidt, E. E. Vaughan, T. Abee, and W. M. de Vos. 2005. Genetic diversity of viable, injured, and dead fecal bacteria assessed by fluorescence-activated cell sorting and 16S rRNA gene analysis. Appl. Environ. Microbiol. 71:4679-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bensch, S., and M. Akesson. 2005. Ten years of AFLP in ecology and evolution: why so few animals? Mol. Ecol. 14:2899-2914. [DOI] [PubMed] [Google Scholar]
- 7.Dennis, P., E. A. Edwards, S. N. Liss, and R. Fulthorpe. 2003. Monitoring gene expression in mixed microbial communities by using DNA microarrays. Appl. Environ. Microbiol. 69:769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ekkapongpisit, M., T. Wannatung, T. Susantad, K. Triwitayakorn, and D. R. Smith. 2007. cDNA-AFLP analysis of differential gene expression in human hepatoma cells (HepG2) upon dengue virus infection. J. Med. Virol. 79:552-561. [DOI] [PubMed] [Google Scholar]
- 9.Frias-Lopez, J., Y. Shi, G. W. Tyson, M. L. Coleman, S. C. Schuster, S. W. Chisholm, and E. F. DeLong. 2008. Microbial community gene expression in ocean surface waters. Proc. Natl. Acad. Sci. U. S. A. 105:3805-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert, J. A., D. Field, Y. Huang, R. Edwards, W. Li, P. Gilna, and I. Joint. 2008. Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS One 3:e3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert, J. A., S. Thomas, N. A. Cooley, A. Kulakova, D. Field, T. Booth, J. W. McGrath, J. P. Quinn, and I. Joint. 2009. Potential for phosphonoacetate utilization by marine bacteria in temperate coastal waters. Environ. Microbiol. 11:111-125. [DOI] [PubMed] [Google Scholar]
- 12.Gill, S. R., M. Pop, R. T. Deboy, P. B. Eckburg, P. J. Turnbaugh, B. S. Samuel, J. I. Gordon, D. A. Relman, C. M. Fraser-Liggett, and K. E. Nelson. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooper, L. V., M. H. Wong, A. Thelin, L. Hansson, P. G. Falk, and J. I. Gordon. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881-884. [DOI] [PubMed] [Google Scholar]
- 14.Klaassens, E. M., W. M. De Vos, and E. E. Vaughan. 2007. Metaproteomics approach to study the functionality of the microbiota in the human infant gastrointestinal tract. Appl. Environ. Microbiol. 73:1388-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klaassens, E. S., R. J. Boesten, M. Haarman, J. Knol, F. H. Schuren, E. E. Vaughan, and W. M. de Vos. 2009. Mixed-species genomic microarray analysis of fecal samples reveals differential transcriptional responses of bifidobacteria in breast- and formula-fed infants. Appl. Environ. Microbiol. 75:2668-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurokawa, K., T. Itoh, T. Kuwahara, K. Oshima, H. Toh, A. Toyoda, H. Takami, H. Morita, V. K. Sharma, T. P. Srivastava, T. D. Taylor, H. Noguchi, H. Mori, Y. Ogura, D. S. Ehrlich, K. Itoh, T. Takagi, Y. Sakaki, T. Hayashi, and M. Hattori. 2007. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 14:169-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liebich, J., C. W. Schadt, S. C. Chong, Z. He, S. K. Rhee, and J. Zhou. 2006. Improvement of oligonucleotide probe design criteria for functional gene microarrays in environmental applications. Appl. Environ. Microbiol. 72:1688-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macfarlane, G. T., and S. Macfarlane. 2007. Models for intestinal fermentation: association between food components, delivery systems, bioavailability and functional interactions in the gut. Curr. Opin. Biotechnol. 18:156-162. [DOI] [PubMed] [Google Scholar]
- 19.Macfarlane, S., E. J. Woodmansey, and G. T. Macfarlane. 2005. Colonization of mucin by human intestinal bacteria and establishment of biofilm communities in a two-stage continuous culture system. Appl. Environ. Microbiol. 71:7483-7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackie, R. I., A. Sghir, and H. R. Gaskins. 1999. Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 69:S1035-S1045. [DOI] [PubMed] [Google Scholar]
- 21.Majima, S., K. Kajino, T. Fukuda, F. Otsuka, and O. Hino. 2000. A novel gene “Niban” upregulated in renal carcinogenesis: cloning by the cDNA-amplified fragment length polymorphism approach. Jpn. J. Cancer Res. 91:869-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neveu, C., C. Charvet, A. Fauvin, J. Cortet, P. Castagnone-Sereno, and J. Cabaret. 2007. Identification of levamisole resistance markers in the parasitic nematode Haemonchus contortus using a cDNA-AFLP approach. Parasitology 134:1105-1110. [DOI] [PubMed] [Google Scholar]
- 23.Pieterse, B., R. H. Jellema, and M. J. van der Werf. 2006. Quenching of microbial samples for increased reliability of microarray data. J. Microbiol. Methods 64:207-216. [DOI] [PubMed] [Google Scholar]
- 24.Poretsky, R. S., N. Bano, A. Buchan, G. LeCleir, J. Kleikemper, M. Pickering, W. M. Pate, M. A. Moran, and J. T. Hollibaugh. 2005. Analysis of microbial gene transcripts in environmental samples. Appl. Environ. Microbiol. 71:4121-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin, J., R. Li, J. Raes, M. Arumugam, K. S. Burgdorf, C. Manichanh, T. Nielsen, N. Pons, F. Levenez, T. Yamada, D. R. Mende, J. Li, J. Xu, S. Li, D. Li, J. Cao, B. Wang, H. Liang, H. Zheng, Y. Xie, J. Tap, P. Lepage, M. Bertalan, J. M. Batto, T. Hansen, D. Le Paslier, A. Linneberg, H. B. Nielsen, E. Pelletier, P. Renault, T. Sicheritz-Ponten, K. Turner, H. Zhu, C. Yu, S. Li, M. Jian, Y. Zhou, Y. Li, X. Zhang, S. Li, N. Qin, H. Yang, J. Wang, S. Brunak, J. Dore, F. Guarner, K. Kristiansen, O. Pedersen, J. Parkhill, J. Weissenbach, P. Bork, S. D. Ehrlich, and J. Wang. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajilić-Stojanović, M., H. G. H. J. Heilig, D. Molenaar, K. Kajander, A. Surakka, H. Smidt, and W. M. de Vos. 2009. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ. Microbiol. 11:1736-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reijans, M., R. Lascaris, A. O. Groeneger, A. Wittenberg, E. Wesselink, J. van Oeveren, E. de Wit, A. Boorsma, B. Voetdijk, H. van der Spek, L. A. Grivell, and G. Simons. 2003. Quantitative comparison of cDNA-AFLP, microarrays, and GeneChip expression data in Saccharomyces cerevisiae. Genomics 82:606-618. [DOI] [PubMed] [Google Scholar]
- 28.Rhee, S. K., X. Liu, L. Wu, S. C. Chong, X. Wan, and J. Zhou. 2004. Detection of genes involved in biodegradation and biotransformation in microbial communities by using 50-mer oligonucleotide microarrays. Appl. Environ. Microbiol. 70:4303-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rizos, D., P. Lonergan, M. P. Boland, R. Arroyo-Garcia, B. Pintado, J. de la Fuente, and A. Gutierrez-Adan. 2002. Analysis of differential messenger RNA expression between bovine blastocysts produced in different culture systems: implications for blastocyst quality. Biol. Reprod. 66:589-595. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 31.Samuel, B. S., and J. I. Gordon. 2006. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc. Natl. Acad. Sci. U. S. A. 103:10011-10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarosh, B. R., and J. Meijer. 2007. Transcriptional profiling by cDNA-AFLP reveals novel insights during methyl jasmonate, wounding and insect attack in Brassica napus. Plant Mol. Biol. 64:425-438. [DOI] [PubMed] [Google Scholar]
- 33.Snel, B., P. Bork, and M. A. Huynen. 2002. The identification of functional modules from the genomic association of genes. Proc. Natl. Acad. Sci. U. S. A. 99:5890-5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taroncher-Oldenburg, G., E. M. Griner, C. A. Francis, and B. B. Ward. 2003. Oligonucleotide microarray for the study of functional gene diversity in the nitrogen cycle in the environment. Appl. Environ. Microbiol. 69:1159-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tatusov, R. L., N. D. Fedorova, J. D. Jackson, A. R. Jacobs, B. Kiryutin, E. V. Koonin, D. M. Krylov, R. Mazumder, S. L. Mekhedov, A. N. Nikolskaya, B. S. Rao, S. Smirnov, A. V. Sverdlov, S. Vasudevan, Y. I. Wolf, J. J. Yin, and D. A. Natale. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turnbaugh, P. J., F. Backhed, L. Fulton, and J. I. Gordon. 2008. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3:213-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turnbaugh, P. J., M. Hamady, T. Yatsunenko, B. L. Cantarel, A. Duncan, R. E. Ley, M. L. Sogin, W. J. Jones, B. A. Roe, J. P. Affourtit, M. Egholm, B. Henrissat, A. C. Heath, R. Knight, and J. I. Gordon. 2009. A core gut microbiome in obese and lean twins. Nature 457:480-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urich, T., A. Lanzen, J. Qi, D. H. Huson, C. Schleper, and S. C. Schuster. 2008. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One 3:e2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaughan, E. E., H. G. Heilig, K. Ben-Amor, and W. M. de Vos. 2005. Diversity, vitality and activities of intestinal lactic acid bacteria and bifidobacteria assessed by molecular approaches. FEMS Microbiol. Rev. 29:477-490. [DOI] [PubMed] [Google Scholar]
- 41.Verberkmoes, N. C., A. L. Russell, M. Shah, A. Godzik, M. Rosenquist, J. Halfvarson, M. G. Lefsrud, J. Apajalahti, C. Tysk, R. L. Hettich, and J. K. Jansson. 2009. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 3:179-189. [DOI] [PubMed] [Google Scholar]
- 42.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman, and M. Kuiper. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner, M., H. Smidt, A. Loy, and J. Zhou. 2007. Unravelling microbial communities with DNA-microarrays: challenges and future directions. Microb. Ecol. 53:498-506. [DOI] [PubMed] [Google Scholar]
- 44.Wu, L., D. K. Thompson, G. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zoetendal, E. G., A. D. L. Akkermans, and W. M. de Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zoetendal, E. G., C. C. G. M. Booijink, E. M. Klaassens, H. G. H. J. Heilig, M. Kleerebezem, H. Smidt, and W. M. de Vos. 2006. Isolation of RNA from bacterial samples of the human gastrointestinal tract. Nat. Protoc. 1:954-959. [DOI] [PubMed] [Google Scholar]
- 47.Zoetendal, E. G., H. G. H. J. Heilig, E. M. Klaassens, C. C. G. M. Booijink, M. Kleerebezem, H. Smidt, and W. M. de Vos. 2006. Isolation of DNA from bacterial samples of the human gastrointestinal tract. Nat. Protoc. 1:870-873. [DOI] [PubMed] [Google Scholar]
- 48.Zoetendal, E. G., M. Rajilić-Stojanović, and W. M. de Vos. 2008. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57:1605-1615. [DOI] [PubMed] [Google Scholar]
- 49.Zoetendal, E. G., E. E. Vaughan, and W. M. de Vos. 2006. A microbial world within us. Mol. Microbiol. 59:1639-1650. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.