Abstract
Pharmaceutical products are released at low concentrations into aquatic environments following domestic wastewater treatment. Such low concentrations have been shown to induce transcriptional responses in microorganisms, which could have consequences on aquatic ecosystem dynamics. In order to test if these transcriptional responses could also be observed in complex river microbial communities, biofilm reactors were inoculated with water from two rivers of differing trophic statuses and subsequently treated with environmentally relevant doses (ng/liter to μg/liter range) of four pharmaceuticals (erythromycin [ER], gemfibrozil [GM], sulfamethazine [SN], and sulfamethoxazole [SL]). To monitor functional gene expression, we constructed a 9,600-feature anonymous DNA microarray platform onto which cDNA from the biofilms was hybridized. Pharmaceutical treatments induced both positive and negative transcriptional responses from biofilm microorganisms. For instance, ER induced the transcription of several stress, transcription, and replication genes, while GM, a lipid regulator, induced transcriptional responses from several genes involved in lipid metabolism. SN caused shifts in genes involved in energy production and conversion, and SL induced responses from a range of cell membrane and outer envelope genes, which in turn could affect biofilm formation. The results presented here demonstrate for the first time that low concentrations of small molecules can induce transcriptional changes in a complex microbial community. The relevance of these results also demonstrates the usefulness of anonymous DNA microarrays for large-scale metatranscriptomic studies of communities from differing aquatic ecosystems.
Surface waters worldwide are often contaminated with treated sewage effluent containing pharmaceutical and personal-care products (PPCP) in the ng/liter to μg/liter range. Their presence reflects the fact that despite variation in excretion rates and types of metabolites, some drugs exit the human body relatively unchanged and are subsequently not fully removed during sewage treatment. Concern is growing regarding potential environmental effects of PPCP pollution for a number of reasons: (i) many drugs interact with a biological target shared by humans, other animals, and even plants and microorganisms; (ii) most act at relatively low concentrations; (iii) pharmaceuticals need time to achieve the desired effect and, thus, are designed specifically to resist degradation in the body (15); (iv) pharmaceuticals are constantly added to aquatic ecosystems, and their rate of addition often exceeds transformation or degradation rates; and (v) chronic exposure has been linked to the development of antibiotic-resistant bacteria (7).
Current ecological assessment approaches for PPCP monitoring (using conventional aquatic toxicity tests) are performed in isolation and out of the context of a larger and more relevant ecological structure (8). Biofilms, for both their basal role in aquatic food webs and their importance in fundamental processes such as biodegradation and biogeochemical cycling, are ideal candidates for monitoring the ecological effects of pharmaceutical pollution on aquatic environments. Other advantages of biofilms for such studies include their continual exposure to contaminants, abundance, ubiquity, and stationary state (21). Although the subinhibitory pollutant concentrations involved preclude the use of marker genes or species, since subtle and complex effects are expected, an approach quantifying the expression of a range of genes might be appropriate.
Anonymous DNA microarrays are frequently used for transcriptomic studies of organisms whose genomes have not been sequenced (5, 14, 23, 26, 31, 32). Using this approach, cDNA is hybridized to unsequenced DNA or cDNA fragments that are printed on a microarray, and only the probes that display significant responses to the treatment of interest are sequenced. Similarly, anonymous DNA microarrays could be an interesting alternative for the comparative metatranscriptomic analysis of environmental samples. The advantages of such an approach are 2-fold: first, there is no need for previous environmental genomic knowledge; second, a large number of samples can be examined without tedious metatranscriptomic sequencing each time a new sample is analyzed. Previously, anonymous microarrays for analyses of mixed microbial communities were used mainly at the genomic level to detect or differentiate particular species (6, 16-18, 27). More recently, a 2,000-probe microarray made of 2.0-kb anonymous fragments was successfully used to fingerprint a simple sludge sample (40). Those previous studies were directed at genomic information, but a similar approach could be devised to observe the metatranscriptome of microbial communities.
The objectives of the present study were (i) to demonstrate the utility of anonymous microarrays in large-scale metatranscriptomic studies and (ii) to identify the transcriptional responses of river biofilms to environmentally relevant doses of different types of pharmaceutical products. In order to do so, we constructed a 9,600-feature anonymous microarray platform using total DNA extracted from environmental biofilm samples. We then hybridized the cDNA obtained from individual samples separately to this microarray and used the resulting hybridization patterns to obtain a high-definition expression fingerprint of the microbial community and to identify the genes that were most differentially expressed between the treatments.
MATERIALS AND METHODS
Study sites.
The city of Regina, Saskatchewan, Canada, treats its sewage at a modern sewage treatment plant located on Wascana Creek (WC). From late October through March, most flow in the creek is tertiary-treated sewage effluent, while during the remaining months, treated sewage effluent is diluted by natural flow a maximum of only 9.4 times. Downstream of the sewage treatment plant, WC is N hypersaturated and hypertrophic with respect to total phosphorus (M. J. Waiser, V. Tumber, and J. Holm, submitted for publication). In such systems, dilution is low, and effects on aquatic organisms may be more severe than in those systems where sewage effluents are highly diluted, as in the South Saskatchewan River (SSR). The SSR receives tertiary-treated sewage that is highly diluted (in the 200:1 range during winter flows and in the 33:1 range during low summer flows), and the river is nutrient (nitrogen and phosphorus) and carbon limited and therefore oligotrophic (19, 24). Recent seasonal field surveys of WC (Waiser et al., submitted) and SSR detected the presence of a number of antibiotics (erythromycin [ER], clindamycin, trimethoprim, sulfamethazine [SN], norfloxacin, and sulfamethoxazole [SL]) downstream of the sewage treatment plants for Regina and Saskatoon. These antibiotics were detected in concentrations of ng liter−1. Also detected at higher concentrations (in μg liter−1) in WC was the lipid regulator gemfibrozil (GM).
Rotating annular reactors for biofilm growth.
The rotating annular reactor design for biofilm development was previously described (19, 20). Two duplicate experiments were carried out, with reactors being inoculated with water from WC (nutrient rich) or the SSR (nutrient poor). Treatments consisted of the addition of various pharmaceutical compounds: 1 μg liter−1 ER, 1 μg liter−1 GM, 0.5 μg liter−1 SN, and 0.5 μg liter−1 SL. Nothing was added to the control reactors (CO). Six different mixed-treatment reactor experiments were also carried out (for a total of 33 samples per river) and used to construct the anonymous microarray, but results of these treatments are not discussed in the present contribution. All treatments were replicated independently three times. WC biofilms were used to construct the anonymous microarray and to observe the biofilm response to pharmaceuticals, while the SSR biofilms were used to test whether the anonymous microarray could be used with biofilm samples other than the ones used to construct it.
Nucleic acid extraction, amplification, and labeling.
Total DNA and RNA were simultaneously extracted from 250- to 500-mg biofilm subsamples using bead beating in a CTAB (hexadecyl-trimethyl-ammonium bromide) buffer followed by phenol-chloroform purification (35, 38). Nucleic acids were resuspended in 100 μl diethyl pyrocarbonate (DEPC)-water and separated into two 50-μl aliquots. One aliquot was treated with RNase A to give DNA, and the other was treated with DNase to give RNA. mRNA was enriched by using the MicrobExpress kit (Ambion, Austin, TX), amplified by using the MessageAmp II kit (Ambion), and labeled with Cy3 dyes (GE Healthcare, Piscataway, NJ) by using an amino allyl cDNA labeling kit (Ambion). The composite DNA sample (see below) was labeled with Cy5 by using the BioPrime Array CGH genomic labeling kit (Invitrogen, Carlsbad, CA).
Anonymous microarray platform.
Ten microliters (3.4 ± 1.1 μg) of DNA extracted from each of the 33 samples from WC biofilms was pooled together to form one composite DNA sample. This composite sample was fragmented by sonication at a power of 8 W for 5 s and size selected (400 to 600 bp) on a 2% agarose gel. Gel-extracted, sonicated DNA was end repaired by using an End-It kit (Epicentre Biotechnologies, Madison, WI) and incubated with Taq DNA polymerase and ATP for 1 h at 72°C to create an “A” overhang compatible with UA ligation. End-repaired DNA was then ligated into a pDrive vector (Qiagen, La Jolla, CA) and transformed into XL1-Blue competent cells (Stratagene, La Jolla, CA). The 9,600 clones were picked by a VersArray colony picker (Bio-Rad, Hercules, CA) at the Concordia University Centre for Structural and Functional Genomics (Montreal, QC, Canada). DNA was extracted from clones by boiling lysis (5 min at 95°C) and then amplified by PCR using vector-specific primers. PCR products were purified by using Multi-Screen FB 96-well purification plates (Millipore, Billerica, MA). Following gel analysis and DNA UV quantification, 6,351 PCR products were classified as “good” (66.2%), and 917 PCR products were classified as “weak” (9.6%). The remaining reactions (2,332; 24.3%) showed either no product or multiple bands and were classified as “failed” reactions. PCR-amplified full-length 16S rRNA genes from an uncultured bacterium (closest match to GenBank accession no. GQ397043 with 95% identity) were added to be printed twice in the last row of each of the 48 subarrays. All PCR products were then printed in duplicate on amino-silane-coated glass slides with a VersArray Chip Writer Pro printer (Bio-Rad).
Microarray hybridization, scanning, and data processing.
Prehybridization was carried out at 50°C for 1 h by submerging slides in a solution containing 1% bovine serum albumin (BSA), 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and 0.1% SDS. The product of the whole cDNA labeling reaction was dried in a Speed-Vac apparatus, resuspended in 45 μl DIG Easy Hyb hybridization buffer (Roche Diagnostics, Laval, QC, Canada), and mixed with 750 ng of Cy5-labeled composite DNA (in 45 μl of DIG buffer). This mixture was denatured at 95°C for 2 min and hybridized to the microarray slides for 16 h at 50°C on a Slide Booster apparatus (Advalytix-Beckman Coulter, Munich, Germany). Slides were washed three times in 0.1× SSC and 0.1% SDS for 5 min at 42°C, rinsed three times in 0.1× SSC, and dried with filtered nitrogen. Slides were then scanned with a ScanArray Lite scanner (Perkin-Elmer, Boston, MA) at a 10-μm resolution. Resulting image files were gridded and quantified by using ScanArray Express software. Text files containing quantitative data were imported into the R package (v. 2.9.0; R Foundation for Statistical Computing) and normalized by using the “limma” package. Spot intensities were corrected for background by using the “backgroundCorrect” function with the “normexp” method. Intensities were then normalized within each array by using the “normalizeWithinArrays” function with the “loess” method. Spot log ratios were normalized between arrays by using the “normalizeBetweenArrays” function with the “Gquantile” method. The resulting normalized A matrix (average log-2 expression values) was then exported to Excel, where duplicate probe values were averaged. This matrix was used for further statistical analyses. Microarray data discussed here have been deposited in the NCBI Gene Expression Omnibus (GEO) (10).
Sequencing and sequence analysis.
Following correspondence analyses (see “Statistical analyses” below), probes that showed the highest scores (in absolute values) on the first and second ordination axes were identified, retrieved from the plates used to construct the microarray, amplified, and sent for Sanger sequencing at the Laval University Bio-Molecular Analysis Platform (Quebec City, QC, Canada). Sequences were compared against the “nr” database using Blastx at the NCBI website. For each sequence, the best match showing an E value below 1 × 10−5 was retrieved.
qRT-PCR.
For some of the sequenced probes identified as being particularly responsive to treatments, PCR primers were designed by using the NCBI Primer-BLAST tool (www.ncbi.nlm.nih.gov/tools/primer-blast/). Primer sequences are listed in Table S1 in the supplemental material. These primers were then used in real-time reverse transcriptase (RT) PCRs (qRT-PCRs) to confirm trends observed on the microarray. qRT-PCRs were carried out at an annealing temperature of 55°C using an iScript One-Step RT-PCR kit with Sybr green (Bio-Rad) as previously described (34, 36). Genes used to design the primers were amplified from plasmids, and their concentration was adjusted from 1 × 106 to 1 × 102 using 10-fold serial dilutions. The 16S rRNA was quantified by using primers and conditions described previously by Fierer et al. (12) against a serial dilution of linearized plasmids containing full-length 16S rRNA genes. For all reactions, several no-RT and no-template controls were carried out and yielded no detectable signals. Phage lambda DNA was used to correct for potential PCR inhibitors in biofilm extracts (1). When the recovery of phage lambda DNA was below 100%, quantification values for all other genes were corrected accordingly. PCR inhibition ranged from 0.0% to 69.3%, within previously reported values for environmental samples (34).
Statistical analyses.
All statistical analyses were carried out with the R package (R Foundation for Statistical Computing, Vienna, Austria). Correspondence analyses were carried out by using the “cca” function of the “vegan” package. The microarray printing batch was included as a covariable for which the effect was removed before performing the analysis. Correspondence analysis was first carried out with all replicates, but a large variation was observed between some replicates. Upon careful visual inspection of microarray pictures, several problems were detected with outlier replicates (large scratches on slides and abnormal background for some subarrays, etc.), and they were therefore removed from the data set before further analyses. For analysis of variance (ANOVA), normality was tested by using the “shapiro.test” function. When necessary, data were log or square root transformed to meet parametric ANOVA assumptions. ANOVA and post hoc Tukey honestly significant difference (HSD) tests were then carried out by using the “aov” and “TukeyHSD” functions, respectively. When transformations failed to normalize data, Kruskal-Wallis and associated multiple-comparison tests were carried out by using the “kruskal.test” and the “kruskalmc” functions of the “pgirmess” library, respectively.
Accession numbers.
Detailed microarray designs were deposited in the NCBI GEO and can be accessed through GEO accession numbers GSE20501 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE20501) and GPL10021. Sequence data produced in the current study were deposited in the GenBank database under accession numbers GS901815 to GS901924 (WC) and GS926646 to GS926705 (SSR).
RESULTS
Normalized hybridization patterns were used for two purposes: (i) to provide a fingerprint of the metatranscriptome of each sample and (ii) to identify the probes that were most strongly linked to the experimental treatments. The latter analysis was compared for selected probes with qRT-PCR results.
Microarray analysis of Wascana Creek.
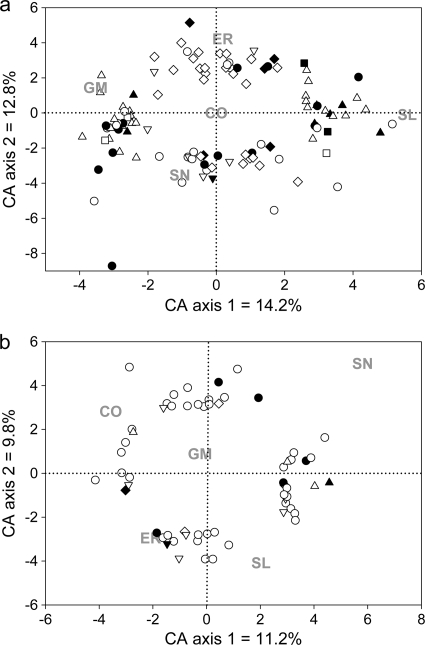
Correspondence analysis ordinations indicated that biofilms from Wascana Creek (WC) replicate reactors clustered mostly together and that the different drug treatments were clearly separated on the first and second axes, with the control located right in the ordination center (Fig. 1 a). Individual probe scores on the first and second axes were thus used as proxies for association strength with treatment. The 96 highest positive and negative scores for each axis were selected and sent for sequencing. Approximately 30 high-quality sequences had significant matches in GenBank (E value below 1 × 10−5) for each side of each axis. These probes are depicted in Fig. 1a and are listed in Tables S2 to S5 in the supplemental material. Probes were determined to be either positively or negatively associated with a treatment (sometimes both) based on their average normalized intensity for that treatment compared to the other treatments (Tables S2 to S5 and Fig. 2). It should be noted that the two first axes accounted for only 27% of the variation in the data set, and even though they represent the strongest variation in the data, other important trends might be missed by the ordination procedure.
FIG. 1.
Correspondence analysis biplot of anonymous microarray hybridization patterns for cDNA extracted from biofilms growing in water from Wascana Creek (a) and the South Saskatchewan River (b) and subjected to environmentally relevant doses of pharmaceutical compounds. Positions of the treatments are the mean values for the replicates (n = 2 to 3). Only the probes listed in Tables S2 to S9 in the supplemental material are depicted. ▴, probes associated exclusively with SL; ▾, probes associated exclusively with SN; ⧫, probes associated exclusively with ER; ▪, probes associated exclusively with GM; •, probes associated with multiple treatments. In a, probes in black were used in Fig. 2 or for real-time PCR quantification. In b, probes in black are the ones mentioned in Table 1. CO, control; CA, correspondence analysis.
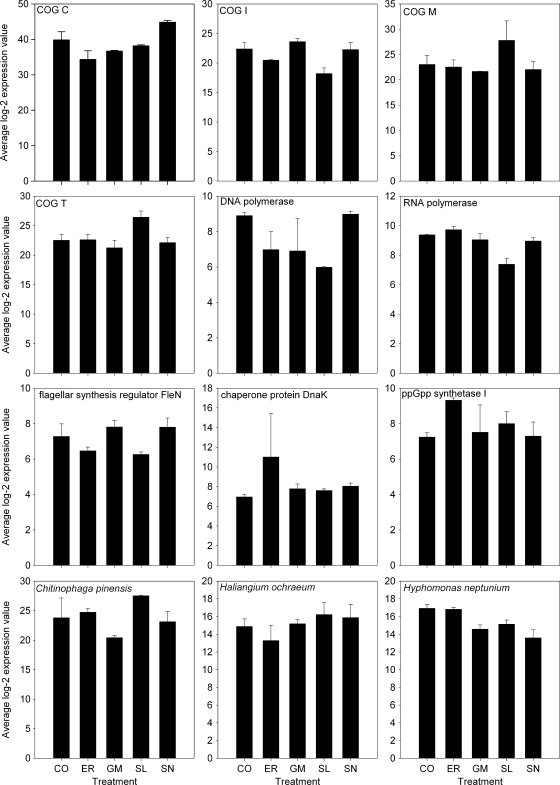
FIG. 2.
Average log-2 expression values for selected probes following exposure of Wascana Creek biofilms to different pharmaceutical products. COG C (energy production and conversion), sum of probes 2003, 5271, 13407, 13905, and 16399; COG I (lipid transport and metabolism), sum of probes 6171, 8775, and 19275; COG M (cell envelope biogenesis, outer membrane), sum of probes 847, 14137, and 14385; COG T (signal transduction), sum of probes 1849, 5589, and 8271. DNA polymerase (COG L [replication, recombination, and repair]), probe 2027; RNA polymerase (COG K [transcription]), probe 13807; the flagellar synthesis regulator FleN (COG D [cell division and chromosome partitioning]), probe 18099; the chaperone protein DnaK (COG O [posttranslational modification, protein turnover, and chaperones]), probe 13925; ppGpp synthetase I (COG K [transcription] and COG T [signal transduction]), probe 8047; Chitinophaga pinensis, sum of probes 605, 14393, and 19439; Haliangium ochraeum, sum of probes 739 and 8467; Hyphomonas neptunium, sum of probes 8129 and 14253. See Tables S2 to S5 in the supplemental material for more details about the probes. CO, control. Error bars represent the standard deviations.
Microarray analysis of the South Saskatchewan River.
The South Saskatchewan River (SSR) was chosen because it differed from WC in a key environmental parameter (i.e., nutrient availability or trophic status). Successful hybridization patterns were obtained from biofilms grown with water from the SSR. In correspondence analyses, different replicates of a single treatment showed more variability than for WC, but average positions for each treatment were clearly separated from each other on the two ordination axes (Fig. 1b). Relative treatment positions were not exactly as seen for WC samples, but some similarities were evident (e.g., opposition of ER and SN treatments). The 48 highest positive and negative scores for each axis were selected and sent for sequencing. Approximately 15 high-quality sequences had significant matches in GenBank (E value below 1 × 10−5) for each side of each axis. These probes are depicted in Fig. 1b and are listed in Tables S6 to S9 in the supplemental material. Probes were determined to be either positively or negatively associated with a treatment (sometimes both) based on their average normalized intensity for that treatment compared to the other treatments (Tables S6 to S9).
Most-responsive probes in the WC biofilms.
The best matches in GenBank and the associated COG (Cluster of Orthologous Groups of proteins) categories for sequenced probes are listed in Tables S2 to S5 in the supplemental material. Some of the potentially most relevant probes for each treatment are mentioned below and are represented in Fig. 2. The significance of the differences could not be formally tested by ANOVA because after removing outliers, most treatments only had 2 replicates (see Materials and Methods). Several probes related to the cell envelope and outer membrane (COG M) or involved in signal transduction (COG T) showed a positive response to sulfamethoxazole (SL) (Table S2 and Fig. 2). In contrast, probes related to DNA-directed DNA and RNA polymerases were negatively affected by SL. Three probes related to Chitinophaga pinensis showed a negative response to gemfibrozil (GM) (Table S3 and Fig. 2), indicating that this organism might be inhibited by GM. Three probes related to lipid transport and metabolism (COG I) and one probe related to a flagellar synthesis regulator showed positive responses to GM. A probe related to guanosine tetraphosphate (ppGpp) synthetase showed a positive response to erythromycin (ER) (Table S4 and Fig. 2). The probe that was the most positively associated with ER was linked to the chaperone protein DnaK (heat shock protein 70). Similarly, several probes potentially related to stress response genes showed a positive or negative response to ER: the UmuD protein, type II restriction enzyme, and the RecA protein. Several probes related to energy production and conversion (COG C), to carbohydrate transport and metabolism (COG G), and to Haliangium ochraeum responded negatively to ER treatment. In contrast, five genes related to energy production and conversion (COG C) were positively affected by sulfamethazine (SN) (Table S5 and Fig. 2). Two probes related to Hyphomonas neptunium were negatively associated with SN, while several replication-, recombination-, and repair (COG L)-related genes were either negatively or positively affected by SN.
Most-responsive probes in the SSR biofilms.
The best matches in GenBank and the associated COG category for sequenced probes are listed in Tables S6 to S9 in the supplemental material. Interestingly, 11 of the 192 probes having the highest scores on each side of the two first axes for the SSR biofilms were also among the 384 probes that had the highest scores for the WC biofilms. Among these 11 probes, 3 had significant matches in databases and are reported in Table 1. These probes showed almost-identical treatment associations (Table 1). Careful screening of results from BLAST analyses showed interesting similarities between several probes from the SSR and WC (Table 1).
TABLE 1.
Probes selected that were among those most influenced by the treatments and that were identical or highly similar for Wascana Creek and South Saskatchewan River
| Probe | Gene | Organism | Positive association(s) (location[s]) | Negative association(s) (location[s]) |
|---|---|---|---|---|
| Identical | ||||
| 13925 | Chaperone protein DnaK | Spirosoma linguale DSM 74 | ER (WC, SSR), SN (SSR) | GM (SSR) |
| 19493 | Acyl-CoA dehydrogenase | Alcanivorax sp. strain DG881 | —a | ER (WC, SSR), GM (SSR) |
| 10047 | Conserved hypothetical protein | Arthrospira maxima CS-328 | SL (WC, SSR) | —a |
| Similar | ||||
| WC | ||||
| 14393 | TonB-dependant receptor | Chitinophaga pinensis DSM 2588 | SL | GM |
| 605 | RNA polymerase | Chitinophaga pinensis DSM 2588 | — | GM |
| 19439 | TonB-dependent receptor | Chitinophaga pinensis DSM 2588 | — | GM |
| SSR | ||||
| 9611 | 3-Isopropylmalate dehydratase | Chitinophaga pinensis DSM 2588 | SN | ER |
| 35 | protein of unknown function | Chitinophaga pinensis DSM 2588 | GM | SN |
| 13463 | DEAD/DEAH box helicase | Chitinophaga pinensis DSM 2588 | — | SN |
| WC | ||||
| 12659 | DEAD/DEAH box helicase | Desulfovibrio salexigens DSM 2638 | ER | — |
| SSR | ||||
| 13463 | DEAD/DEAH box helicase | Chitinophaga pinensis DSM 2588 | — | SN |
| WC | ||||
| 4917 | RecA protein | Prevotella veroralis F0319 | — | ER |
| SSR | ||||
| 6609 | RecD/TraA family helicase | Methylobacterium sp. strain 4-46 | SN | ER, GM, SL |
—, no association.
qRT-PCR.
Some probes identified as being particularly interesting (in boldface type in Tables S2 to S5 in the supplemental material) were used to design qRT-PCR primers. These primers were then employed to confirm and quantify the expression patterns observed on microarrays for WC biofilms. Values for the different assays are given in Table 2. While responses were quite variable between different replicates of a single treatment, differences observed were not significant (P > 0.05 by ANOVA). Because of this lack of significance, only general trends are mentioned below. Interestingly, all treatments decreased the amount of 16S rRNA by more than half compared to controls. Furthermore, ER generally caused quite large increases in the levels of expression of all the different genes, while GM produced a general decrease in the gene expression level. Some trends observed in the qRT-PCR assays were highly concordant with microarray results. For instance, the expression levels of ppGpp synthetase and the chaperone protein DnaK measured by qRT-PCR were higher for the ER treatments. Similarly, the level of TonB-dependent receptor (needed for high-affinity binding and energy-dependent uptake of specific substrates) expression was higher for SL treatments and lower for GM treatments, just as on the microarray. Other genes also showed trends in the qRT-PCR assays that provided information complementary to the microarray data. For instance, DNA-directed DNA polymerase B and the UDP-N-acetylglucosamine carboxyvinyltransferase genes (involved in cell wall formation) showed increased expression levels following all treatments compared to those of controls. Trends in some qRT-PCR assays, however, were not similar to those of the microarrays. In fact, both photosystem I reaction center subunit X and acetyl coenzyme A (CoA) carboxylase indicated trends opposite to the microarray.
TABLE 2.
qRT-PCR quantification of different genes among those most affected by the different treatments in the biofilm of Wascana Creek (n = 3)
| Treatment | Avg level (SD) |
|||||||
|---|---|---|---|---|---|---|---|---|
| ppGpp synthetase I (105 copies g−1 biofilm) | TonB-dependent receptor (106 copies g−1 biofilm) | DNA-directed DNA polymerase B (106 copies g−1 biofilm) | Chaperone protein DnaK (105 copies g−1 biofilm) | Photosystem I reaction center subunit X (106 copies g−1 biofilm) | Acetyl-CoA carboxylase (105 copies g−1 biofilm) | UDP-N-acetylglucosamine 1-carboxyvinyltransferase (104 copies g−1 biofilm) | 16S rRNA gene (109 copies g−1 biofilm) | |
| CO | 4.21 (3.34) | 2.78 (1.28) | 1.69 (1.36) | 2.84 (2.21) | 0.48 (0.57) | 1.00 (0.66) | 0.17 (0.26) | 7.88 (6.42) |
| ER | 11.53 (15.4) | 10.82 (14.8) | 4.20 (5.92) | 8.71 (10.7) | 21.39 (36.9) | 9.86 (15.8) | 109.77 (190) | 2.37 (2.06) |
| GM | 3.85 (1.84) | 1.92 (1.45) | 1.96 (2.46) | 1.76 (1.6) | 0.10 (0.09) | 0.57 (0.52) | 0.72 (1.23) | 2.43 (1.73) |
| SL | 9.99 (3.66) | 17.1 (20.9) | 4.95 (3.07) | 4.20 (2.88) | 4.75 (4.46) | 6.92 (4.97) | 5.94 (5.87) | 2.38 (2.08) |
| SN | 4.84 (1.15) | 8.41 (7.22) | 2.37 (1.79) | 1.21 (1.28) | 2.71 (3.57) | 2.22 (2.11) | 0.66 (0.64) | 1.06 (0.33) |
DISCUSSION
The present study tracked the transcriptional responses of complex microbial communities to environmentally relevant concentrations of pharmaceutical compounds. Although several studies previously observed transcriptional responses of isolated bacteria to subinhibitory levels of antibiotics, a set of specific signature genes responsive to each antibiotic has yet to be clearly identified (9). Although the exposure of complex microbial communities in this study resulted in an intricate molecular response, there were striking similarities to the responses of isolated bacterial strains reported in previous studies (9, 22). For instance, the pharmaceutical compounds tested in this study caused clear shifts in the expression patterns of several genes related to various functions, including outer membrane formation, lipid metabolism, and energy production.
Erythromycin.
Many small bioactive molecules exhibit contrasting properties when tested at low concentrations compared to high concentrations, a phenomenon called hormesis (9). For instance, at high concentrations, erythromycin inhibits translation, but at subinhibitory concentrations, erythromycin was shown to affect a wide range of cellular functions not necessarily related to translation (9). One key molecule in linking the response to antibiotics and transcription is guanosine tetraphosphate (ppGpp) (2). The levels of ppGpp are related to a large number of cellular functions (3), and the transcription of the gene involved in its synthesis was positively affected by ER in the present study. SL also increased the level of transcription of ppGpp synthetase, indicating that antibiotics with different modes of action influence ppGpp levels, further highlighting the potentially central role of this messenger in the response to subinhibitory antibiotic concentrations. Furthermore, several studies have shown that translation inhibitors often induced the expression of bacterial heat shock and cold shock genes (25, 29). Accordingly, in this study a heat shock protein (chaperone DnaK) was overexpressed in the ER treatments. Other genes potentially related to stress response that were influenced by ER included a type II restriction enzyme, the UmuD protein, and the RecA protein, some of which were previously associated with antibiotic exposure (22). ppGpp synthetase was also previously associated with stress response in bacteria (4, 33).
Sulfonamides (sulfamethoxazole and sulfamethazine).
To the best of our knowledge, microbial transcriptional responses to subinhibitory concentrations of sulfonamide antibiotics have not yet been addressed. At high concentrations, these antibiotics inhibit folic acid synthesis. Folic acid is needed in purine metabolism, and consequently, sulfonamide antibiotics inhibit DNA and RNA synthesis. In the present study, low concentrations of both SN and SL induced transcriptional changes in several genes related to replication and transcription (e.g., DNA and RNA polymerases). Other effects included changes in gene expression related to the cell envelope and outer membrane (SL treatment). Such changes might be related to the negative and positive relationships previously observed between biofilm formation and low doses of antibiotics (2, 21, 22, 30). SN also elicited a positive response of several energy production and conversion genes, potentially indicating changes in the central metabolism of some biofilm microorganisms. Interestingly, the two sulfonamide antibiotics tested here did not elicit the same responses from study biofilms even though their molecular structures and modes of action are almost identical. This further demonstrates the high specificity of microbial responses to antibiotics (11).
Gemfibrozil.
We are not aware of any study reporting the transcriptomic response of microorganisms to low doses of the lipid regulator gemfibrozil. Recently, however, it was shown that high doses of GM inhibited bacterial fatty acid synthesis (28). A variety of responses, including an increase in acetylcholinesterase, succinate dehydrogenase, and glucose-6-P dehydrogenase activities, has also been observed for fish cells at low GM doses (41). In our study, genes that showed a positive response to GM included several genes related to lipid metabolism, indicating that GM might affect this process at low concentrations. Another potentially important gene responding positively to GM was a flagellar synthesis regulator. For some microorganisms this response could indicate a shift from sessile to planktonic forms, i.e., dispersal from the biofilm. Such a shift has been shown for bacteria in response to subinhibitory doses of antibiotics (22), and it could have repercussions on biofilm formation by reducing attachment and/or increasing dispersal. It could also influence the bacterial transcriptional response, as this response differed in bacteria grown in liquid versus solid media (13). From our results, it also appeared that GM could cause some changes in community structure by inhibiting specific organisms (e.g., Chitinophaga pinensis). The other pharmaceutical products (ER, SL, and SN) also inhibited or stimulated some microorganisms, indicating that the small molecules tested all have the potential to cause shifts in community structure and/or function. Interestingly, all the products tested caused a relatively large decrease in the levels of 16S rRNA, indicating that these pharmaceutical products either decreased microbial biomass or decreased general activity within the biofilm. In fact, the bacterial biomass within the biofilms as measured by confocal laser microscopy decreased when exposed to the pharmaceutical products (J. R. Lawrence, M. J. Waiser, and D. R. Korber, unpublished results). Alternatively, pharmaceutical products at low doses could cause an inhibition of more sensitive microorganisms, or perhaps, the concentration of different compounds reached inhibitory levels locally within the biofilm.
Anonymous microarrays.
Although the construction of anonymous microarray platforms is initially labor-intensive, there are several advantages to using this tool compared to other transcriptomic and microarray approaches. First, we can now interrogate the same set of transcripts for numerous samples from similar environments without having to carry out in-depth transcriptomic sequencing. Second, the complete separation of mRNA from rRNA in microbial metatranscriptomic studies is difficult, and the process reduces the amount of data that can be effectively analyzed. This problem was circumvented in the present study by building an anonymous microarray platform using genomic DNA. Since the gene density of bacteria is high (on average, one open reading frame per 1,000 bp), most of the 400- to 600-bp probes contained a large portion of a least one gene. Third, another strength of the anonymous microarray approach is that it solves issues related to not-yet-sequenced genes in environmental studies. Indeed, by basing construction on genomic material retrieved from the environment, anonymous microarrays can reveal unknown sequences and, importantly, provide a tool for novel gene discovery. Such novel genes, not showing any significant matches in databases or matching only hypothetical proteins, were detected here. Further refinement of the microarray platform could be achieved by eliminating probes that did not show significant differences between treatments and replacing them with another set of anonymous probes. Alternatively, sequenced probes identified as particularly interesting and responsive to the treatments could be printed together on a smaller microarray platform.
Another important advantage of the anonymous microarray platform is that it can be used with extracted RNA not originating from the biofilms used in actual platform construction. This approach therefore could be useful for monitoring PPCP effects on reactor-grown biofilms from differing aquatic environments and probably also biofilms grown in situ. Out of several hundred screened probes that showed the strongest treatment response, 11 showed overlap between the two rivers. The probability of independently selecting the same 11 probes in random data sets of the size mentioned above is (384/9,600 × 192/9,600)11 = 8.59 × 10−35, which is extremely unlikely. The selection of identical probes is therefore not a result of chance but an indication that the small molecules tested caused reproducible changes in biofilm communities for both rivers. Furthermore, several other probes showed similar patterns in the SSR and WC. These probes were either similar genes from different organisms or different genes from identical organisms. Although the association with treatments differed in most cases, similarities suggest that, independently of the aquatic ecosystem, the small molecules tested are affecting a defined set of genes and organisms. Differences between the responses of the remaining probes could have been caused by the relatively small number of probes selected but may also reflect differences in biofilm nutritional status. Indeed, some changes caused by pharmaceuticals in river biofilms have been linked to a nutrient effect (21), which could have been more prominent in the SSR (oligotrophic) than in WC (eutrophic). Furthermore, biofilm community compositions from the two ecosystems were probably different, and accordingly, similar genes from different organisms were affected.
The discrepancies between qPCR and microarray data reported here are not necessarily indicative of a lack of quantitative power for the microarray platform. Such divergences were previously found for a range of different genes (37, 39). In our case, incongruities could be due to differences in probe specificities (400 to 600 bp) compared to the primers (∼20 bp). Primers were designed to be specific to the probe sequenced and to exclude all bacterial sequences present in the “nr” database of GenBank. It is possible that not-yet-sequenced bacteria or eukaryotes present in environmental samples have RNA that matched the primers used here.
Concluding remarks.
The exposure of complex microbial communities to environmentally relevant concentrations of pharmaceuticals induced transcriptional responses that mirrored those reported previously for isolated organisms in the literature (for the better-studied erythromycin) or that agreed with general knowledge about the effects of low concentrations of small molecules on transcriptional activities in microorganisms (for the less-well-studied sulfonamide antibiotics and gemfibrozil). The relevance of genes identified as being responsive to treatments also indicated that the anonymous microarray approach undertaken here provides useful transcriptomic information from complex environmental samples. The microarray was successfully used with biofilms grown in an environment that differed in a key parameter (nutrient content) compared to the environment from which the microarray was constructed. The anonymous microarray platform, or a reduced version with relevant probes, could be useful to rapidly fingerprint the metatranscriptome of river microbial communities. With further developments, the microarray described here could help monitor effects of different PPCPs on river ecosystems, as the response of microbial communities to pharmaceutical products are complex and beyond the capacity of a single marker gene or species.
Supplementary Material
Acknowledgments
This study was supported by Environment Canada's Strategic Applications of Genomics in the Environment (STAGE) program. E.Y. was supported by an NSERC postdoctoral fellowship.
We acknowledge Sylvie Sanschagrin, Mélanie Arbour, Diane Labbé, and the staff of NRC-BRI's Microarray Laboratory for technical assistance.
Footnotes
Published ahead of print on 18 June 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Beller, H. R., S. R. Kane, T. C. Legler, and P. J. J. Alvarez. 2002. A real-time polymerase chain reaction method for monitoring anaerobic, hydrocarbon-degrading bacteria based on a catabolic gene. Environ. Sci. Technol. 36:3977-3984. [DOI] [PubMed] [Google Scholar]
- 2.Boehm, A., S. Steiner, F. Zaehringer, A. Casanova, F. Hamburger, D. Ritz, W. Keck, M. Ackermann, T. Schirmer, and U. Jenal. 2009. Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress. Mol. Microbiol. 72:1500-1516. [DOI] [PubMed] [Google Scholar]
- 3.Braeken, K., M. Moris, R. Daniels, J. Vanderleyden, and J. Michiels. 2006. New horizons for (p)ppGpp in bacterial and plant physiology. Trends Microbiol. 14:45-54. [DOI] [PubMed] [Google Scholar]
- 4.Bugrysheva, J. V., A. V. Bryksin, H. P. Godfrey, and F. C. Cabello. 2005. Borrelia burgdorferi rel is responsible for generation of guanosine-3′-diphosphate-5′-triphosphate and growth control. Infect. Immun. 73:4972-4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannon, C. H., C. S. Kua, E. K. Lobenhofer, and P. Hurban. 2006. Capturing genomic signatures of DNA sequence variation using a standard anonymous microarray platform. Nucleic Acids Res. 34:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho, J. C., and J. M. Tiedje. 2001. Bacterial species determination from DNA-DNA hybridization by using genome fragments and DNA microarrays. Appl. Environ. Microbiol. 67:3677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costanzo, S. D., J. Murby, and J. Bates. 2005. Ecosystem response to antibiotics entering the aquatic environment. Mar. Pollut. Bull. 51:218-223. [DOI] [PubMed] [Google Scholar]
- 8.Daughton, C. G., and T. A. Ternes. 1999. Pharmaceuticals and personal care products in the environment: agents of subtle change? Environ. Health Perspect. 107:907-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies, J., G. B. Spiegelman, and G. Yim. 2006. The world of subinhibitory antibiotic concentrations. Curr. Opin. Microbiol. 9:445-453. [DOI] [PubMed] [Google Scholar]
- 10.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fajardo, A., and J. L. Martínez. 2008. Antibiotics as signals that trigger specific bacterial responses. Curr. Opin. Microbiol. 11:161-167. [DOI] [PubMed] [Google Scholar]
- 12.Fierer, N., J. A. Jackson, R. Vilgalys, and R. B. Jackson. 2005. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 71:4117-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goh, E.-B., G. Yim, W. Tsui, J. McClure, M. G. Surette, and J. Davies. 2002. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc. Natl. Acad. Sci. U. S. A. 99:17025-17030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hegarty, M. J., J. M. Jones, I. D. Wilson, G. L. Barker, J. A. Coghill, P. Sanchez-Baracaldo, G. Q. Liu, R. J. A. Buggs, R. J. Abbott, K. J. Edwards, and S. J. Hiscock. 2005. Development of anonymous cDNA microarrays to study changes to the Senecio floral transcriptome during hybrid speciation. Mol. Ecol. 14:2493-2510. [DOI] [PubMed] [Google Scholar]
- 15.Jones, O. A. H., N. Voulvoulis, and J. N. Lester. 2004. Potential ecological and human health risks associated with the presence of pharmaceutically active compounds in the aquatic environment. Crit. Rev. Toxicol. 34:335-350. [DOI] [PubMed] [Google Scholar]
- 16.Kim, B. C., J. H. Park, and M. B. Gu. 2004. Development of a DNA microarray chip for the identification of sludge bacteria using an unsequenced random genomic DNA hybridization method. Environ. Sci. Technol. 38:6767-6774. [DOI] [PubMed] [Google Scholar]
- 17.Kim, B. C., J. H. Park, and M. B. Gu. 2005. Multiple and simultaneous detection of specific bacteria in enriched bacterial communities using a DNA microarray chip with randomly generated genomic DNA probes. Anal. Chem. 77:2311-2317. [DOI] [PubMed] [Google Scholar]
- 18.Kingsley, M. T., T. A. Straub, D. R. Call, D. S. Daly, S. C. Wunschel, and D. P. Chandler. 2002. Fingerprinting closely related Xanthomonas pathovars with random nonamer oligonucleotide microarrays. Appl. Environ. Microbiol. 68:6361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawrence, J. R., M. R. Chenier, R. Roy, D. Beaumier, N. Fortin, G. D. W. Swerhone, T. R. Neu, and C. W. Greer. 2004. Microscale and molecular assessment of impacts of nickel, nutrients, and oxygen level on structure and function of river biofilm communities. Appl. Environ. Microbiol. 70:4326-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawrence, J. R., G. D. W. Swerhone, and T. R. Neu. 2000. A simple rotating annular reactor for replicated biofilm studies. J. Microbiol. Methods 42:215-224. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence, J. R., G. D. W. Swerhone, L. I. Wassenaar, and T. R. Neu. 2005. Effects of selected pharmaceuticals on riverine biofilm communities. Can. J. Microbiol. 51:655-669. [DOI] [PubMed] [Google Scholar]
- 22.Linares, J. F., I. Gustafsson, F. Baquero, and J. L. Martinez. 2006. Antibiotics as intermicrobial signaling agents instead of weapons. Proc. Natl. Acad. Sci. U. S. A. 103:19484-19489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Micheletto, S., L. Rodriguez-Uribe, R. Hernandez, R. D. Richins, J. Curry, and M. A. O'Connell. 2007. Comparative transcript profiling in roots of Phaseolus acutifolius and P. vulgaris under water deficit stress. Plant Sci. 173:510-520. [Google Scholar]
- 24.Neu, T. R., G. D. W. Swerhone, U. Bockelmann, and J. R. Lawrence. 2005. Effect of CNP on composition and structure of lotic biofilms as detected with lectin-specific glycoconjugates. Aquat. Microb. Ecol. 38:283-294. [Google Scholar]
- 25.Ng, W.-L., K. M. Kazmierczak, G. T. Robertson, R. Gilmour, and M. E. Winkler. 2003. Transcriptional regulation and signature patterns revealed by microarray analyses of Streptococcus pneumoniae R6 challenged with sublethal concentrations of translation inhibitors. J. Bacteriol. 185:359-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parro, V., and M. Moreno-Paz. 2003. Gene function analysis in environmental isolates: the nif regulon of the strict iron oxidizing bacterium Leptospirillum ferrooxidans. Proc. Natl. Acad. Sci. U. S. A. 100:7883-7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasquer, F., C. Pelludat, B. Duffy, and J. E. Frey. 2010. Broad spectrum microarray for fingerprint-based bacterial species identification. BMC Biotechnol. 10:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reich-Slotky, R., C. A. Kabbash, P. Della-Latta, J. S. Blanchard, S. J. Feinmark, S. Freeman, G. Kaplan, H. A. Shuman, and S. C. Silverstein. 2009. Gemfibrozil inhibits Legionella pneumophila and Mycobacterium tuberculosis enoyl coenzyme A reductases and blocks intracellular growth of these bacteria in macrophages. J. Bacteriol. 191:5262-5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sabina, J., N. Dover, L. J. Templeton, D. R. Smulski, D. Soll, and R. A. LaRossa. 2003. Interfering with different steps of protein synthesis explored by transcriptional profiling of Escherichia coli K-12. J. Bacteriol. 185:6158-6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schreiber, F., and U. Szewzyk. 2008. Environmentally relevant concentrations of pharmaceuticals influence the initial adhesion of bacteria. Aquat. Toxicol. 87:227-233. [DOI] [PubMed] [Google Scholar]
- 31.Wintz, H., L. J. Yoo, A. Loguinov, Y. Y. Wu, J. A. Steevens, R. D. Holland, R. D. Beger, E. J. Perkins, O. Hughes, and C. D. Vulpe. 2006. Gene expression profiles in fathead minnow exposed to 2,4-DNT: correlation with toxicity in mammals. Toxicol. Sci. 94:71-82. [DOI] [PubMed] [Google Scholar]
- 32.Wu, Y. G., S. Rozenfeld, A. Defferrard, K. Ruggiero, J. A. Udall, H. Kim, D. J. Llewellyn, and E. S. Dennis. 2005. Cycloheximide treatment of cotton ovules alters the abundance of specific classes of mRNAs and generates novel ESTs for microarray expression profiling. Mol. Genet. Genomics 274:477-493. [DOI] [PubMed] [Google Scholar]
- 33.Yan, X., C. Zhao, A. Budin-Verneuil, A. Hartke, A. Rince, M. S. Gilmore, Y. Auffray, and V. Pichereau. 2009. The(p)ppGpp synthetase RelA contributes to stress adaptation and virulence in Enterococcus faecalis V583. Microbiology 155:3226-3237. doi: 10.1099/mic.0.026146-0. [DOI] [PubMed] [Google Scholar]
- 34.Yergeau, E., M. Arbour, R. Brousseau, D. Juck, J. R. Lawrence, L. Masson, L. G. Whyte, and C. W. Greer. 2009. Microarray and real-time PCR analyses of the responses of high Arctic soil bacteria to hydrocarbon pollution and bioremediation treatments. Appl. Environ. Microbiol. 75:6258-6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yergeau, E., S. Bokhorst, A. H. L. Huiskes, H. T. S. Boschker, R. Aerts, and G. A. Kowalchuk. 2007. Size and structure of bacterial, fungal and nematode communities along an Antarctic environmental gradient. FEMS Microbiol. Ecol. 59:436-451. [DOI] [PubMed] [Google Scholar]
- 36.Yergeau, E., H. Hogues, L. G. Whyte, and C. W. Greer. 15 April 2010. The functional potential of high Arctic permafrost revealed by metagenomic sequencing, qPCR, and microarray analyses. ISME J. [Epub ahead of print.] doi: 10.1038/ismej.2010.41. [DOI] [PubMed]
- 37.Yergeau, E., S. Kang, Z. He, J. Zhou, and G. A. Kowalchuk. 2007. Functional microarray analysis of nitrogen and carbon cycling genes across an Antarctic latitudinal transect. ISME J. 1:163-179. [DOI] [PubMed] [Google Scholar]
- 38.Yergeau, E., and G. A. Kowalchuk. 2008. Responses of Antarctic soil microbial communities and associated functions to temperature and freeze-thaw cycle frequency. Environ. Microbiol. 10:2223-2235. [DOI] [PubMed] [Google Scholar]
- 39.Yergeau, E., S. A. Schoondermark-Stolk, E. L. Brodie, S. Déjean, T. Z. DeSantis, O. Gonçalves, Y. M. Piceno, G. L. Andersen, and G. A. Kowalchuk. 2009. Environmental microarray analyses of Antarctic soil microbial communities. ISME J. 3:340-351. [DOI] [PubMed] [Google Scholar]
- 40.Yokoi, T., Y. Kaku, H. Suzuki, M. Ohta, H. Ikuta, K. Isaka, T. Sumino, and M. Wagatsuma. 2007. ‘FloraArray’ for screening of specific DNA probes representing the characteristics of a certain microbial community. FEMS Microbiol. Lett. 273:166-171. [DOI] [PubMed] [Google Scholar]
- 41.Zurita, J. L., G. Repetto, Á. Jos, M. Salguero, M. López-Artíguez, and A. M. Cameán. 2007. Toxicological effects of the lipid regulator gemfibrozil in four aquatic systems. Aquat. Toxicol. 81:106-115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.