Abstract
The model rhizobacterium Pseudomonas putida KT2440 and other fluorescent pseudomonads possess two bacterioferritins, Bfrα and Bfrβ. However, the regulatory systems controlling the expression of these genes and the roles of these proteins in iron homeostasis are ill defined. Our studies show that both bfrα and bfrβ were monocistronic: promoter motifs and transcriptional start sites were identified, and Fur boxes and σS-dependent regulatory motifs were absent. The expressions of bfrα and bfrβ were enhanced by iron exposure and were maximal in cells rapidly growing in a high-iron environment. Both bfrα and bfrβ were positively regulated by Fur, and both were expressed independently of adjoining, functionally related genes. The loss of Bfrα or Bfrβ individually resulted in a significant reduction (ca. 17%) in cellular iron levels, and the deletion of both bfrα and bfrβ reduced cellular iron levels by 38% relative to those of the wild type. The mutants varied in their abilities to grow in low-iron medium; while growths (rate and final cell density) of single mutants and the wild type were similar, that of the double mutant was reduced significantly. Mutants lacking Bfrα and/or Bfrβ showed no change relative to the wild type in sensitivity to reactive oxygen species toxicity. Collectively, the data show that while Bfrα and Bfrβ could function independently of each other, an interaction-dependent function cannot be ruled out. Furthermore, regardless of the mechanism, a primary benefit of the bacterioferritins to P. putida KT2440 appears to be the enhancement of its survival in the environment by strengthening its tolerance to iron starvation.
Fluorescent pseudomonads such as Pseudomonas putida and P. aeruginosa are important in human health and environmental processes, and a key element for their activity and survival is iron (4, 6, 15, 27, 35, 39). While essential, levels of free iron in cells must be controlled to mitigate reactions in which it participates that generate reactive oxygen species (ROS) (32). To this end, one iron management strategy is the funneling of the element into ferritins (9). In addition to scavenging free iron to defend against ROS generation, ferritins could also serve as a reserve from which an iron-starved cell could draw (1, 8). Thus, ferritins could be viewed as having dual functionality: one serving an acute, immediate need (toxicity mitigation) and the other addressing a potentially longer-term condition (starvation tolerance).
Ferritins are omnipresent in bacteria and exist in two general types, bacterial ferritin (Ftn) and bacterioferritin (Bfr) (37). Both possess a dinuclear ferroxidase center, but Ftn differs from Bfr in that it lacks heme-binding-site motifs and has a third metal-binding site at the ferroxidase center, which Bfr lacks (2, 3). Three forms of Bfr have been described: Bfr, Bfrα, and Bfrβ (1, 10, 11, 23, 26). Bfrα differs from Bfr and Bfrβ in that it lacks a conserved Met-52 residue involved in heme binding, which is present in Bfr and Bfrβ (5). Bfr and Bfrβ have the same array of functional motifs and are distinguished only by the phylogenetic divergence of their primary structure (23). Bacterial genomes typically show one of two patterns of bfr and ftn distribution (37). The most common pattern is a combination of ftn (one or two copies) with bfr (single copy) and is exemplified by Escherichia coli (1), Salmonella enterica (40), and Erwinia chrysanthemi (7). A second pattern is the possession of both bfrα and bfrβ and the absence of ftn(s); this pattern is exhibited by fluorescent pseudomonads (11, 23, 26), Synechocystis sp. strain PCC6803 (21), and Neisseria gonorrhoeae (10).
Despite their widespread occurrence, much is still unknown about the function(s) of ferritins in bacteria, particularly the bacterioferritins. In this regard, for organisms that possess ftn and bfr, there is good evidence showing Ftn, but not Bfr, as functioning in ROS toxicity mitigation and/or starvation tolerance (1, 7, 40). Furthermore, the amounts of total cellular iron bound by Bfr compared to those bound by Ftn are highly variable (7, 14, 40). The situation is similarly ill defined for the bfrα-bfrβ combination in fluorescent pseudomonads, where functional analysis has focused on bfrα (23, 26). In P. aeruginosa, Bfrα appeared to make no significant contribution to the total cellular iron pool (1, 23). However, it was suggested previously that Bfrα could participate in ROS defense by enhancing catalase (KatA) activity by serving as a heme donor to that enzyme (23). In contrast, compared to the wild type (WT), mutants of P. putida Corvallis (23, 26) lacking bfrα showed no difference in katA expression or growth in iron-depleted medium. Thus, if Bfrα functions in ROS defense and/or starvation tolerance, that activity has not yet been clearly established for fluorescent pseudomonads. Studies of Bfrβ have been more limited than those of Bfrα, and the role(s) that Bfrβ may play in ROS defense or starvation tolerance has, to the best of our knowledge, not yet been determined for fluorescent pseudomonads.
In fluorescent pseudomonads, the regulation and expression of bfrα and bfrβ are also in need of clarification. In this regard, there are conflicting data on the responsiveness of bfrα to environmental iron levels: while analysis with gene reporters and Northern blotting show that bfrα expression is induced by iron exposure (23), a previously reported transcriptomic study indicated that iron exposure induced the expression of bfrβ but not bfrα (33). The transcriptional organizations of bfrα and bfrβ are also not fully defined: while for bfrα, transcription is known to be independent of its downstream neighbor katA (23, 26), the transcriptional organization of bfrβ in relation to its upstream neighbor bfd has not been established. The latter has been determined for E. coli (17), Yersinia pestis (16), and E. chrysanthemi (7), where bfr is cotranscribed with its upstream neighbor bfd. This organization has been proposed to represent a means to regulate the production of two functionally linked genes (Bfd facilitates iron mobilization from Bfr). It is unknown whether fluorescent pseudomonads employ a similar strategy for the regulation of bfd and bfrβ.
The model rhizobacterium P. putida KT2440 is like other fluorescent pseudomonads in possessing single copies of bfrα and bfrβ. In the present study, strain KT2440 was used to gain further insights into the ill-defined areas of molecular biology and physiology regarding these genes and proteins. Our aims were 2-fold. First, we sought to delineate mechanisms that control the production of Bfrα and Bfrβ, including regulatory motifs, transcriptional structure, interaction with the ferric uptake regulator (Fur) protein, and effects of iron exposure on gene expression. Second, we sought to determine the extent to which these proteins may contribute to ROS defense and/or iron nutrition.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Cloning was done with E. coli JM109; E. coli S17(λpir) was used for conjugal transfer as the donor strain and as the host for maintaining engineered constructs. E. coli strains (Table 1) were routinely grown in LB (Luria-Bertani) broth at 37°C. P. putida strain KT2440 and all its derivatives (Table 1) were grown in either LB broth or mineral salts medium (19) supplemented with 10 mM benzoic acid (MSMB) at 28°C. Iron levels in MSMB were adjusted by the addition of appropriate volumes of FeCl3 (12 mM in 6 N HCl). The medium used for culturing under “high-iron conditions” was either LB broth or MSMB containing 15 μM FeCl3. For “low-iron conditions” cells were grown in MSMB containing 200 μM 2,2′-dipyridyl. When needed, antibiotics were added as follows: rifampin (Rif) at 50 μg/ml, ampicillin (Amp) at 100 μg/ml, and gentamicin (Gm) at 60 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics and/or plasmid construction | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| S17-1 | hsdR17 (rK− mK−) recA RP4-2 (Tcr::Mu-Kmr::Tn7 Strr) | 36 |
| JM109 | F′ [traD36 proAB+lacIqlacZΔM15] recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi-1 mcrA Δ(lac-proAB) | Promega |
| Pseudomonas putida | ||
| SCH65 | Δbfrβ | This study |
| SCH88 | Δbfrα | This study |
| SCH89 | Δbfrα and Δbfrβ | This study |
| SCH99 | bfrα::rluc in KT2440R; WT | 11 |
| SCH109 | bfrβ::rluc in KT2440R; WT | 11 |
| SCH140 | Δfur bfrα::rluc in KT2440R | This study |
| SCH144 | Δfur bfrβ::rluc in KT2440R | This study |
| Plasmids | ||
| pGEM-T Easy | Cloning vector; Ampr | Promega |
| pRL-SV40 | Renilla luciferase gene (rluc) | Promega |
| pEX18Gm | Allelic exchange vector; Gmr | 20 |
| pSCH56 | bfrβ downstream region in pGEM-T Easy | This study |
| pSCH58 | bfrβ upstream region in pGEM-T Easy | This study |
| pSCH59 | bfrα upstream region in pGEM-T Easy | This study |
| pSCH60 | bfrα upstream region in pEX18-Gm | This study |
| pSCH62 | bfrβ upstream and downstream regions in pEX18-Gm | This study |
| pSCH77 | bfrα downstream region in pGEM-T Easy | This study |
| pSCH83 | bfrα upstream and downstream regions in pEX18-Gm | This study |
Nucleic acid manipulations.
Genomic DNA extractions were done with a genomic DNA extraction kit (Promega, Madison, WI), and plasmid DNA was purified with the QIAprep spin miniprep kit (Qiagen, Germantown, MD). DNA ligations, restriction endonuclease digestions, and agarose gel electrophoresis were performed according to standard methods (34). Transformation was done by the calcium chloride method for E. coli and by conjugation or electroporation for P. putida strains. PCR amplifications were done with the Failsafe PCR system (Epicenter Technology, Madison, WI). PCR products were separated on 0.7 to 1.0% (wt/vol) agarose gels, and the bands were purified with the QIAquick gel extraction system (Qiagen). RNA was extracted with an RNeasy kit (Qiagen).
RT-PCR analysis.
Total RNA was isolated from P. putida KT2440 cells grown in high-iron MSMB and digested with DNase I. After 1 h of incubation, DNase I was inactivated by a 15-min incubation at 72°C, and RNA was then purified by phenol-chloroform extraction. Reverse transcription (RT)-PCR was done by using SuperScript II (Invitrogen, Carlsbad, CA) at 42°C for 1.5 h with random hexamer primers (Gibco, Cambridge, MA). PCR amplification of the fragments spanning the upstream and downstream regions of bfrα was done by using primers pp19/pp20, pp20/pp21, and pp22/pp23 (Table 2). For bfrβ, upstream and downstream fragments were amplified by using primers pp9/pp24, pp16/pp25, and pp7/pp26 (Table 2). Genomic DNA and total RNA extracts (not reverse transcribed) were used for RT-PCR as the positive and negative controls, respectively. RT-PCR products were analyzed by electrophoresis on a 2% agarose gel stained with ethidium bromide; amplicons were then purified and sequenced to confirm their identity.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′)a |
|---|---|
| pp1 | 5′-(p)CATCTGCGACTGCAGG |
| pp2 | GGCATCGAGCTGTGCGAACTGCACAAGG |
| pp3 | CCTGCGTGCGCAGCTGGCCGACAC |
| pp4 | CGCACGCATGTCAGGGGTGCCTTCG |
| pp5 | CGCTCGTAGAGCTTGGACAGGCCCC |
| pp6 | 5′-(p)GCCCATCTGCGACTG |
| pp7 | CATTGTGAAACCGCTGGCGACTTCGG |
| pp8 | CGAGGAAGAGCACATCGACTGGCTGG |
| pp9 | GCCCAGGTCCTGAACGTTAGGAATACC |
| pp10 | CGTGCTTGCCGAGTTTGTTCAGCCC |
| pp11 | CGGAATTCCGAGCCGCGACGAGAACCCATTCGAC |
| pp12 | TGCTCTAGAGCAGGTCGCGTGCCGCCAGTTCGCCC |
| pp13 | TGCTCTAGAGCACCACAGACTCAAGTAAAAGCCCC |
| pp14 | CCCAAGCTTGGGCCTTTACCCCTTGGCCGCCACC |
| pp15 | CGGAATTCCGCTCATGACAGGCCGACTCCGTCG |
| pp16 | TGCTCTAGAGCACCAATCTTCGTACATGCGTGCG |
| pp17 | TGCTCTAGAGCACGCCCCATAAAAAAACCCGCTGATCG |
| pp18 | CCCAAGCTTGGGCGCGTACTGGCTCACTGGAGGTCG |
| pp19 | GCGGATCCGGCTTACGGCG |
| pp20 | CGGTGCTACCAACCTCCAGG |
| pp21 | TTCACATGGAGAATCAGGGCG |
| pp22 | CCTGGAGGTTGGTAGCACCG |
| pp23 | GCAATACCGTGGTGGTGATCG |
| pp24 | CGAGCCGCCACCAATGTCGCC |
| pp25 | GGGCTGAACAAACTCGGCAAGCACG |
| pp26 | ACGCTGCGCTCAATGGCGTACCC |
| pp27 | ATGAAAGGCGACGTAAGCG |
| pp28 | CTGCAAACCCTGATCAAGG |
Restriction sites on the primers are underlined. EcoRI was engineered on the 5′ ends of pp11 and pp15; XbaI was engineered on the 5′ ends of pp12 and pp13; HindIII was engineered on the 5′ ends of pp14 and pp18. 5′-(p), 5′ phosphorylated.
TSS determination.
Analysis by 5′ rapid amplification of cDNA ends (5′-RACE) was done by using the TaKaRa 5′-Full RACE core set under the conditions recommended by the supplier. For bfrα, the RT reaction was done with the 5′-phosphorylated RT primer pp1 (Table 2). After RT, mRNA was digested with RNase H, and cDNA was then concatenated by using T4 RNA ligase. The region of interest was then amplified by using two sets of primers for regions of bfrα in a nested PCR. In the first PCR, RT products were used as a template and amplified with primers pp2 and pp4 (Table 2). In the second PCR, the template was a 10-fold dilution of the round 1 PCR product, and amplification was done by using primers pp3 and pp5 (Table 2). The 5′-RACE products were isolated, purified, ligated into the pGEM-T Easy vector, and sequenced. The same strategy for 5′-RACE was used to identify the transcription start site (TSS) of bfrβ except for the following oligonucleotides: pp6 was used for the initiation of cDNA synthesis, and the primers for the first and second rounds of PCR were pp7/pp9 and pp8/pp10 (Table 2), respectively.
Mutant construction.
To knock out bfrα, upstream (P. putida genome positions 565323 to 566191) and downstream (genome positions 566592 to 567540) fragments of ca. 900 bp were PCR amplified from chromosomal DNA by using primers pp11/pp12 and pp13/pp14, respectively (Table 2). Amplicons were gel purified, cloned into pGEM-T Easy (pSCH59 and pSCH77) (Table 1), and sequenced to confirm their identity. The upstream fragment was released from pSCH59 by EcoRI/XbaI digestion and then inserted into the same sites on pEX18-Gm (pSCH60). For the bfrα downstream fragment, pSCH77 was XbaI/HindIII digested and inserted into the same sites on pSCH60, yielding the bfrα knockout vector pSCH83. Similarly, the bfrβ upstream (genome positions 1243017 to 1242360) and downstream (genome positions 1241985 to 1241280) fragments were amplified by using primers pp15/pp16 and pp17/pp18, respectively, and cloned into pGEM-T Easy (pSCH58 and pSCH56). The inserted fragments were released and subcloned into pEX18-Gm, giving the bfrβ knockout vector pSCH62.
Vector pSCH83 was introduced into P. putida KT2440R by conjugation, and merodiploids were selected on LB plates supplemented with Gm and Rif. The Gm-resistant merodiploids were resolved by plating single colonies onto MSMB medium containing 5% (wt/vol) sucrose according to a method described previously by Chen et al. (11). Putative Δbfrα clones were identified by screening with PCR and checked for susceptibility to Gm and sucrose; one confirmed Δbfrα clone (strain SCH88) was chosen for further analysis. A Δbfrβ strain (SCH65) was created by using the same procedures described above. A Δbfrα Δbfrβ mutant (SCH89) was constructed by repeating the procedures described above with strain SCH88.
Reporter strains SCH99 (bfrα::rluc) and SCH109 (bfrβ::rluc) were constructed as described in our previous work (11) by the insertion of Renilla luciferase (rluc) immediately downstream of bfrα and bfrβ individually. The fur-deleted reporter strains SCH140 (Δfur bfrα::rluc) and SCH144 (Δfur bfrβ::rluc) were created by using vector pSCH113 as described previously (11). Reporter assays were done as described in our prior work (11) by using a commercially available kit (Promega) according to the manufacturer's protocol.
DNA sequence analysis.
Nucleotide sequencing was done through the University of Wisconsin—Madison Biotechnology Center. GenBank database searches were done by using the NCBI Web server (http://www.ncbi.nlm.nih.gov/BLAST). Multiple-sequence alignments were carried out with the ClustalW program and later adjusted manually. Genome sequence data for P. putida KT2440 (GenBank accession number AE015451) was obtained from The Institute for Genomic Research (29), and terminator prediction was done by using the “Terminator List Tool.” RNA folding analysis was conducted with the RNAfold Web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi).
Total iron determination.
Cells (triplicate 20-ml aliquots) were grown in LB broth for 10 h (mid-log phase) and 20 h (stationary phase), collected by centrifugation, washed twice in 50 ml of phosphate-buffered saline (PBS) (1 mM EDTA, pH 7.4), and then resuspended in 2 ml PBS. A 1-ml aliquot was then diluted in 10 ml of 1% nitric acid and analyzed by using inductively coupled plasma-mass spectrometry through the University of Wisconsin—Madison Soil and Plant Analysis Laboratory. The nitric acid solution was analyzed to correct for background. The remaining aliquots (1 ml) were used to measure cell mass and were dried at 100°C until reaching a constant weight.
Statistical analysis.
The Wilcoxon rank sum test was used for all data analyses and applied via the SAS statistical software package (version 9.0; SAS Institute Inc., Cary, NC).
RESULTS AND DISCUSSION
Transcriptional organization of bfrα and bfrβ.
The organization of the bfrα and bfrβ neighborhoods is conserved in genomes of fluorescent pseudomonads (see Fig. S1 in the supplemental material). In P. putida KT2440, the start codon of bfrα and the stop codon of the upstream neighbor (katA) are separated by 163 bp; computational analysis of the region downstream of katA identified a potential transcriptional terminator in the form of a stable (ΔG0 = −36.82 kJ/mol) stem-and-loop structure (CCGGACCTGGTGTCCGG). The start and stop codons of bfrβ and the adjoining upstream gene (bfd) are separated by 199 nucleotides. Like katA, a potential stem-loop element (CATAAAAAAACCCGCTGATCGCGGGTTTTTTTATG) (ΔG0 = −84.94 kJ/mol) is located 10 bp downstream of the bfd stop codon. These G+C-rich regions of hyphenated dyad symmetry could function as transcriptional terminators, indicating that bfrα and bfrβ could be monocistronic and transcribed independently of neighboring genes.
For experimental analyses of transcriptional organization, RT-PCR analysis of total RNA isolated from cells grown under high-iron conditions was done. The bfrα primers yielded a product from the cDNA template similar in size to that amplified from genomic DNA (Fig. 1). In contrast, no product was obtained from a cDNA template with primers targeting the intergenic region between katA and bfrα. Thus, bfrα was transcribed independently of katA. Similar results were obtained from analyses of bfd and bfrβ. As expected, for both bfrα and bfrβ, primers targeting the downstream intergenic regions yielded no product. Thus, bfrα and bfrβ were transcribed as monocistrons.
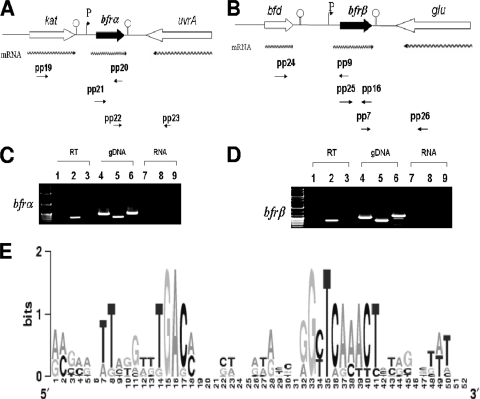
FIG. 1.
Transcriptional organization analysis and transcription start site determination for bfrα and bfrβ in P. putida KT2440. (A) Organization of the bfrα neighborhood in P. putida KT2440. Wavy arrows indicate predicted transcripts, and straight arrows show primer targets used for RT-PCR. (B) Organization of the bfrβ neighborhood in P. putida KT2440. Wavy arrows indicate predicted transcripts, and straight arrows show primer targets used for RT-PCR. (C) PCR detection of the upstream region of bfrα (spanning the kat-bfrα) by using primer pair pp19/pp20 (see Table 2 for details on all primers) (lanes 1, 4, and 7), bfrα by using primer pair pp21/pp20 (lanes 2, 5, and 8), and the bfrα-uvrA junction by using primer pair pp22/pp23 (lanes 3, 6, and 9). (D) PCR detection of the upstream region of bfrβ (spanning the bfd-bfrβ junction) by using primer pair pp24/pp9 (lanes 1, 4, and 7), bfrβ by using primer pair pp25/pp16 (lanes 2, 5, and 8), and the bfrβ-glu junction by using primer pair pp7/pp26 (lanes 3, 6, and 9). For C and D, PCR templates were genomic DNA (gDNA) (positive control), cDNA (RT), or non-reverse-transcribed RNA (RNA) (negative control). (E) Logo of putative promoter motifs for the bfrα and bfrβ genes in selected fluorescent pseudomonads. The nonencoding regions within 100 bp upstream of the bfrα and bfrβ translation start codons were first manually scanned, aligned with ClustalW, and preceded with WebLogo (13). The selected bfrα and bfrβ genes are from P. putida KT2440 (GenBank accession number AE015451), P. aeruginosa PAO1 (accession number AE004091), Pseudomonas fluorescens PfO-1 (accession number CP000094), and Pseudomonas syringae pv. syringae B728a (accession number CP000075) (see Fig. S2 in the supplemental material).
The present finding of bfrα as being monocistronic is consistent with previously described Northern blot analyses of fluorescent pseudomonads (23, 26). Collectively, the data show that while bfrα and katA may be functionally linked, their expression is not. The transcriptional organization of bfrβ in fluorescent pseudomonads has not previously been established. However, the current finding of bfrβ as being monocistronic presents a situation similar to that of bfrα-katA in that the expression of bfrβ can be regulated independently of bfd, the adjoining upstream gene to which it may be functionally linked. The transcriptional organization of bfrα and bfrβ differs from that of bfr in E. coli (17), Y. pestis (16), and E. chrysanthemi (7), where bfr is cotranscribed with its upstream neighbor, bfd. In the case of the latter three organisms, the transcriptional linkage of bfd and bfrβ was previously postulated to represent a mechanism for the efficient production of proteins that may work together (7, 17). However, for organisms in which bfd and bfrβ are cotranscribed, any alteration of Bfd and Bfrβ levels (e.g., for rebalancing of intracellular iron) would rely on posttranscriptional/posttranslational mechanisms. In contrast, strain KT2440 and other fluorescent pseudomonads could modulate levels of Bfrα and Bfrβ (as well as KatA and Bfd) at the transcriptional level and possibly apply posttranscriptional/posttranslational mechanisms for additional tuning (24, 31, 42).
To the best of our knowledge, promoter motifs and TSS for bacterioferritin genes have not been documented for pseudomonads. For bfrα, of six randomly chosen colonies, four had A and two had G as the TSS. The separations of the TSS and the translational start site were 28 bp for A and 24 bp for G; this may indicate that alternative TSSs are used for the regulation of bfrα transcription. For bfrβ, there was a sole G TSS 33 bp upstream of the translational start site. An alignment of intergenic regions within 100 bp upstream of bfr from several fluorescent Pseudomonas strains shows that the putative conserved motifs of a −10 box (TCAAACT) and a −35 box (TGAC) exist upstream of the TSS in both bfrα and bfrβ (Fig. 1). The extended −10 regions for both bfrα (TCACACT) and bfrβ (TCAAACT) had no homology to the canonical TATAAT E. coli promoter consensus sequence, while −35 regions (TGAC) were relatively short compared to that conserved in E. coli (TTGACA). Furthermore, the putative −10 regions of bfrα and bfrβ did not resemble σS-dependent −10 (TGN0-2CYATAMT) motifs (22). Thus, while σS is implicated in the regulation of a variety of iron uptake and storage functions in the stationary phase (12), it is not likely to directly affect the expression of bfrα and bfrβ in P. putida KT2440 (see below). Sequence motifs indicative of Fur-binding sites (GATAATGATAATCATTATC) (30) were not detected in regions upstream of bfrα or bfrβ in P. putida KT2440. Thus, any Fur-mediated regulation of these genes is likely to be indirect and transmitted through the interaction of Fur with other molecules.
Transcriptional analysis of bfrα and bfrβ expression as a function of growth phase and environmental iron levels.
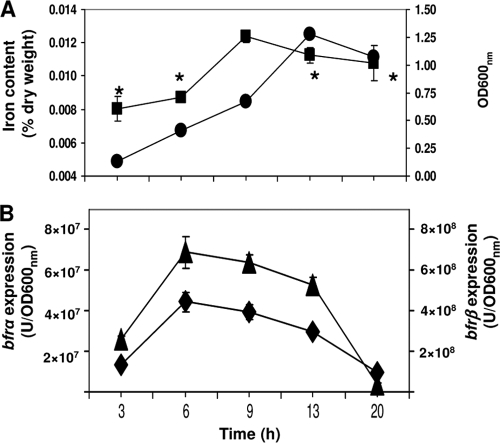
The expression levels of bfrα and bfrβ varied in a similar fashion during the growth cycle (Fig. 2): both increased with time, reaching a maximum at mid-log phase (6 to 9 h), and then decreased into stationary phase (13 to 20 h). Cellular iron levels also varied as a function of growth phase and mirrored the expression patterns of bfrα and bfrβ (Fig. 2): maximum levels were reached in mid-log phase and then slightly decreased as the cells entered stationary phase.
FIG. 2.
Analysis of growth, total cellular iron, and bacterioferritin gene expression. (A) Cell growth (•) in LB medium and total cellular iron (▪). (B) Expression of bfrα (▴) and bfrβ (⧫) in cells grown in LB broth. Luciferase activity was normalized to cell density; values are means of single measurements from triplicate cultures (± standard deviations). The asterisk indicates time points where the cellular iron content was significantly lower (P < 0.05) than that at 9 h.
It is noteworthy that in P. putida KT2440, the growth phase-related variation in the expressions of bfrα and bfrβ contrasts with those of E. coli (17) and E. chrysanthemi (7). In the latter two organisms, the level of bfr expression increased at least 5-fold in the transition from exponential growth to stationary phase, indicating that bfr expression was σS regulated. These divergent expression patterns are consistent with the occurrence of σS regulation motifs in the promoter regions of bfd-bfr in E. coli and E. chrysanthemi (7) and their absence in promoter regions of bfrα and bfrβ in P. putida KT2440.
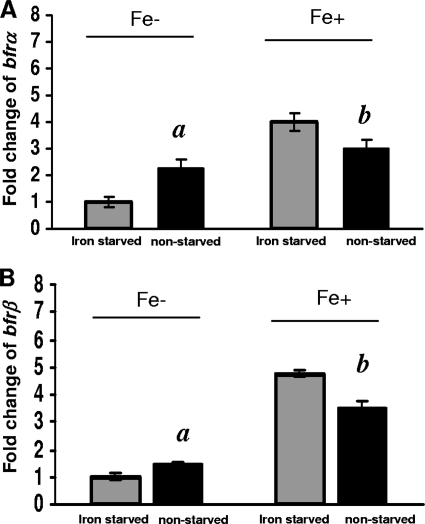
The responses of bfrα and bfrβ to high versus low environmental iron levels were examined in cells that were first precultured either under high-iron conditions (nonstarved, iron-sufficient cells) or low-iron conditions (starved, iron-deficient cells) and then grown under high- versus low-iron conditions (Fig. 3). Under low-iron conditions, in nonstarved cells, the expression levels of bfrα and bfrβ were ca. 2.3- and 1.5-fold higher, respectively, than those in starved cells (significant at a P value of 0.04). In contrast, under high-iron conditions, bfrα and bfrβ expression levels in nonstarved cells were 25% and 29% lower, respectively, than those in starved cells (significant at a P value of 0.04), indicating that expressions of these genes were affected by both the environmental iron level and the initial intracellular iron status.
FIG. 3.
Comparison of bfrα and bfrβ expression levels in iron-starved and nonstarved cells in high- and low-iron medium. (A) Expression of bfrα. (B) Expression of bfrβ. For both genes, reporter activity is expressed as the fold change relative to iron-starved cells in low-iron (Fe−) medium. Values are means of single measurements from triplicate cultures (± standard deviations). Comparisons that were significantly different (P < 0.05) were nonstarved cells versus iron-starved cells in low-iron medium (indicated by “a”) and nonstarved cells versus iron-starved cells in high-iron medium (indicated by “b”).
The relationship between growth phase, environmental iron level, and relative expression levels of bfrα and bfrβ further indicated that the proteins are utilized primarily to store iron during periods of rapid growth under high-iron conditions (Fig. 2). Results of our prior work (11) as well as those of the present study are consistent in showing the upregulation of bfrα in response to iron exposure (Fig. 3), and a similar effect of iron on bfrα expression was documented previously for P. putida Corvallis (26) and P. aeruginosa PAO (23). However, a divergent finding was reported for a transcriptome analysis of P. aeruginosa PAO, where a shift from low- to high-iron conditions yielded a 24-fold increase in the level of bfrβ expression but no change in that of bfrα (33). The latter investigators reasoned that their divergent finding with bfrα was attributable to differences in experimental protocols (microarray versus LacZ reporters and Northern blotting).
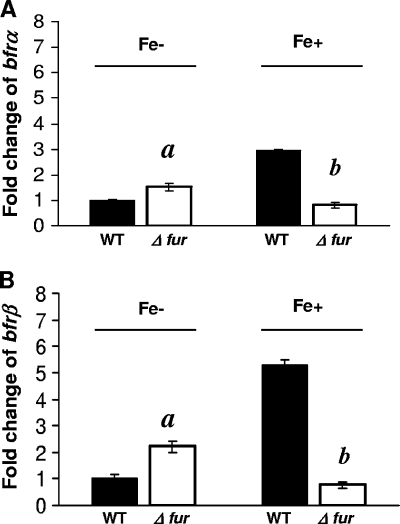
Responses of bfrα and bfrβ to high versus low iron in Fur+ and Fur− backgrounds.
In wild-type (WT) cells (Fur+ background) grown under high-iron conditions, the levels of expression of bfrα and bfrβ were ca. 3- and 6-fold higher, respectively, than levels measured in the Fur− mutant (Fig. 4). In contrast, under low-iron conditions, the levels of expression of bfrα and bfrβ in the Fur− background were ca. 0.5- and 1.2-fold higher, respectively, than those in the WT background. Thus, P. putida KT2440 bfrα and bfrβ were both positively regulated by iron in a Fur-dependent manner.
FIG. 4.
Effect of Fur on expression of bfrα and bfrβ. (A) Expression of bfrα in the wild type (WT) and the Δfur mutant in high- and low-iron medium. (B) Expression of bfrβ in the WT and the Δfur mutant in high- and low-iron medium. For both genes, reporter activity is expressed as the fold change relative to WT cells in low-iron medium. Luciferase activity was normalized to cell density; values are means of single measurements from triplicate cultures (± standard deviations). Cells were cultured in MSMB, and a low-iron condition was created by the addition of 2,2′-dipyridyl (200 μM); a high-iron condition was established by the addition of FeCl3 (15 μM). Comparisons that were significantly different (P < 0.05) were Δfur mutant versus WT cells in low-iron medium (indicated by “a”) and Δfur mutant versus WT cells in high-iron medium (indicated by “b”).
The effects of iron and Fur on the expression of bfrB in P. aeruginosa were investigated previously by Wilderman et al. (42) by using a nonsense mutant. In this case, although the mutant still expressed Fur and retained partial repressor activity, those investigators were able to demonstrate a positive effect of Fur on bfrB expression. The results of the present study with a Fur deletion mutant were consistent with those reported by Wilderman et al. (42) and provided further evidence for Fur as a positive regulator in fluorescent pseudomonads. In E. coli and other bacteria (25), positive regulatory mechanisms involving Fur are mediated via a small antisense RNA, RhyB, in a posttranscriptional manner. Strain KT2440 and other fluorescent pseudomonads have two small RNAs, PrrF1 and PrrF2, which have RhyB-like functionality, but these do not affect bacterioferritin expression (42). Thus, the mechanism by which Fur exerts positive regulatory control on bfrα and bfrβ remains unknown.
Physiological roles of Bfrα and Bfrβ.
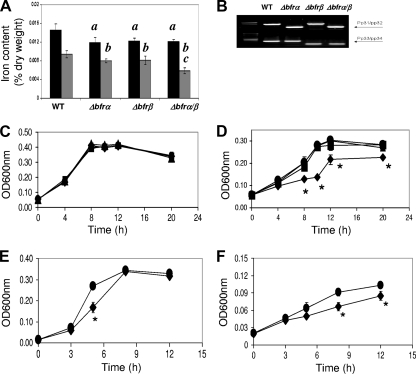
At log phase (10 h), the single mutants (Δbfrα and Δbfrβ) and the double mutant (Δbfrα Δbfrβ) all showed similar reductions in levels of total cellular iron of 16 to 18% relative to that of the wild type. In stationary phase (20 h), levels of total iron in single mutants were reduced by ca. 15%, while that of the double mutant was reduced by 38%, which was significantly (P < 0.05) lower than those of the wild type and single mutants (Fig. 5 A). These data show that while the loss of Bfrα or Bfrβ alone can upset iron homeostasis, the greatest impact was seen in nongrowing cells devoid of bacterioferritin. Possibly, Bfrα and Bfrβ may interact, as suggested previously by Moore et al. (28), to form a hetero-oligomer, which is the primary iron-binding structure for these molecules. Alternatively, these proteins could function independently in iron binding (41) and provide some redundancy in iron management; consistent with this hypothesis are the data showing that the iron reduction in the nongrowing double mutant roughly approximated the sum of the iron loss in the individual mutants. Investigations into the physical properties of Bfrα and Bfrβ are needed to resolve the underlying mechanism(s).
FIG. 5.
Effects of bacterioferritin deletion on cell iron content and growth. (A) Total iron content in cells grown in LB broth. The black bars represent the iron content at log phase, and the gray bars represent the iron content at the stationary phase; values are means of single measurements from triplicate cultures (± standard deviations). For log-phase measurements, data indicated by “a” are significantly different from the WT (P < 0.05). For stationary-phase measures, data indicated by “b” are significantly different from the WT (P < 0.05), and those indicated by “c” are significantly different from the WT and single mutants (P < 0.05). (B) Confirmation of bfrα and/or bfrβ deletion. Screening for bfrα and bfrβ was done with the indicated primers (see Fig. 1A and B and Table 2). (C) Growth of the WT (•), the Δbfrα (▴) and Δbfrβ (▪) single mutants, and the Δbfrα Δbfrβ double mutant (⧫) in high-iron medium (MSMB plus 15 μM FeCl3). Inocula were nonstarved. (D) Growth of the WT, the Δbfrα and Δbfrβ single mutants, and the Δbfrα Δbfrβ double mutant in low-iron medium (MSMB plus 200 μM 2,2′-dipyridyl). Inocula were nonstarved. (E) Growth of the WT and the Δbfrα Δbfrβ double mutant in high-iron medium (MSMB plus 15 μM FeCl3). Inocula were iron starved. (F) Growth of the WT and the Δbfrα Δbfrβ double mutant in low-iron medium (MSMB plus 200 μM 2,2′-dipyridyl). Inocula were iron starved. For C to F, data points are means of single measurements from triplicate cultures (± standard deviations), and an asterisk indicates points where double mutant growth was significantly different (P < 0.05) from that of the WT. There was no significant difference in growth between the WT and single mutants in any test.
When grown in high-iron medium, there was no significant difference in growth between the wild type and any of the three mutant strains (Fig. 5C). In low-iron medium, the growth of the single mutants was similar to that of the wild type (Fig. 5D), but the growth of the double mutant was reduced significantly (Fig. 5D). The behavior of the single mutants was consistent with redundancy in the iron management system proposed above. For the double mutant, impaired growth suggested that having at least one of the bacterioferritins is advantageous in supporting growth and could do so in two ways: by functioning as an iron-scavenging system that mitigates ROS generation and/or by serving as an intracellular reservoir of iron from which the cell can draw to meet nutritional needs.
To explore the potential function of Bfrα and Bfrβ in ROS defense, the wild type and mutants were compared for their sensitivity to H2O2 by using the disk method described previously by Ma et al. (23). The wild type and all bacterioferritin mutants (single and double mutants) showed similar sensitivities to H2O2 (zone of growth inhibition for all averaging 3.5 ± 0.2 cm). In contrast, for the Fur mutant, the zone of growth inhibition was significantly larger (5.6 cm), reflecting a heightened sensitivity to H2O2. These data indicate that while P. putida KT2440 has an ROS defense system that utilizes Fur-regulated proteins (18, 38), bacterioferritin is probably not of primary importance in this response. Similar results were also reported previously for S. enterica serovar Typhimurium and Synechocystis sp. PCC6803, where Bfr contributed to iron storage but did not play a critical role in ROS defense (21, 40).
The potential for Bfrα and Bfrβ to serve as the intracellular reservoir of nutritional iron was examined in two ways: first by comparing wild type and mutant growth in serial passage through low-iron medium (see Fig. S3 in the supplemental material) and second by preculturing cells either under high-iron conditions (nonstarved, iron-sufficient cells) or under low-iron conditions (starved, iron-deficient cells) and comparing the growth of starved cells to that of nonstarved cells in high- and low-iron medium (Fig. 5C and D). In the serial passage experiment, the growth of all cultures decreased with each transfer, but only the double mutant showed a reduction in growth that was greater than that of the wild type (Fig. S3). In the second experiment, when nonstarved cells were grown in high-iron medium, there was, as expected, no difference between any of the cultures (Fig. 5C). However, in low-iron medium, the growth of the nonstarved double mutant was significantly reduced relative to that of the wild type, while that of the single mutants was not (Fig. 5D). Thus, in the subsequent experiments with starved cells that are described below, only the double mutant and the wild type were compared (Fig. 5E and F).
In high-iron medium, the growth of the starved double mutant was significantly reduced in log phase, but the final culture density was similar to that of the wild type (Fig. 5E). This finding suggests that the mutant was impaired in its ability to recover from iron starvation. In low-iron medium, compared to the wild type, the growth of the mutant culture was significantly slower and achieved a final optical density at 600 nm (OD600) that was significantly lower (Fig. 5F). The above-described data indicate that the possession of least one bacterioferritin strengthens the tolerance of P. putida KT2400 to iron starvation. However, the loss of both Bfrα and Bfrβ has a far greater effect and may reflect an interaction between these proteins similar to that described above for iron storage. The bacterioferritins improve starvation tolerance, presumably by serving as an iron source. The potential for the recycling of iron from Bfrα or Bfrβ is supported by the work of Expert et al. (14), who, by using 59Fe-labeling methods, showed previously that a portion of Bfr-bound iron was redistributed to other proteins in E. chrysanthemi. The effect that the loss of bacterioferritin formation has on strain KT2440 (and, consequently, the importance of Bfrα and Bfrβ) is perhaps best appreciated in the context of the potential impact on the growth and/or survival of strain KT2440 in soil. In this case, the mutant's weakened competitive fitness would be manifested over a time frame expanded by orders of magnitude (e.g., months, years, or decades) beyond that displayed in Fig. 5F, and over such periods, the mutant could be effectively displaced from the microbial community.
This study has provided insights into the regulation and physiological function of Bfrα and Bfrβ in P. putida KT2440. Collectively, the data show that the expressions of bfrα and bfrβ are maximal during periods of rapid growth in a high-iron environment and involved positive regulation by Fur. The proteins can be expressed independently of each other and of adjoining, functionally related genes. While Bfrα or Bfrβ may be able to function independently, interaction-dependent function cannot be ruled out. Finally, a primary benefit of the bacterioferritins to P. putida KT2440 appears to be an enhancement of its survival in the environment by strengthening its tolerance to iron starvation.
Supplementary Material
Acknowledgments
These studies were funded by a grant from the USDA NRI-CGP (award number 2006-02624 to W.F.B. and W.J.H.).
We thank M. Bagdasarian for constructive advice and for kindly providing P. putida strain KT2440R.
Footnotes
Published ahead of print on 18 June 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abdul-Tehrani, H., A. J. Hudson, Y.-S. Chang, A. R. Timms, C. Hawkins, J. M. Williams, P. M. Harrison, J. R. Guest, and S. C. Andrews. 1999. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J. Bacteriol. 181:1415-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, S. C. 1998. Iron storage in bacteria. Adv. Microb. Physiol. 40:281-351. [DOI] [PubMed] [Google Scholar]
- 3.Baaghil, S., A. Lewin, G. R. Moore, and N. E. LeBrun. 2003. Core formation in Escherichia coli bacterioferritin requires a functional ferroxidase center. Biochemistry 42:14047-14056. [DOI] [PubMed] [Google Scholar]
- 4.Banin, E., M. L. Vasil, and E. P. Greenberg. 2005. Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 102:11076-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertani, E. L., J. S. Huang, B. A. Weir, and J. L. Kirschvink. 1997. Evidence for two types of subunits in the bacterioferritin of Magnetospirillum magnetotacticum. Gene 201:31-36. [DOI] [PubMed] [Google Scholar]
- 6.Bodey, G. P., R. Bolivar, V. Fainstein, and L. Jadeja. 1983. Infections caused by Pseudomonas aeruginosa. Rev. Infect. Dis. 5:279-313. [DOI] [PubMed] [Google Scholar]
- 7.Boughammoura, A., B. F. Matzanke, L. Bottger, S. Reverchon, E. Lesuisse, D. Expert, and T. Franza. 2008. Differential role of ferritins in iron metabolism and virulence of the plant-pathogenic bacterium Erwinia chrysanthemi 3937. J. Bacteriol. 190:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrondo, M. A. 2003. Ferritins, iron uptake and storage from the bacterioferritin viewpoint. EMBO J. 22:1959-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chasteen, N. D., and P. M. Harrison. 1999. Mineralization in ferritin: an efficient means of iron storage. J. Struct. Biol. 126:182-194. [DOI] [PubMed] [Google Scholar]
- 10.Chen, C.-Y., and S. A. Morse. 1999. Neisseria gonorrhoeae bacterioferritin: structural heterogeneity, involvement in iron storage and protection against oxidative stress. Microbiology 145:2967-2975. [DOI] [PubMed] [Google Scholar]
- 11.Chen, S., W. F. Bleam, and W. J. Hickey. 2009. Simultaneous analysis of bacterioferritin gene expression and intracellular iron status in Pseudomonas putida KT2440 by using a rapid dual luciferase reporter assay. Appl. Environ. Microbiol. 75:866-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornelis, P., S. Matthijs, and L. Van Oeffelen. 2009. Iron uptake regulation in Pseudomonas aeruginosa. BioMetals 22:15-22. [DOI] [PubMed] [Google Scholar]
- 13.Crooks, G. E., G. Hon, J.-M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Expert, D., A. Boughammoura, and T. Franza. 2008. Siderophore-controlled iron assimilation in the enterobacterium Erwinia chrysanthemi. J. Biol. Chem. 283:36564-36572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Expert, D., C. Enard, and C. Masclaux. 1996. The role of iron in plant host-pathogen interactions. Trends Microbiol. 4:232-237. [DOI] [PubMed] [Google Scholar]
- 16.Gao, H., D. Zhou, Y. Li, Z. Guo, Y. Han, Y. Song, J. Zhai, Z. Du, X. Wang, J. Lu, and R. Yang. 2008. The iron-responsive Fur regulon in Yersinia pestis. J. Bacteriol. 190:3063-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garg, R. P., C. J. Vargo, X. Cui, and D. M. Kurtz. 1996. A [2Fe-2S] protein encoded by an open reading frame upstream of the Escherichia coli bacterioferritin gene. Biochemistry 35:6297-6301. [DOI] [PubMed] [Google Scholar]
- 18.Heim, S., M. Ferrer, H. Heuer, D. Regenhardt, M. Nimtz, and K. N. Timmis. 2003. Proteome reference map of Pseudomonas putida strain KT2440 for genome expression profiling: distinct responses of KT2440 and Pseudomonas aeruginosa strain PAO1 to iron deprivation and a new form of superoxide dismutase. Environ. Microbiol. 5:1257-1269. [DOI] [PubMed] [Google Scholar]
- 19.Hickey, W. J., and D. D. Focht. 1990. Degradation of mono-, di-, and trihalogenated benzoic acids by Pseudomonas aeruginosa JB2. Appl. Environ. Microbiol. 56:3842-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 21.Keren, N., R. Aurora, and H. B. Pakrasi. 2004. Critical roles of bacterioferritins in iron storage and proliferation of cyanobacteria. Plant Physiol. 135:1666-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lacour, S., and P. Landini. 2004. Sigma S-dependent gene expression at the onset of stationary phase in Escherichia coli: function of sigma S-dependent genes and identification of their promoter sequences. J. Bacteriol. 186:7186-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma, J.-F., U. A. Ochsner, M. G. Klotz, V. K. Nanayakkara, M. L. Howell, Z. Johnson, J. E. Posey, M. L. Vasil, J. J. Monaco, and D. J. Hassett. 1999. Bacterioferritin A modulates catalase A (KatA) activity and resistance to hydrogen peroxide in Pseudomonas aeruginosa. J. Bacteriol. 181:3730-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mass, E., H. Salvail, G. Desnoyers, and M. Arguin. 2007. Small RNAs controlling iron metabolism. Curr. Opin. Microbiol. 10:140-145. [DOI] [PubMed] [Google Scholar]
- 25.Masse, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, C. D., Y. C. Kim, M. K. Walsh, and A. J. Anderson. 2000. Characterization and expression of the Pseudomonas putida bacterioferritin alpha subunit gene. Gene 247:199-207. [DOI] [PubMed] [Google Scholar]
- 27.Molina, L., C. Ramos, E. Duque, M. C. Ronchel, J. M. García, L. Wyke, and J. L. Ramos. 2000. Survival of Pseudomonas putida KT2440 in soil and in the rhizosphere of plants under greenhouse and environmental conditions. Soil Biol. Biochem. 32:315-321. [Google Scholar]
- 28.Moore, G. R., F. H. Kadir, F. K. al-Massad, N. E. Le Brun, A. J. Thomson, C. Greenwood, J. N. Keen, and J. B. Findlay. 1994. Structural heterogeneity of Pseudomonas aeruginosa bacterioferritin. Biochem. J. 304:493-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. Martin dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. C. Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. Eisen, K. N. Timmis, A. Düsterhöft, B. Tümmler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 30.Ochsner, U. A., and M. L. Vasil. 1996. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc. Natl. Acad. Sci. U. S. A. 93:4409-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oglesby, A. G., J. M. Farrow, J.-H. Lee, A. P. Tomaras, E. P. Greenberg, E. C. Pesci, and M. L. Vasil. 2008. The influence of iron on Pseudomonas aeruginosa physiology. J. Biol. Chem. 283:15558-15567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orino, K., L. Lehman, Y. Tsuji, H. Ayaki, S. V. Torti, and F. M. Torti. 2001. Ferritin and the response to oxidative stress. Biochem. J. 357:241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palma, M., S. Worgall, and L. Quadri. 2003. Transcriptome analysis of the Pseudomonas aeruginosa response to iron. Arch. Microbiol. 180:374-379. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 35.Schmidt, K. D., B. Tummler, and U. Romling. 1996. Comparative genome mapping of Pseudomonas aeruginosa PAO with P. aeruginosa C, which belongs to a major clone in cystic fibrosis patients and aquatic habitats. J. Bacteriol. 178:85-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology (NY) 1:784-791. [Google Scholar]
- 37.Smith, J. L. 2004. The physiological role of ferritin-like compounds in bacteria. Crit. Rev. Microbiol. 30:173-185. [DOI] [PubMed] [Google Scholar]
- 38.Tarassova, K., R. Tegova, A. Tover, R. Teras, M. Tark, S. Saumaa, and M. Kivisaar. 2009. Elevated mutation frequency in surviving populations of carbon-starved rpoS-deficient Pseudomonas putida is caused by reduced expression of superoxide dismutase and catalase. J. Bacteriol. 191:3604-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Temple, L. M., A. E. Sage, H. P. Schweizer, and P. V. Phibbs, Jr. 1998. Carbohydrate catabolism in Pseudomonas aeruginosa, p. 35-72. In T. C. Montie (ed.), Pseudomonas. Plenum Press, New York, NY.
- 40.Velayudhan, J., M. Castor, A. Richardson, K. L. Main-Hester, and F. C. Fang. 2007. The role of ferritins in the physiology of Salmonella enterica sv. Typhimurium: a unique role for ferritin B in iron-sulphur cluster repair and virulence. Mol. Microbiol. 63:1495-1507. [DOI] [PubMed] [Google Scholar]
- 41.Weeratunga, S. K., C. E. Gee, S. Lovell, Y. Zeng, C. L. Woodin, and M. Rivera. 2009. Binding of Pseudomonas aeruginosa apobacterioferritin-associated ferredoxin to bacterioferritin B promotes heme mediation of electron delivery and mobilization of core mineral iron. Biochemistry 48:7420-7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilderman, P. J., N. A. Sowa, D. J. FitzGerald, P. C. FitzGerald, S. Gottesman, U. A. Ochsner, and M. L. Vasil. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. U. S. A. 101:9792-9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.