Abstract
Oxalic acid occurs extensively in nature and plays diverse roles, especially in pathological processes. Due to its highly oxidizing effects, hyperabsorption or abnormal synthesis of oxalate can cause serious acute disorders in mammals and can be lethal in extreme cases. Intestinal oxalate-degrading bacteria could therefore be pivotal in maintaining oxalate homeostasis and reducing the risk of kidney stone development. In this study, the oxalate-degrading activities of 14 bifidobacterial strains were measured by a capillary electrophoresis technique. The oxc gene, encoding oxalyl-coenzyme A (CoA) decarboxylase, a key enzyme in oxalate catabolism, was isolated by probing a genomic library of Bifidobacterium animalis subsp. lactis BI07, which was one of the most active strains in the preliminary screening. The genetic and transcriptional organization of oxc flanking regions was determined, unraveling the presence of two other independently transcribed open reading frames, potentially responsible for the ability of B. animalis subsp. lactis to degrade oxalate. pH-controlled batch fermentations revealed that acidic conditions were a prerequisite for a significant oxalate degradation rate, which dramatically increased in cells first adapted to subinhibitory concentrations of oxalate and then exposed to pH 4.5. Oxalate-preadapted cells also showed a strong induction of the genes potentially involved in oxalate catabolism, as demonstrated by a transcriptional analysis using quantitative real-time reverse transcription-PCR. These findings provide new insights into the characterization of oxalate-degrading probiotic bacteria and may support the use of B. animalis subsp. lactis as a promising adjunct for the prophylaxis and management of oxalate-related kidney disease.
Oxalate is a normal end product of amino acid metabolism and must be excreted, predominantly via the kidney, to maintain homeostasis (22). Oxalate is also present in a wide range of foods and drinks, and the normal dietary intake is variable, ranging from 70 to 920 mg per day (23). Because of its highly oxidizing effects and the capability to combine with cations to form insoluble salts, this organic dicarboxylate is extremely toxic for most forms of life. In humans, oxalate can cause a variety of pathological disorders, including hyperoxaluria, urolithiasis, cardiomyopathy, and renal failure (29, 41, 59). Hyperoxaluria is the single strongest promoter of kidney stone formation, whose medical management represents a burden to the individual patient as well as the health care system (49). The lack of new medications and the continued poor compliance with drug therapy have led to a growing interest in dietary manipulation and novel therapies aimed at preventing recurrent stone formation. Unfortunately, an oxalate-free diet is difficult to achieve and would probably be deficient in essential nutrients. Hence, other approaches to reducing urinary oxalate for management of stone disease have been explored.
The discovery of oxalate-degrading bacteria within the human gastrointestinal tract has opened the way to a flurry of research regarding their potential role in reducing urinary excretion of oxalic acid (12, 20, 21, 25, 26, 30, 31, 35, 40, 44, 45). The first intestinal oxalate-degrading bacterium to be described was Oxalobacter formigenes, an obligate anaerobe which relies exclusively on oxalate metabolism for energy (2). In O. formigenes, oxalate is decarboxylated to give CO2 and formate in a two-step pathway that is mediated by the coupled action of the enzymes formyl-coenzyme A (CoA) transferase (Frc) (6) and oxalyl-CoA decarboxylase (Oxc) (7). Frc catalyzes the transfer of CoA from formate to oxalate, thus activating the oxalyl moiety for the thiamine-dependent decarboxylation mediated by Oxc. Several studies have established a direct correlation between the disappearance of O. formigenes from the intestinal microbiota and the appearance of hyperoxaluria symptoms, confirming its major role in maintaining oxalate homeostasis and making it the leading probiotic candidate for the management of kidney stone disease (21, 25, 26, 30, 44, 45, 49). However, the complicated growth requirements of O. formigenes, with the need to determine the optimal dietary oxalate level required to maintain its colonization, severely limited its administration (47).
Probiotic use of Lactobacillus and Bifidobacterium strains that metabolize oxalate might provide a valid alternative to O. formigenes. Bifidobacteria and lactobacilli are common inhabitants of mammalian guts, including that of humans (55). Some strains of these genera are known to have health-promoting effects and have been added for decades as functional ingredients in food products (48). In particular, strains of Bifidobacterium animalis subsp. lactis are widely used in dairy products in North America and Europe (8) due to several benefits associated with their administration, such as improvement of various physiological conditions, modulation of the host immune response, and reduction of certain risks that impact health (37).
Increasing reports documented a significant reduction in urinary oxalate levels associated with probiotic administration in both animals and humans (12, 31, 35, 39, 40, 58) and suggested the existence of oxalate-degrading activity in Bifidobacterium and Lactobacillus species. Azcarate-Peril et al. (5) identified a cluster of genes encoding the Oxc and Frc proteins in Lactobacillus acidophilus NCFM and showed that mildly acidic conditions were a prerequisite for their transcription. Similarly, an oxalate-dependent induction of the oxc gene in cells first adapted to subinhibitory concentrations of oxalate and then exposed to pH 5.5 was also observed in Lactobacillus gasseri ATCC 33323 (34). Moreover, Turroni et al. (51) screened the oxalate-degrading ability of several lactobacilli isolated from functional foods and pharmaceutical preparations and demonstrated the functionality of L. acidophilus LA14 Oxc and Frc in oxalate catabolism.
To the best of our knowledge, only one study has so far investigated the potential role of Bifidobacterium in the human intestinal degradation of oxalate (19). In that study, the oxalate-degrading capacity of 12 bifidobacterial strains was evaluated and the first in-depth genetic and functional characterization of Oxc from B. animalis subsp. lactis DSM 10140 was provided. However, until now no information has existed about other bifidobacterial genes potentially involved in oxalate degradation and their regulation at the transcriptional level under different culture conditions.
In the present work, a sensitive and selective capillary electrophoretic method was applied to determine the oxalate-degrading activities in several Bifidobacterium strains. Among the most active strains, B. animalis subsp. lactis BI07 was selected for further analysis. Putative genes encoding Oxc and Frc were identified and characterized. The impact of acidic conditions on oxalate degradation and the expression levels of these predicted genes were evaluated.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains used in this study were the following: Bifidobacterium adolescentis ATCC 15703, B. animalis subsp. lactis DSM 10140, BA30, Bb12, BI07, and L15, B. bifidum S16, B. breve ATCC 15700 and BBSF, B. catenulatum B665, B. longum biotype longum S123, ATCC 15707, and W11, and B. longum biotype suis ATCC 27533. The B, L, S, and W strains belonged to the collection of our laboratory. In particular, B. bifidum S16 (15), B. catenulatum B665, B. longum biotype longum S123 (13), and B. animalis subsp. lactis BA30 (14) had been isolated from human feces, whereas B. breve BBSF and B. animalis subsp. lactis BI07 and L15 had been isolated from commercial probiotic products (15). B. longum biotype longum W11 and B. animalis subsp. lactis Bb12 were kindly provided by Alfa Wassermann (Bologna, Italy) and Christian Hansen A/S (Hørsholm, Denmark), respectively. All strains were verified for genus and species by 16S rRNA gene sequence typing.
Bifidobacteria were grown anaerobically at 37°C in the nutritionally complex medium (NCM) described by Mlobeli et al. (38), containing (in grams per liter) tryptone (Oxoid, Hampshire, United Kingdom), 10.0; peptone water, 5.0; yeast extract (Oxoid), 5.0; Tween 80, 0.5; NaCl, 4.5; KCl, 0.5; MgCl2·6H2O, 0.15; KH2PO4, 0.4; NH4Cl, 1.0; and cysteine-HCl, 1.2. Peptone water was prepared as follows (in g/liter): bacteriological peptone (Oxoid), 10.0; NaCl, 5.0; and Tween 80, 1.0 (pH 7.2). The pH of the medium was adjusted to 6.5 before autoclaving. Solid medium was prepared by the addition of 1.5% (wt/vol) agar (Oxoid) to NCM. Sucrose (20 g/liter) was used as an energy source, sterilized separately, and aseptically added to the culture medium. Anaerobic conditions were achieved in anaerobic jars supplemented with a pad of Anaerocult A (Merck, Milan, Italy).
Capillary electrophoresis analysis of oxalate-degrading activity and sucrose consumption in Bifidobacterium strains.
Bifidobacterium strains were cultured for 5 days with 5 mM sodium oxalate. B. animalis subsp. lactis DSM 10140, for which oxalate-degrading activity had already been demonstrated (19), was used as a positive control. As a negative control, B. adolescentis ATCC 15703, whose complete genome sequence does not harbor oxc-related genes (NCBI accession no. NC-008618), was chosen. Uninoculated broth medium was utilized as the experimental control. Residual oxalate in bacterial culture supernatants and control broth was measured at the end of the incubation period using the capillary electrophoresis (CE) method with indirect UV absorbance detection as described by Holmes (24), slightly modified as described below.
Electrophoretic experiments were performed on a Biofocus 2000 apparatus (Bio-Rad, Hercules, CA) in a polyethylenimine-coated capillary (50-μm internal diameter; 31-cm effective length). The capillary was provided by Composite Metal Service (Ilkley, United Kingdom). The coating procedure followed the method proposed by Erim et al. (18). The applied voltage was maintained at 20 kV (anodic detection) at a controlled temperature of 25°C. The samples, diluted 1:10 and filtered (0.45-μm filters), were injected hydrodynamically using a pressure of 2 lb/in2 s−1. Ten mM sodium chromate, pH 7.7, was used as the background electrolyte; 0.2 g/liter sodium sulfate was chosen as the internal standard. The detection wavelength was 254 nm.
Culture broth samples of B. animalis subsp. lactis BI07 were taken over time and further analyzed for formate production by using the CE conditions described above for oxalate. On these cultural supernatants, the residual concentration of sucrose was also measured. For the carbon source determination, electrophoretic separations were carried out at a constant voltage of 7 kV (anodic detection) at a controlled temperature of 25°C on a capillary with a 500μm internal diameter and an effective length of 31 cm (Composite Metal Service). The electrolyte solution developed by Jager et al. (28) for carbohydrate determinations in dairy products, comprising 15 mM potassium sorbate, 0.3 mM cetyl trimethyl ammonium bromide, and 50 mM NaOH, pH 12, was chosen as the background electrolyte; 1.0 g/liter lactose was used as the internal standard.
Identification of the analytes was performed by comparison of the migration times obtained in actual samples with those of standard solutions. Spiking experiments (standard addition method) confirmed the peak identity. The quantification of oxalate, formate, and sucrose was carried out by determining the corrected peak area A′, defined as the ratio between the analyte peak area and the corresponding migration time, and by comparing this ratio to that of the internal standard (sodium sulfate for oxalate and formate and lactose for sucrose). The sensitivity data at the detection wavelength of 254 nm were estimated as the limit of detection (LOD) (signal/noise ratio [S/N] = 3) and limit of quantitation (LOQ) (S/N = 10), and they were found to be 0.05 and 0.15 mM for oxalate and formate and 0.03 and 0.10 mg/ml for sucrose. The repeatability of the separation system for oxalate and formate was evaluated by replicated analysis of a reaction mixture containing oxalate and formate at the concentration of 0.25 mM. The intraday (n = 5) relative standard deviation percentages (RSD%) of migration time of oxalate (tm = 2.10) and formate (tm = 3.99) were 1.24% and 1.04%, respectively. The interday (n = 10) RSD% of migration time were 2.14% for oxalate and 2.66% for formate. The RSD% of the corrected peak area ratio (analyte to internal standard) were 2.06% and 1.83% (intraday) for oxalate and formate, respectively, and they were found to be less than 4% over a two-consecutive-day experiment. Replicated injections of 0.15 mg/ml sucrose were performed to determine the reproducibility of the electrophoretic system for the carbon source. The RSD% of migration time (tm = 8.74) were 1.15% (intraday) and 2.49% (interday). The intra- and interday (2 days) RSD% of the corrected peak area ratio were found to be 1.49% and 3.37%, respectively.
B. animalis subsp. lactis BI07 growth was monitored by following changes in optical density at 600 nm (OD600).
Detection of the oxc gene in Bifidobacterium strains.
Genomic DNA of Bifidobacterium strains was isolated using the DNeasy blood and tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
PCR was carried out in a Biometra thermal cycler T gradient (Biometra, Göttingen, Germany) with AmpliTaq Gold (Applied Biosystems, Foster City, CA) as the DNA polymerase. The primer set oxc-L/oxc-R (Table 1), used to amplify the chromosomal DNAs, was designed by aligning the oxc genes from O. formigenes (accession no. M77128) and probiotic bacteria whose complete or partial genome sequences were available at the beginning of this study: L. acidophilus NCFM (3), L. gasseri ATCC 33323 (36), and B. animalis subsp. lactis DSM 10140 (8).
TABLE 1.
Oligonucleotides used in this study
| Primer | Nucleotide sequence (5′-3′) | Purposea |
|---|---|---|
| oxc-L | GCTACCACGAACTGCTTCCC | PCR |
| oxc-R | CCCATAACGCCCCAAGTACC | PCR |
| EMBL3-F | TTATGCCCGAGAAGATGTTGA | Sequencing |
| EMBL3-R | TATACATGGTTCTCTCCAGAG | Sequencing |
| A | TCGCTTTCGCGCACCTGTATT | Primer walking |
| B | TCGCGAACTGCGCATTCCGT | Primer walking |
| C | TCATGCGCGCAATCACCGATT | Primer walking |
| D | TGGGTGGTTGTCGGTGGTCT | Primer walking |
| E | ATGAGCGTCTCGGCGAGGTA | Primer walking |
| F | GGTCACTGATTTCGCACGTAT | Primer walking and RT-qPCR |
| G | TCGGTCTTCTCGACGAATTCA | Primer walking and RT-qPCR |
| H | ACCGCGAACCCCTACCTCAA | Primer walking |
| I | AAGCGGCCGTATGCGCGTTGAT | Primer walking and RT-qPCR |
| J | GAGATGATGATGCCGGAGAATGC | Primer walking and RT-qPCR |
| K | CGCCTGCGTAATTTGGGACA | Primer walking |
| L | TACGCAGCGACAGGCACCAT | Primer walking |
| M | ACCAGCTCGATCGGGGATAT | Primer walking |
| N | CGACTTGCACTCGTTGACGAT | Primer walking |
| O | CGACGATCATGAGCTACACCAA | Primer walking |
| P | GAAGCTCGGCGAGAACAA | Primer walking |
| Q | ATCGGTCTCCCAGCCCTT | Primer walking and RT-qPCR |
| R | CGTGTACCAGTCCATGCAGAA | Primer walking and RT-qPCR |
| S | GGCCAACAAGATACGGGAAA | Primer walking |
| T | GTGTGGACGGTGACGTAGATT | Primer walking |
| Bif164b | GGGTGGTAATGCCGGATG | RT-qPCR |
| Bif662b | CCACCGTTACACCGGGAA | RT-qPCR |
| GAPDH-L | CTCGAGTGCACCGGCTTCTA | RT-qPCR |
| GAPDH-R | TGAAGCCGTACTCGTTGTCGTA | RT-qPCR |
| gmk-L | TGGGTTTCGGTCTCCGCCAC | RT-qPCR |
| gmk-R | AGCTCCACCTTCGCGGTCTC | RT-qPCR |
| recA-L | ACGGCCATCTTCATCAACCAG | RT-qPCR |
| recA-R | GGTGAACCAAGAACCGGACTT | RT-qPCR |
| rpoB-Lc | CCAGGTCGGCGAGCTGAT | RT-qPCR |
| rpoB-Rc | TACGGGGTCTCGATGAAGC | RT-qPCR |
Construction and screening of B. animalis subsp. lactis BI07 genomic library for oxc gene.
The isolated chromosomal DNA from B. animalis subsp. lactis BI07 was partially digested with the restriction enzyme MboI. DNA fragments ranging from 9 to 23 kb were isolated using a continuous sucrose gradient (10 to 40%) and then ligated to the lambda EMBL3 vector BamHI arms using the lambda EMBL3/BamHI vector kit (Stratagene, La Jolla, CA) as per the manufacturer's instructions. Ligated DNA was packed in vitro using Gigapack III Gold-11 packaging extract provided with the kit, and the library was plated on the XL1-Blue MRA (P2) host strain.
The screening of the B. animalis subsp. lactis BI07 genome library was performed by using the 1,001-bp amplicon, obtained by amplifying the B. animalis subsp. lactis BI07 genome with the oxc-L and oxc-R primers, digoxigenin-dUTP labeled (Roche, Mannheim, Germany) following the supplier's instructions. The plaque hybridization was carried out according to standard procedures (43).
Sequencing and analysis of the genomic DNA fragment from oxc-positive clones.
The B. animalis subsp. lactis BI07 genome fragments from positive clones were amplified by using the primers EMBL3-F and EMBL3-R (Table 1), based on the cloning site flanking regions on the lambda EMBL3 vector BamHI arms. To determine the nucleotide sequence of the oxc gene and its up- and downstream regions, a primer walking sequencing strategy was employed (Table 1). Nucleotide sequencing was performed by the dideoxy chain termination method using BigDye terminators and the ABI 3730 automated DNA sequencer (Applied Biosystems) for analysis. The primary DNA sequence data were assembled using the GCG software program, version 10 (Wisconsin package; Genetics Computer Group, Madison, WI).
DNA sequence analysis and similarity searches of nucleotide and protein databases were carried out using the BLAST network service (BlastN, BlastX, and BlastP) (4) at the NCBI website (http://www.ncbi.nlm.nih.gov/).
Conserved domains in the potential proteins encoded by the open reading frames (ORFs) of interest were inferred from the amino acid sequences by using the Protein Families Database of Alignments and HMMs (http://www.sanger.ac.uk/Software/Pfam/) and clusters of orthologous groups of proteins (http://www.ncbi.nlm.nih.gov/COG/). Membrane-spanning regions of the translated gene products were predicted by the TMpred software program (http://www.ch.embnet.org/software/TMPRED_form.html).
Fermentation experiments.
Batch fermentations were performed in 1-liter stirred-tank bioreactors (ADI 1025 Bio Console and ADI 1010 Bio Controller; Applikon Biotechnology, Schiedam, Netherlands) using 500 ml NCM supplemented with 20 g/liter sucrose. For inoculum buildup, B. animalis subsp. lactis BI07 was incubated anaerobically at 37°C for 18 h in the same culture medium and propagated twice using 2% (vol/vol) inoculum. All fermentations were carried out anaerobically by sparging the medium with filter-sterilized nitrogen at a rate of 0.1 liters/min. The temperature was kept at 37°C, and constant stirring (300 rpm) was applied. The initial pH value was adjusted to 6.5 and controlled through automatic addition of 4 M NaOH. Six parallel fermentation processes were performed, in which, 24 h after the inoculum, when the culture was in exponential phase with an average specific growth rate of 0.23 h−1 and a biomass concentration of approximately 9 × 108 CFU/ml, the medium composition and the culture pH were modified with the following: (i) lowering of pH to 5.5 with or without addition of 5 mM sodium oxalate, (ii) lowering of pH to 4.5 with or without addition of 5 mM sodium oxalate, (iii) addition of 5 mM sodium oxalate, and (iv) no variation. Each treatment was applied for 24 h. Finally, a further fermentation experiment was carried out under the following conditions: 24 h of growth at pH 6.5 in the presence of 5 mM sodium oxalate and then addition of 50 mM sodium oxalate and lowering and maintenance of pH 4.5 for 24 h. As a control, B. animalis subsp. lactis BI07 was grown at pH 6.5 in the presence of 5 mM sodium oxalate without a change in the experimental conditions. All the fermentative parameters were controlled online (BioXpert Lite software; Applikon Biotechnology). Samples were withdrawn at appropriate intervals and analyzed for bacterial growth, oxalate degradation, and transcriptional activity. Each fermentation was carried out in triplicate.
Cellular growth was followed by determination of the OD600 and plate counting on NCM agar.
Concentrations of oxalate and formate were measured on the cultural supernatants by CE as reported above.
Data concerning cellular growth and oxalate consumption in the different pH-controlled batch cultures were compared using Student's t test on paired samples (SigmaStat v. 3.5; Systat Software Inc., San Jose, CA).
RNA extraction and cDNA synthesis.
Total RNA was extracted from 1.5 ml of bacterial culture using the Illustra RNAspin Mini RNA isolation kit (GE Healthcare, Uppsala, Sweden), starting with digestion of bacterial cell wall in 100 μl of 0.1% diethyl pyrocarbonate (DEPC)-treated Tris-EDTA (TE) buffer containing 15 mg/ml lysozyme for 3 h at 37°C. The purified RNA was treated with DNase I on RNAspin Mini columns (GE Healthcare) and eluted in 60 μl of RNase-free water. The RNA integrity was confirmed by formaldehyde agarose gel electrophoresis according to standard procedures (43). RNA quantity (A260) and quality (A260/280) were assessed by analysis with the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE).
First-strand cDNA was synthesized in a total volume of 20 μl using random hexamers and Transcriptor reverse transcriptase according to the manufacturer's instructions (Transcriptor First Strand cDNA synthesis kit; Roche). To check for residual DNA, each RNA sample was also subjected to a cDNA synthesis reaction without addition of reverse transcriptase enzyme (NoRT). The reaction mixtures were incubated in a Biometra thermal cycler T gradient (Biometra) under the following cycling conditions: 65°C for 10 min, 25°C for 10 min, 55°C for 30 min, and 85°C for 5 min to inactivate the enzyme.
Quantitative real-time PCR.
Quantitative real-time PCR (qPCR) was carried out with a LightCycler system (Roche). Amplifications were performed in a 20-μl reaction volume containing 1 μl of cDNA template, equivalent to 50 ng RNA starting material, 4 μl of SYBR green PCR master mix, and a 500-nM concentration of the appropriate gene-specific primers (Table 1). The following cycle profile was used: one cycle at 95°C for 10 min, 40 cycles at 95°C for 15 s, 58°C or 60°C (only for the Q/R primer set) for 25 s, 72°C for 30 s, and an additional incubation step at 89°C for 5 s for fluorescence acquisition. The cycle threshold (CT) was defined as the first PCR cycle in which the generated fluorescence is recorded as statistically significant above the background. All sample and primer combinations were assessed in triplicate. To check for background contamination or residual chromosomal DNA, each run included a negative control and a NoRT cDNA reaction. When a ΔCT value of >4 between the sample and its respective NoRT was obtained, the DNA contamination level was considered negligible. Product detection and PCR specificity were checked postamplification by examining the temperature-dependent melting curves of the PCR products.
cDNA products were subsequently amplified by conventional PCR using primers internal to the genes of interest. PCR products were analyzed by agarose gel electrophoresis.
Analysis of the relative expression levels of the oxc, ORF-1, and ORF-4 genes.
To quantify the gene expression, relative quantification by means of a standard curve was used. Standard curves of the same RNA sample were registered in each qPCR run for each gene by analyzing 2-fold serial dilutions of the cDNA. The amplification efficiency (E) was calculated for each primer pair using the equation E = [10(−1/slope) − 1] × 100.
A panel of five housekeeping genes, one gene encoding rRNA (the 16S rRNA gene) and four genes encoding mRNA (glyceraldehyde 3-phosphate dehydrogenase [GAPDH] gene, gmk, recA, and rpoB), was evaluated for gene expression stability. CT values from the LightCycler system (Roche) were converted into relative quantities and imported into the geNorm v. 3.5 software program, a Visual Basic application (VBA) for Microsoft Excel (53). For each housekeeping gene, the gene stability M, defined as the average pairwise variation for a particular reference gene with all the other tested control genes, was measured. Ideally, M should be lower than or equal to 1.5, and the highest M value reflects the least stable expression level. A normalization factor (NF), based on the geometric mean of the best-performing housekeeping genes, was calculated by geNorm and applied for data processing. To determine the optimal number of genes (n) to include in NF, the pairwise variation Vn/n + 1 was calculated between two sequential NFs (NFn and NFn + 1). According to Vandesompele et al. (53), a cutoff value of 0.15, below which the inclusion of an additional control gene is not required, was considered. The relative oxc, ORF-1, and ORF-4 expression levels were reported as relative quantities of the target gene against NF.
Differences between experimental conditions were assessed by Student's t test, using SigmaStat v. 3.5 (Systat Software Inc.), with P values of <0.05 considered to be significant.
Nucleotide sequence accession numbers.
The sequence of the DNA fragment containing the oxc gene from B. animalis subsp. lactis BI07 was deposited in the GenBank database under the following accession numbers: AB488699, AB488700, AB488701, AB488702, and AB488703.
RESULTS
Oxalate-degrading activity and oxc gene detection in Bifidobacterium strains.
A CE technique was used for the determination of the oxalate-degrading activities in 14 Bifidobacterium strains belonging to the human species B. adolescentis, B. bifidum, B. breve, B. catenulatum and B. longum biotype longum, the B. animalis subsp. lactis, widely used in dairy and pharmaceutical products, and the swine species B. longum biotype suis.
Only the five B. animalis subsp. lactis strains showed oxalate-degrading activity with 100% of oxalate consumption after 5 days of incubation, whereas no degrading activity was exhibited by all the other bifidobacterial strains tested. As expected, 0 and 100% oxalate degradation values were obtained for the negative (B. adolescentis ATCC 15703) and positive (B. animalis subsp. lactis DSM 10140) controls, respectively. The 14 Bifidobacterium strains were subsequently analyzed by PCR for the detection of the oxc gene by using the oxc-L/oxc-R primer set. Again, only the five B. animalis subsp. lactis strains gave the specific amplicon of 1,001 bp (data not shown).
Kinetic analysis of oxalate and sucrose consumption by B. animalis subsp. lactis BI07.
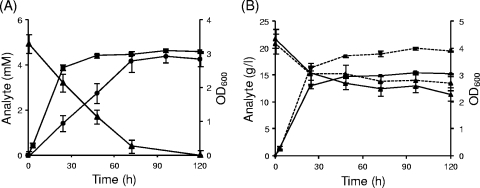
The time course of oxalate consumption by B. animalis subsp. lactis BI07 was determined. As shown in Fig. 1 A, the degradation profile was characterized by a linear, growth-phase-independent disappearance of 5 mM oxalate from the culture supernatants. Thirty-three percent of oxalate was consumed during the first 24 h of incubation, corresponding to the active growth phase, whereas stationary-phase cells showed a linear consumption until 72 h, when a residual oxalate concentration of 0.4 mM, equivalent to 92% degradation, was reached. At the same time, an increase in the concentration of formic acid as a result of oxalate decarboxylation was observed (Fig. 1A). Formate production started immediately and simultaneously with oxalate consumption. Within 72 h of growth, a concentration of 4.2 mM, corresponding to a conversion of 84% of the oxalate added, was produced.
FIG. 1.
Oxalate-degrading activity and sucrose consumption by B. animalis subsp. lactis BI07. (A) Kinetics of 5 mM oxalate degradation (triangles) and formate production (circles) and growth curve (squares) in the presence of 20 g/liter sucrose in the culture medium. For formate, the concentration values were corrected by subtracting at each time point the corresponding values obtained when B. animalis subsp. lactis BI07 was grown in the absence of oxalate. (B) Time course of 20 g/liter sucrose consumption (triangles) and growth curves (squares) in the presence (solid lines) or absence (dashed lines) of 5 mM sodium oxalate. Each point represents the mean of data from three independent experiments. The error bars indicate standard deviations.
B. animalis subsp. lactis BI07 oxalate-degrading activity was also evaluated under the same experimental conditions except for the absence of sucrose in the culture medium composition. In this case, bifidobacteria were unable to grow and to utilize oxalate, as demonstrated by the recovery of almost all the oxalate added to the cultural broth at the fifth day of incubation (data not shown). Lack of growth was also observed when neither oxalate nor sucrose was added to NCM (data not shown).
Consumption kinetics of sucrose was also analyzed (Fig. 1B). Exponentially dividing cells rapidly consumed the carbon source (30% degradation, 0.25-g/liter/h consumption rate) during the first 24 h. From 24 to 120 h of incubation, after the exponential growth phase, the remaining sucrose was slowly degraded to half the initial concentration (0.07-g/liter/h consumption rate). When oxalate was not added to the culture medium, a similar profile for sucrose consumption was obtained, whereas the growth patterns showed statistically significant differences in terms of biomass yields at all time points (Fig. 1B).
Construction of a gene bank for B. animalis subsp. lactis BI07 and isolation of the oxc gene.
To identify the genes potentially involved in oxalate consumption, an MboI-digested genomic library for B. animalis subsp. lactis BI07 was created in lambda EMBL3 and screened in Escherichia coli XL1-Blue MRA (P2) cells. Two positive clones were identified by plaque hybridization with the digoxigenin-dUTP-labeled oxc gene probe and were designated F and S. Preliminary restriction enzyme analysis revealed that F contained a 10.2-kb insert and S contained a 9.5-kb insert (data not shown).
Analysis of the 10.2-kb insert of clone F.
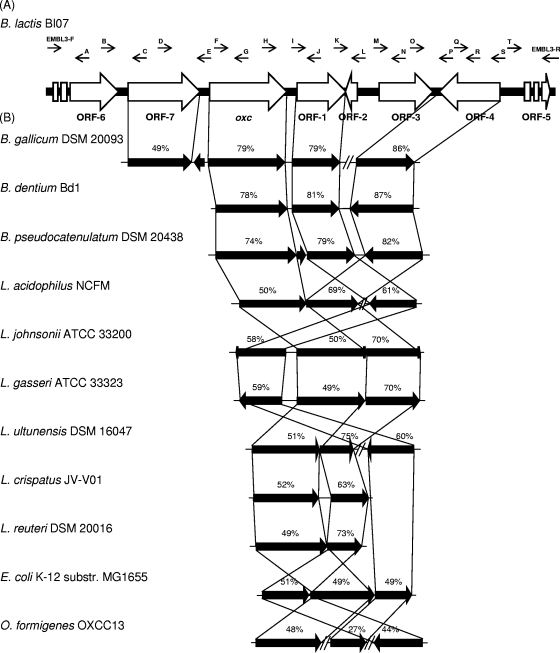
A total of 10,208 bp was sequenced from clone F by a primer walking strategy, unraveling the presence of five complete and two partial ORFs around the putative oxc gene (Fig. 2 A). Apart from ORF-2 and -4, all the other ORFs were oriented in the same direction as oxc. Bioinformatic analysis of the 1,782-bp oxc gene, encoding a 593-amino-acid (aa) protein (Mr of 63,710), revealed 100% homology with the corresponding gene from B. animalis subsp. lactis DSM 10140, whose complete genome has been recently sequenced (8). The function and significance of B. animalis subsp. lactis DSM 10140 Oxc in oxalate catabolism have already been demonstrated (19). BlastX analysis of the B. animalis subsp. lactis oxc gene revealed the highest similarities to the putative oxalyl-CoA decarboxylases of Bifidobacterium gallicum, B. dentium, and B. pseudocatenulatum. High similarity scores were also found to the Oxc enzymes of several Lactobacillus strains, as well as E. coli and O. formigenes (Fig. 2B). The protein encoded by oxc has a conserved domain that is present in thiamine pyrophosphate (TPP)-requiring enzymes (acetolactate synthase, pyruvate dehydrogenase [cytochrome], glyoxylate carboligase, and phosphonopyruvate decarboxylase) (COG0028). In the oxc product, the N-terminal TPP-binding domain (pfam02776) starts at residue 23 and spans 165 aa, the central domain (pfam00205) starts at residue 212 and spans 130 aa, and the C-terminal TPP-binding domain (pfam02775) starts at residue 418 and spans 135 aa.
FIG. 2.
Genetic organization around the oxc gene in B. animalis subsp. lactis BI07 (A) and comparison with the corresponding loci in bifidobacteria, lactobacilli, E. coli, and O. formigenes (B). A through T, primers A through T used for primer walking (Table 1). Each arrow indicates an ORF, and the length of the arrow is proportional to the length of the predicted ORF. Levels of amino acid identity, expressed as percentages, are shown. The organisms used were B. gallicum DSM 20093 (accession no. NZ_ABXB00000000), B. dentium Bd1 (54), B. pseudocatenulatum DSM 20438 (accession no. NZ_ABXX00000000), L. acidophilus NCFM (3), Lactobacillus johnsonii ATCC 33200 (accession no. NZ_ACGR00000000), Lactobacillus gasseri ATCC 33323 (36), Lactobacillus ultunensis DSM 16047 (accession no. NZ_ACGU00000000), Lactobacillus crispatus JV-V01 (accession no. NZ_ACKR00000000), Lactobacillus reuteri DSM 20016 (accession no. NC_009513), E. coli K-12 MG1655 (accession no. NC_000913), and O. formigenes OXCC13 (accession no. NZ_ACDQ00000000).
ORF-1 begins on the same strand, 104 nucleotides downstream of the oxc gene. The deduced protein has a theoretical Mr of 39,350 and a pI of 8.99. BLAST analysis of the 367 aa encoded by ORF-1 revealed similarities to a number of permeases, containing the conserved COG0679 and pfam03547 domains (Fig. 2B). The hydrophobicity plot of the predicted protein encoded by ORF-1 revealed the presence of 10 transmembrane segments, composed of several hydrophobic residues. The transmembrane topology was predicted as N terminus outside, with the N-terminal portion of the protein likely to be located in the extracellular space.
ORF-2 was identified 7 nucleotides downstream of ORF-1 in the opposite orientation to oxc. The gene was predicted to encode a protein containing 43 aa and having a calculated Mr of 4,740. Protein comparison showed a significant similarity only to a non-pfam hypothetical protein of B. animalis subsp. lactis DSM 10140 (accession no. gb|ACS48365).
The third complete ORF, located 863 nucleotides downstream of ORF-1 on the same strand, is 1,137 bp in length, encoding 378 aa (Mr of 40,030). At the protein level, the ORF-3 product showed the highest overall identities to the secreted beta-mannosidases of B. animalis subsp. lactis (100% identity) (8), Streptomyces coelicolor, and Streptomyces lividans (50% identity; accession no. emb|CAA20610 and gb|AAA26710). A conserved cellulose or protein binding domain (pfam02013) is present in the C-terminal end of these proteins.
ORF-4, which reads divergently from the oxc gene, consists of 1,332 bp encoding a protein of 443 aa, with a deduced Mr of 48,900 and a calculated pI of 5.03. Comparison of the predicted amino acid sequence to the database revealed the highest similarities to the putative formyl-CoA transferases of B. animalis subsp. lactis, B. dentium, B. gallicum, and B. pseudocatenulatum. Levels of identity of >63% were observed with the Frc enzymes from lactobacilli which harbor the oxc gene. Additionally, the ORF-4 product exhibited 44 and 49% identity (62 and 67% similarity) with the proteins encoded by the O. formigenes frc and E. coli yfdW genes, respectively (Fig. 2B). Analysis of the predicted protein sequence for pfam matches revealed the presence of the conserved pfam02515 domain, belonging to the third family of CoA transferases, which starts at residue 74 and spans 207 aa. As for Oxc, no regions involved in membrane sorting or anchoring were detected in the ORF-4 amino acid chain, suggesting its cytoplasmic localization.
The last complete ORF, identified upstream of oxc and designated ORF-7, contains 552 codons. The deduced amino acid sequence, with theoretical Mr and pI values of 60,260 and 8.98, displayed 100% identity with the voltage gated chloride channel family protein from B. animalis subsp. lactis DSM 10140 (8) and significant similarities to the putative chloride channel EriC proteins from B. gallicum (Fig. 2B) and Eggerthella lenta (44% identity; accession no. ZP_03895186). In the predicted protein sequence, a conserved domain, present in EriC proteins and involved in inorganic ion transport and metabolism, was detected (COG0038). The hydropathy plot of the ORF-7 product revealed the presence of 11 membrane-spanning regions with an inside-outside orientation.
Two incomplete ORFs were also identified, both on the same strand of the oxc gene. The first, ORF-5, contains 8 codons for the N-terminal end of a protein, which showed the highest similarity to a putative integrase of B. animalis subsp. lactis DSM 10140 (83% identity; accession no. YP_002970424). The second, ORF-6, located upstream of ORF-7, contains 345 aa, which exhibited 100% identity to a predicted outer membrane protein involved in collagen adhesion of B. animalis subsp. lactis DSM 10140 (accession no. YP_002970431).
The comparison with other bifidobacteria revealed that the genetic organizations around the oxc gene are similar (Fig. 2B). Indeed, oxc, ORF-1, and ORF-4 are present in the same gene order in B. gallicum, B. dentium, and B. pseudocatenulatum. Interestingly, in the B. gallicum genome, the gene encoding the putative Frc protein is located 14 ORFs downstream on the same strand of oxc. The gene inventory in unrelated taxa (e.g., Lactobacillus, Escherichia, and Oxalobacter) was shown to be different in terms of gene order and transcriptional orientation, which is in accordance with their distant phylogenetic positions. The only exception is represented by O. formigenes, where, more than 500 kbp are inserted between oxc and ORF-4 (Fig. 2B).
Characterization of the oxc transcript.
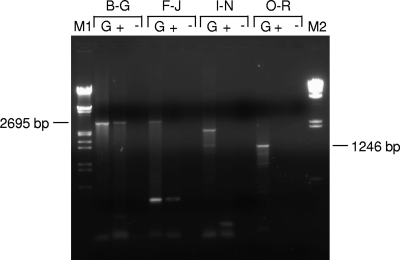
To examine if the oxc gene is transcribed polycistronically, cDNA synthesized from B. animalis subsp. lactis BI07 total RNA was amplified by PCR with several primer pairs that probed four different regions (Fig. 2A). Primers B and G were used to detect transcripts running from ORF-7 to oxc, primers F and J for transcripts running from oxc to ORF-1, primers I and N for transcripts running from ORF-1 to ORF-3, and the primer set O/R to determine if ORF-3 and ORF-4 were coordinately expressed on the same RNA transcript. A clear signal with the predicted product size was obtained only with primers B and G, whereas the PCRs amplifying the intergenic regions between oxc and ORF-1 and between ORF-1 and ORF-3 did not yield amplicons (Fig. 3). No PCR product was obtained even with the primer pair O/R, as expected on the basis of the opposite transcriptional directions of ORF-3 and ORF-4. These results indicated that the ORF-7 and oxc genes were present on the same mRNA transcript, but the transcript did not extend downstream of oxc. In contrast, ORF-1, ORF-3, and ORF-4 were most likely transcribed independently.
FIG. 3.
RT-PCR analysis of the transcriptional organization around the oxc gene in B. animalis subsp. lactis BI07. G, positive genomic DNA control; +, first-strand cDNA generated by RT reaction; −, negative-control reaction lacking RT enzyme. For cDNA synthesis, total RNA extracted from B. animalis subsp. lactis BI07 cells grown in a batch fermentor at pH 6.5 for 24 h was used as a template. B, F, G, I, J, N, O, and R, primers used for RT-PCR (see Table 1); M, molecular weight marker (M1, lambda DNA/EcoRI + HindIII; M2, lambda DNA/HindIII; M-Medical Fermentas, Milan, Italy). The expected size of amplicons is indicated on each side in bp.
Fermentation experiments.
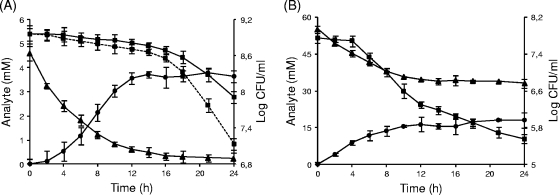
To evaluate the impact of acidic conditions on the oxalate-degrading activity of B. animalis subsp. lactis BI07, different pH-controlled batch cultures were carried out as previously performed by Azcarate-Peril et al. (5) for L. acidophilus NCFM. As reported in Table 2, significant oxalate consumption (82.6%) was measured when B. animalis subsp. lactis was challenged with pH 4.5, whereas negligible values of degradation were exhibited by the pH 6.5- and 5.5-controlled cultures. At pH 4.5, B. animalis subsp. lactis BI07 consumed most of the oxalate added (70%) within 10 h of fermentation, with an accompanying viable cell decline of 0.1 log unit (Fig. 4 A). During the last 14 h, both the cell viability and the oxalate consumption strongly decreased. However, statistically significant differences were found between the pH 4.5-controlled culture and the other pH-controlled cultures in terms of oxalate consumption rates throughout the whole fermentation course (Table 2). Formic acid was produced simultaneously with the oxalate degradation (Fig. 4A). After 24 h of fermentation, a formate concentration of 3.7 mM, corresponding to a conversion of 90% of the degraded oxalate, was measured. Interestingly, at the end of the fermentation process, a cell concentration value 1 log unit higher than that found in the absence of oxalate was reached (Fig. 4A). When comparing pH 6.5- and 5.5-controlled batch cultures with and without oxalate addition, no statistically significant differences in terms of cell viability were found (data not shown).
TABLE 2.
B. animalis subsp. lactis BI07 oxalate-degrading activities in different pH-controlled batch fermentationsa
| Exptl conditionsb | Oxalate degraded, mmol/liter (%) | Mean oxalate degradation rate (mmol/CFU/h × 10−13) in time periodc: |
|
|---|---|---|---|
| 1 | 2 | ||
| pH 6.5 + 5 mM oxa | 0.28 ± 0.02 (5.6) | 0.71 ± 0.09 | 0.34 ± 0.08 |
| pH 5.5 + 5 mM oxa | 0.59 ± 0.03 (11.9) | 1.10 ± 0.09 | 0.10 ± 0.07 |
| pH 4.5 + 5 mM oxa | 4.13 ± 0.2 (82.6) | 4.64 ± 0.93 | 1.62 ± 0.15 |
| pH 4.5 + 50 mM oxa | 23.81 ± 1.7 (43.3) | 989.87 ± 62.42 | 2,099.06 ± 222.01 |
Data are the means ± standard deviations of data from three independent experiments.
Variations in terms of culture pH and oxalate (oxa) concentration applied after 24 h of growth at pH 6.5. In the last batch fermentation, B. animalis subsp. lactis BI07 was preadapted to 5 mM sodium oxalate during the first 24 h and then exposed to pH 4.5 and a 10-times-higher concentration of oxalic acid.
Mean oxalate degradation rate, expressed as mmol of oxalate degraded per CFU per h and calculated during the first 10 h (1) or in the last 14 h (2) of fermentation after changing the experimental conditions.
FIG. 4.
Oxalate-degrading activity of B. animalis subsp. lactis BI07 in pH 4.5-controlled batch fermentations after changing the experimental conditions. (A) After 24 h of growth at pH 6.5, the fermentation pH was lowered to 4.5 with the simultaneous addition of 5 mM sodium oxalate. (B) Bifidobacterium was preadapted to 5 mM sodium oxalate during the first 24 h and then exposed to pH 4.5 and a 10-times-higher concentration of oxalic acid. Kinetics of oxalate degradation (triangles) and formate production (circles) were determined by capillary electrophoresis as described in Materials and Methods. Cell viability (squares) is shown. In panel A, the data on B. animalis subsp. lactis viability at pH 4.5 without oxalate addition (dashed lines) were also reported. Each point represents the mean of data from three independent experiments. The error bars indicate standard deviations.
Based on these results, a further fermentation experiment was performed by preadapting Bifidobacterium to a noninhibiting concentration of oxalate (5 mM) prior to the pH shift to 4.5. The acid challenge was combined with the addition of a higher concentration of oxalate (50 mM), as reported by Azcarate-Peril et al. (5). Similarly to what occurred at pH 4.5 without preadaptation, oxalate was degraded mainly at the beginning of fermentation but to a much greater extent (Fig. 4B and Table 2). Although, after a short lag phase, the cell viability began to decline sharply, B. animalis subsp. lactis BI07 preadapted cells degraded oxalic acid with a statistically higher rate, approaching 200-fold the corresponding value measured in the absence of preadaptation during the first 10 h of fermentation after the pH shift (Table 2). Formic acid was produced at a concentration of 18.1 mM, which was equivalent to a conversion of 76% of the oxalate degraded. In the pH 6.5 control culture preadapted to 5 mM oxalate, only 5% of oxalic acid was degraded (data not shown).
Relative expression of the oxc, ORF-1, and ORF-4 genes in response to pH shift and oxalate addition.
To investigate the effect of pH shift and oxalate addition on the transcriptional levels of the B. animalis subsp. lactis BI07 oxc, ORF-1, and ORF-4 genes, the relative abundances of gene transcripts were determined following the normalization strategy outlined by Vandesompele et al. (53).
In each qPCR run, a standard curve was used to calculate the amplification efficiency (E) for all the genes analyzed (oxc, ORF-1, ORF-4, 16S rRNA, GAPDH gene, gmk, recA, and rpoB). E was >85% for the GAPDH, gmk, and recA reference genes, whereas a slope of the standard curve of less than −4.1, indicative of an E value of <75%, was obtained for 16S rRNA, rpoB, and the target genes. The correlation coefficients were >0.99. The reproducibility and reliability of the assay were assessed by repeating the cDNA synthesis and qPCR three times under identical conditions. The intra-assay coefficients of variation (CV) for cDNA synthesis and qPCR were about 5% and 3%, respectively. The interassay CVs ranged between 10% and 5%. The interreplicate (between fermentations) CVs were lower than 10%.
The gene expression stability measure M calculated by geNorm was 0.739 for the GAPDH gene, 0.773 for gmk, 0.828 for recA, and 0.841 for rpoB. The 16S rRNA gene was the worst-scoring housekeeping gene, with an M value of 1.717; thus, it was eliminated from the calculation of a suitable normalization factor (NF). Since the pairwise variation between two sequential NFs containing an increasing number of genes was above the cutoff value of 0.15 proposed by Vandesompele et al. (53), the remaining four genes were needed as internal controls for geometric averaging.
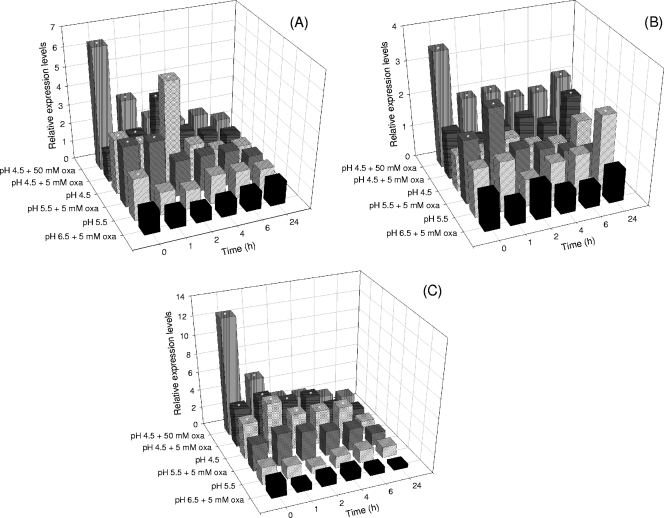
As shown in Fig. 5, when 5 mM sodium oxalate was provided, maintaining a pH of 6.5, all the three genes evaluated, oxc, ORF-1, and ORF-4, were expressed without considerable differences, indicating the absence of a specific effect of oxalate without lowering the pH. In contrast, after exposure to pH 4.5, for oxc (Fig. 5A) and ORF-4 (Fig. 5C), the relative expression immediately and consistently increased independently of oxalate addition. The highest expression levels, with a statistically significant average increase of 5.5-fold with respect to the basal expression, were observed after 1 and 2 h in the absence of oxalate, respectively, for ORF-4 and oxc. At pH 4.5, ORF-1 transcripts were detected but their amounts were not significantly higher than those found in B. animalis subsp. lactis BI07 control cells (Fig. 5B). Modest levels of induction of the gene encoding the putative permease were exhibited after the pH shift to 5.5 and addition of oxalate, with a mean of 2.7 at 1 h after the end of log-phase growth (t1). In pH 5.5-controlled batch cultures, statistically larger amounts of the oxc gene transcript were also observed (with 3.2 as the maximal fold change). In almost all the pH-controlled batch cultures, the gene transcript levels tended to decrease over time.
FIG. 5.
Relative expression levels of the oxc (A), ORF-1 (B), or ORF-4 (C) gene of B. animalis subsp. lactis BI07 in different pH-controlled batch cultures at 0, 1, 2, 4, 6, and 24 h after the variation of the experimental conditions. The gene transcript amounts were normalized against four housekeeping genes (GAPDH gene, gmk, recA, and rpoB) and compared to the control culture represented by exponentially dividing cells at pH 6.5. Along the y axis, the variations in terms of culture pH and oxalate concentration applied after 24 h of growth at pH 6.5 are indicated. In the last batch fermentation, Bifidobacterium was preadapted to 5 mM sodium oxalate during the first 24 h and then exposed to pH 4.5 and a 10-times-higher concentration of oxalic acid. * , P < 0.05.
When Bifidobacterium was preadapted to 5 mM oxalate during the first 24 h of growth, all the loci examined were highly induced. Their expression showed a maximum level (6.5 for oxc, 3.5 for ORF-1, and 12 for ORF-4) immediately after the variation in the experimental conditions and had already started to decrease after 1 h of treatment. However, the relative amounts of all transcripts remained consistently higher than the basal expression at all time points. Statistically significant differences were observed at t0 for all genes between preadapted cultures and pH 4.5-controlled cultures without preadaptation. In all the other samples analyzed, no significant induction or repression of the oxc, ORF-1, and ORF-4 genes was observed.
DISCUSSION
In this work, a preliminary screening concerning oxalate-degrading activity in 14 Bifidobacterium strains comprising 4 species and 3 subspecies originating from humans, animals or probiotic products was carried out by capillary electrophoresis (CE). In a difference from previous reports (19), no oxalate consumption was observed for B. adolescentis, B. bifidum, B. breve, B. catenulatum, B. longum biotype longum, and B. longum biotype suis, whereas all the B. animalis subsp. lactis strains tested proved to be equally active, with 100% oxalate degradation. It is noteworthy that Federici et al. (19) measured the oxalate concentration using a commercial enzymatic kit with low sensitivity and specificity of analysis due to the complexity of the reaction mixture. Therefore, the differences registered in the Bifidobacterium oxalate degradation values can be easily explained by considering the high separation efficiency and sensitivity which characterize CE. Furthermore, only small amounts of sample and a short analysis time are required, making CE a much more suitable and reliable analytical approach than the conventional spectrophotometric methods. Confirming our findings and in accordance with the in silico screening for oxalyl-CoA decarboxylase (oxc)-related genes in members of Bifidobacterium whose complete or partial genome sequences are available, among all the bifidobacteria tested, the oxc gene was detected by PCR only in the B. animalis subsp. lactis strains.
Owing to the high level of oxalate-degrading activity and the wide use in dairy and pharmaceutical products, B. animalis subsp. lactis BI07 was selected as a model strain for further investigation. About 3.4 kbp downstream of the B. animalis subsp. lactis oxc gene, the bioinformatic analysis identified the presence of a formyl-CoA transferase (frc) ortholog (ORF-4), strengthening the hypothesis that B. animalis subsp. lactis has a mechanism of oxalate degradation similar to that of other intestinal anaerobic bacteria, such as the “specialist” O. formigenes, Eubacterium lentum, and Providencia sp. (42). As demonstrated for O. formigenes (46), the genetic analysis showed that oxc and frc in B. animalis subsp. lactis BI07 are not part of a polycistronic operon. Similarity searches of the oxc and frc gene sequences from B. animalis subsp. lactis BI07 against the available partial or complete Bifidobacterium genomes showed that besides B. animalis subsp. lactis, only B. gallicum, B. dentium, and B. pseudocatenulatum harbor putative oxc and frc genes. Interestingly, apart from B. gallicum, which appears to be exclusively associated with animal feces (32), the other Bifidobacterium species with a potential oxalate-degrading activity seem to prefer a cosmopolitan lifestyle without a clear ecological specialization (32, 50, 56). As reported for all these bifidobacterial genome sequences, an ORF encoding a putative permease (ORF-1) was identified flanking the oxc gene on the same strand in B. animalis subsp. lactis BI07. Although this gene product has no similarity to the O. formigenes oxalate:formate antiporter protein OxlT (1), nor to the ABC transporter of L. acidophilus NCFM, which is associated with oxc (5), its presence in the oxc surrounding regions suggests its potential role as an oxalate transporter protein. However, the arrangement of oxc, frc, and the gene encoding the putative oxalate transporter in the B. gallicum, B. dentium, and B. pseudocatenulatum genomes is somewhat different from B. animalis subsp. lactis with regard to gene order and orientation. In particular, B. animalis subsp. lactis BI07 ORF-2 and ORF-3, located between oxc and frc orthologs, did not display a significant sequence similarity to any known genes of Bifidobacterium, indicating that they are either specific to B. animalis subsp. lactis or are yet to be determined in the other incomplete genomes. In contrast, the complete B. animalis subsp. lactis ORF (ORF-7) identified upstream of oxc showed 49% predicted amino acid identity to the B. gallicum putative chloride channel protein EriC gene. It has been proposed that these channel proteins may function as an electrical shunt for an outwardly directed virtual proton pump that is linked to amino acid decarboxylation, allowing enteric bacteria to survive in the strongly acidic environment of the stomach (27). In this way, the persistent proton influx that arises from high extracellular acidity could be continually counteracted in the cytoplasm by the decarboxylation-linked proton utilization. Since oxalate is decarboxylated to formyl-CoA in a reaction that consumes a proton after intracellular uptake and activation to oxalyl-CoA, the concerted action of permease, Oxc, Frc, and EriC could allow Bifidobacterium to withstand acid challenges.
To investigate whether B. animalis subsp. lactis BI07 could rely on oxalate metabolism as a major energy source, its oxalate-degrading activity was determined in the absence of the sugar source. Under these conditions, neither B. animalis subsp. lactis growth nor oxalate consumption was observed, providing evidence that oxalate cannot be used as the sole carbon source and that its metabolism is strictly dependent on the cell physiological state. Conversely, according to our data, no impact of oxalate on B. animalis subsp. lactis sugar metabolism was determined.
The role of an acidic environment in the B. animalis subsp. lactis BI07 oxalate degradation pattern was evaluated in pH-controlled batch cultures analogously to what has already been reported for L. acidophilus (5) and L. gasseri (34). Fermentation experiments indicated that only at pH 4.5 is there a significant oxalate consumption and that the degradation rate dramatically increases in oxalate-preadapted cells. Confirming that oxalate degradation by B. animalis subsp. lactis proceeds via a decarboxylation step, formate production was demonstrated. Our results indicated that approximately 0.9 mol of formate were produced per mol of oxalate degraded, as reported for O. formigenes (2). Interestingly, when oxalate was supplied, the ability of B. animalis subsp. lactis BI07 to resist the detrimental effects of acidic pH was improved.
For the first time, using quantitative real-time reverse transcription-PCR (RT-qPCR), the regulation of B. animalis subsp. lactis oxc, frc, and the gene encoding the putative permease was proven. To quantify accurately the cDNA levels and to identify real gene-specific variations, the normalization strategy proposed by Vandesompele et al. (53) was chosen. Since there is no consensus for prokaryotic endogenous control (52), five presumed housekeeping genes belonging to different functional classes, the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene, gmk (encoding guanylate kinase), recA (encoding recombinase A), rpoB (encoding subunit beta of DNA-directed RNA polymerase), and 16S rRNA (encoding small subunit [SSU] ribosome, four copies), were tested. Although 16S rRNA is among the most commonly used cellular products for transcript normalization (11), in our study the metabolic housekeeping gene transcripts were found to work best.
After normalization of the expression data, the B. animalis subsp. lactis BI07 ORF-4, encoding the putative Frc enzyme, was the most highly induced gene, exhibiting between 2.5- and 12-fold-higher levels of expression in the pH 4.5-controlled batch cultures. Acidic conditions were also a prerequisite for oxc transcription, which was most active within 2 h after the pH shift from 6.5 to 4.5. As for L. acidophilus NCFM (5) and L. gasseri ATCC 33323 (34), when bifidobacterial cells were preadapted to subinhibitory concentrations of oxalate and subsequently exposed to lower pH values plus a higher concentration of oxalic acid, the relative oxc and frc mRNA abundances were significantly increased. However, with respect to lactobacilli, in B. animalis subsp. lactis BI07 overall, higher transcriptional levels were observed, and more importantly, a pH value of 4.5 instead of 5.5 was fundamental for gene expression as well as oxalate-degrading activity. After oxalate preadaptation, a lower but significant magnitude of upregulation was also detected for ORF-1. For all the three genes, no specific effect of oxalate was observed without a lowering of the pH. Further, decreased transcript levels were detected over time, mainly for oxc and frc, suggesting a progressive reduction of the transcriptional and metabolic activities of B. animalis subsp. lactis BI07.
In conclusion, our results indicate that in B. animalis subsp. lactis BI07, the pattern of oxalate degradation and the transcriptional activities of the genes potentially involved in oxalate catabolism are pH dependent. The discovery of genes whose expression responds to changes in pH is particularly important. The pH of the lumen of the proximal colon is reported to be somewhat lower (5.5 to 6.5) than that of the distal colon (6.5 to 7.0), mainly as a result of the higher fermentation rate of saccharolytic microbial groups within the proximal region (17). The pH of both regions may be transiently and locally reduced following ingestion of nondigestible but fermentable carbohydrates (9, 10, 16). The oxalate-degrading activity which uniquely characterizes the B. animalis subsp. lactis species over the other bifidobacteria most commonly encountered in probiotic products may justify the use of this probiotic species as a promising adjunct for the prevention and treatment of oxalate-associated disorders.
Acknowledgments
This work was financed by the project “Mechanisms of interaction and cell-cell communication in lactic acid bacteria from food ecosystems: transcriptomics, proteomics and metabolomics,” PRIN 2007 funds, from the Ministry of University and Scientific Research of Italy.
Footnotes
Published ahead of print on 2 July 2010.
REFERENCES
- 1.Abe, K., Z. S. Ruan, and P. C. Maloney. 1996. Cloning, sequencing, and expression in Escherichia coli of OxlT, the oxalate:formate exchange protein of Oxalobacter formigenes. J. Biol. Chem. 271:6789-6793. [DOI] [PubMed] [Google Scholar]
- 2.Allison, M. J., K. A. Dawson, W. R. Mayberry, and J. G. Foss. 1985. Oxalobacter formigenes gen. nov., sp. nov.: oxalate-degrading anaerobes that inhabit the gastrointestinal tract. Arch. Microbiol. 141:1-7. [DOI] [PubMed] [Google Scholar]
- 3.Altermann, E., W. M. Russell, M. A. Azcarate-Peril, R. Barrangou, B. L. Buck, O. McAuliffe, N. Souther, A. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. R. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. U. S. A. 102:3906-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azcarate-Peril, M. A., J. M. Bruno-Bárcena, H. M. Hassan, and T. R. Klaenhammer. 2006. Transcriptional and functional analysis of oxalyl-coenzyme A (CoA) decarboxylase and formyl-CoA transferase genes from Lactobacillus acidophilus. Appl. Environ. Microbiol. 72:1891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baetz, A. L., and M. J. Allison. 1990. Purification and characterization of formyl-coenzyme A transferase from Oxalobacter formigenes. J. Bacteriol. 172:3537-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baetz, A. L., and M. J. Allison. 1989. Purification and characterization of oxalyl-coenzyme A decarboxylase from Oxalobacter formigenes. J. Bacteriol. 171:2605-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrangou, R., E. P. Briczinski, L. L. Traeger, J. R. Loquasto, M. Richards, P. Horvath, A. C. Coute-Monvoisin, G. Leyer, S. Rendulic, J. L. Steele, J. R. Broadbent, T. Oberg, E. G. Dudley, S. Schuster, D. A. Romero, and R. F. Roberts. 2009. Comparison of the complete genome sequences of Bifidobacterium animalis subsp. lactis DSM 10140 and Bl-04. J. Bacteriol. 191:4144-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bird, A. R., M. S. Vuaran, R. A. King, M. Noakes, J. Keogh, M. K. Morell, and D. L. Topping. 2008. Wholegrain foods made from a novel high-amylose barley variety (Himalaya 292) improve indices of bowel health in human subjects. Br. J. Nutr. 99:1032-1040. [DOI] [PubMed] [Google Scholar]
- 10.Bown, R. L., J. A. Gibson, G. E. Sladen, B. Hicks, and A. M. Dawson. 1974. Effects of lactulose and other laxatives on ileal and colonic pH as measured by a radiotelemetry device. Gut 15:999-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bustin, S. A. 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25:169-193. [DOI] [PubMed] [Google Scholar]
- 12.Campieri, C., M. Campieri, V. Bertuzzi, E. Swennen, D. Matteuzzi, S. Stefoni, F. Pirovano, C. Centi, S. Ulisse, G. Famularo, and C. De Simone. 2001. Reduction of oxaluria after an oral course of lactic acid bacteria at high concentration. Kidney Int. 60:1097-1105. [DOI] [PubMed] [Google Scholar]
- 13.Candela, M., E. Biagi, M. Centanni, S. Turroni, M. Vici, F. Musiani, B. Vitali, S. Bergmann, S. Hammerschmidt, and P. Brigidi. 2009. Bifidobacterial enolase, a cell surface receptor for human plasminogen involved in the interaction with the host. Microbiology 155:3294-3303. [DOI] [PubMed] [Google Scholar]
- 14.Candela, M., F. Perna, P. Carnevali, B. Vitali, R. Ciati, P. Gionchetti, F. Rizzello, M. Campieri, and P. Brigidi. 2008. Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int. J. Food Microbiol. 125:286-292. [DOI] [PubMed] [Google Scholar]
- 15.Candela, M., G. Seibold, B. Vitali, S. Lachenmaier, B. J. Eikmanns, and P. Brigidi. 2005. Real-time PCR quantification of bacterial adhesion to Caco-2 cells: competition between bifidobacteria and enteropathogens. Res. Microbiol. 156:887-895. [DOI] [PubMed] [Google Scholar]
- 16.Chung, Y.-C., C.-K. Hsu, C.-Y. Ko, and Y.-C. Chan. 2007. Dietary intake of xylooligosaccharides improves the intestinal microbiota, fecal moisture, and pH value in the elderly. Nutr. Res. 27:756-761. [Google Scholar]
- 17.Cummings, J. H., and G. T. Macfarlane. 1991. The control and consequences of bacterial fermentation in the human colon. J. Appl. Bacteriol. 70:443-459. [DOI] [PubMed] [Google Scholar]
- 18.Erim, F. B., A. Cifuentes, H. Poppe, and J. C. Kraak. 1995. Performance of a physically adsorbed high-molecular-mass polyethyleneimine layer as coating for the separation of basic proteins and peptides by capillary electrophoresis. J. Chromatogr. A 708:356-361. [DOI] [PubMed] [Google Scholar]
- 19.Federici, F., B. Vitali, R. Gotti, M. R. Pasca, S. Gobbi, A. B. Peck, and P. Brigidi. 2004. Characterization and heterologous expression of the oxalyl coenzyme A decarboxylase gene from Bifidobacterium lactis. Appl. Environ. Microbiol. 70:5066-5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldfarb, D. S., F. Modersitzki, and J. R. Asplin. 2007. A randomized, controlled trial of lactic acid bacteria for idiopathic hyperoxaluria. Clin. J. Am. Soc. Nephrol. 2:745-749. [DOI] [PubMed] [Google Scholar]
- 21.Hatch, M., J. Cornelius, M. Allison, H. Sidhu, A. Peck, and R. W. Freel. 2006. Oxalobacter sp. reduces urinary oxalate excretion by promoting enteric oxalate secretion. Kidney Int. 69:691-698. [DOI] [PubMed] [Google Scholar]
- 22.Holmes, R. P. 2000. Oxalate synthesis in humans: assumptions, problems, and unresolved issues. Mol. Urol. 4:329-332. [PubMed] [Google Scholar]
- 23.Holmes, R. P., and M. Kennedy. 2000. Estimation of oxalate content of foods and daily oxalate intake. Kidney Int. 57:1662-1667. [DOI] [PubMed] [Google Scholar]
- 24.Holmes, R. P. 1995. Measurement of urinary oxalate and citrate by capillary electrophoresis and indirect ultraviolet absorbance. Clin. Chem. 41:1297-1301. [PubMed] [Google Scholar]
- 25.Hoppe, B., B. Beck, N. Gatter, G. von Unruh, A. Tischer, A. Hesse, N. Laube, P. Kaul, and H. Sidhu. 2006. Oxalobacter formigenes: a potential tool for the treatment of primary hyperoxaluria type 1. Kidney Int. 70:1305-1311. [DOI] [PubMed] [Google Scholar]
- 26.Hoppe, B., G. von Unruh, N. Laube, A. Hesse, and H. Sidhu. 2005. Oxalate degrading bacteria: new treatment option for patients with primary and secondary hyperoxaluria? Urol. Res. 33:372-375. [DOI] [PubMed] [Google Scholar]
- 27.Iyer, R., T. M. Iverson, A. Accardi, and C. Miller. 2002. A biological role for prokaryotic ClC chloride channels. Nature 419:715-718. [DOI] [PubMed] [Google Scholar]
- 28.Jager, A. V., F. G. Tonin, and M. F. Tavares. 2007. Comparative evaluation of extraction procedures and method validation for determination of carbohydrates in cereals and dairy products by capillary electrophoresis. J. Sep. Sci. 30:586-594. [DOI] [PubMed] [Google Scholar]
- 29.James, L. F. 1972. Oxalate toxicosis. Clin. Toxicol. 5:231-243. [DOI] [PubMed] [Google Scholar]
- 30.Kaufman, D. W., J. P. Kelly, G. C. Curhan, T. E. Anderson, S. P. Dretler, G. M. Preminger, and D. R. Cave. 2008. Oxalobacter formigenes may reduce the risk of calcium oxalate kidney stones. J. Am. Soc. Nephrol. 19:1197-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwak, C., B. C. Jeong, J. H. Ku, H. H. Kim, J. J. Lee, C. S. Huh, Y. J. Baek, and S. E. Lee. 2006. Prevention of nephrolithiasis by Lactobacillus in stone-forming rats: a preliminary study. Urol. Res. 34:265-270. [DOI] [PubMed] [Google Scholar]
- 32.Lamendella, R., J. W. Santo Domingo, C. Kelty, and D. B. Oerther. 2008. Bifidobacteria in feces and environmental waters. Appl. Environ. Microbiol. 74:575-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewanika, T. R., S. J. Reid, V. R. Abratt, G. T. Macfarlane, and S. Macfarlane. 2007. Lactobacillus gasseri Gasser AM63(T) degrades oxalate in a multistage continuous culture simulator of the human colonic microbiota. FEMS Microbiol. Ecol. 61:110-120. [DOI] [PubMed] [Google Scholar]
- 35.Lieske, J. C., D. S. Goldfarb, C. De Simone, and C. Regnier. 2005. Use of a probiotic to decrease enteric hyperoxaluria. Kidney Int. 68:1244-1249. [DOI] [PubMed] [Google Scholar]
- 36.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavolv, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goldstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J.-H. Lee, I. Diaz-Muniz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sulllivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U. S. A. 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marco, M. L., S. Pavan, and M. Kleerebezem. 2006. Towards understanding molecular modes of probiotic action. Curr. Opin. Biotechnol. 17:204-210. [DOI] [PubMed] [Google Scholar]
- 38.Mlobeli, N. T., N. A. Gutierrez, and I. S. Maddox. 1998. Physiology and kinetics of Bifidobacterium bifidum during growth on different sugars. Appl. Microbiol. Biotechnol. 50:125-128. [Google Scholar]
- 39.Murphy, C., S. Murphy, F. O'Brien, M. O'Donoghue, T. Boileau, G. Sunvold, G. Reinhart, B. Kiely, F. Shanahan, and L. O'Mahony. 2009. Metabolic activity of probiotics-oxalate degradation. Vet. Microbiol. 136:100-107. [DOI] [PubMed] [Google Scholar]
- 40.Okombo, J., and M. Liebman. 2010. Probiotic-induced reduction of gastrointestinal oxalate absorption in healthy subjects. Urol. Res. Doi: 10.1007/s00240-010-0262-9. [DOI] [PubMed]
- 41.Rodby, R. A., T. S. Tyazka, and J. W. Williams. 1991. Reversal of cardiac dysfunction secondary to type I primary hyperoxaluria after combined liver-kidney transplantation. Am. J. Med. 90:498-504. [PubMed] [Google Scholar]
- 42.Sahin, N. 2003. Oxalotrophic bacteria. Res. Microbiol. 154:399-407. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J. E., F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 44.Sidhu, H., M. J. Allison, J. M. Chow, A. Clark, and A. B. Peck. 2001. Rapid reversal of hyperoxaluria in a rat model after probiotic administration of Oxalobacter formigenes. J. Urol. 166:1487-1491. [PubMed] [Google Scholar]
- 45.Sidhu, H., M. E. Schmidt, J. G. Cornelius, S. Thamilselvan, S. R. Khan, A. Hesse, and A. B. Peck. 1999. Direct correlation between hyperoxaluria/oxalate stone disease and the absence of the gastrointestinal tract-dwelling bacterium Oxalobacter formigenes: possible prevention by gut recolonization or enzyme replacement therapy. J. Am. Soc. Nephrol. 10:S334-S340. [PubMed] [Google Scholar]
- 46.Sidhu, H., S. D. Ogden, H. Y. Lung, B. G. Luttge, A. L. Baetz, and A. B. Peck. 1997. DNA sequencing and expression of the formyl coenzyme A transferase gene, frc, from Oxalobacter formigenes. J. Bacteriol. 179:3378-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siva, S., E. R. Barrack, G. P. Reddy, V. Thamilselvan, S. Thamilselvan, M. Menon, and M. Bhandari. 2009. A critical analysis of the role of gut Oxalobacter formigenes in oxalate stone disease. BJU Int. 103:18-21. [DOI] [PubMed] [Google Scholar]
- 48.Stanton, C., R. P. Ross, G. F. Fitzgerald, and D. van Sinderen. 2005. Fermented functional foods based on probiotics and their biogenic metabolites. Curr. Opin. Biotechnol. 16:198-203. [DOI] [PubMed] [Google Scholar]
- 49.Tracy, C. R., and M. S. Pearle. 2009. Update on the medical management of stone disease. Curr. Opin. Urol. 19:200-204. [DOI] [PubMed] [Google Scholar]
- 50.Turroni, F., E. Foroni, P. Pizzetti, V. Giubellini, A. Ribbera, P. Merusi, P. Cagnasso, B. Bizzarri, G. L. de'Angelis, F. Shanahan, D. van Sinderen, and M. Ventura. 2009. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl. Environ. Microbiol. 75:1534-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turroni, S., B. Vitali, C. Bendazzoli, M. Candela, R. Gotti, F. Federici, F. Pirovano, and P. Brigidi. 2007. Oxalate consumption by lactobacilli: evaluation of oxalyl-CoA decarboxylase and formyl-CoA transferase activity in Lactobacillus acidophilus. J. Appl. Microbiol. 103:1600-1609. [DOI] [PubMed] [Google Scholar]
- 52.Vandecasteele, S. J., W. E. Peetermans, R. Merckx, and J. Van Eldere. 2001. Quantification of expression of Staphylococcus epidermidis housekeeping genes with Taqman quantitative PCR during in vitro growth and under different conditions. J. Bacteriol. 183:7094-7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:research0034.1-0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ventura, M., F. Turroni, A. Zomer, E. Foroni, V. Giubellini, F. Bottacini, C. Canchaya, M. J. Claesson, F. He, M. Mantzourani, L. Mulas, A. Ferrarini, B. Gao, M. Delledonne, B. Henrissat, P. Coutinho, M. Oggioni, R. S. Gupta, Z. Zhang, D. Beighton, G. F. Fitzgerald, P. W. O'Toole, and D. van Sinderen. 2009. The Bifidobacterium dentium Bd1 genome sequence reflects its genetic adaptation to the human oral cavity. PLoS Genet. 5:e1000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ventura, M., S. O'Flaherty, M. J. Claesson, F. Turroni, T. R. Klaenhammer, D. van Sinderen, and P. W. O'Toole. 2009. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat. Rev. Microbiol. 7:61-71. [DOI] [PubMed] [Google Scholar]
- 56.Ventura, M., M. O'Connell-Motherway, S. Leahy, J. A. Moreno-Munoz, G. F. Fitzgerald, and D. van Sinderen. 2007. From bacterial genome to functionality; case bifidobacteria. Int. J. Food Microbiol. 120:2-12. [DOI] [PubMed] [Google Scholar]
- 57.Vitali, B., S. Turroni, F. Dal Piaz, M. Candela, V. Wasinger, and P. Brigidi. 2007. Genetic and proteomic characterization of rifaximin resistance in Bifidobacterium infantis BI07. Res. Microbiol. 158:355-362. [DOI] [PubMed] [Google Scholar]
- 58.Weese, J. S., H. E. Weese, L. Yuricek, and J. Rousseau. 2004. Oxalate degradation by intestinal lactic acid bacteria in dogs and cats. Vet. Microbiol. 101:161-166. [DOI] [PubMed] [Google Scholar]
- 59.Williams, H. E., and L. H. Smith. 1968. Disorders of oxalate metabolism. Am. J. Med. 45:715-735. [DOI] [PubMed] [Google Scholar]