Abstract
We detected and characterized two distinct scuticociliate ciliates inside Acropora corals in the South China Sea. One, voraciously foraging on Symbiodinium, resembled the brown band disease of ciliates. The other, which is closely related to Paranophrys magna, grazed on detritus instead of Symbiodinium. These two ciliates may serve contrasting functions (competitor versus “cleaner”) in the coral-ciliate-Symbiodinium triangular relationship.
Various ciliates have been found to dwell in coral reefs (1, 2, 5, 6, 9, 15, 16, 18, 23, 24), but their genetic and physiological diversities remain poorly understood. Best studied were those associated with coral diseases, such as skeleton-eroding band (SEB) (1, 17) and brown band syndrome (BrB) (5, 24). BrB causes a brown zone on the coral, often sandwiched by healthy tissue and exposed white skeleton (24), which occurs when a large mass of a scuticociliate ciliate is present. BrB has been described for three coral families (Acroporidae, Pocilloporidae, and Faviidae) on the Great Barrier Reef, Australia (5, 21, 24), and for Porites astreoides and Montastraea faveolata in Florida (8). The responsible ciliate has been classified in different lineages (4, 8), but a recent molecular analysis based on the 18S rRNA gene (18S rDNA) identified it as a novel species closely related to the scuticociliate genera Uronema and Paranophrys (6). This ciliate was observed to consume the spat of the host coral (8) and to intensively ingest the coral's essential endosymbiont Symbiodinium; the ingested alga could remain intact and photosynthetically active within the ciliate after ingestion (21). Whether other ciliates cohabit with BrB ciliates in the coral and whether they would play different ecological functions in the coral microcosm are of high interest but remain obscure. In an attempt to address these issues, we found two closely related but distinct ciliates from two Acropora corals in the South China Sea. By culturing, microscopic examination, molecular (18S rDNA) analysis of a community DNA extract, and microscopic examination of isolated single cells and cultures, we observed distinct morphologies, phylogenetic positions, and trophic behaviors in these two ciliates.
Sampling of corals and intracavity microbial assemblages.
Coral branches were collected at 18°12′N, 109°28′E (latitude, longitude) at a depth of 1.6 m at Sanya, off Hainan Island in the South China Sea. The temperature was 27.1°C. After collection, each coral branch was transported in ambient seawater and transported to the laboratory in Sanya within 45 min. After the surface was rinsed with 0.45-μm-filtered seawater, the internal content of the coral (endosymbionts) was washed off using a Waterpik Ultra dental water jet (WP70EC; Alibaba, Shenzhen, China). Six 2-ml subsamples were preserved in 1% formaldehyde and immediately frozen in liquid nitrogen. The remainder was kept at ambient temperature for culturing. The corals were identified as Acropora microclados and Acropora hyacinthus as previously described (22). In situ and laboratory observations showed no BrB symptom in these two coral samples.
Microscopic observation, culturing, and characterization of the dominant ciliates.
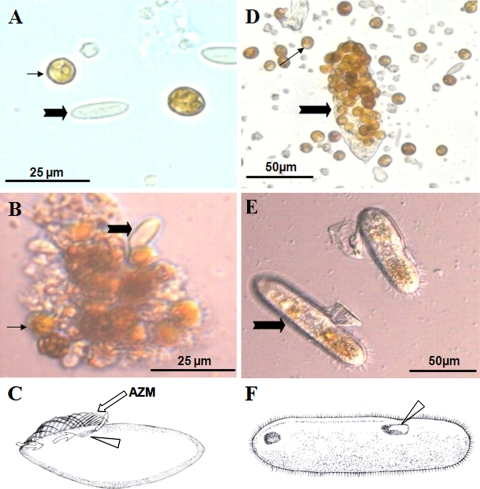
Microscopic examination of the preserved microbial samples revealed abundant Symbiodinium organisms, the endosymbiotic dinoflagellate (zooxanthella) essential for coral growth, and ciliates mainly of two morphotypes (Fig. 1). While both ciliates were detected in each coral, the microbial sample from A. microclados (sample 1) was dominated by a small ciliate species (Cil1) (Fig. 1A and B), about 20.3 to 30.9 (25.1 ± 4.9) μm in length and 8.7 to 11.1 (9.5 ± 0.8) μm in width, whereas that from A. hyacinthus (sample 2) was dominated by a markedly larger species (Cil2) (Fig. 1D and E), about 78.7 to 116.3 (98.0 ± 10.7) μm in length and 21.1 to 31.2 (25.9 ± 2.5) μm in width. Cil1 morphologically resembled Paranophrys magna (7), whereas Cil2 appeared identical to the reported BrB ciliates (21).
FIG. 1.
Micrographs and schematic drawings of the two ciliates detected in this study. (A to C) Samples (Cil1) from Acropora microclados before (A) and after (B) culturing and drawing (C) to show the peristome (arrowhead), adoral zone of membranelles (AZM) (straight arrow), and feeding current (curvy arrows) generated by the vibration of AZM. (D to F) Samples (Cil2) from Acropora hyacinthus before (C) and after (D) culturing and drawing (F) to show the peristome (arrowhead). Thick filled arrows denote the dominant ciliates; thin filled arrows denote Symbiodinium.
The live-cell suspension was cultured at 20°C in f/16 medium (8-fold dilution of f/2 medium [12a]) and with illumination at 90 μE·m−2·s−1 to sustain Symbiodinium growth. The cultures were examined daily. Near the peristome of Cil1 there was a small, well-developed adoral zone of membranelles (AZM), which was not found in Cil2 (Fig. 1C). The Cil1 cells were similar in size to Symbiodinium and exhibited slow movement. Daily microscopic examination of the live cultures showed no consumption of Symbiodinium (Fig. 1A and B); instead, detritus was ingested by rapid filtering of water through the current generated by the movement of the AZM (Fig. 1C). In contrast, Cil2 rapidly moved and voraciously ingested Symbiodinium cells; 6 to 70 Symbiodinium cells were observed per ciliate (in most cases >20) (Table 1; Fig. 1D). The peristome of Cil2 cells is large enough to engulf the cells of Symbiodinium (Fig. 1F).
TABLE 1.
Morphological characteristics of the ciliates and their abundances relative to Symbiodinium
| Ciliate | Source | Avg size (length by width [μm]) (no. of organisms) | Shape | Motility | Feeding | Mean ratio of Symbiodinium organisms to ciliates ± SD (no. of expts) |
|
|---|---|---|---|---|---|---|---|
| Before culture | After culture | ||||||
| Cil1 | Acropora microclados | 25.1 ± 4.9 by 9.5 ± 0.8 (9) | Short spindle | Slow | Filter | 8.5 ± 3.4 (6) | 9.5 ± 10.6 (3) |
| Cil2 | Acropora hyacinthus | 98.0 ± 10.7 by 26.0 ± 2.5 (19) | Elongated oval | Rapid | Engulf | 106.4 ± 44.8 (4) | 0.6 ± 9.4 (3) |
Consistent with the above observations, the Symbiodinium-to-ciliate abundance ratio was 8.5 ± 3.4 (n = 3) in sample 1 (Fig. 1A) and 106.4 ± 44.8 (n = 3) in sample 2 (Fig. 1D) before culturing. After incubation for 5 days, the ratio in sample 1 remained largely unchanged, at 9.5 ± 10.6 (n = 3), and Symbiodinium cells aggregated, showing signs of cell senescence and degradation, with scavenging Cil1 ciliates noticeable (Fig. 1B). In sample 2, the postincubation ratio decreased dramatically, to 0.6 ± 9.4 (n = 3), and Symbiodinium was almost completely depleted (Table 1; Fig. 1E).
Molecular identification of the two dominant ciliates.
When ciliates became abundant, the cultures were moved to darkness for 2 days to inhibit the growth of Symbiodinium. Ciliate cells were harvested by centrifuging a 10-ml sample of each culture at 10,000 × g for 10 min. Cell pellets were resuspended in 1 ml DNA lysis buffer (0.1 M EDTA, pH 8.0, 1% SDS, 200 μg ml−1 proteinase K), and DNA was extracted as described previously (26). PCR was carried out with a pair of 18S rDNA universal primers (25) to detect as many ciliate species as possible. Reaction conditions were 94°C for 30 s, 58°C for 30 s, and 72°C for 45 s for 35 cycles, preceded by a denaturing step at 94°C for 3 min and followed by an extension step at 72°C for 7 min. PCR products were cloned using a TA vector, and 20 clones were randomly picked for sequencing of both strands using the BigDye sequencing kit. Molecular cloning and sequencing of 18S rDNA yielded one sequence from Cil1 and two slightly different sequences from Cil2. The difference between the two sequences from Cil2 was only 0.60%, suggesting that they likely represented intragenomic gene copies or intraspecific ciliate populations. A chimera check (14) showed that these sequences were not chimeric, which is consistent with the result of full-length hit coverage in BLAST.
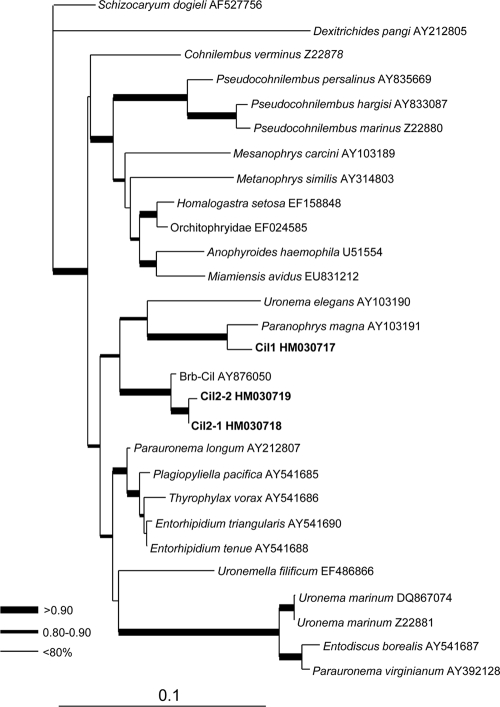
BLAST analysis showed that the 18S rDNA sequence of Cil1 was 97% identical to that of P. magna and that the two Cil2 sequences were 99% identical to that of the BrB ciliate. To further verify the relationships between these and other ciliates, a maximum likelihood phylogenetic tree was inferred from the sequences obtained in our study and in GenBank (hits in the BLAST analysis). ModelTest was run to select the most appropriate evolutionary model. The selected general time-reversible (GTR) model with gamma distribution was employed for maximum likelihood analysis using PhyML3.0 aLRT with 4 categories of substitution rates, the proportion of invariable sites at 0.521, and the gamma shape parameter at 0.413 as estimated from the data set (10). In the resultant tree, the Cil1 sequence was allied with P. magna with very strong bootstrap support (100%), and the Cil2 sequences formed a strongly supported (99%) cluster with the BrB ciliate (Fig. 2). These two types of ciliates were in the same clade and hence phylogenetically related. Neither of the two ciliates, however, showed significant similarity to Maristentor dinoferus, the ciliate that was previously reported to ingest and harbor Symbiodinium, because when the 18S rDNA of this species was included in the phylogenetic analysis, it formed a separate clade at the basal position with very long branches (not shown).
FIG. 2.
Maximum likelihood phylogenetic tree inferred from the 18S rDNA sequences retrieved in this study and those from related organisms available in GenBank. Each terminal branch is shown by species names (or lowest taxonomic level possible) followed by the GenBank accession number. Branch support is categorized as >90% (thickest line), 80 to 90% (thick line), and <80% (thin line). The scale bar denotes the number of substitutions per site.
Linking the genotypes to the morphotypes via single-cell PCR.
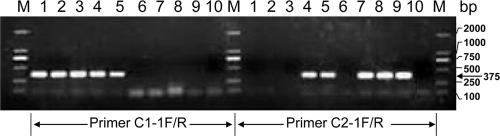
With the ciliate sequences obtained from the community DNA, a multialignment was conducted to identify specific regions from which to design genotype-specific PCR primers. These primers were then used to detect the specific genotypes from the original preserved samples and the cultures to verify that both genotypes occurred in both corals and each genotype specifically corresponded to one of the two morphotypes observed. Single cells from each morphotype were isolated from the initially preserved samples under an inverted microscope. The isolated cells (three cells from each morphotype) were rinsed carefully with 0.45-μm-filtered seawater and individually subjected to PCR analyses. In order to cross-check both genotypes for each cell, PCR was run first using the universal 18S rDNA primers mentioned above. Then the product was diluted and used as the template for a second PCR using the primers C1-1F, 5′-CCGACTCGGAATCGGGCAGGCTAC-3′ (forward), and C1-1R, 3′-TGCATAGCTTCGCATACCTTC-5′ (reverse), which are specific for ciliate Cil1, and primers C2-1F, 5′-CCGACTCGGAATCGGACAGGCCTT-3′, and C2-1R, 3′-CACCAAACTTTGCGTACCTTC-5′), which are specific for ciliate Cil2. Performed with 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min, PCR showed that the C1-1F/C1-1R primer set amplified only Cil1 cells and not Cil2 cells and that the C2-1F/C2-1R set amplified only Cil2 cells and not Cil1 cells (Fig. 3). PCR products were purified (26) and directly sequenced; results confirmed their corresponding identities, as expected from the primers. When used in PCR for DNA isolated from the original two samples (i.e., before culturing) where both morphotypes were found, both primer sets produced positive results from each sample. However, they did not show amplification for DNA isolated from the ambient water sample collected at the same time with the coral samples (Fig. 3). These results unambiguously linked the Cil1 morphotype with the Cil1 genotype and the Cil2 morphotype with the Cil2 genotype and further indicated that both ciliates occurred in each of the two coral species. The lack of PCR amplification for the ambient water suggests that these ciliates probably do not occur in substantial abundances as free-living organisms, which is consistent with the outcome of no significant matches when we used the 18S rDNA sequences that we obtained in this study in a BLAST search of the CAMERA database (43,240,119 entries, 16,900,401,306 bp), where data are typically derived from samples of tens to hundreds of liters of water.
FIG. 3.
PCR results with Cil1- and Cil2-specific primers. Shown are the results of three PCRs with a single cell of morphotype Cil1 (lanes 1, 2, and 3) or Cil2 (lanes 7, 8, and 9); reactions for the original (i.e., before culturing) samples 1 (lane 4) and 2 (lane 5) and reactions for the ambient water sample (lane 6) are also shown. Autoclaved distilled water was used as a negative control for each primer set (lane 10). Lanes M on the left, middle, and the right contain molecular size standards, with the molecular sizes of the bands indicated on the right. The molecular size of the PCR products, essentially the same as for the Cil1- and Cil2-specific primers, is marked on the right with an arrow.
Apparent differential functions of the two types of ciliates.
Previous observations tend to support the view that corals ingest zooplankton (e.g., ciliates) and detritus (containing a whole microbial food chain) as major nutritive sources (3, 11, 13, 19, 20). It has been reported that the total number of ciliates ingested by coral and the ingestion rates decrease when the light intensity increases (12), suggesting functional complementarity between heterotrophy and phototropy in the coral. Our observations in the present study suggest that the functional association between the coral and the ciliates may be more complex. The two types of ciliates detected here exhibit different trophic modes and appear to have totally different relationships with the coral host. Cil1 moved slowly in the culture and rapidly filtered ambient water by the movement of the AZM, apparently feeding on detritus and bacteria in the water. This ciliate can be a competitor for the nutritive substances (particulate matter and microbes) but also can serve as a “cleaner” for the host corals especially if high abundances of the photosynthetic Symbiodinium organisms die as a result of external or internal environmental stress. However, this ciliate does not ingest Symbiodinium and thus probably does not have much influence on the autotrophy of the host coral. In contrast, the BrB-associated ciliate moves rapidly and voraciously ingests Symbiodinium. Therefore, the BrB-associated ciliate may be the host coral's rival, competing for the “common garden.” In agreement with the previous report showing that ingested Symbiodinium cells remained photosynthetically competent inside this ciliate (21), we observed the deep colors of the pigments in the intracellular Symbiodinium cells. This observation suggests that ciliates may temporarily exploit the photosynthetic capability of Symbiodinium after ingestion. However, the disappearance of Symbiodinium in the culture over a 5-day period indicates that this ciliate is more of a grazer rather than a “farmer.” This grazing tendency may be the cause of the BrB disease in the host coral, but the lack of noticeable BrB or other syndromes in the corals that we sampled suggests that the causative ciliates may need to reach a certain level of abundance (as a result of active growth or depressed digestion by the host coral) before triggering the brown band symptom. In this case, the health of the coral may hinge on a delicate balance between ingestion and digestion of ciliates and Symbiodinium growth. Much remains to be investigated to gain a good understanding on the coral-ciliate-Symbiodinium relationship.
Nucleotide sequence accession numbers.
The gene sequences obtained in this study have been deposited in the GenBank database under accession numbers HM030717 to HM030719.
Acknowledgments
We thank George McManus from the Department of Marine Sciences, University of Connecticut, for help with the English in our paper.
This project was supported by the CAS International Partnership Project for the Innovational Group on Tropical Marine Ecological Process Studies (grant KZCX2-YW-T001), the NSFC-Overseas Collaboration Fund (grant 40828006), and the U.S. National Science Foundation (grant EF-0629624).
Footnotes
Published ahead of print on 25 June 2010.
REFERENCES
- 1.Antonius, A. 1999. Halofolliculina corallasia, a new coral-killer ciliate on Indo-Pacific reefs. Coral Reefs 18:300. [Google Scholar]
- 2.Antonius, A., and A. D. Lipscomb. 2000. First protozoan coral-killer in the Indo-Pacific. Atoll Res. Bull. 481:1-21. [Google Scholar]
- 3.Ayukai, T. 1995. Retention of phytoplankton and planktonic microbes on coral reefs within the Great Barrier Reef, Australia. Coral Reefs. 14:141-147. [Google Scholar]
- 4.Borneman, E. H. (ed.). 2001. Aquarium corals: selection, husbandry, and natural history. T. F. H. Publications, Inc., Neptune City, NJ.
- 5.Bourne, D. G., H. Boyett, and B. Willis. 2004. Microbiology of brown band disease (BRB) affecting Acropora corals of the Great Barrier Reef, abstr. 874. Abstr. 10th Int. Symp. Microb. Ecol. Cancun, Mexico, 22 to 27 August 2004.
- 6.Bourne, D. G., H. V. Boyett, M. E. Henderson, A. Muirhead, and B. L. Willis. 2008. Identification of a ciliate (Oligohymenophora: Scuticociliatia) associated with brown band disease (BrB) on corals of the Great Barrier Reef. Appl. Environ. Microbiol. 74:883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carey, P. G. 1992. Marine interstitial ciliates—an illustrated key. Chapman and Hall, London, United Kingdom.
- 8.Cooper, W., D. Lirman, M. Schmale, and D. Lipscomb. 2007. Consumption of coral spat by histophagic ciliates. Coral Reefs 26:249-250. [Google Scholar]
- 9.Cróquer, A., C. Bastidas, D. Lipscomp, R. E. Rodríguez-Martínez, E. Jordan-Dahlgren, and H. M. Guzman. 2006. First report of folliculinid ciliates affecting Caribbean scleractinian corals. Coral Reefs 25:187-191. [Google Scholar]
- 10.Dereeper, A., V. Guignon, G. Blanc, S. Audic, S. Buffet, F. Chevenet, J. F. Dufayard, S. Guindon, V. Lefort, M. Lescot, J. M. Claverie, and O. Gascuel. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36(Web Server issue):W465-W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrant, P. A., M. A. Borowitzka, R. Hinde, and R. J. King. 1987. Nutrition of the temperate Australian soft coral Capnella gaboensis. Mar. Biol. 95:575-581. [Google Scholar]
- 12.Ferrier-Pagès, C., A. Allemand, J. P. Gattuso, and J. Jaubert. 1998. Microheterotrophy in the zooxanthellate coral Stylophora pistillata: effects of light and ciliate density. Limnol. Oceanogr. 43:1639-1648. [Google Scholar]
- 12a.Guillard, R. R., and J. H. Ryther. 1962. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt and Detonula confervacea (Cleve.) Gran. Can. J. Microbiol. 18:229-239. [DOI] [PubMed] [Google Scholar]
- 13.Lewis, J. B. 1992. Heterotrophy in corals: zooplankton predation by the hydrocoral Millepora complanata. Mar. Ecol. Prog. Ser. 90:251-256. [Google Scholar]
- 14.Lin, S., H. Zhang, Y. Hou, L. Miranda, and D. Bhattacharya. 2006. Development of a dinoflagellate-oriented PCR primer set leads to the detection of picoplanktonic dinoflagellates from Long Island Sound. Appl. Environ. Microbiol. 72:5626-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lobban, C. S., and M. Schefter. 1996. An abundance of marine Stentor (Ciliophora: Spirotrichea) epiphytic on Padina (Phaeophyta). Micronesica 19:99-100. [Google Scholar]
- 16.Lobban, C. S., M. Schefter, A. G. B. Simpson, X. Pochon, J. Pawlowski, and W. Foissner. 2002. Maristentor dinoferus n. gen., n. sp., a giant heterotrich ciliate (Spirotrichea: Heterotrichida) with zooxanthellae, from coral reefs on Guam, Mariana Islands. Mar. Biol. 140:411-423. [Google Scholar]
- 17.Riegl, B., and A. Antonius. 2003. Halofolliculina skeleton eroding band (SEB): a coral disease with fossilization potential? Coral Reefs 22:48. [Google Scholar]
- 18.Rodríguez, S., A. Cróquer, H. M. Guzmán, and C. Bastidas. 2008. A mechanism of transmission and factors affecting coral susceptibility to Halofolliculina sp. infection. Coral Reefs 28:67-77. [Google Scholar]
- 19.Sebens, K. P., K. S. Vandersall, L. A. Savina, and K. R. Graham. 1996. Zooplankton capture by two scleractinian corals, Madracis mirabilis and Montastrea cavernosa in a field enclosure. Mar. Biol. 127:303-317. [Google Scholar]
- 20.Sorokin, Y. I. 1991. Biomass, metabolic rates and feeding of some common reef zoantharians and octocorals. Aust. J. Mar. Freshwat. Res. 42:729-741. [Google Scholar]
- 21.Ulstrup, K. E., M. Kühl, and D. G. Bourne. 2007. Zooxanthellae harvested by ciliates associated with brown band syndrome of corals remain photosynthetically competent. Appl. Environ. Microbiol. 73:1968-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wallace, C. C. (ed.). 1999. Staghorn coral of the world—a revision of the coral genus Acropora. CSIRO Publishing, Melbourne, Australia.
- 23.Wichterman, R. 1942. A new ciliate from a coral of Tortugas and its symbiotic algae. Pap. Tottugas Lab. 33:107-111. [Google Scholar]
- 24.Willis, B. L., C. A. Page, and E. A. Dinsdale. 2004. Coral disease in the Great Barrier Reef, p. 69-104. In E. Rosenberg and Y. Loya (ed.), Coral health and disease. Springer-Verlag, New York, NY.
- 25.Zhang, H., and S. Lin. 2002. Detection and quantification of Pfiesteria piscicida by using the mitochondrial cytochrome b gene. Appl. Environ. Microbiol. 68:989-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, H., and S. Lin. 2005. Development of a cob-18S rDNA real-time PCR assay for quantifying Pfiesteria shumwayae in the natural environment. Appl. Environ. Microbiol. 71:7053-7063. [DOI] [PMC free article] [PubMed] [Google Scholar]