Abstract
Environmental waters are an important reservoir for Vibrio cholerae, and effective surveillance of the pathogen can help to warn of and prevent infection with this potentially fatal pathogen. An immunofluorescent-aggregation (IFAG) assay to detect V. cholerae O1 and O139 was established and evaluated with estuarine water samples. The practical application of this assay was compared with the conventional culture method and real-time PCR. The IFAG method had a sensitivity of 103 CFU/ml for detection of V. cholerae O1 and O139 strains in a suspension containing 10 different species of enterobacterial strains (total, 105 CFU/ml). Ten fluorescent bacterial aggregate colonies were randomly picked and tested positive in serum agglutination tests for the V. cholerae O1 and O139 strains, showing a high specificity. The enrichment broths of 146 samples of estuarine water were tested, and the percentage positive by the IFAG assay was 19.9% (29/146), which was significantly higher than that of the conventional culture method (10.3%, 15/146; P < 0.01) but lower than that of real-time PCR (29.5%, 43/146; P < 0.01). The coincidence rates of real-time PCR and IFAG detection were decreased with the reduction of the V. cholerae concentration. The IFAG method, with a high specificity and a relatively high sensitivity, may be used for detection and isolation of V. cholerae in environmental water samples.
Vibrio cholerae causes disease by colonizing, proliferating, and secreting toxins in the intestine of the host. The resulting profuse diarrhea and vomiting can be fatal if left untreated. Among the more than 200 known Vibrio cholerae serogroups, only the O1 and O139 strains are pathogenic, causing epidemics and pandemics throughout history (24). The El Tor biotype of the O1 serogroup first emerged in Indonesia at the beginning of the seventh pandemic in 1961 (16), became prevalent in the Ganges delta of the Indian subcontinent, and then spread into other countries (13, 26). The first non-O1 serogroup epidemic, which emerged in southern India and then spread into many Asian countries, was caused by an O139 strain of V. cholerae (3, 5). Currently, cholera is still a major threat to public health in many countries of Asia, Africa, and South America (4).
Many cholera epidemics originated in the coastal areas (21), where water habitats are important niches contributing to the prevalence of V. cholerae (15, 21). Surveillance studies in the Bay of Bengal found that cholera epidemics correlated with the propagation of V. cholerae in estuarine waters (14, 21). Remote sensing data analysis showed that sea surface temperature (SST) and sea surface height (SSH) are also associated with the cholera outbreaks, suggesting that the epidemics are climate linked (11, 23). Although V. cholerae was once thought to inhabit only the human small intestine, it is known that the pathogen can persist in environmental waters during nonepidemic periods (12). This bacterium attaches to the bodies of zooplankton, crustaceans, and aquatic plants in seawater and estuarine water and survives by forming a biofilm on the surface of these organisms (1) or as a biofilm in the water (2).
Therefore, monitoring of V. cholerae O1 and O139 in the estuarine waters in the interim period between cholera epidemics is of great significance for early warning of an outbreak, as well as prevention and control of cholera epidemics when they occur. To date, a number of methods for detection of V. cholerae in environmental water have been developed. Due to the general low level of V. cholerae in water and to reduce the impact of other bacteria that can interfere with its detection, selective enrichment of the bacterial population from water samples, followed by selective isolation and culture in selective medium, is generally required. This method is time-consuming, especially in handling a large number of water samples. Current rapid detection methods include the use of immunomagnetic beads (25) and nucleic acid-based methods such as PCR (9, 22) and DNA probe hybridization (10). The sensitivity of immunomagnetic bead detection is low, and there is a greater possibility of false positives. The PCR and DNA probe hybridization techniques require designing of specific primers or probes that can detect only nucleic acids rather than live bacterial cells. The recently developed real-time PCR technique has high sensitivity in detecting V. cholerae (7, 17), although it also detects nucleic acids, and it still requires isolation and culture of sample after PCR detection.
The fluorescent labeling technique uses antibodies to specifically identify pathogens, and some studies have applied this method to detection of V. cholerae in environmental water samples. Brayton et al. established a fluorescent-antibody (FA) direct viable count (FA-DVC) assay (8) in Bangladesh which yields relatively high specificity and sensitivity and can rapidly and easily detect culturable and nonculturable V. cholerae O1. The direct fluorescent antibody (DFA) assay has also been applied to detect viable but nonculturable V. cholerae O1 in environmental water (6) and in studies on the role of V. cholerae biofilm on the transmission of cholera (2). In addition, Hasan et al. (18) developed two monoclonal antibody-based methods, the coagglutination test and the direct fluorescent-antibody test, which has a high specificity, and the detection limit for V. cholerae O139 in environmental water samples reached 2.0 × 103 CFU/ml and 1.5 × 102 CFU/ml, respectively. Therefore, use of FA may be an effective method to directly detect V. cholerae in samples, followed by isolation and culture.
In this study, we established an immunofluorescent-aggregation (IFAG) assay using fluorescence-labeled monoclonal antibodies against V. cholerae O1 and O139, respectively. After enrichment for bacterial populations in estuarine water samples, V. cholerae was detected in the samples, and then the observed fluorescent bacterium aggregates were picked for further isolation and culture. We also evaluated the feasibility of our IFAG assay by comparison with the direct enrichment, isolation, and culture method, as well as real-time PCR detection. The combination of IFAG and real-time PCR for environmental monitoring of V. cholerae can provide rapid and efficient detection of V. cholerae O1 and O139.
MATERIALS AND METHODS
Experimental strains.
V. cholerae N16961 (O1 strain) and MO45 (O139 strain) were used in this study to establish our IFAG assay. Additionally, we selected 20 isolates of the O1 El Tor strain from China collected during the years between 1964 and 2005 (10 isolates of toxigenic strains from patients and 10 isolates of nontoxigenic strains from environmental sources) and 20 isolates of V. cholerae O139 from China collected during 1994 to 2005 (10 isolates of toxigenic strains from patients and 10 isolates of nontoxigenic strains from environmental sources). Other Vibrio bacteria that were isolated from environmental waters were used to determine specificity of our detections methods; these included 8 strains of V. parahaemolyticus, 5 of V. mimicus, 5 of V. metschnikovii, 5 of V. vulnificus, 3 of V. alginolyticus, 3 of V. furnissii, 3 of V. fluvialis, 2 of non-O1/non-O139 V. cholerae), other enteric bacteria (including 10 strains [each] of Escherichia coli, Shigella, Salmonella, Klebsiella, Citrobacter, and Proteus), and 5 of Aeromonas hydrophila. All of the test strains had undergone serum or biochemical identification and were preserved in our laboratories.
Fluorescent antibody preparation.
V. cholerae O1 strain Wujiang-2 (serotype Inaba, isolated from a patient; our laboratory store) and O139 strain MO45 were used for generation of monoclonal antibodies (MAbs), and the MAbs were prepared in our laboratory. Cells were grown on nutrient agar at 37°C for 12 to 14 h, killed by formaldehyde (Formalin) at a concentration of 1.0% for 8 h, and then washed three times with 0.9% NaCl, with 109 CFU/ml of suspension used as an antigen, to immunize mice (female BALB/c; age, 6 to 8 weeks). All other methods of immunization, hybridization, and selection of hybridoma cell lines were as described previously (19). IgG antibodies were purified by the octanoic acid-ammonium sulfate method, and the purity and content were determined by SDS-PAGE. The concentration of IgG was adjusted to 10 mg/ml, and after incubation in 1 mg/ml dichlorotriazinyl aminofluorescein (DTAF) (Sigma) solution (pH 8.6) at 37°C for 1 h, the purified fluorescent antibodies were collected by a Sephadex G-50 purification column. The fluorescent-antibody solution was prepared with antibodies at concentrations of 0.5 mg/ml in peptone solution (water with 1% peptone, 1% NaCl, pH 8.6).
IFAG.
One hundred microliters (each) of peptone solution with fluorescent antibodies specific for V. cholerae O1 and O139 in peptone water was dropped in two points on glass slides, which were boiled in paraffin oil for 10 min before use. Ten microliters of the sample to be analyzed was mixed with the O1 fluorescent-antibody solution droplet, and another 10 μl mixed solution was mixed with the O139 fluorescent antibody solution droplet. After culturing in a wet disc at 37°C for 6 h, the samples were observed under a fluorescence microscope (Nikon TS100) equipped with a 200-V high-voltage mercury lamp and a light filter (ELWD0.3T1-SNCP). If fluorescent masses with irregular edges (bacterial aggregates) were observed, it was judged to be a positive IFAG detection. For IFAG-positive samples, fluorescent bacterial aggregates were drawn with an aggregate picker (a manually made glass capillary pipette with an about 0.5-mm inner diameter) under the microscope and transferred to one of three drops of sterile peptone water on a slide. With the use of a microscope to locate the bacteria, the labeled aggregates were “washed” by serial transfer with the capillary pipette, each time expelling the aggregate into one droplet and then drawing it up again to transfer to the next drop. After the final “wash” in the last droplet, the fluorescent bacterial aggregate was dropped onto a gentamicin agar plate (Shanghai KeMaJa Biotech), streaked, and cultured at 37°C for 8 to 12 h. Suspected colonies of V. cholerae were selected to be tested with diagnostic serum for V. cholerae O1 and O139 (National Institute for the Control of Pharmaceutical and Biological Products) and monoclonal antibodies to O1 and O139 in slide agglutination tests.
Sensitivity and specificity of detection of IFAG.
Single colonies of the O1 N16961 and O139 MO45 strains were cultured in LB enriched broth at 37°C overnight. The pure culture media were serially diluted 10-fold and plated for colony counts. Ten microliters of the serially diluted samples were detected by IFAG to assess its sensitivity; the LB broth samples with no V. cholerae strains were used as the blank controls. In the specificity experiments, each of the serially diluted samples (including the blank samples) was added with an equal volume of interfering bacteria mixture (the final concentration of each of the interfering bacteria in the mixture was 105 CFU/ml, including non-V. cholerae, V. cholerae non-O1/non-O139, and other enteric bacteria listed under “Experimental strains”). Each of the serially diluted samples was detected by IFAG for either V. cholerae O1 or O139 in separate diluted samples. The bacterial aggregates detected as fluorescence positive by the IFAG assay were picked under the microscope using the aggregate picker, streak inoculated onto a gentamicin agar plate, and cultured at 37°C for 8 to 12 h. Suspected V. cholerae colonies on the plates were selected, and slide agglutination tests were performed using V. cholerae O1 and O139 diagnostic serum for verification.
Collection of estuarine water samples.
Water samples were collected from the Pearl River in the Guangzhou urban area and from its estuary, and V. cholerae was detected. The sites selected were watercourses associated with past outbreaks, according to past monitoring data, and areas around terminals for seawater products. During July 2008 to October 2008, a total of 146 water samples were collected in the 4 consecutive months. At each sampling point, three samples of 450 ml surface water (more than 30 cm below the surface) were collected using sterile water sample bottles. After sealing, the sample bottles were sent at room temperature to the laboratories on the same day for isolation and culture of V. cholerae.
Conventional culture.
The enrichment of cultures was done by adding 50 ml of 10× alkaline peptone water to 450 ml water sample, which was adjusted to a pH of 8.4 by NaOH. The enriched cultures were incubated at 37°C for 8 h and then streak inoculated on a gentamicin agar plate and a thiosulfate citrate bile salts sucrose (TCBS) agar plate for V. cholerae screening. The plates were cultured at 37°C for 8 to 12 h, and the suspected colonies on the TCBS agar plate were inoculated on a nutrient agar plate for pure culture at 37°C for 8 to 12 h. The culture was tested in slide agglutination tests with diagnostic sera for V. cholerae O1 and O139. Suspected colonies on the gentamicin agar plate were directly assayed in the slide agglutination tests with diagnostic sera. At least five suspected colonies in each sample were picked (all were picked when fewer than 5 colonies were in the plate) for slide agglutination tests. The culture was performed independently along with other detection methods.
Real-time PCR.
This study applied the established SYBR green I real-time PCR method (27). The primers O1-P2F (5′-GCG TAA ATA TCT AAA CGA TTG CAT TG-3′) and O1-P2R (5′ AAA CTC AGT TTC GAA GCG ATC AA 3′) amplify a fragment of 83 bp from the O1 rfb gene. The primers O139-P1F (5′-GCG GTG TAG CGG GTT TTA TTA G-3′) and O139-P1R (5′-TGC ATA ATA CTT TCG ACC ATG GA-3′) amplify a fragment of 76 bp from O139 rfb. The real-time reaction conditions in the LightCycler (Roche) were as follows: stage 1, 95°C for 10 s; stage 2, 95°C for 5 s and 60°C for 20 s, 40 cycles; stage 3, dissolution curve analysis, 95°C for 0 s, 65°C for 15 s, and 95°C for 0 s. DNA was extracted using a kit (Tiangen Biotech) from 1 ml enrichment broth of water samples as the template. The real-time PCR was performed independently along with other detection methods.
PCR detection of the toxin gene ctxAB.
Primers for detection of the ctxAB gene isolated from water samples were CT1 (5′-ATTTTGAGGTGTTCCATGTG-3′) and CT2 (5′-ATAAAGCAGTCAGGTGGTCT-3′), with a product length of 749 bp; PCR conditions were at 94°C for 5 min; 94°C for 30 s, 58°C for 40 s, and 72°C for 50 s for 30 cycles; and 72°C for 7 min.
Statistical analysis.
The percent-positive results of different detection methods were compared by the χ2 test using the row × column table (McNemar's test).
RESULTS
Specificity of monoclonal IgG antibodies to V. cholerae O1 and O139.
High-purity IgG antibody bands were observed by protein electrophoresis with the DTAF-labeled monoclonal antibodies to V. cholerae O1 and O139 (data not shown). In determining the specificities of IgG antibodies to V. cholerae O1 and O139, the two MAbs were found to agglutinate only with the corresponding V. cholerae serogroup, and no cross-agglutination was detected. The two antibodies also did not cross-agglutinate with non-O1/non-O139 V. cholerae strains, nor with those of other Vibrio spp., including V. parahaemolyticus (n = 8), V. mimicus (n = 5), V. metschnikovii (n = 5), V. vulnificus (n = 5), V. alginolyticus (n = 3), V. furnissii (n = 3), and V. fluvialis (n = 3), and enteric bacteria, including E. coli, Shigella, Salmonella, Klebsiella, Citrobacter, and Proteus (10 strains each) and A. hydrophila (n = 5). The results showed that the monoclonal antibodies used in this study were specific to V. cholerae O1 and O139.
Sensitivity of IFAG detection.
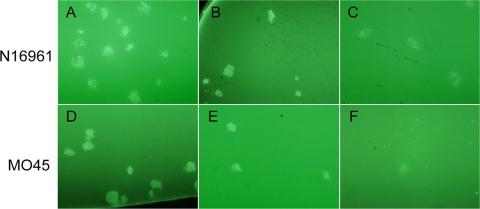
The pure culture media of the O1 N16961 and O139 MO45 strains were serially diluted 10-fold in LB medium. Ten microliters of these cultures were used as test samples to determine the sensitivity of the IFAG assay. Plate colony counts determined the concentration of LB culture of N16961 to be 6.5 × 107 CFU/ml, and that of MO45 was 2.8 × 107 CFU/ml. The serially diluted samples were then detected by the IFAG assay in triplicate for each sample. When fluorescent bacterial aggregates were detected by IFAG one or more times, the sample was determined to be positive. Fluorescent aggregates of bacteria formed by bacterial agglutination were visible under the microscope (Fig. 1). We found that the detection limit of the IFAG assay for both the O1 and O139 strains reached 103 CFU/ml. At this concentration, each of the N16961 and MO45 isolates had one out of the triplicate samples test positive by IFAG. In fact, when the concentration was 103 CFU/ml, the number of V. cholerae CFU in the tested samples was only 101 in the 10 μl used for IFAG detection. After these bacteria were agglutinated with the fluorescent antibodies, the formed fluorescent aggregates of bacteria were still easily identified in the dark background.
FIG. 1.
The fluorescent aggregates of bacteria of V. cholerae O1 strain N16961 (upper) and O139 strain MO45 (lower) in LB culture. The aggregates were observed under the fluorescence microscope (20 × 40). The concentration in LB samples of N16961 was 6.5 × 105 CFU/ml (A), 6.5 × 104 CFU/ml (B), or 6.5 × 103 CFU/ml (C), and that for MO45 was 2.8 × 105 CFU/ml (D), 2.8 × 104 CFU/ml (E), or 2.8 × 103 CFU/ml (F), respectively.
Determination of IFAG specificity.
To evaluate IFAG specificity in the laboratory, LB cultures of N16961 and MO45 were serially diluted 10-fold and then added to an equal volume of mixed solution of interfering bacteria. In the serially diluted samples containing V. cholerae, the content of N16961 or MO45 was 1.75 × 106 to 1.75 × 101 CFU/ml or 3.0 × 106 to 3.0 × 101 CFU/ml, respectively. When the final concentrations of strains N16961 and MO45 were >103 CFU/ml, fluorescent aggregates were observed in the mixed bacterial solution with the 10 types of interfering bacteria with the use of IFAG (see Fig. 2). The fluorescent aggregates were randomly drawn and transferred into gentamicin plates for culture, and the cultured suspected colonies were assayed by serum agglutination tests for the diagnosis of V. cholerae O1 and O139, which was matched with the original V. cholerae serogroups in the samples. No fluorescent aggregates were observed in the samples with only the mixture of the interfering bacteria.
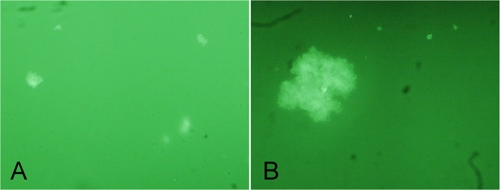
FIG. 2.
IFAG detection for fluorescent aggregates of V. cholerae under the fluorescence microscope. (A) O1 strain N16961, 1.75 × 103 CFU/ml, 10 × 40; (B) O139 strain MO45, 3.0 × 103 CFU/ml, 40 × 40.
Detection of water samples.
To evaluate the sensitivity of IFAG for V. cholerae in environmental water samples, we compared the IFAG method by using the samples collected from estuarine waters with the conventional culture and real-time PCR methods, carried out simultaneously. All these different methods were performed independently. Among the 146 samples collected from the Pearl River in the Guangzhou urban area and from its estuary, the percent-positive results of IFAG and real-time PCR were 19.9% (29/146) and 29.5% (43/146), respectively, both of which were significantly higher than 10.3% (15/146), attained by the conventional method (Table 1). There was also a significant difference between the percent-positive result of IFAG and that of real-time PCR detection (Table 1).
TABLE 1.
Positive results among conventional culture, IFAG, IFAG plus culture, and real-time PCRa
| Parameter for results | Value for method |
|||
|---|---|---|---|---|
| Conventional culture | IFAG | IFAG + culture | Real-time PCR | |
| No. positive | 15 | 29 | 22 | 43 |
| No. negative | 129 | 115 | 124 | 103 |
| % Positive | 10.3 | 19.9 | 15.1 | 29.5 |
Total sample, n = 146. Statistical tests: between conventional culture and IFAG, χ2 = 12.1 and P < 0.01; between IFAG and real-time PCR, χ2 = 12.1 and P < 0.01.
In the detection of water samples by IFAG, an example of the bacterial aggregates observed under the fluorescence microscope is shown in Fig. 3. Of the 29 samples positive by IFAG detection (19 samples of V. cholerae O1 and 10 of V. cholerae O139), 22 isolates were obtained by drawing the fluorescent bacterial aggregates to the gentamicin agar plates for culture (75.9% positive) (Table 1). Among them, 16 strains were O1 and 6 strains were V. cholerae O139 (no samples obtained tested positive for both serogroups). The serotypes of V. cholerae O1 and O139 by initial IFAG assay detection were consistent with those of the strains determined after culture of the IFAG-positive samples. The strains which were initially obtained by conventional culture were also positive by IFAG and IFAG plus culture detection, and the serotypes were consistent with those of isolated strains obtained by culture of IFAG-positive samples. This showed that the isolation rate of culture after detection with IFAG was higher than that by conventional isolation.
FIG. 3.
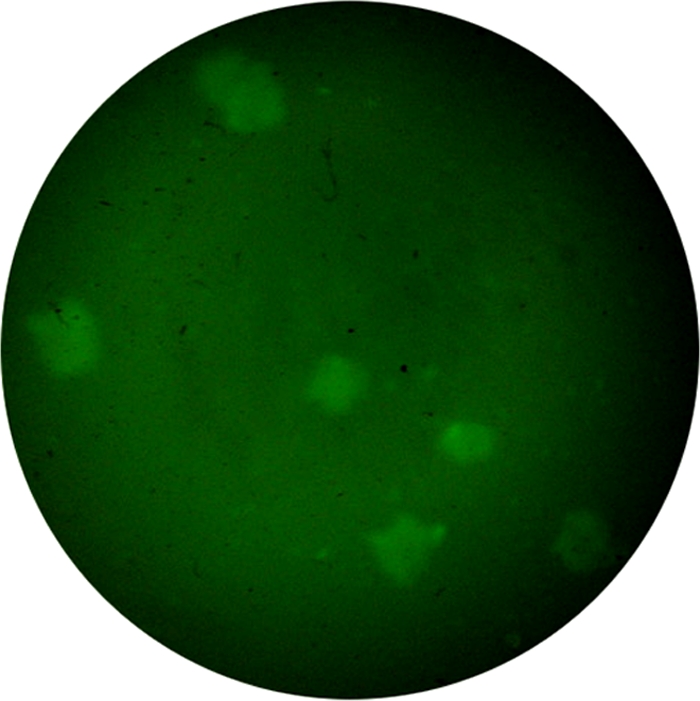
Fluorescent aggregates of V. cholerae (serogroup O1) by IFAG detection of a water sample.
We simultaneously used real-time PCR to detect V. cholerae O1 and O139 in the water samples. It was found that all the samples positive by the conventional culture, IFAG, and IFAG-plus-culture methods were consistent with the samples positive by real-time PCR, including the serotypes of isolates. We grouped the 43 real-time-PCR-positive samples according to the threshold cycle (CT) values of PCR detection and compared the percent-positive results of IFAG, IFAG plus culture, and conventional culture within these samples (Table 2). In the lower-CT groups, the coincidence rate between the results of conventional culture, IFAG, IFAG plus culture, and real-time PCR was high, and V. cholerae was more easily isolated by the conventional method. With the CT values increasing, the percent-positive results of IFAG and culture decreased. Therefore, when the concentration of V. cholerae in the enrichment broth was low, the detection abilities of conventional culture methods and IFAG were decreased.
TABLE 2.
Comparison of samples positive by IFAG, IFAG plus culture, and conventional culture methods with real-time PCR
| CT groupa | No. (%b) of samples positive by: |
|||
|---|---|---|---|---|
| Real-time PCR | IFAG | IFAG + culture | Conventional culture | |
| Serogroup O1 | ||||
| ≤20.00 | 8 | 8 (100) | 8 (100) | 7 (87.5) |
| 20.01-23.00 | 10 | 8 (80) | 7 (70) | 3 (30) |
| 23.01-26.00 | 4 | 3 (75) | 0 (0) | 0 (0) |
| 26.01-29.00 | 1 | 0 (0) | 0 (0) | 0 (0) |
| ≥29.01 | 1 | 0 (0) | 0 (0) | 0 (0) |
| Total | 24 | 19 | 15 | 10 |
| Serogroup O139 | ||||
| ≤20.00 | 7 | 7 (100) | 7 (100) | 5 (71.4) |
| 20.01-23.00 | 3 | 0 (0) | 0 (0) | 0 (0) |
| 23.01-26.00 | 6 | 3 (50) | 0 (0) | 0 (0) |
| 26.01-29.00 | 1 | 0 (0) | 0 (0) | 0 (0) |
| ≥29.01 | 2 | 0 (0) | 0 (0) | 0 (0) |
| Total | 19 | 10 | 7 | 5 |
The CT value increases with a decreasing amount of template or a high template concentration.
Values within parentheses are percentages relative to the number of real-time PCR-positive samples.
We also detected the virulence gene ctxAB for V. cholerae strains isolated from water samples in this study. The ctxAB genes in all 22 of the V. cholerae isolates were negative, indicating that the V. cholerae strains in these waters were predominantly nontoxigenic.
DISCUSSION
In this study, the sensitivity of the established IFAG method for detection of V. cholerae O1 and O139 in laboratory culture was 103 CFU/ml; the method was also highly specific. In the mixed bacterial solution containing 10 species of interfering bacteria and the target V. cholerae O1 and O139 strains, IFAG was able to specifically detect fluorescent aggregates of V. cholerae O1 and O139 without cross-reactivity. Furthermore, the aggregates were verified to be V. cholerae O1 and O139 in the subsequent culture and serum agglutination tests, showing high specificity. Hasan et al (18). used DFA to detect V. cholerae O139; because the water samples were filtered and concentrated in that study, the sensitivity of V. cholerae O139 detection reached 1.5 × 102 CFU/ml and the specificity was 100%. In this study, because neither the pure cultures nor on-site water samples were filtered or concentrated, the sensitivity of detection was one log lower than that of Hasan et al., but our assay had the same specificity.
A number of methods have been evaluated for detection of V. cholerae O1 and O139 in samples from environmental waters. The colloidal gold technique was used to easily screen V. cholerae, but the sensitivity is low (requiring the bacteria concentration in samples to be more than 105 CFU/ml). Conventional culture requires a larger sample size, for which preprocessing steps for isolation and detection are laborious and cumbersome. The real-time PCR technique has high sensitivity in detecting V. cholerae, but it still requires culture and isolation of sample after PCR detection when the strain is needed. The IFAG assay has high specificity and a higher isolation rate than the conventional culture and should be a good choice for environmental surveillance of V. cholerae, especially surveillance and study based on obtaining isolates from environmental samples. We previously performed studies on the use of IFAG for detection of V. cholerae by using antisera to V. cholerae O1 and O139 (28, 29). Here we analyzed the enriched broth of estuarine water samples and used monoclonal antibodies to serogroups O1 and O139 in IFAG. We determined that the IFAG percentage positive was higher than that of conventional culture while the assay time was less than that of conventional culture methods. Additionally, with its high specificity, IFAG can be used to improve the isolation rate for V. cholerae and efficiency when dealing with a large number of samples, since only IFAG-positive samples are forwarded for further isolation. Also, the isolation rate could be increased by drawing the fluorescent aggregates of bacteria in the IFAG-positive samples, compared to that for conventional culture.
In this study, 22 out of 29 IFAG-positive water samples were positive by subsequent culture of fluorescent aggregates, but for 7 samples, there was a failure to culture V. cholerae from the fluorescent aggregates (Table 1). Among possible reasons for the inability to obtain culture from some aggregates, one may be the presence of viable but nonculturable (VBNC) V. cholerae in the waters. Studies by Binsztein et al (6) suggested that in the interim period between cholera epidemics, V. cholerae is still present in the environment in a VBNC state, indicating that even if fluorescent aggregates of bacteria are detected, no viable bacteria can be obtained by culture. The second reason is that even if fluorescent aggregates were obtained, they may have been suppressed in the gentamicin plate of subsequent culture or no colony could be formed in the gentamicin agar by a single colony due to interference from other bacteria. The third reason may be that the colonies were false positive due to incomplete washings and the presence of nonspecific fluorescence.
The IFAG percentage positive (19.9%) was lower than the real-time PCR positive rate (29.5%). The discordance between the two methods (where the real-time PCR gave a positive result and the IFAG result was negative) was found mainly in samples with higher CT values by real-time PCR detection. At low V. cholerae concentrations, the sensitivity of the assay or the formation of fluorescent aggregates of bacteria may have been affected by the composition of estuarine water after enrichment. Moreover, this study used a small amount (10 μl) of enrichment broth for IFAG detection, and this small sample could possibility result in weak detection.
In environmental monitoring of cholera, a large number of samples is often required. Enrichment, isolation and identification, and conventional culture methods are labor intensive, with many time-consuming steps. This study demonstrated that during handling of samples for real-time fluorescent PCR screening, the real-time PCR-positive samples can be used for isolation with the use of IFAG. This process not only reduces the workload of detection but also improves the isolation rate.
Nontoxigenic V. cholerae was isolated from the estuary water of the Pearl River in this study. Currently China has a smaller number of reported cholera cases each year than before, less than 200 cases during the most recent 3 years. In the year we conducted this study, fewer than 10 cases of cholera were found in the Guangdong province. In the interim period between cholera outbreaks, V. cholerae bacteria present mainly as nontoxigenic strains (20). However, if nontoxigenic strains abnormally increase during environmental monitoring, it would suggest that toxigenic strains may also be increasing under the same environmental conditions, which are suitable for propagation of the pathogen. Therefore, the method described in our study would help to more efficiently accumulate monitoring data for V. cholerae in environmental waters, which can provide a basis for early warnings of cholera outbreaks.
Acknowledgments
This work was supported by grants from the National High Technology Research and Development Program of China (863 Program) (no. 2006AA02Z425), the National Natural Science Foundation of China (no. 30872260), and the National Priority Program for Prevention and Control of Infectious Diseases (no. 2008ZX10004-012).
Footnotes
Published ahead of print on 25 June 2010.
REFERENCES
- 1.Alam, M., M. Sultana, G. B. Nair, R. B. Sack, D. A. Sack, A. K. Siddique, A. Ali, A. Huq, and R. R. Colwell. 2006. Toxigenic Vibrio cholerae in the aquatic environment of Mathbaria, Bangladesh. Appl. Environ. Microbiol. 72:2849-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alam, M., M. Sultana, G. B. Nair, A. K. Siddique, N. A. Hasan, R. B. Sack, D. A. Sack, K. U. Ahmed, A. Sadique, H. Watanabe, C. J. Grim, A. Huq, and R. R. Colwell. 2007. Viable but nonculturable Vibrio cholerae O1 in biofilms in the aquatic environment and their role in cholera transmission. Proc. Natl. Acad. Sci. U. S. A. 104:17801-17806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albert, M. J., A. K. Siddique, M. S. Islam, A. S. Faruque, M. Ansaruzzaman, S. M. Faruque, and R. B. Sack. 1993. Large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet 341:704. [DOI] [PubMed] [Google Scholar]
- 4.Barua, D. 1992. History of cholera, p. 1-5. In D. Barua and W. B. Greenough III (ed.), Cholera. Plenum, New York, NY.
- 5.Bhattacharya, M. K., S. K. Bhattacharya, S. Garg, P. K. Saha, D. Dutta, G. B. Nair, B. C. Deb, and K. P. Das. 1993. Outbreak of Vibrio cholerae non-O1 in India and Bangladesh. Lancet 341:1346-1347. [DOI] [PubMed] [Google Scholar]
- 6.Binsztein, N., M. C. Costagliola, M. Pichel, V. Jurquiza, F. C. Ramirez, R. Akselman, M. Vacchino, A. Huq, and R. Colwell. 2004. Viable but nonculturable Vibrio cholerae O1 in the aquatic environment of Argentina. Appl. Environ. Microbiol. 70:7481-7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackstone, G. M., J. L. Nordstrom, M. D. Bowen, R. F. Meyer, P. Imbro, and A. DePaola. 2007. Use of a real time PCR assay for detection of the ctxA gene of Vibrio cholerae in an environmental survey of Mobile Bay. J. Microbiol. Methods 68:254-259. [DOI] [PubMed] [Google Scholar]
- 8.Brayton, P. R., M. L. Tamplin, A. Huq, and R. R. Colwell. 1987. Enumeration of Vibrio cholerae O1 in Bangladesh waters by fluorescent-antibody direct viable count. Appl. Environ. Microbiol. 53:2862-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakraborty, S., J. Khanam, Y. Takeda, and G. B. Nair. 1999. Application of PCR for detection of toxigenic Vibrio cholerae O1 in water samples during an outbreak of cholera. Trans. R. Soc. Trop. Med. Hyg. 93:527-528. [DOI] [PubMed] [Google Scholar]
- 10.Chun, J., A. Huq, and R. R. Colwell. 1999. Analysis of 16S-23S rRNA intergenic spacer regions of Vibrio cholerae and Vibrio mimicus. Appl. Environ. Microbiol. 65:2202-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colwell, R. R. 1996. Global climate and infectious disease: the cholera paradigm. Science 274:2025-2031. [DOI] [PubMed] [Google Scholar]
- 12.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faruque, S. M., Asadulghani, A. R. Alim, M. J. Albert, K. M. Islam, and J. J. Mekalanos. 1998. Induction of the lysogenic phage encoding cholera toxin in naturally occurring strains of toxigenic Vibrio cholerae O1 and O139. Infect. Immun. 66:3752-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faruque, S. M., M. J. Islam, Q. S. Ahmad, A. S. Faruque, D. A. Sack, G. B. Nair, and J. J. Mekalanos. 2005. Self-limiting nature of seasonal cholera epidemics: role of host-mediated amplification of phage. Proc. Natl. Acad. Sci. U. S. A. 102:6119-6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gil, A. I., V. R. Louis, I. N. Rivera, E. Lipp, A. Huq, C. F. Lanata, D. N. Taylor, E. Russek-Cohen, N. Choopun, R. B. Sack, and R. R. Colwell. 2004. Occurrence and distribution of Vibrio cholerae in the coastal environment of Peru. Environ. Microbiol. 6:699-706. [DOI] [PubMed] [Google Scholar]
- 16.Glass, R. I., and R. E. Black. 1992. The epidemiology of cholera, p. 129-154. In D. Barua and W. B. Greenough III (ed.), Cholera. Plenum, New York, NY.
- 17.Gubala, A. J. 2006. Multiplex real-time PCR detection of Vibrio cholerae. J. Microbiol. Methods 65:278-293. [DOI] [PubMed] [Google Scholar]
- 18.Hasan, J. A., A. Huq, G. B. Nair, S. Garg, A. K. Mukhopadhyay, L. Loomis, D. Bernstein, and R. R. Colwell. 1995. Development and testing of monoclonal antibody-based rapid immunodiagnostic test kits for direct detection of Vibrio cholerae O139 synonym Bengal. J. Clin. Microbiol. 33:2935-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendriksen, C. F., and W. de Leeuw. 1998. Production of monoclonal antibodies by the ascites method in laboratory animals. Res. Immunol. 149:535-542. [PubMed] [Google Scholar]
- 20.Huq, A., R. R. Colwell, M. A. Chowdhury, B. Xu, S. M. Moniruzzaman, M. S. Islam, M. Yunus, and M. J. Albert. 1995. Coexistence of Vibrio cholerae O1 and O139 Bengal in plankton in Bangladesh. Lancet 345:1249. [DOI] [PubMed] [Google Scholar]
- 21.Huq, A., R. B. Sack, A. Nizam, I. M. Longini, G. B. Nair, A. Ali, J. G. Morris, Jr., M. N. Khan, A. K. Siddique, M. Yunus, M. J. Albert, D. A. Sack, and R. R. Colwell. 2005. Critical factors influencing the occurrence of Vibrio cholerae in the environment of Bangladesh. Appl. Environ. Microbiol. 71:4645-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lipp, E. K., I. N. Rivera, A. I. Gil, E. M. Espeland, N. Choopun, V. R. Louis, E. Russek-Cohen, A. Huq, and R. R. Colwell. 2003. Direct detection of Vibrio cholerae and ctxA in Peruvian coastal water and plankton by PCR. Appl. Environ. Microbiol. 69:3676-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lobitz, B., L. Beck, A. Huq, B. Wood, G. Fuchs, A. S. Faruque, and R. Colwell. 2000. Climate and infectious disease: use of remote sensing for detection of Vibrio cholerae by indirect measurement. Proc. Natl. Acad. Sci. U. S. A. 97:1438-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sack, D. A., R. B. Sack, G. B. Nair, and A. K. Siddique. 2004. Cholera. Lancet 363:223-233. [DOI] [PubMed] [Google Scholar]
- 25.Seo, K. H., R. E. Brackett, and J. F. Frank. 1998. Rapid detection of Escherichia coli O157:H7 using immuno-magnetic flow cytometry in ground beef, apple juice, and milk. Int. J. Food Microbiol. 44:115-123. [DOI] [PubMed] [Google Scholar]
- 26.Siddique, A. K., K. Zaman, A. H. Baqui, K. Akram, P. Mutsuddy, A. Eusof, K. Haider, S. Islam, and R. B. Sack. 1992. Cholera epidemics in Bangladesh: 1985-1991. J. Diarrhoeal Dis. Res. 10:79-86. [PubMed] [Google Scholar]
- 27.Wang, X. M., D. C. Wang, H. L. Tan, H. J. Zhong, J. D. Chen, B. S. Li, C. W. Ke, M. Y. Yan, J. Zhang, and B. Kan. 2007. Development and application of real-time polymerase chain reaction to detect Vibrio cholerae O1 and O139 in river water. Chin. J. Epidemiol. 28:768-771. [PubMed] [Google Scholar]
- 28.Xu, X., B. Gu, P. Hu, M. Xi, H. Jin, and C. Chen. 2003. Improved immunofluorescence agglomerate assay in detection of O1 and O139 Vibrio cholerae. Chin. Prev. Med. 4:204-206. [Google Scholar]
- 29.Xu, X., B. Gu, H. Jin, Y. Zheng, M. Chen, and P. Hu. 2007. Application of modified immunofluorescence colonies method to detect Vibrio cholerae in natural river and antibiotic resistance analysis. Chin. J. Health Lab. Technol. 17:320-322. [Google Scholar]