Abstract
A novel strategy for in vivo immobilization of enzymes on the surfaces of inclusion bodies has been established. It relies on expression in Escherichia coli of the polyhydroxybutyrate synthase PhaC from Cupriavidus necator, which carries at its amino terminus an engineered negatively charged α-helical coil (Ecoil) and forms inclusion bodies upon high-level expression. Coexpression in the same cell of galactose oxidase (GOase) from Fusarium spp. carrying a carboxy-terminal positively charged coil (lysine-rich coil [Kcoil]) sequence results in heterodimeric coiled-coil formation in vivo and in the capture of the enzyme in active form on the surface of the inclusion body particle. These round-shaped enzyme-decorated microparticles, with sizes of approximately 0.7 μm, can be isolated from lysed cells simply by centrifugation. The cost-effective one-step generation and isolation of enzymes immobilized on inclusion body particles may become useful for various applications in bioprocessing and biotransformation.
In recent years, significant advances were made in enzyme-mediated synthesis of bulk and fine chemicals. In particular, for the synthesis of compounds having one or more stereo centers, enzymes are the catalysts of choice due to their unprecedented stereoselectivity and substrate specificity (14, 30, 35). For numerous biotransformations, immobilized enzymes that are conjugated to inert carrier particles are used. This strategy has several advantages over direct usage of the biocatalyst, as follows. Rather often, but not always, the enzyme becomes stabilized by surface immobilization, and the activity is retained. Moreover, the enzyme can be removed after the biotransformation reaction and reused. However, enzyme purification and enzyme immobilization remain the main technological hurdles, also in terms of cost efficiency for the development of commercially viable biocatalysts (23). Whole-cell biotransformation is considered to be a viable alternative (13) but has some other intrinsic disadvantages, such as, for example, unwanted side reactions performed by enzymes of the microbial producer, the need to purify the product from microbial metabolites and macromolecules, or permeability issues for the substrate or reaction product with respect to the microbial cell membrane (7).
As a consequence, it would be desirable to have a method available in which the enzyme of interest is synthesized by a microbial host and becomes intracellularly immobilized directly to the surface of a microparticle that is produced by the same cell. The enzyme-decorated particle could then be isolated simply by cell lysis and centrifugation and used for a desired biotransformation process.
Several examples which put that concept into practice that rely on the synthesis of proteins as in-frame fusions to phasins, which are attached to the surfaces of intracellular polymer polyhydroxybutyrate (PHB) granules, have been described recently. By this approach, model proteins could be sequestered to the surfaces of PHB granules (2, 8, 10). A similar approach that also relies on PHB particle synthesis is the C-terminal fusion of the protein of interest to the enzyme PHB synthase PhaC from Cupriavidus necator that is responsible for the polymerization of 3-hydroxyacyl-coenzyme A (CoA) to PHB granules, a process that requires coexpression of two other enzymes, PhaA and PhaB (21). PhaC has been found not only to catalyze polymer synthesis but also to remain covalently attached to the polymer core. It has been demonstrated that various functional proteins can be displayed on the polymer surface and that these protein-decorated polymer beads can be isolated from Escherichia coli and stably maintained outside the cell (1, 11, 15, 17-19, 21-22). However, isolation of PHB particles from E. coli cell lysates is somewhat technically demanding since it requires ultracentrifugation through a glycerol gradient (20) to strip the surface layers of the granules of contaminating proteins and phospholipids.
Here we describe an alternative route to the intracellular production of enzyme-decorated microparticles that relies on the simultaneous synthesis of insoluble protein inclusion bodies and of the enzyme of choice, which becomes tightly attached to the particle surface in multiple copies.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown in double yeast tryptone (dYT) medium at 37°C, with chloramphenicol (25 μg/ml) and/or ampicillin (100 μg/ml) added to preserve maintenance of plasmids. Cells were grown for 3 h in the presence of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for the induction of inclusion body synthesis. Then, enzyme synthesis was induced by addition of 2 μg/ml anhydrotetracycline, and cell growth was continued at 28°C for another 12 to 16 h.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| E. coli strains | ||
| BL21(DE3) | F−ompT gal dcm lon hsdSB(rB− mB−) λ(DE [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) | 26 |
| BL21(DE3)(pACYC-MEP) | Myc-Ecoil-PhaC (cat) | This study |
| BL21(DE3)(pASK75-GKE) | His10-GOase-Kcoil-Etag (bla) | This study |
| BL21(DE3)(pACYC-MEP, pASK75-GKE) | Myc-Ecoil-PhaC His10-GOase-Kcoil-Etag (cat bla) | This study |
| BL21(DE3)(pJM9131) | PhaA PhaB PhaC (Kanr) | This study |
| BL21(DE3)(pET22b-MycPhaC) | Myc-PhaC (bla) | This study |
| Plasmids | ||
| pJM9131 | ColE1 ori δ70 prom PhaA PhaB PhaC Kmr | 31 |
| pET22b-MycPhaC | ColE1 ori T7 prom Myc-PhaC Apr | This study |
| pACYC-MEP | P15A ori T7 prom Myc-Ecoil-PhaC Cmr | This study |
| pASK75-GKE | ColE1 ori tetA prom His10-GOase-Kcoil-Etag Apr | This study |
Vector constructions.
The PhaC coding sequence was amplified from plasmid pJM9131 (2), using PCR primers that also introduce a myc epitope coding sequence, and introduced into pET-22b (Novagen) to give pET22b-MycPhaC. The resulting Myc-PhaC fusion was extended by an engineered negatively charged α-helical coil (Ecoil) coding sequence by PCR and introduced into pACYCDuet-1 (Novagen) to give pACYC-MEP. Vector pASK75-GKE is a derivate of pASK75 (24), which carries the gene for the galactose oxidase (GOase) variant M1 (28), amino-terminally fused to a decahistidine tag and carboxy-terminally fused to an E epitope (GAPVPYPDPLEPR) and a lysine-rich coil (Kcoil) coding sequence. Vector sequences are available upon request.
Isolation of inclusion bodies.
Inclusion bodies were isolated from 1 liter of bacterial liquid culture after mechanical French press disruption of cells and centrifugation at 18,000 × g for 30 min. The precipitated inclusion bodies were resuspended in 30 ml phosphate-buffered saline (PBS) and, after recentrifugation, again suspended in 10 ml phosphate-buffered saline.
Determination of galactose oxidase activity.
An aliquot of 10 μl inclusion body suspension was added to 140 μl of reaction buffer [phosphate-buffered saline, 100 mM α-D-galactose, 1 U/ml horseradish peroxidase, and 10 mM 2,2′-azinobis(3-ethylbenzthiazoline)-6-sulfoninc acid (ABTS)]. Development of the green color was followed by measurement of absorption at 405 nm (9).
Flow cytometry.
For flow cytometric analysis, inclusion bodies were pelleted by centrifugation in a tabletop centrifuge for 3 min and, after resuspension in 10 μl PBS containing 0.1 mg/ml of the respective antibody (anti-myc antibody [Abcam] or anti-E antibody [Abcam]), they were incubated for 10 min on ice. After addition of 190 μl of PBS, inclusion bodies were centrifuged for 3 min and resuspended in 10 μl PBS containing 1:10-diluted biotinylated goat anti-mouse antibody (for myc epitope staining) or fluorescein-coupled goat-anti rabbit antibody (for E epitope staining), followed by incubation for 10 min on ice. After addition of 190 μl of PBS, inclusion bodies were pelleted by centrifugation and resuspended in PBS buffer for flow cytometry. For myc epitope staining, inclusion bodies were resuspended in 10 μl PBS containing streptavidin R-phycoerythrin conjugate (0.1 mg/ml; Invitrogen), followed by incubation for 10 min on ice, addition of 190 μl of PBS, and recentrifugation. Finally, inclusion bodies were resuspended in PBS for flow cytometric analysis. A total of 50,000 to 100,000 events were collected on a Cytomation MoFlo cell sorter. Parameters were set as follows: forward scatter, side scatter, 600 (LIN mode, amplification factor 6); FL1 (fluorescein), 600 (LOG mode); FL2 (phycoerythrin), 600 (LOG mode); trigger parameter, side scatter. The sample flow rate was adjusted to an event rate of approximately 10,000 s−1.
Nile red staining of inclusion bodies and PHB particles.
E. coli BL21(DE3) strains harboring pJM9131 or pACYC-MEP were grown at 37°C in dYT containing 780 μM Nile red (3, 25) Gene expression from the pha operon in pJM9131 and the myc-Ecoil gene-phaC fusion in pACYC-MEP was induced by addition of 1 mM IPTG at an optical density (OD) of 0.5. After 16 h of growth, cells from a 100-μl aliquot were harvested by centrifugation and washed three times by addition of 200 μl PBS. Cells were analyzed by flow cytometry. A total of 100,000 events were collected in a Cytomation MoFlo cell sorter.
SDS-PAGE.
Protein samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). To this end, 12-μl samples were mixed with 5 μl solubilization buffer (8 M urea, 200 mM Tris, 200 mM dithiothreitol [DTT], 2% [wt/vol] SDS) and incubated at 99°C for 10 min. Samples were loaded onto a 12.5% SDS-polyacrylamide gel. Gels were stained with Coomassie brilliant blue R-250.
Transmission electron microscopy.
PHB synthase inclusion bodies were resuspended in phosphate-buffered saline (PBS). Copper grids (400-mesh carbon coated or Formvar coated) were dipped in the inclusion body suspension for 1 min. Excess liquid was removed with filter paper. Samples were observed in a Zeiss EM109 electron microscope. Images were recorded on 70-mm film using a trans-fiber-optic photography (TFP) camera or a Gatan MultiScan 600W camera attached to the microscope. The microscope was operated at an 80-kV accelerating voltage and a total magnification of ×7,000 to 200,000 on the camera.
Fluorescence microscopy.
E. coli BL21(DE3) harboring pACYC-MEP and pASK75-GKE was grown at 37°C in dYT containing 780 μM Nile red. Gene expression of Myc-Ecoil-PhaC was induced by addition of 1 mM IPTG at an optical density of 0.5. After 3 h of growth, gene expression of GOase-Kcoil-(E epitope tag) Etag was induced by addition of 0.2 μg/ml anhydrotetracycline. After 16 h of growth, cells were washed with PBS and used for fluorescence microscopy analysis that was performed with an iMic fluorescence microscope (TILL Photonics).
Production and purification of GOase.
His-tagged GOase-Kcoil-Etag was isolated from 1 liter of bacterial liquid culture after mechanical French press disruption of cells and centrifugation at 18,000 × g for 30 min. GOase enzyme was purified from the supernatant via immobilized metal-ion affinity chromatography and dialyzed in phosphate-buffered saline (PBS).
Determination of inclusion body loading capacity.
Inclusion bodies (0.25 mg and 0.125 mg [wet weight]) were incubated with 120 μl GOase (1.43 μM) in a final volume of 150 μl. After 2-fold centrifugation and being washed, the enzyme load was determined by measuring the GOase activity of 10 μl of a 1:100 dilution of the inclusion body suspension in a total of 150 μl PBS. The specific activity of soluble GOase was determined by measuring the velocity of generation of H2O2 in a coupled assay, using peroxidase and ABTS as substrates (9) and various concentrations of GOase, and compared to the velocity of ABTS oxidation mediated by immobilized GOase. The relative specific activity, expressed as ΔOD405/min·μg enzyme, where OD405 is the optical density at 405 nm, was determined to be 2.5 for soluble GOase. The 0.25-mg and 0.125-mg samples of inclusion bodies displayed the same total volume activity, measured to be 130 ΔOD405/min, which corresponds to approximately 50 μg enzyme. The overall surface load with GOase was calculated to be in the range of 0.2 mg/mg inclusion bodies for the 0.25-mg sample of inclusion bodies and 0.4 mg/mg for the 0.125-mg sample. Since identical GOase activities were measured for both amounts of inclusion bodies, it was concluded that saturation of enzyme binding to inclusion body particles has not been reached yet.
RESULTS
PhaC inclusion body formation.
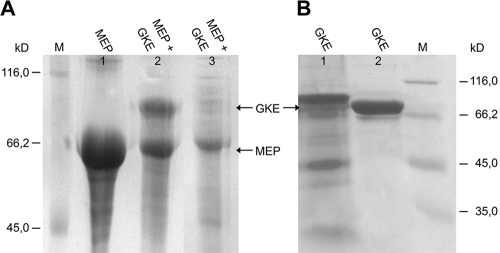
In a attempt to immobilize galactose oxidase on the surface of PHB granules via fusion to PHB synthase PhaC, we made the serendipitous finding that expression under the control of the strong T7 promoter of a modified PhaC that contained an amino-terminal myc epitope tag resulted in the formation of inclusion bodies. These intracellular deposits were undistinguishable by light microscopy from PHB granules generated by simultaneous expression of the entire pha operon encoding PhaA, PhaB, and PhaC from plasmid pJM9131 (data not shown). IPTG-induced E. coli cells transformed with pJM9131 or pET22b-MycPhaC (which encodes a PHB synthase with an amino-terminal myc epitope tag) as well as cells transformed with both plasmids were lysed, and particles were collected by centrifugation and subjected to SDS-gel electrophoresis (Fig. 1). Particles resulting from Myc-PhaC expression alone could be completely dissolved in 8 M urea, which was not the case for PHB granules that remained largely refractive to urea solubilization. While no observable amounts of PhaC protein were detected upon urea-SDS solubilization of PHB granules, large amounts of protein with the expected molecular mass of 65,000 for PhaC accumulated upon expression of Myc-PhaC from pET22b-MycPhaC, both in the presence and in the absence of pJM9131.
FIG. 1.
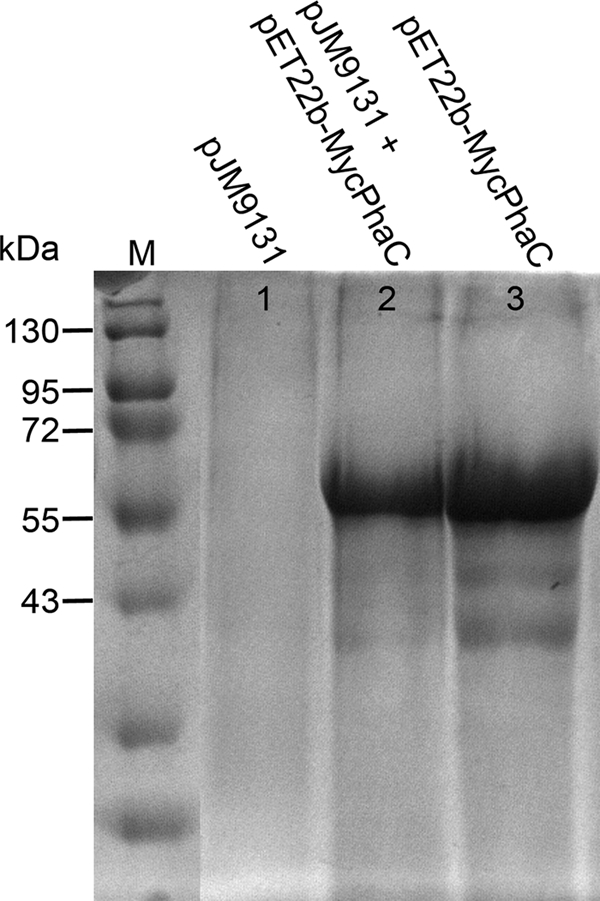
SDS-PAGE of intracellular particles. Particles from overnight cultures of E. coli BL21(DE3) harboring plasmid pJM9131 (lane 1), pET22b-MycPhaC and pJM9131 (lane 2), or pET22b-MycPhaC (lane 3) were solubilized in urea-SDS buffer and analyzed by SDS-PAGE. M, molecular mass standard, with sizes indicated.
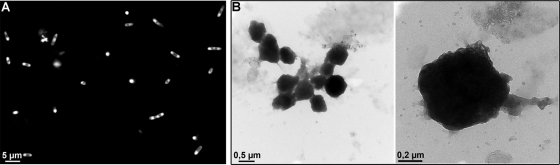
Expression in BL21(DE3) of a Myc-PhaC variant with an amino-terminal extension by a glutamic acid-rich coil sequence from medium-copy-number plasmid pACYC-MEP also resulted in the intracellular accumulation of spherical particles with narrow size distribution and diameters of approximately 0.7 μm (Fig. 2).
FIG. 2.
Fluorescence and electron microscopy of PhaC particles. (A) Fluorescence microscopy of IPTG-induced cells of BL21(DE3) harboring pACYC-MEP and pASK75-GKE stained with Nile red. (B) Electron microscopy of isolated PhaC particles.
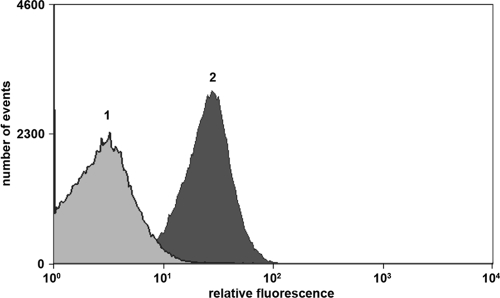
These data indicate that the particles formed upon expression of Myc-PhaC are the result of inclusion body formation rather than of the production of PHB granules staining. E. coli cells producing Myc-PhaC-derived inclusion bodies are indistinguishable from cells containing PHB granules by light microscopy and hard to distinguish by fluorescence microscopy after Nile red staining. However, flow cytometric analysis revealed that the red fluorescent dye Nile red that is commonly used for the staining of PHB granules accumulated to a higher extent in E. coli cells synthesizing PHB granules than in cells producing Myc-PhaC (Fig. 3). Moreover, a PhaC variant was generated that contained a cysteine-to-alanine exchange of the active-site cysteine that is known to be required for catalytic function of the enzyme (22). Compared to wild-type PhaC, this variant displayed no changes in particle accumulation in E. coli, neither with respect to the number of particles per cell nor to particle size or morphology (data not shown). Hence, we conclude that the observed particles are the result of the formation of insoluble inclusion bodies that consist mainly of the PhaC protein.
FIG. 3.
Fluorescence-activated cell sorter (FACS) histogram of Nile red-stained BL21(DE3) cells harboring pACYC-MEP (peak 1) or pJM9131 (peak 2).
Enzyme immobilization on inclusion body particles.
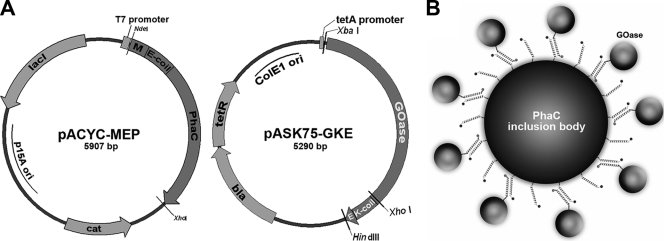
To investigate whether an enzyme can be immobilized on the surface of PhaC inclusion bodies via coiled-coil formation through association with charged α-helical extensions, experiments were conducted using galactose oxidase from Fusarium as a model enzyme. Galactose oxidase M1 has been widely used by others for genetic engineering purposes aimed at optimizing stability and substrate specificity (9, 27-28). To immobilize the enzyme on the surface of the PhaC protein, the protein was endowed with a coiled-coil sequence containing glutamic acid (Ecoil) or lysine residues (Kcoil), which are known to form tight heterodimers with estimated KD (equilibrium dissociation constant) values of 1 nM (6). The Ecoil sequence (EVSALEK)5 was fused to the amino terminus of PhaC, and the Kcoil sequence (KVSALKE)5 was fused to the carboxy terminus of galactose oxidase. Both variant genes were placed under the control of independently inducible promoters and introduced into two different, mutually compatible plasmids to ensure maximal experimental flexibility (Fig. 4). Vector pACYC-MEP contains a medium-copy-number p15A origin and the modified PhaC-encoding sequence fused to an Ecoil under T7 promoter control. Plasmid pASK75-GKE contains a ColE1 replication origin and encodes galactose oxidase M1 with a C-terminally fused E epitope tag and a Kcoil sequence under tetA promoter control. Both plasmids can be maintained simultaneously in E. coli due to compatible origins and different resistance markers.
FIG. 4.
Galactose oxidase display on PhaC inclusion bodies. (A) Schematic outline of expression vectors pACYC-MEP and pASK75-GKE. lacI, gene encoding the lac repressor; M, myc epitope encoding sequence (EQKLISEEDL); PhaC, gene encoding phasin synthase; cat, chloramphenicol resistance gene; GOase, galactose oxidase M1 gene; K, Kcoil-encoding sequence (KVSALKE)5; E, E epitope tag (GAPVPYPDPLEPR); bla, ampicillin resistance gene; tetR, gene encoding the tet repressor. Restriction sites that were used for cloning purposes are indicated. (B) Scheme for immobilization of GOase on the surface of PhaC inclusion bodies via intracellular coexpression and leucine zipper formation.
Expression of Myc-Ecoil-PhaC was induced with IPTG to allow for inclusion body formation, and 3 h later, anhydrotetracycline was added to induce synthesis of the GOase-Kcoil-Etag fusion protein, followed by overnight growth in the presence of both inducers. Cells continued to grow after induction of PhaC and GOase expression. After cell lysis, inclusion bodies and supernatant were investigated by SDS-PAGE. As shown in Fig. 5A, in the inclusion body, a fraction of cells coexpressing two PhaC and GOase protein bands appeared that corresponded to the expected molecular weights of both proteins. The band with an Mr of 78,000, indicative of the expression and accumulation of GOase on inclusion bodies, was absent in cells lacking the GOase expression plasmid. Only minor amounts of PhaC and no GOase were detected in the soluble fraction, which indicated that GOase was nearly completely captured into the PhaC inclusion bodies. However, when produced from pASK75-GKE without coexpression of Myc-Ecoil-PhaC, GOase was found exclusively in the soluble fraction, and no inclusion body formation was detected (Fig. 5B).
FIG. 5.
GOase immobilization on PhaC particles. (A) SDS-PAGE analysis of inclusion body fractions and soluble fraction of BL21(DE3) containing expression plasmids, as follows: lane 1, inclusion body fraction of BL21(DE3) containing pACYC-MEP; lane 2, inclusion body fraction of BL21(DE3) containing both pACYC-1MEP and pASK75-GKE; lane 3, soluble fraction of BL21(DE3) harboring both pACYC-MEP and pASK75-GKE. (B) SDS-PAGE analysis of the inclusion body fraction and soluble fraction of BL21(DE3) containing pASK75-GKE, as follows: lane 1, inclusion body fraction; lane 2, soluble fraction.
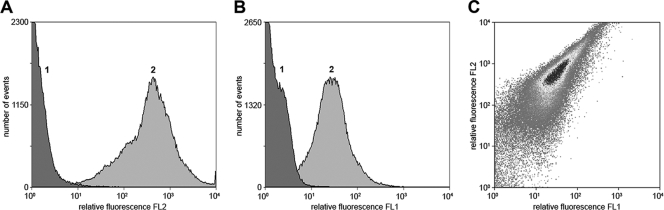
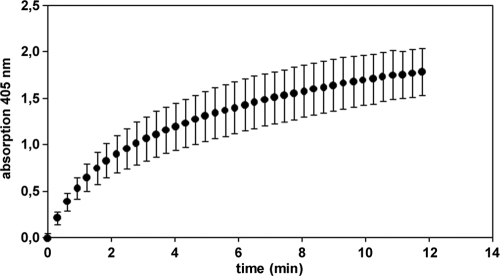
Inclusion bodies from cells containing both expression plasmids were separately stained with anti-myc antibody and anti-E antibody to confirm coexpression of both proteins and accumulation of GOase on the particle surface. Biotinylated anti-myc antibody binding to the myc epitope that was fused to the PhaC N terminus was detected by incubation with streptavidin R-phycoerythrin conjugate (Fig. 6A). Likewise, the E epitope, which is indicative for the presence of surface-exposed GOase that is accessible to the antibody, was detected by successive particle staining using an anti-E antibody and a fluorescein-labeled second antibody, followed by detection by flow cytometry (Fig. 6B). Since a mouse monoclonal antibody was used for myc epitope staining and a rabbit polyclonal antibody was used for E epitope staining, both epitopes could be detected simultaneously without cross-staining, corroborating the colocalization of the PhaC N-terminal Ecoil and of GOase Kcoil fusion on the inclusion body surface (Fig. 6C). Moreover, the isolated inclusion bodies displayed GOase activity (Fig. 7).
FIG. 6.
FACS histograms of inclusion bodies produced by recombinant BL21(DE3) harboring pACYC-MEP and pASK75-GKE. (A) Cell staining without (peak 1) and with (peak 2) anti-myc antibody, followed by incubation with biotinylated anti-mouse immunoglobulin and streptavidin R-phycoerythrin conjugate. (B) Cell staining without (peak 1) and with (peak 2) anti-E antibody, followed by incubation with fluorescein-conjugated anti-rabbit immunoglobulin. (C) Double staining with both antibodies.
FIG. 7.
GOase activity testing of 6 μg (dry weight) inclusion bodies produced by recombinant BL21(DE3) harboring pACYC-MEP and pASK75-GKE. Formation of hydrogen peroxide was detected after addition of galactose in a coupled reaction followed by the horseradish peroxidase-mediated oxidation at 405 nm of ABTS by H2O2. Error bars show the standard deviations from three independent measurements.
GOase expression and particle loading capacity.
Enzyme activity of cells producing GOase alone was compared to the activity of cells synthesizing the enzyme together with inclusion bodies. Simultaneous synthesis of PhaC inclusion bodies and GOase resulted in a 35% reduction of GOase activity (data not shown), which indicates that target enzyme synthesis was slightly impaired. By measuring the GOase activity of a defined amount of inclusion bodies that were loaded with saturating amounts of GOase and under the assumption that the specific activity of the enzyme remains unchanged upon particle binding, one can estimate the binding capacity of the particles for GOase to be at least 200 mg (wet weight) of inclusion bodies per g.
DISCUSSION
We have established a method for the direct immobilization of an enzyme to inclusion body particles that are formed in E. coli. Inclusion bodies are widely used for recombinant protein production. Little is known about the structural features of proteins that form intracellular deposits. They have been found to bind amyloid-diagnostic dyes, which indicates a high level of organized β-sheet structure (5, 32-33). Bacterial inclusion bodies have been found to have some amyloid-like properties, suggesting that they might contain structures similar to those contained in amyloid-like fibrils (32).
Inclusion bodies are produced by many different proteins and differ from each other with respect to size, shape, and morphology (4, 32). The inclusion bodies described here consist of the PhaC protein from Cupriavidus necator; they are round shaped and have a narrow size distribution. They can be easily purified from E. coli cell lysates via centrifugation and have a low content of contaminating host proteins. They can be dispersed in aqueous buffers and show no unwanted aggregation behavior or stickiness to surfaces, which may at least in part be due to the presence of a large number of acidic coils that have been introduced via N-terminal fusion to PhaC and give rise to a charged particle surface. Antibody titration to detect surface-exposed myc epitopes (34) flanking the coil region indicates that several hundred thousand coils reside on the surface of a single particle (data not shown). This acidic helical structure can be used to capture an enzyme of interest that is coexpressed in the same host cells and contains a basic coil fused to its C terminus (6, 29), as exemplified in this study with the enzyme galactose oxidase. Since both coils of the heterodimeric α-helical coiled coil are held together both by hydrophobic interactions of a leucine zipper and by electrostatic interaction of the lysine and glutamate residues of the helical backbone, a tight interaction results, with a KD in the nanomolar range (6). Moreover, cysteine residues were placed at the C terminus of each coil, aimed at allowing the formation of an intermolecular disulfide bond in an environment where oxidizing conditions prevail (36). To what extent these disulfide bonds are formed during isolation and storage of inclusion bodies remains to be determined.
Expression of a protein of interest in a cellular environment, where large amounts of inclusion bodies are produced, carries two inherent possible risks. First, the yield of synthesis of the soluble protein might be strongly reduced in the presence of concomitant large-scale production of inclusion bodies due to overload of the cellular protein synthesis machinery. Second, protein synthesis in the presence of inclusion bodies may result in unwanted inclusion body formation of the enzyme under consideration, since the inclusion body-forming unfolded protein might interact with the newly synthesized enzyme via intermolecular hydrophobic interaction and lead to formation of denatured and aggregated target enzyme. Only a slight reduction of cellular GOase activity was observed upon coexpression of PhaC inclusion bodies, which indicates that the enzyme is produced in active form in the presence of inclusion bodies. This finding may at least in part be due to the fact that inclusion body production and enzyme synthesis are regulated separately, since expression of PhaC is under T7 promoter control and synthesis of GOase is under the control of the tetA promoter/operator. Hence, synthesis of the target enzyme could be induced several hours after the start of inclusion body production. No efforts have been made yet to optimize the time points of induction or the offset of induction of expression of PhaC inclusion bodies and the target enzyme to be captured.
To our knowledge, this is the first description of the usage of inclusion bodies for intracellular capture of a coexpressed target enzyme. Other strategies for enzyme display on intracellular particles, such as, e.g., that used by polyhydroxyalkanoates, rely on the synthesis of the protein of interest as a fusion protein with proteins that are members of the PHB synthesis machinery or attach to the particle surface, which may result in comparably low surface loads (1, 11, 16). In contrast, the loading capacity of the proteinaceous inclusion body particles used here for GOase exceeds 200 mg (wet weight) of inclusion bodies per g. Hence, the formation of particle-immobilized enzymes may be limited by the expression yield of the enzyme moiety itself rather than by the binding capacity of the inclusion bodies.
Simple cell disruption and centrifugation are sufficient to obtain relatively pure enzyme-decorated particle preparations that may become a cost-efficient alternative to whole-cell biotransformation. We have experimental evidence that this enzyme-capturing strategy can be extended to other enzymes of biotechnological relevance, such as, e.g., monoamine oxidases. It will be interesting to see whether this scheme can be further expanded to the simultaneous in vivo immobilization of multiple different enzymes that are commonly being used in whole-cell biotransformation, such as, e.g., cofactor dependent enzymes like ketoreductases, together with enzymes that are required for cofactor regeneration (12).
Acknowledgments
This work was supported in part by Deutsche Forschungsgemeinschaft through grant SPP1170.
We thank Karin Faist, Institute for Microbiology and Genetics, TU Darmstadt, for help with transmission electron microscopy studies; Nico Herdergott, Institute for Microbiology and Genetics, TU Darmstadt, for help with fluorescence microscopy; and Nicholas J. Turner, School of Chemistry, University of Manchester, for providing the GOase M1 coding sequence.
Footnotes
Published ahead of print on 25 June 2010.
This paper is dedicated to Professor Hans-Joachim Fritz on the occasion of his retirement.
REFERENCES
- 1.Atwood, J. A., and B. H. Rehm. 2009. Protein engineering towards biotechnological production of bifunctional polyester beads. Biotechnol. Lett. 31:131-137. [DOI] [PubMed] [Google Scholar]
- 2.Barnard, G. C., J. D. McCool, D. W. Wood, and T. U. Gerngross. 2005. Integrated recombinant protein expression and purification platform based on Ralstonia eutropha. Appl. Environ. Microbiol. 71:5735-5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Betscheider, D., and J. Jose. 2009. Nile blue A for staining Escherichia coli in flow cytometer experiments. Anal. Biochem. 384:194-196. [DOI] [PubMed] [Google Scholar]
- 4.Bowden, G. A., A. M. Paredes, and G. Georgiou. 1991. Structure and morphology of protein inclusion bodies in Escherichia coli. Biotechnology (NY) 9:725-730. [DOI] [PubMed] [Google Scholar]
- 5.Carrio, M., N. Gonzalez-Montalban, A. Vera, A. Villaverde, and S. Ventura. 2005. Amyloid-like properties of bacterial inclusion bodies. J. Mol. Biol. 347:1025-1037. [DOI] [PubMed] [Google Scholar]
- 6.Chao, H., D. L. Bautista, J. Litowski, R. T. Irvin, and R. S. Hodges. 1998. Use of a heterodimeric coiled-coil system for biosensor application and affinity purification. J. Chromatogr. B Biomed. Sci. Appl. 715:307-329. [DOI] [PubMed] [Google Scholar]
- 7.Chen, R. R. Z. 2007. Permeability issues in whole-cell bioprocesses and cellular membrane engineering. Appl. Microbiol. Biotechnol. 74:730-738. [DOI] [PubMed] [Google Scholar]
- 8.de Almeida, A., P. I. Nikel, A. M. Giordano, and M. J. Pettinari. 2007. Effects of granule-associated protein PhaP on glycerol-dependent growth and polymer production in poly(3-hydroxybutyrate)-producing Escherichia coli. Appl. Environ. Microbiol. 73:7912-7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Escalettes, F., and N. J. Turner. 2008. Directed evolution of galactose oxidase: generation of enantioselective secondary alcohol oxidases. Chembiochem 9:857-860. [DOI] [PubMed] [Google Scholar]
- 10.Gillies, A. R., R. B. Mahmoud, and D. W. Wood. 2009. PHB-intein-mediated protein purification strategy. Methods Mol. Biol. 498:173-183. [DOI] [PubMed] [Google Scholar]
- 11.Grage, K., A. C. Jahns, N. Parlane, R. Palanisamy, I. A. Rasiah, J. A. Atwood, and B. H. Rehm. 2009. Bacterial polyhydroxyalkanoate granules: biogenesis, structure, and potential use as nano-/micro-beads in biotechnological and biomedical applications. Biomacromolecules 10:660-669. [DOI] [PubMed] [Google Scholar]
- 12.Huisman, G. W., J. Liang, and A. Krebber. 2010. Practical chiral alcohol manufacture using ketoreductases. Curr. Opin. Chem. Biol. 14:122-129. [DOI] [PubMed] [Google Scholar]
- 13.Ishige, T., K. Honda, and S. Shimizu. 2005. Whole organism biocatalysis. Curr. Opin. Chem. Biol. 9:174-180. [DOI] [PubMed] [Google Scholar]
- 14.Meyer, H. P., and N. J. Turner. 2009. Biotechnological manufacturing options for organic chemistry. Mini-Rev. Org. Chem. 6:300-306. [Google Scholar]
- 15.Mifune, J., K. Grage, and B. H. Rehm. 2009. Production of functionalized biopolyester granules by recombinant Lactococcus lactis. Appl. Environ. Microbiol. 75:4668-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moldes, C., P. Garcia, J. L. Garcia, and M. A. Prieto. 2004. In vivo immobilization of fusion proteins on bioplastics by the novel tag BioF. Appl. Environ. Microbiol. 70:3205-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parlane, N. A., D. N. Wedlock, B. M. Buddle, and B. H. Rehm. 2009. Bacterial polyester inclusions engineered to display vaccine candidate antigens for use as a novel class of safe and efficient vaccine delivery agents. Appl. Environ. Microbiol. 75:7739-7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peters, V., D. Becher, and B. H. Rehm. 2007. The inherent property of polyhydroxyalkanoate synthase to form spherical PHA granules at the cell poles: the core region is required for polar localization. J. Biotechnol. 132:238-245. [DOI] [PubMed] [Google Scholar]
- 19.Peters, V., and B. H. Rehm. 2006. In vivo enzyme immobilization by use of engineered polyhydroxyalkanoate synthase. Appl. Environ. Microbiol. 72:1777-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Potter, M., H. Muller, F. Reinecke, R. Wieczorek, F. Fricke, B. Bowien, B. Friedrich, and A. Steinbuchel. 2004. The complex structure of polyhydroxybutyrate (PHB) granules: four orthologous and paralogous phasins occur in Ralstonia eutropha. Microbiology 150:2301-2311. [DOI] [PubMed] [Google Scholar]
- 21.Rehm, B. H. 2007. Biogenesis of microbial polyhydroxyalkanoate granules: a platform technology for the production of tailor-made bioparticles. Curr. Issues Mol. Biol. 9:41-62. [PubMed] [Google Scholar]
- 22.Rehm, B. H., R. V. Antonio, P. Spiekermann, A. A. Amara, and A. Steinbuchel. 2002. Molecular characterization of the poly(3-hydroxybutyrate) (PHB) synthase from Ralstonia eutropha: in vitro evolution, site-specific mutagenesis and development of a PHB synthase protein model. Biochim. Biophys. Acta 1594:178-190. [DOI] [PubMed] [Google Scholar]
- 23.Sheldon, R. A. 2007. Enzyme immobilization: the quest for optimum performance. Adv. Synth. Catal. 349:1289-1307. [Google Scholar]
- 24.Skerra, A. 1994. Use of the tetracycline promoter for the tightly regulated production of a murine antibody fragment in Escherichia coli. Gene 151:131-135. [DOI] [PubMed] [Google Scholar]
- 25.Spiekermann, P., B. H. A. Rehm, R. Kalscheuer, D. Baumeister, and A. Steinbuchel. 1999. A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch. Microbiol. 171:73-80. [DOI] [PubMed] [Google Scholar]
- 26.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 27.Sun, L. H., T. Bulter, M. Alcalde, I. P. Petrounia, and F. H. Arnold. 2002. Modification of galactose oxidase to introduce glucose 6-oxidase activity. Chembiochem 3:781-783. [DOI] [PubMed] [Google Scholar]
- 28.Sun, L. H., I. P. Petrounia, M. Yagasaki, G. Bandara, and F. H. Arnold. 2001. Expression and stabilization of galactose oxidase in Escherichia coli by directed evolution. Protein Eng. 14:699-704. [DOI] [PubMed] [Google Scholar]
- 29.Tripet, B., L. Yu, D. L. Bautista, W. Y. Wong, R. T. Irvin, and R. S. Hodges. 1996. Engineering a de novo-designed coiled-coil heterodimerization domain off the rapid detection, purification and characterization of recombinantly expressed peptides and proteins. Protein Eng. 9:1029-1042. [DOI] [PubMed] [Google Scholar]
- 30.Turner, N. J. 2009. Directed evolution drives the next generation of biocatalysts. Nat. Chem. Biol. 5:567-573. [DOI] [PubMed] [Google Scholar]
- 31.Valentin, H. E., and A. Steinbuchel. 1994. Application of enzymatically synthesized short-chain-length hydroxy fatty acid coenzyme A thioesters for assay of polyhydroxyalkanoic acid synthases. Appl. Microbiol. Biotechnol. 40:699-709. [Google Scholar]
- 32.Wang, L., S. K. Maji, M. R. Sawaya, D. Eisenberg, and R. Riek. 2008. Bacterial inclusion bodies contain amyloid-like structure. PLoS Biol. 6:1791-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wasmer, C., L. Benkemoun, R. Sabate, M. O. Steinmetz, B. Coulary-Salin, L. Wang, R. Riek, S. J. Saupe, and B. H. Meier. 2009. Solid-state NMR spectroscopy reveals that E. coli inclusion bodies of HET-s(218-289) are amyloids. Angew. Chem. Int. Ed. Engl. 48:4858-4860. [DOI] [PubMed] [Google Scholar]
- 34.Wentzel, A., A. Christmann, T. Adams, and H. Kolmar. 2001. Display of passenger proteins on the surface of Escherichia coli K-12 by the enterohemorrhagic E. coli intimin EaeA. J. Bacteriol. 183:7273-7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wohlgemuth, R. 2009. The locks and keys to industrial biotechnology. New Biotechnol. 25:204-213. [DOI] [PubMed] [Google Scholar]
- 36.Zhou, N. E., C. M. Kay, and R. S. Hodges. 1993. Disulfide bond contribution to protein stability: positional effects of substitution in the hydrophobic core of the two-stranded alpha-helical coiled-coil. Biochemistry 32:3178-3187. [DOI] [PubMed] [Google Scholar]