Abstract
Acidilobus saccharovorans is an anaerobic, organotrophic, thermoacidophilic crenarchaeon isolated from a terrestrial hot spring. We report the complete genome sequence of A. saccharovorans, which has permitted the prediction of genes for Embden-Meyerhof and Entner-Doudoroff pathways and genes associated with the oxidative tricarboxylic acid cycle. The electron transfer chain is branched with two sites of proton translocation and is linked to the reduction of elemental sulfur and thiosulfate. The genomic data suggest an important role of the order Acidilobales in thermoacidophilic ecosystems whereby its members can perform a complete oxidation of organic substrates, closing the anaerobic carbon cycle.
Acidophilic microorganisms are widely dispersed in natural acidic environments, including volcanic hot springs, and are, in the majority, aerobes (14). However, such anoxic, high-temperature, acidic environments are inhabited by metabolically versatile anaerobic thermoacidophiles of the archaeal phylum Crenarchaeota. Lithoautotrophic thermoacidophiles oxidize molecular hydrogen in the course of elemental sulfur (S0) respiration. Organotrophs couple the oxidation of organic substrates to the reduction of S0 or thiosulfate. They all belong to the genus Acidilobus in the family Acidilobaceae and to the genus Caldisphaera in the family Caldisphaeraceae (4, 13, 22, 24). Acidilobaceae and Caldisphaeraceae form the crenarchaeal order Acidilobales (24). Acidilobus saccharovorans was isolated from an acidic hot spring of Uzon Caldera, Kamchatka, Russia (24). It is an obligately anaerobic acidophile with a range of growth from pH 2.5 to 5.8 (optimum at pH 3.5 to 4) and a temperature range from 60 to 90°C (optimum at 80 to 85°C). It utilizes a wide range of proteinaceous and carbohydrate substrates and cannot grow lithoautotrophically on H2 and CO2 (24). S0 and thiosulfate stimulate growth and are reduced to H2S. Protons cannot serve as electron acceptors, since no H2 is produced during growth in the absence of S0 (24). Genomic sequences of aerobic, thermoacidophilic euryarchaea Thermoplasma acidophilum (26) and Picrophilus torridus (8) give an insight into the thermoacidophilic survival strategy. However, no genomes of obligately anaerobic, thermoacidophilic archaea were available until now. Here we present the genome of A. saccharovorans and show that it encodes numerous hydrolytic enzymes and metabolic pathways necessary for the utilization and complete mineralization of organic substrates in its natural habitat, acidic hot springs.
Genome sequencing and annotation.
A. saccharovorans 345-15T (DSM 16705) was obtained from the culture collection of the Laboratory of Hyperthermophilic Microbial Communities, the Winogradsky Institute of Microbiology of the Russian Academy of Sciences, and grown as described previously (24). The A. saccharovorans genome was sequenced on a Roche GS FLX genome sequencer by the standard protocol for a shotgun genome library. The GS FLX run (1/4 plate) resulted in the generation of about 22 Mb of sequences with an average read length of 230 bp. The GS FLX reads were assembled into 11 contigs by a GS De Novo Assembler (Roche). The contigs were oriented into scaffolds, and the complete genome sequence was obtained upon the generation and sequencing of appropriate PCR fragments. The rRNA genes were identified with RNAmmer (17). tRNA genes were located with tRNAscan-SE (19). Protein-encoding genes were identified with the GLIMMER gene finder (5), followed by a round of manual curation. Genome annotation was performed with the AutoFACT tool (16). Protein similarity was analyzed by BLASTP search (1). Signal peptides were predicted with SignalP version 3.0 (3).
General features of the genome.
A. saccharovorans has a single circular chromosome of 1,496,453 bp with no extrachromosomal elements (see Fig. S1 in the supplemental material). There is a single copy of the 16S-23S rRNA operon and a single distantly located 5S rRNA gene. A total of 45 tRNA genes coding for all 20 amino acids are scattered over the genome. The cumulative GC skew profile (9, 18) displays two local minima that likely correspond to the DNA replication origins (see Fig. S1 in the supplemental material). Genome annotation reveals 1,499 potential protein-encoding genes, 972 (65%) of which can be functionally assigned. The functions of the remaining 527 genes cannot be predicted from the deduced amino acid sequences; 246 of them are unique to A. saccharovorans.
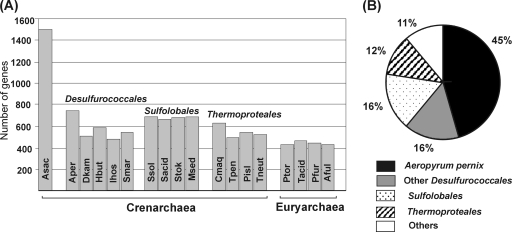
The numbers of predicted protein-encoding genes of A. saccharovorans that have homologs in other archaeal genomes are shown in Fig. 1 A. The closest relative of A. saccharovorans is the aerobic, neutrophilic crenarchaeon Aeropyrum pernix; they share about half of their proteomes (∼740 proteins). Fewer A. saccharovorans proteins have homologs in the genomes of other members of the crenarchaeal orders Desulfurococcales and Thermoproteales (Fig. 1B). Notably, A. saccharovorans shares higher numbers of proteins (661 to 687, 16% of best BLASTP hits) with the phylogenetically distant aerobic thermoacidophiles of the order Sulfolobales (24). Predicted functions of most of the 199 proteins of A. saccharovorans showing the best BLASTP hits in Sulfolobales proteomes are related to transport (54 proteins) or are unknown (86 proteins). A. saccharovorans and Sulfolobales often coexist in acidic hot springs, and lateral gene transfer may be responsible for the relatively high similarity between their proteomes. Extensive lateral gene transfer was suggested to explain the high fraction of genes shared by Sulfolobales and aerobic euryarchaeal thermoacidophiles P. torridus and T. acidophilum (8, 26). However, it is unlikely that extensive gene transfer occurred between A. saccharovorans and euryarchaeal thermoacidophiles, because the numbers of proteins shared with P. torridus (428) and T. acidophilum (459), as well as with nonacidophilic euryarchaea Pyrococcus furiosus (440) and Archaeoglobus fulgidus (427), are similar.
FIG. 1.
Comparisons of proteomes of A. saccharovorans and other archaea. (A) The numbers of A. saccharovorans protein-encoding genes present in the genomes of other archaea. Genes were considered present in a pair of genomes if the region of similarity covered >70% of the corresponding protein with an E value of <10−10. Abbreviations: Asac, A. saccharovorans; Aper, Aeropyrum pernix; Dkam, Desulfurococcus kamchatkensis; Hbut, Hyperthermus butylicus; Ihos, Ignicoccus hospitalis; Smar, Staphylothermus marinus; Ssol, Sulfolobus solfataricus; Sacid, Sulfolobus acidocaldarius; Stok, Sulfolobus tokodaii; Msed, Metallosphaera sedula; Cmaq, Caldivirga maquilingensis; Tpen, Thermofilum pendens; Pisl, Pyrobaculum islandicum; Tneut, Thermoproteus neutrophilus; Ptor, Picrophilus torridus; Tacid, Thermoplasma acidophilum; Pfur, Pyrococcus furiosus; Aful, Archaeoglobus fulgidus. (B) The best BLASTP hits of A. saccharovorans proteins. Significant BLASTP hits were detected for 1,218 of 1,499 proteins found in the A. saccharovorans genome. Proteins unique to A. saccharovorans were excluded from this analysis.
Metabolism of proteins, carbohydrates, and lipids.
Growth of A. saccharovorans on proteins and peptides may be enabled by the function of several extracellular proteolytic enzymes. The imported peptides can be fermented with concomitant substrate-level phosphorylation to generate ATP (Fig. 2; see Table S1 in the supplemental material). The presence of genes for extra- and intracellular glycoside hydrolases and sugar transporters (Fig. 2 and Table S1) correlates with the growth of A. saccharovorans on carbohydrates (24). Notably, the A. saccharovorans genome encodes at least two primary ABC-type sugar transport systems (Table S1) and more than 20 secondary transporters. A similar high ratio of secondary transport systems to ATP-consuming primary transport systems was found in the thermoacidophile P. torridus (8), suggesting that the high external proton concentration is extensively used by A. saccharovorans for transport processes.
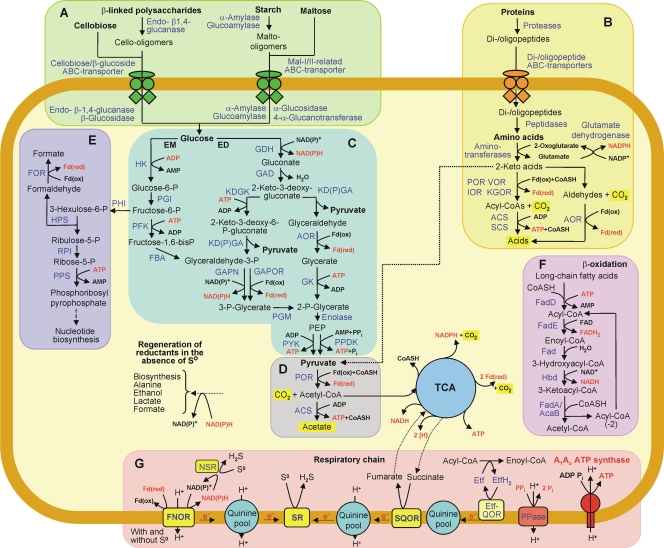
FIG. 2.
Overview of catabolic pathways encoded by the A. saccharovorans genome. Substrates utilized are in boldface, enzymes and proteins encoded by genes identified on the genome are in blue, and energy-rich intermediate compounds are in red. Panels: A, utilization of carbohydrates; B, utilization of proteins; C, glycolysis (EM and ED pathways); D, pyruvate degradation; E, pentose phosphate synthesis; F, β-oxidation of long-chain fatty acids; G, formation of proton motive force coupled with ATP generation. TCA, oxidative tricarboxylic acid cycle. Abbreviations: POR, pyruvate:ferredoxin oxidoreductase; VOR, 2-ketoisovalerate:ferredoxin oxidoreductase; IOR, indolepyruvate:ferredoxin oxidoreductase; KGOR, 2-ketoglutarate:ferredoxin oxidoreductase; ACS, acetyl-CoA synthetase; SCS, succinyl-CoA synthetase; AOR, aldehyde:ferredoxin oxidoreductases; HK, ADP-dependent hexokinase; PGI, phosphoglucose isomerase; PFK, ADP-dependent phosphofructokinase; FBA, fructose-1,6-bisphosphate aldolase; GAPOR, glyceraldehyde-3-phosphate:ferredoxin oxidoreductase; GAPN, nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase; PGM, phosphoglycerate mutase; PYK, pyruvate kinase; PPDK, pyruvate phosphate dikinase; PEP, phosphoenolpyruvate; GDH, glucose dehydrogenase; GAD, gluconate dehydratase; KDGK, 2-keto-3-deoxy-gluconate kinase; KD(P)GA, 2-keto-3-deoxy-(6-phospho)gluconate aldolase; GK, glycerate kinase; PHI, 6-phospho-3-hexuloisomerase; HPS, 3-hexulose-6-phosphate synthase; RPI, ribose-5-phosphate isomerase; PPS, phosphoribosyl pyrophosphate synthase; FOR, formaldehyde:ferredoxin oxidoreductase; Fd(ox), oxidized ferredoxin; Fd(red), reduced ferredoxin; FNOR, ferredoxin:NAD(P)+ oxidoreductase complex; NSR, NAD(P)H:elemental sulfur oxidoreductase; PPase, H+-translocating pyrophosphatase; SQOR, succinate:quinone oxidoreductase complex; SR, sulfur reductase complex; Etf, electron transfer flavoprotein; Etf-QOR, electron transfer protein-quinone oxidoreductase; CoASH, coenzyme A; FAD, flavin adenine dinucleotide; FADH2, reduced FAD.
Glucose in archaea is metabolized to pyruvate via either the Embden-Meyerhof (EM) pathway or the Entner-Doudoroff (ED) pathway. Both modified EM and ED pathways are encoded by A. saccharovorans genome (Fig. 2), suggesting their operation in parallel as found in Thermoproteus tenax (32). The ED pathway can be beneficial at the upper temperature range of growth, since it avoids the most heat-labile intermediates (31). The EM pathway may account for the synthesis of fructose-6-phosphate used in the ribulose monophosphate pathway (Fig. 2).
The genome contains 14 genes encoding esterases and genes encoding a complete β-oxidation pathway, suggesting the ability of A. saccharovorans to utilize triacylglycerides and long-chain fatty acids available from the environment (Fig. 2 and Table S1). To our knowledge, besides A. saccharovorans, A. fulgidus is the only archaeon to encode the enzymes of β-oxidation (15).
Conservation of energy.
A. saccharovorans may conserve energy by substrate-level phosphorylation as well as by oxidative phosphorylation. During growth in the presence of S0 as a terminal electron acceptor, H2S is produced and acetate is formed (24). It is likely that in the presence of S0, acetyl coenzyme A (acetyl-CoA) generated from the oxidation of substrates is concomitantly converted to acetate by fermentation pathway as well as oxidized to CO2 by anaerobic respiration (Fig. 2). The A. saccharovorans genome encodes all eight enzymes of the oxidative tricarboxylic acid (TCA) cycle (Fig. 2 and Table S1). This cycle operates in T. tenax and Pyrobaculum islandicum and was proposed to function in both oxidative and reductive directions, enabling the complete oxidation of organic substrates to CO2 and H2S, as well as CO2 fixation during autotrophic growth (12, 30, 31, 32). In A. saccharovorans, which cannot grow autotrophically (24), the TCA cycle appears to operate only in the oxidative direction. Indeed, citrate lyase, the key enzyme of the reductive TCA cycle, is not encoded.
During growth of A. saccharovorans by anaerobic respiration in the presence of S0, reduced ferredoxin, NAD(P)H, and H produced in the TCA cycle can be oxidized by a concerted action of a set of membrane-bound protein complexes and cytoplasmic proteins, resulting in the generation of transmembrane proton gradient. The genome analysis reveals a putative membrane-bound protein complex (ASAC_0373 to ASAC_0383), which is partially homologous to bacterial NADH:quinone oxidoreductase (respiratory complex I). From the 14 subunits (NuoA to NuoN) present in Escherichia coli complex I (7), this putative A. saccharovorans complex contains 11 subunits (NuoA to NuoD and NuoH to NuoN), while three subunits (NuoEFG) known to form the dehydrogenase domain involved in NADH binding and oxidation (28) are missing. Thus, the A. saccharovorans complex most likely accepts electrons from a donor other than NADH. ASAC_0380 is also homologous to the MbxL subunit of the membrane-bound oxidoreductase (MBX) complex from P. furiosus (29). It has been shown that in the MBX complex, the oxidation of ferredoxin and the reduction of NAD(P)+ are coupled to the generation of a proton motive force in the presence of S0 (29). A similar function of the MBX complex in the crenarchaea D. kamchatkensis (25) and S. marinus (2), as well as in the euryarchaeon T. sibiricus (20), has been proposed. Based on these data, we propose that ASAC_0373-ASAC_0383 encodes a proton-translocating ferredoxin:NAD(P)+ oxidoreductase complex (FNOR) which accepts electrons from reduced ferredoxin and transfers them to NAD(P)+, thereby translocating protons and establishing a proton gradient (Fig. 2). NAD(P)H is then oxidized by the encoded NAD(P)H:elemental sulfur oxidoreductase (NSR) (ASAC_1028), forming H2S, or enters other processes linked to NAD(P)H reoxidation (e.g., thiosulfate reduction). In addition, the NuoB (Nqo6) subunit, which transfers electrons to menaquinone in Thermus thermophilus complex I (27), is present in the A. saccharovorans FNOR complex (ASAC_0382), suggesting the possibility of electron transfer not only to NAD(P)+ but also to quinones in the respiratory chain and further to membrane-bound sulfur reductase (SR; ASAC_1394 to ASAC_1397) (Fig. 2; see Table S1 in the supplemental material).
The transmembrane proton gradient could also be generated by the function of the predicted integral membrane protein H+-translocating pyrophosphatase (PPase; ASAC_1013) closely related to functionally characterized enzyme from Pyrobaculum aerophilum (6). The pyrophosphatase may work in concert with H+-ATP synthase to scavenge energy from biosynthetic waste pyrophosphate in order to maintain the proton gradient, especially under energy-limiting conditions.
Besides involving the proposed FNOR complex, the respiratory chain of A. saccharovorans presumably involves the succinate:quinone oxidoreductase complex (SQOR; ASAC_1440 to ASAC_1443), which is the membrane-bound component of the oxidative TCA cycle. Electrons from the oxidation of succinate in the TCA cycle would be delivered by SQOR to the quinones in the membrane.
The genome contains a gene cluster (corresponding to ASAC_0264 to ASAC_0267) encoding two subunits of electron transfer flavoprotein (EtfAB; ASAC_0264 and ASAC_0265, respectively), a ferredoxin-like protein (ASAC_0266), and an electron transfer flavoprotein-quinone oxidoreductase (Etf-QOR; ASAC_0267). In analogy to the mitochondrial enzymes, ASAC_0264 to ASAC_0267 might be involved in the oxidation of fatty acids and some amino acids (10). These substrates would be oxidized by acyl-CoA dehydrogenases (FadE) with the Etf as an acceptor, which would pass the electrons to Etf-QOR and further to quinone in the respiratory chain of A. saccharovorans.
The genome analysis also suggests a second site of S0 reduction in A. saccharovorans. It involves a predicted membrane-bound SR complex, which would function as the terminal oxidase of S0-dependent respiration. Based on the sequence analyses, we propose that the SR is composed of the hydrophilic catalytic subunit ASAC_1397, the electron transfer subunit ASAC_1396, and the membrane-bound subunits ASAC_1395 and ASAC_1394.
Summarizing the data on the electron transfer in A. saccharovorans, we propose that its respiratory chain functioning during the growth with S0 is branched (Fig. 2). First, the electrons from reduced ferredoxin could be utilized by FNOR and NSR. It would be linked to proton translocation and the first site of S0 reduction. Second, a part of the electrons from FNOR, as well as the electrons from SQOR and Etf-QOR, would be transferred to the quinone pool. Oxidation of quinol by the SR would also be linked to proton translocation and the second site of reduction of S0. During growth in the absence of S0, the TCA cycle is not functional and the respiratory chain involving the SQOR complex, quinones, and SR seems not to operate. However, a proton gradient still could be established by the oxidation of reduced ferredoxin in the reaction of the proposed FNOR complex linked to reoxidation of NAD(P)H in reactions yielding reduced fermentation products.
Some anaerobic Crenarchaeota were reported to use in addition to S0 other terminal electron acceptors, including sulfate, thiosulfate, and nitrate. The presence of thiosulfate stimulated the growth of A. saccharovorans and led to production of H2S (24). We have predicted the function of ASAC_1397 as a component of membrane-bound SR complex (see above). Nevertheless, in analogy with thiosulfate reductase of Salmonella enterica involved also in the reduction of S0 (11), the dual function of ASAC_1397 in the reduction of S0 as well as of thiosulfate can be suggested.
The established proton gradient can be used by ATP synthase for the synthesis of ATP. The genome of A. saccharovorans encodes a single A1Ao ATP synthase composed of nine subunits (Table S1). The subunit K (c) (ASAC_0393) of the membrane-embedded motor does not contain conserved residues involved in Na+ binding in Na+-ATP synthases (21), strongly suggesting that H+ is the coupling ion in the A1Ao ATP synthase. During growth of A. saccharovorans in the absence of S0, the required proton gradient could be established only partially by the function of the FNOR complex and H+-translocating PPase. An additional proton gradient required for maintenance of normal intracellular pH could be generated by the reverse activity of A1Ao ATP synthase.
Possible ecological role of Acidilobales.
Organic substances reaching terrestrial acidic hot springs from surrounding areas as well as produced by lithoautotrophic microorganisms serve as substrates for organotrophic thermoacidophiles. Metabolic activity of these organisms is supported by fast anaerobic oxidation of [14C]sucrose to CO2 in acidic hot springs (23). Catabolic pathways and energy generation mechanisms predicted from the genome of A. saccharovorans explain its growth on proteins, carbohydrates, and lipids (Fig. 2). Though phylogenetically Acidilobales are most closely related to Desulfurococcales, they metabolically resemble Thermoproteales. Similarly to T. tenax, A. saccharovorans possesses both the modified EM and ED pathways of glucose catabolism. Another feature common between A. saccharovorans and Thermoproteales is the presence of the TCA cycle, which enables a complete oxidation of organic substances in the presence of external anaerobic electron acceptors. Adaptation of A. saccharovorans to acidic conditions is reflected by a high ratio of secondary transporters to ATP-dependent primary transporters, the suggested reversibility of H+-ATP synthase, the function of the encoded H+-translocating PPase, and the proposed two proton translocation sites in the anaerobic respiratory chain.
Physiological functions predicted by A. saccharovorans genome analysis (Fig. 2) and the wide distribution of Acidilobus spp. in acidic hot springs (23, 24) suggest an important role of Acidilobales in microbial communities of such habitats, closing the anaerobic carbon cycle through complete mineralization of organic substrates via S0 respiration. Contrary to mesophilic microbial communities, in which the process of anaerobic destruction of organic substances consists of several stages performed by different groups of microorganisms (hydrolysis, fermentation, syntrophic reactions, and terminal oxidation), in thermal environments this could be achieved by a single group of organisms, Thermoproteales in neutral hot springs and Acidilobales in acidic ones.
Nucleotide sequence accession number.
The annotated genome sequence of A. saccharovorans 345-15T has been deposited in the GenBank database under accession number CP001742.
Supplementary Material
Acknowledgments
This work was supported by the Federal Agency for Science and Innovations of Russia (contract 02.512.11.2201).
The work of M.I.P. and E.A.B.-O. on the analysis of growth characteristics of A. saccharovorans was supported by the Molecular and Cellular Biology program of RAS.
Footnotes
Published ahead of print on 25 June 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, I. J., L. Dharmarajan, J. Rodriguez, S. Hooper, I. Porat, L. E. Ulrich, J. G. Elkins, K. Mavromatis, H. Sun, M. Land, A. Lapidus, S. Lucas, K. Barry, H. Huber, I. B. Zhulin, W. B. Whitman, B. Mukhopadhyay, C. Woese, J. Bristow, and N. Kyrpides. 2009. The complete genome sequence of Staphylothermus marinus reveals differences in sulfur metabolism among heterotrophic Crenarchaeota. BMC Genomics 10:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. J. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, E. S., R. A. Jackson, G. Encarnasion, J. A. Zahn, T. Beard, W. D. Leavitt, Y. Pi, C. L. Zhang, A. Pearson, and G. G. Geesey. 2007. Isolation, characterization, and ecology of sulfur-respiring crenarchaea inhabiting acid-sulfate-chloride-containing geothermal spring in Yellowstone National Park. Appl. Environ. Microbiol. 73:6669-6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drozdowicz, Y. M., Y. P. Lu, V. Patel, S. Fitz-Gibbon, J. H. Miller, and P. A. Rea. 1999. A thermostable vacuolar-type membrane pyrophosphatase from the archaeon Pyrobaculum aerophilum: implications for the origins of pyrophosphate-energized pumps. FEBS Lett. 460:505-512. [DOI] [PubMed] [Google Scholar]
- 7.Friedrich, T., and D. Scheide. 2000. The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett. 479:1-5. [DOI] [PubMed] [Google Scholar]
- 8.Fütterer, O., A. Angelov, H. Liesegang, G. Gottschalk, C. Schleper, B. Schepers, C. Dock, G. Antranikian, and W. Liebl. 2004. Genome sequence of Picrophilus torridus and its implications for life around pH 0. Proc. Natl. Acad. Sci. U. S. A. 101:9091-9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grigoriev, A. 1998. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 26:2286-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrmann, G., E. Jayamani, G. Mai, and W. Buckel. 2008. Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J. Bacteriol. 190:784-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinsley, A. P., and B. C. Berks. 2002. Specificity of respiratory pathways involved in the reduction of sulfur compounds by Salmonella enterica. Microbiology 148:3631-3638. [DOI] [PubMed] [Google Scholar]
- 12.Hu, Y., and J. F. Holden. 2006. Citric acid cycle in the hyperthermophilic archaeon Pyrobaculum islandicum grown autotrophically, heterotrophically, and mixotrophically with acetate. J. Bacteriol. 188:4350-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itoh, T., K. Suzuki, P. C. Sanchez, and T. Nakase. 2003. Caldisphaera lagunensis gen. nov., sp. nov., a novel thermoacidophilic crenarchaeote isolated from a hot spring at Mt Maquiling, Philippines. Int. J. Syst. Evol. Microbiol. 53:1149-1154. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, D. B., and K. B. Hallberg. 2009. Carbon, iron and sulfur metabolism in acidophilic microorganisms. Adv. Microb. Physiol. 54:201-255. [DOI] [PubMed] [Google Scholar]
- 15.Klenk, H. P., R. A. Clayton, J. F. Tomb, O. White, K. E. Nelson, K. A. Ketchum, R. J. Dodson, M. Gwinn, E. K. Hickey, J. D. Peterson, D. L. Richardson, A. R. Kerlavage, D. E. Graham, N. C. Kyrpides, R. D. Fleischmann, J. Quackenbush, N. H. Lee, G. G. Sutton, S. Gill, E. F. Kirkness, B. A. Dougherty, K. McKenney, M. D. Adams, B. Loftus, S. Peterson, C. I. Reich, L. K. McNeil, J. H. Badger, A. Glodek, L. Zhou, R. Overbeek, J. D. Gocayne, J. F. Weidman, L. McDonald, T. Utterback, M. D. Cotton, T. Spriggs, P. Artiach, B. P. Kaine, S. M. Sykes, P. W. Sadow, K. P. D'Andrea, C. Bowman, C. Fujii, S. A. Garland, T. M. Mason, G. J. Olsen, C. M. Fraser, H. O. Smith, C. R. Woese, and J. C. Venter. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364-370. [DOI] [PubMed] [Google Scholar]
- 16.Koski, L. B., M. W. Gray, B. F. Langi, and G. Burger. 2005. AutoFACT: an automatic functional annotation and classification tool. BMC Bioinformatics 6:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagesen, K., P. Hallin, E. A. Rødland, H. H. Staerfeldt, T. Rognes, and D. W. Ussery. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35:3100-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lobry, J. R. 1996. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 13:660-665. [DOI] [PubMed] [Google Scholar]
- 19.Lowe, T., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mardanov, A. V., N. V. Ravin, V. A. Svetlitchnyi, A. V. Beletsky, M. L. Miroshnichenko, E. A. Bonch-Osmolovskaya, and K. G. Skryabin. 2009. Metabolic versatility and indigenous origin of the archaeon Thermococcus sibiricus, isolated from a Siberian oil reservoir, as revealed by genome analysis. Appl. Environ. Microbiol. 75:4580-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pisa, K. Y., H. Huber, M. Thomm, and V. Müller. 2007. A sodium ion-dependent A1AO ATP synthase from the hyperthermophilic archaeon Pyrococcus furiosus. FEBS J. 274:3928-3938. [DOI] [PubMed] [Google Scholar]
- 22.Prokofeva, M. I., M. L. Miroshnichenko, N. A. Kostrikina, N. A. Chernyh, B. B. Kuznetsov, T. P. Tourova, and E. A. Bonch-Osmolovskaya. 2000. Acidilobus aceticus gen. nov., sp. nov., a novel anaerobic thermoacidophilic archaeon from continental hot vents in Kamchatka. Int. J. Syst. Evol. Microbiol. 50:2001-2008. [DOI] [PubMed] [Google Scholar]
- 23.Prokofeva, M. I., I. I. Rusanov, N. V. Pimenov, and E. A. Bonch-Osmolovskaya. 2006. Organotrophic activity in Kamchatka hot springs with low pH. Mikrobiologiia 75:284-286. [PubMed] [Google Scholar]
- 24.Prokofeva, M. I., N. A. Kostrikina, T. V. Kolganova, T. P. Tourova, A. M. Lysenko, A. V. Lebedinsky, and E. A. Bonch-Osmolovskaya. 2009. Isolation of the anaerobic thermoacidophilic crenarchaeote Acidilobus saccharovorans sp. nov. and proposal of Acidilobales ord. nov., including Acidilobaceae fam. nov. and Caldisphaeraceae fam. nov. Int. J. Syst. Evol. Microbiol. 59:3116-3122. [DOI] [PubMed] [Google Scholar]
- 25.Ravin, N. V., A. V. Mardanov, A. V. Beletsky, I. V. Kublanov, T. V. Kolganova, A. V. Lebedinsky, N. A. Chernyh, E. A. Bonch-Osmolovskaya, and K. G. Skryabin. 2009. Complete genome sequence of the anaerobic, peptide-fermenting hyperthermophilic crenarchaeon Desulfurococcus kamchatkensis. J. Bacteriol. 191:2371-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruepp, A., W. Graml, M. L. Santos-Martinez, K. K. Koretke, C. Volker, H. W. Mewes, D. Frishman, S. Stocker, A. N. Lupas, and W. Baumeister. 2000. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 407:508-513. [DOI] [PubMed] [Google Scholar]
- 27.Sazanov, L. A., and P. Hinchliffe. 2006. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 311:1430-1436. [DOI] [PubMed] [Google Scholar]
- 28.Sazanov, L. A. 2007. Respiratory complex I: mechanistic and structural insights provided by the crystal structure of the hydrophilic domain. Biochemistry 46:2275-2288. [DOI] [PubMed] [Google Scholar]
- 29.Schut, G. J., S. L. Bridger, and M. W. Adams. 2007. Insights into the metabolism of elemental sulfur by the hyperthermophilic archaeon Pyrococcus furiosus: characterization of a coenzyme A-dependent NAD(P)H sulfur oxidoreductase. J. Bacteriol. 189:4431-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selig, M., and P. Schönheit. 1994. Oxidation of organic compounds to CO2 with sulfur or thiosulfate as electron acceptor in the anaerobic hyperthermophilic archaea Thermoproteus tenax and Pyrobaculum islandicum proceeds via the citric acid cycle. Arch. Microbiol. 162:286-294. [Google Scholar]
- 31.Siebers, B., B. Tjaden, K. Michalke, C. Dörr, H. Ahmed, M. Zaparty, P. Gordon, C. W. Sensen, A. Zibat, H. P. Klenk, S. C. Schuster, and R. Hensel. 2004. Reconstruction of the central carbohydrate metabolism of Thermoproteus tenax by use of genomic and biochemical data. J. Bacteriol. 186:2179-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaparty, M., B. Tjaden, R. Hensel, and B. Siebers. 2008. The central carbohydrate metabolism of the hyperthermophilic crenarchaeote Thermoproteus tenax: pathways and insights into their regulation. Arch. Microbiol. 190:231-245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.