Abstract
Since enterohemorrhagic Escherichia coli (EHEC) isolates of serogroup O156 have been obtained from human diarrhea patients and asymptomatic carriers, we studied cattle as a potential reservoir for these bacteria. E. coli isolates serotyped by agglutination as O156:H25/H−/Hnt strains (n = 32) were isolated from three cattle farms during a period of 21 months and characterized by rapid microarray-based genotyping. The serotyping by agglutination of the O156 isolates was not confirmed in some cases by the results of DNA-based serotyping as only 25 of the 32 isolates were conclusively identified as O156:H25. In the multilocus sequence typing (MLST) analysis, all EHEC O156:H25 isolates were characterized as sequence type 300 (ST300) and ST688, which differ by a single-nucleotide exchange in the purA gene. Oligonucleotide microarrays allow simultaneous detection of a wider range of EHEC-associated and other E. coli virulence markers than other methods. All O156:H25 isolates showed a wide spectrum of virulence factors typical for EHEC. The stx1 genes combined with the EHEC hlyA (hlyAEHEC) gene, the eae gene of the ζ subtype, as well as numerous other virulence markers were present in all EHEC O156:H25 strains. The behavior of eight different cluster groups, including four that were EHEC O156:H25, was monitored in space and time. Variations in the O156 cluster groups were detected. The results of the cluster analysis suggest that some O156:H25 strains had the genetic potential for a long persistence in the host and on the farm, while other strains did not. As judged by their pattern of virulence markers, E. coli O156:H25 isolates of bovine origin may represent a considerable risk for human infection. Our results showed that the miniaturized E. coli oligonucleotide arrays are an excellent tool for the rapid detection of a large number of virulence markers.
Shiga toxin-producing Escherichia coli (STEC) strains comprise a group of zoonotic enteric pathogens (45). In humans, infections with some STEC serotypes may result in hemorrhagic or nonhemorrhagic diarrhea, which can be complicated by the hemolytic uremic syndrome (HUS) (32). These STEC strains are also designated enterohemorrhagic Escherichia coli (EHEC). Consequently, EHEC strains represent a subgroup of STEC with high pathogenic potential for humans. Although E. coli O157:H7 is the most frequent EHEC serotype implicated in HUS, other serotypes can also cause this complication. Non-O157:H7 EHEC strains including serotypes O26:H11/H−, O103:H2/H−, O111:H8/H10/H−, and O145:H28/H25/H− and sorbitol-fermenting E. coli O157:H− isolates are present in about 50% of stool cultures from German HUS patients (10, 42). However, STEC strains that cause human infection belong to a large number of E. coli serotypes, although a small number of STEC isolates of serogroup O156 were associated with human disease (7). Strains of the serotypes O156:H1/H8/H21/H25 were found in human cases of diarrhea or asymptomatic infections (9, 22, 25, 26). The detection of STEC of serogroup O156 from healthy and diseased ruminants such as cattle, sheep, and goats was reported by several authors (1, 11-13, 21, 39, 46, 50, 52). Additional EHEC-associated virulence genes such as stx, eae, hlyAEHEC, or nlaA were found preferentially in the serotypes O156:H25 and O156:H− (11-13, 21, 22, 50, 52).
Numerous methods exist for the detection of pathogenic E. coli, including genotypic and phenotypic marker assays for the detection of virulence genes and their products (19, 47, 55, 57). All of these methods have the common drawback of screening a relatively small number of determinants simultaneously. A diagnostic DNA microarray based on the ArrayTube format of CLONDIAG GmbH was developed as a viable alternative due to its ability to screen multiple virulence markers simultaneously (2). Further microarray layouts working with the same principle but different gene targets were developed for the rapid identification of antimicrobial resistance genes in Gram-negative bacteria (5) and for the rapid DNA-based serotyping of E. coli (4). In addition, a protein microarray for E. coli O serotyping based on the ArrayTube format was described by Anjum et al. (3).
The aim of our study was the molecular genotyping of bovine E. coli field isolates of serogroup O156 based on miniaturized E. coli oligonucleotide arrays in the ArrayStrip format and to combine the screening of E. coli virulence markers, antimicrobial resistance genes, and DNA serotyping targets, some of which were partially described previously for separate arrays (2, 4, 5). The epidemiological situation in the beef herds from which the isolates were obtained and the spatial and temporal behavior of the clonal distribution of E. coli serogroup O156 were analyzed during the observation period. The potential risk of the isolates inducing disease in humans was assessed.
MATERIALS AND METHODS
Bacterial strains and phenotyping methods.
A total of 32 E. coli isolates of serogroup O156 were isolated from three cattle farms (farms B, C, and D) during 21 months of monitoring in four German cattle farms. No isolates of this serogroup were found on farm A. Details on the monitoring program, the participating farms, their management practices, and the sampling procedure were reported elsewhere (30). On farm D, two groups of cattle (groups 1 and 2) were investigated. The cattle of group 2 were only born after the animals of group 1 had been slaughtered. E. coli O156 isolates were obtained from both groups. All 32 isolates were identified primarily as STEC. The serotypes were determined by National Reference Centre for Salmonella and Other Enterics (Robert Koch-Institut, Wernigerode, Germany). Fermentation of sorbitol was detected on sorbitol McConkey agar (SIFIN GmbH, Berlin, Germany) and the production of EHEC hemolysin was detected on enterohemolysin agar (Haipha GmbH, Eppelheim, Germany). The expression of Shiga toxins was monitored by an enzyme-linked immunosorbent assay (ELISA) using monoclonal antibodies specific for Stx1 or by the Vero cell neutralization assay described previously (54).
Genotype characterization.
RNA-free genomic DNAs of the E. coli O156 isolates were prepared using the Zymo ZR fungal/bacterial DNA kit (Hiss Diagnostic GmbH, Freiburg, Germany) from 2 ml of overnight cultures. The DNA concentration was determined spectrophotometrically at 260 nm and analyzed for fragmentation by agarose gel electrophoresis. Miniaturized E. coli oligonucleotide arrays in the ArrayStrip format (CLONDIAG GmbH, Jena, Germany) containing gene targets for the identification of virulence genes (2), antimicrobial resistance genes (5), and DNA-based serotyping (4) were used for the genetic characterization of the O156 E. coli isolates. A complete list of primers and probes is available upon request from the corresponding author.
For labeling and biotinylation of the genomic DNA, a site-specific labeling approach was used (43). The primer elongation reaction was performed using the primer mixture and a deoxynucleoside triphosphate (dNTP) solution containing 1 mM dATP, 1 mM dCTP, 1 mM dGTP, 0.65 mM dTTP, and 0.35 mM biotin-16-dUTP (Roche Penzberg, Germany). The mixture for the elongation reaction contained 0.3 μl Therminator polymerase (New England Biolabs, Frankfurt am Main, Germany), 3 μl Therminator polymerase buffer (New England Biolabs), 3 μl primer solution, 3 μl dNTP stock solution, and 1 to 1.5 μg unfragmented genomic DNA free of any RNA of the E. coli isolates. The reaction was started with denaturation (5 min at 96°C), and then 45 cycles of 60 s at 96°C, 20 s at 62°C, and 40 s at 72°C followed. The sample was then cooled down to 4°C and hybridized with the DNA array. For hybridization, the HybKit (CLONDIAG GmbH) was used with an adapted protocol. Initially, each ArrayStrip was washed with 200 μl double-distilled water and 150 μl of buffer C1 using a thermomixing device (5 min, 55°C, 550 rpm [Eppendorf GmbH, Hamburg, Germany]). The sample consists of 10 μl labeled sample and 90 μl buffer C1. It was transferred into the ArrayStrip and incubated (60 min, 55°C, 550 rpm). The sample was then removed from the tube, and the array was washed twice (12 min, 40°C, 550 rpm) with buffer C2. Afterwards, 100 μl conjugate solution was added for 15 min at 30°C and 550 rpm, followed by a washing step with 200 μl buffer C5 for 15 min at 30°C and 550 rpm. The ArrayStrip was then stained with buffer D1 (100 μl, 10 min, no agitation), aspirated, photographed using the ArrayMate instrument (CLONDIAG GmbH, Jena, Germany), and automatically analyzed. Mean signal intensity (mean) and local background (lbg) were measured for each probe position, and values were calculated by the formula value = 1 − mean/lbg. Resulting values below 0.1 were considered negative, and those above 0.3 were considered positive. Values between 0.1 and 0.3 were regarded as ambiguous. Validation was performed using a collection of sequenced control strains (GenBank accession no. AE005174, LCL_10009, FM180568, U00096, and LCL_10006). These control strains were tested by the method described above.
In addition, the E. coli O serogroups were characterized by MboII restriction endonuclease digests of amplified O-antigen gene clusters (rfb restriction fragment length polymorphism [RFLP]), as described elsewhere (20). The detection of the eae subtypes and the tir and espB subtypes was performed as described previously (17, 48, 53, 60). The nucleotide sequences of intimin genes of three isolates (WH-02/23/021-2, WH-02/25/010-9, and WH-04/25/005-1) were determined by PCR (Primer-fwd, 5′-GTTTCCCGCTCGATGATGCTAC-3′; Primer-rev, 5′-GCGAATAGAAAAGGTGGCTGGAG-3′) followed by DNA sequencing in a LiCOR 4200 system using the DYEnamic direct cycle sequencing kit (GE Healthcare Europe GmbH, Freiburg, Germany). A plasmid profile analysis was also conducted (28, 29).
MLST analysis.
Internal fragments of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) of all O156 isolates were analyzed, and the genetic relationships between different sequence types (STs) (assigned in accordance with the E. coli MLST website [http://mlst.ucc.ie/mlst/dbs/Ecoli] were determined as described elsewhere (10). Phylogenetic analyses were conducted and the minimum-spanning tree was generated using the appropriate tools in the BioNumerics software v6.0 (Applied Maths NV, Sint-Martens-Latem, Belgium).
CHEF-PFGE and cluster analysis.
Preparation of genomic DNA for contour-clamped homogeneous electric field-pulsed-field gel electrophoresis (CHEF-PFGE) was done by a protocol described previously (28, 29, 38). Slices of the DNA agarose plugs were equilibrated in the respective restriction endonuclease buffer and then digested for 4 h with XbaI, NotI, BlnI (AvrII), or SpeI (New England Biolabs GmbH, Frankfurt am Main, Germany). The resulting fragments were separated in 1.0% agarose gels (Biozyme Gold agarose; Biozyme GmbH, Germany) in 0.5× Tris-borate-EDTA (TBE) at 10°C in a CHEF Mapper XA system. The pulse times for XbaI and NotI digests were increased from 5 to 50 s (gradient of 6 V/cm) during 25 h at a constant angle of 120°. The switch time values for BlnI and SpeI were set using the Auto Algorithm function of the CHEF Mapper XA to separate fragments in the range of 50 to 450 kb (BlnI) or 30 to 350 kb (SpeI), respectively. After electrophoresis, the gels were stained with 500 ml ethidium bromide solution (50 μg/ml), and the banding patterns were recorded under UV illumination. Interpretation of the PFGE patterns was performed by visual inspection and computer analysis (BioNumerics software v6.0; Applied Maths NV, Sint-Martens-Latem, Belgium). All fragments larger than 45 kb (up to 23 fragments per isolate for XbaI, up to 15 for NotI, up to 14 for BlnI, and up to 16 for SpeI) were included in the clonal analysis of the isolates. Distance matrices were calculated by pairwise comparisons of the fragmentation patterns produced by each of the four restriction endonucleases used for the PFGE analysis. To this end, the total character difference was used, which counts the pairwise differences between two given patterns. A cluster analysis was performed with the distance matrices using the neighbor-joining algorithm, an agglomerative cluster method which generates a phylogenetic tree based on distance matrices (51). The cluster analysis was conducted in PAUP* for Windows (version 4.0).
Nucleotide sequence accession numbers.
The DNA sequences of the complete eae genes of the three O156:H25 isolates have been submitted to GenBank under accession no. GU944691, GU944692, and GU944693.
RESULTS
Analyses of EHEC virulence-associated factors.
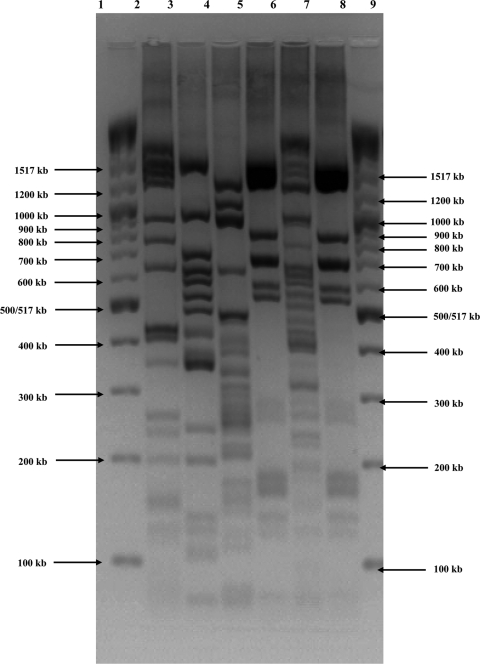
All tested isolates were serotyped by agglutination as O156:H25/H−/Hnt. We detected the fliC gene for the H25 antigen in 26 of the isolates by using the oligonucleotide microarrays. In the remaining 6 isolates, the fliC genes for H8, H11 (found twice), H43, or H46 (found twice) were detected (Table 1). When the O antigens were typed with the oligonucleotide microarrays, only an O103 antigen was detected in the isolate with H43 antigen; all other hybridization results were negative. The number of O antigens detectable by this method is limited. There are no gene probes for the detection of the O156 antigen, for example. All isolates were therefore also characterized by MboII digests of amplified O-antigen gene clusters (rfb RFLP). Twenty-five isolates showed a pattern typical for O156 as described by Coimbra et al. (20). The patterns of some remaining isolates varied, and they also differed from the O156 pattern (Fig. 1). Classification of these isolates using the rfb RFLP database described by Coimbra et al. (20) was not possible. All 25 isolates with the O156 pattern were also positive for the H25 antigen. Consequently, these isolates were designated EHEC O156:H25.
TABLE 1.
Serotyping and phylogenetic characteristics of tested E. coli O156 isolates classified by the time pattern of isolationa
| Sampling day | Source | Isolate | Serotype by agglutination | Result by: |
Plasmid size(s) in kb | Cluster | |||
|---|---|---|---|---|---|---|---|---|---|
| MLST |
DNA-based serotyping |
||||||||
| ST | CC | MboII RFLP | fliC | ||||||
| 211 | Cow 26 (farm B) | WH-02/26/008-10 | O156:H25 | 688 | NA | O156 | H25 | 75, 6.9, and 5.1 | 8 |
| 274 | Cow 25 (farm B) | WH-02/25/010-9 | O156:Hnt | 688 | NA | O156 | H25 | 75, 6.9, and 5.1 | 8 |
| WH-02/25/010-10 | O156:H25 | 688 | NA | O156 | H25 | 75, 6.9, and 5.1 | 8 | ||
| 330 | Cow 16 (farm C) | WH-04/16/007-2 | O156:H− | 58 | 155 | ? | H25 | 155 | 1 |
| 365 | Cow 25 (farm C) | WH-04/25/005-1 | O156:H25 | 300 | NA | O156 | H25 | 75 | 8 |
| 400 | Cow 4 (farm B) | WH-02/04/017-3 | O156:Hnt | 300 | NA | O156 | H25 | 75 | 7 |
| WH-02/04/017-9 | O156:Hnt | 300 | NA | O156 | H25 | 75 | 7 | ||
| 512 | Cow 28 (farm B) | WH-02/28//018-1 | O156:H25 | 300 | NA | O156 | H25 | 75 | 6 |
| WH-02/28//018-3 | O156:H25 | 300 | NA | O156 | H25 | 75 | 6 | ||
| WH-02/28//018-4 | O156:H25 | 300 | NA | O156 | H25 | 75 | 6 | ||
| WH-02/28//018-6 | O156:H− | 300 | NA | O156 | H25 | 75 | 6 | ||
| 533 | Cow 25 (farm B) | WH-02/25/019-1 | O156:H25 | 300 | NA | O156 | H25 | 75 | 7 |
| WH-02/25/019-9 | O156:H25 | 300 | NA | O156 | H25 | 75 | 7 | ||
| 547 | Cow 12 (farm D, group 1) | WH-03/12/016-1 | O156:H− | 300 | NA | O156 | H25 | 75 and 6.9 | 7 |
| WH-03/12/016-2 | O156:H− | 300 | NA | O156 | H25 | 75 and 6.9 | 7 | ||
| WH-03/12/016-8 | O156:Hnt | 300 | NA | O156 | H25 | 75 and 6.9 | 7 | ||
| 554 | Cow 23 (farm B) | WH-02/23/021-2 | O156:Hnt | 300 | NA | O156 | H25 | 75 | 7 |
| WH-02/23/021-3 | O156:Hnt | 300 | NA | O156 | H25 | 75 | 7 | ||
| WH-02/23/021-5 | O156:Hnt | 300 | NA | O156 | H25 | 75 | 7 | ||
| WH-02/23/021-9 | O156:H25 | 300 | NA | O156 | H25 | 75 | 7 | ||
| WH-02/23/021-10 | O156:H25 | 300 | NA | O156 | H25 | 75 | 7 | ||
| 638 | Cow 25 (farm B) | WH-02/25/024-3 | O156:H− | 300 | NA | O156 | H25 | 75 | 7 |
| WH-02/25/024-5 | O156:H25 | 300 | NA | O156 | H25 | 75 | 7 | ||
| 708 | Cow 2 (farm D, group 2) | WH-05/02/001-2 | O156:H25 | 1308 | NA | ? | H46 | 110, 85, and 5.1 | 2 |
| WH-05/02/001-3 | O156:Hnt | 300 | NA | O156 | H25 | 75 and 6.9 | 7 | ||
| WH-05/02/001-8 | O156:H25 | 1308 | NA | ? | H46 | 110, 85, and 5.1 | 2 | ||
| WH-05/02/001-9 | O156:Hnt | 300 | NA | O156 | H25 | 75 and 6.9 | 7 | ||
| 764 | Cow 23 (farm D, group 2) | WH-05/23/003-1 | O156:H25 | 10 | 10 | ? | H11 | 155, 110, 75, 50, 21, 5.9, and 3.1 | 3 |
| Cow 25 (farm D, group 2) | WH-05/25/003-1 | O156:H25 | 48 | 10 | O103 | H43 | 75, 35, 21, 7.5, and 2.9 | 4 | |
| 792 | Cow 25 (farm D, group 2) | WH-05/25/004-1 | O156:H25 | 300 | NA | O156 | H25 | 75 and 6.9 | 5 |
| WH-05/25/004-3 | O156:H25 | 10 | 10 | ? | H11 | 155, 110, 75, 50, 21, 5.9, 3.1, and 2.9 | 3 | ||
| 841 | Cow 12 (farm D, group 2) | WH-05/12/006-2 | O156:Hnt | 327 | NA | ? | H8 | 85 | 4 |
CC, clonal complex; MLST, multilocus sequence typing; ST, sequence type; NA, CC not assigned; H−, nonmotile; Hnt, not typeable; ?, not detectable.
FIG. 1.
Electropherogram of MboII restriction of amplified O-antigen gene clusters (rfb RFLP). Lane 1, 2-log DNA ladder (New England Biolabs GmbH, Germany); lane 2, WH-02/23/021-2; lane 3, WH-04/16/007-2; lane 4, WH-05/02/001-8; lane 5, WH-05/23/003-1; lane 6, WH-05/25/003-1; lane 7, WH-05/25/004-3; lane 8, 2-log DNA ladder.
Numerous EHEC-associated and other E. coli virulence markers were tested using the oligonucleotide microarrays. An stx1 gene characterized as the stx1 subtype was found in all 25 EHEC O156:H25 isolates. The detection of the stx1 genes corresponded with the demonstration of the eae gene, the hlyAEHEC gene and the espP gene in the same isolates (see Table S1 in the supplemental material). For the subtyping of the intimin genes, a PCR product of 2,430 bp was amplified from all EHEC O156:H25 isolates using SK1/LP6B primers (60). The eae genes were therefore considered as members of the eae-ζ subgroup. These results were confirmed by DNA sequencing of the complete eae genes of the three O156:H25 isolates (GenBank accession no. GU944691, GU944692, and GU944693). The nucleotide sequences were identical or nearly identical relative to the published sequences of ζ-intimin genes of several E. coli serotypes (6, 33, 60), but also to that of an O156:H− strain (GenBank accession no. AY520904.1). We detected both locus of enterocyte effacement (LEE)-encoded and non-LEE-encoded genes of the type III secretion system (T3SS) in the EHEC O156:H25 isolates. An espA gene (hybridization with three DNA probes [espA-O103:H2, espA-O127:H7, and espA-O55:H7]) was found in all 25 isolates. Furthermore, the genes espF (hybridization of one variant), tccP (hybridization of both variants in 23 isolates and of one variant in two isolates), espJ (hybridization of one probe), nleA (hybridization of all probes), and nleB (hybridization of one probe) were presented in all EHEC O156:H25 isolates. The nleC gene was found in 16 of the 25 EHEC O156:H25 isolates. Detection of a translocated intimin receptor (tir) gene with the gene probes for tir genes of the oligonucleotide microarrays failed. We detected, however, a tirα gene and an espBα gene in all EHEC O156:H25 isolates by using the PCRs described previously (17), but the expected sizes of the PstI restriction fragments of the tirα amplicons were not obtained. We found two fragments of ca. 170 kb (data not shown). We also detected the etpD gene, which is associated with a type II secretion system, the gene for a heat-stable enterotoxin (astA gene), and the gene for fimbria adhesion (lpfA gene) in all EHEC O156:H25 isolates. The cba gene (coding for a bacteriocin) was found in five of these isolates; the katP gene (15) was found only once (see Table S1 in the supplemental material). The detected virulence markers of the seven non-O156:H25 isolates differed considerably from the EHEC O156:H25 virulence patterns (Table 1; see Table S1 in the supplemental material).
An approximately 75-kb large plasmid was detected in 28 of the tested E. coli isolates. This plasmid was found exclusively in 16 of the 25 EHEC 156:H25 isolates; 1 (6.9 kb) or 2 (6.9 kb or 5.1 kb) additional plasmids were detected in the other nine isolates (Table 1). The plasmid profiles of the other three E. coli isolates carrying the 75-kb plasmid varied substantially. Several larger plasmids and a number of smaller plasmids were present in these isolates (Table 1). In the remaining isolates, single plasmids of 155 kb or 85 kb, respectively, or two plasmids (110 kb and 5.1 kb) were found in addition to the 85-kb plasmid.
The production of the Shiga toxins was tested by ELISA and Vero cell neutralization assay. All isolates with the stx1 gene produced Stx1. The cytotoxicity of the Stx1 was very high for Vero cells upon induction with mitomycin C. The EHEC hemolysin was produced by all EHEC O156:H25 isolates harboring hlyAEHEC. Sorbitol was fermented by all tested E. coli isolates.
MLST analysis.
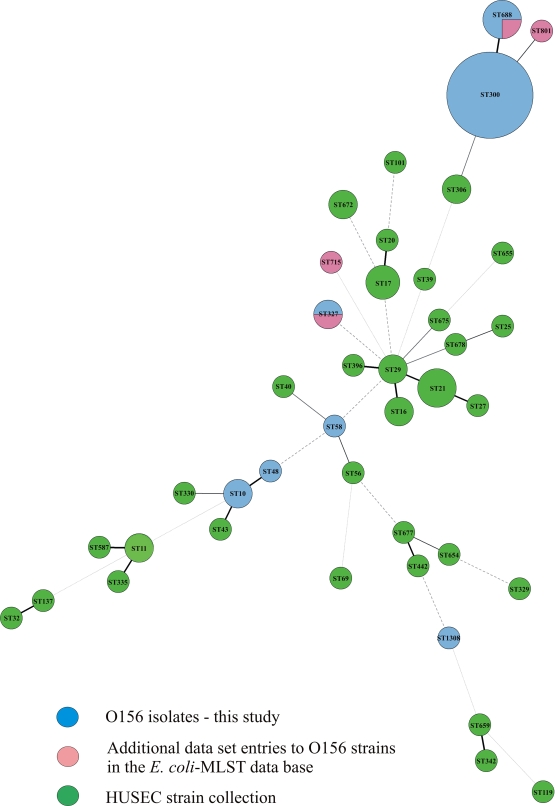
MLST analyses were performed with all 32 E coli isolates (Table 1). Twenty-two isolates grouped in the sequence type ST300 and 3 in ST688. The difference between ST300 and ST688 is a single-nucleotide exchange in the purA gene (Fig. 2). These 25 isolates were previously identified as EHEC O156:H25 in the DNA-based serotyping. The other seven isolates were grouped into different sequence types (ST). Three isolates were members of the clonal complex (CC) 10 (ST10 and ST48), two were characterized as ST1308, and one isolate each was detected as ST58 (CC155) and ST327. A comparison of all O156 STs with all STEC STs associated with HUS (HUSEC collection described by Mellmann et al. [42]) and with the additional data set entries for O156 strains in the E. coli MLST database (http://mlst.ucc.ie/mlst/dbs/Ecoli) is displayed in Fig. 2.
FIG. 2.
Minimum-spanning tree based on the multilocus sequence typing allelic profiles that portray the clonal distribution of the tested 32 E. coli O156 isolates in relation to the HUSEC collection (described by Mellmann et al. [42]) and to the additional data set entries for O156 in the E. coli MLST database (http://mlst.ucc.ie/mlst/dbs/Ecoli). Each circle represents a given sequence type, and the size of each circle is proportional to the number of strains analyzed. Connecting lines show the number of identical alleles between two STs (thick black line, 6 of 7 alleles identical; thinner black lines, 5 alleles identical; thin black lines, 4 alleles identical; thin dashed lines, ≤3 alleles identical).
Analyses of the antimicrobial resistance genes.
Antimicrobial resistance genes were rarely detected. An EHEC O156:H25 isolate hybridized with one of three DNA probes (probe blaMOX-CMY-612) for a plasmid encoding AmpC-like β-lactamase gene (blaCMY gene) and with one of three probes detecting resistance genes against sulfonamides (probe sul2-11). Antimicrobial resistance genes were not detected in the remaining O156:H25 strains. Hybridization with a tetA gene (encoding tetracycline resistance) and with two probes for sulfonamide resistance genes (probes sul1-11 and sul2-11) were observed in two non-O156:H25 isolates.
Clonal relationships.
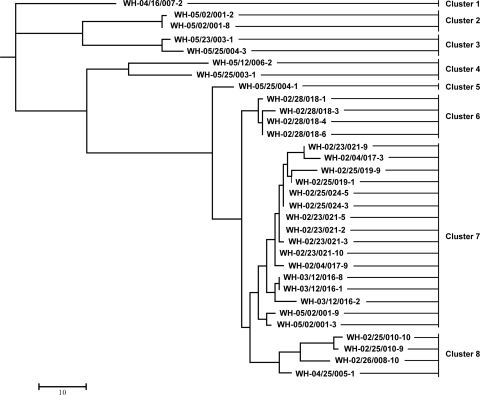
The tested 32 bovine E. coli isolates were found over a period of 21 months in three of four cattle herds monitored at the same time (30). In farm D, we serially investigated two groups of cattle. A total of 8 different clusters were detected. The identified EHEC O156:H25 isolates belonged to four different cluster groups, but the genetic distances to the other four cluster groups comprising the non-O156:H25 isolates were much higher than the distances within the EHEC O156:H25 cluster groups (Fig. 3; see Fig. S1 in the supplemental material).
FIG. 3.
Neighbor-joining tree of bovine E. coli O156 isolates based on the restriction pattern obtained after digestion with XbaI, NotI, BlnI, and SpeI (see Fig. S1 in the supplemental material).
From farm B, all 18 tested isolates were identified as EHEC O156:H25. These isolates were obtained from seven different animals during a period of 12 months. They belonged to three different clusters (clusters 6, 7, and 8) (Fig. 3; see Fig. S1 in the supplemental material). First, EHEC O156:H25 was found in two different animals (cattle 25 and 26) on sampling days 211 and 274. These isolates were typed as ST688 in the MLST analysis and grouped in cluster 8. Isolates of this sequence type were not detected subsequently during the monitoring. We obtained the other 15 isolates during a period of 9 months. This period started half a year after the isolation of the first EHEC O156:H25 isolate. Certain isolates belonging to cluster 7 were found during this period in the same or different animals (Table 1 and Fig. 3; see Fig. S1 in the supplemental material). Exceptions were four EHEC O156:H25 isolates detected in cow 28 on day 512 which grouped in cluster 6.
The three isolates of group 1 from farm D were invariably EHEC O156:H25. In contrast, only three of the nine isolates of group 2 from the same farm were identified as EHEC O156:H25. Interestingly, the isolates of group 1 and two isolates of group 2 belonged to the same cluster, 7, in which most of the isolates from farm B grouped. The EHEC O156:H25 isolate WH-05/25/004-1 represented a separate cluster, 5 (Table 1 and Fig. 3; see Fig. S1 in the supplemental material). The EHEC O156:H25 isolates were detected on 2 days, when non-O156:H25 isolates were also found in the same animals (cow 2, day 708; cow 25, day 792) in group 2. The divergences of the non-O156:H25 isolates were very high relative to all obtained EHEC O156:H25 isolates and particularly among the non-O156:H25 isolates (Table 1 and Fig. 3; see Fig. S1 in the supplemental material).
Two isolates obtained from two different animals on sequential sampling days in farm C were typed as EHEC O156:H25 and non-O156:H25. The EHEC O156:H25 isolate was identified as a member of cluster 8. The non-O156:H25 isolate had the greatest genetic divergence from all other isolates (Table 1 and Fig. 3; see Fig. S1 in the supplemental material).
DISCUSSION
Ruminants, especially cattle, are considered the primary reservoir for human infections with EHEC (34). For EHEC strains of serogroup O156, this fact seems to apply only to a very small number of cases. Several authors have described the detection of isolates of serogroup O156 from healthy and diseased ruminants, particularly cattle and sheep. They are referred to most often in reports from Spain but were also detected in Germany, Canada, and India (1, 8, 12, 13, 21, 30, 36, 39, 46, 50, 52, 59). In contrast, O156 isolates were only sporadically found in cases of human disease. Two isolates associated with bloody diarrhea and asymptomatic disease were reported from Finland (25, 26). Cases of asymptomatic disease and diarrhea associated with O156 were also found in Germany (9, 22, 37). In our study, 32 bovine E. coli O156:H25/H−/Hnt isolates were characterized by molecular genotyping based on miniaturized E. coli oligonucleotide arrays to assess their potential to cause EHEC diseases in humans.
Serotyping results by agglutination of the O156 isolates were not confirmed in some cases by DNA-based serotyping because only 25 of the 32 isolates were conclusively identified as O156:H25. In contrast, one isolate was typed as O103:H43, and the classification of the O groups by DNA-based serotyping was not possible for the other isolates. There may be several reasons for discrepancies between the serotyping by agglutination and the DNA-based methods. First, it is possible that more than one rfb RFLP pattern exists for O156, but not all patterns may be described in the database. Several different patterns were shown for other O groups: for example, for O2, O5, O8, O26, O55, O85, O111, O125, O147, and O149 (20). Second, potential cross-reactions in the agglutination reaction may have led to the false determination of serotypes. Moreover, it is also possible that the originally serotyped isolate has changed and is no longer identical to the isolate genotyped later on.
In the MLST analysis, all EHEC O156:H25 isolates were characterized as ST300 and ST688, which differ by a single nucleotide exchange in the purA gene. Both sequence types were previously found in human EHEC O156:H25 isolates from Germany (9). Other data set entries regarding O156 in the E. coli MLST database (http://mlst.ucc.ie/mlst/dbs/Ecoli) are very rare. However, it is interesting that a bovine O156:K+ isolate from the United Kingdom (http://mlst.ucc.ie/mlst/dbs/Ecoli) and four human O156:H8 isolates from Germany (9) were characterized as ST327. We also found the same sequence type in one of our non-O156:H25 isolates.
Oligonucleotide microarrays allow the simultaneous detection of a much wider range of EHEC-associated and other E. coli virulence markers than other methods. All of our O156:H25 isolates demonstrated a wide spectrum of virulence factors typical for EHEC. The detection of the combination of genes stx1, eae, and hlyAEHEC, as in our isolates, has previously been reported for O156:H21/H25/H− isolates from cattle and sheep (12, 13, 50). To date, 10 distinct variants of eae have been described (17, 33, 48, 49). Some serotypes were closely associated with a particular intimin variant (5, 17, 18, 56). Some authors described an eae-ζ gene in O156:H25/H− strains (12, 13, 22, 50). Our study confirms these associations. All bovine EHEC O156:H25 isolates were typed as members of the eae-ζ subgroup. The DNA sequences of the complete eae genes were identical for both the isolates from the two different MLST ST300 and ST688 and for isolates from different farms (farms B and C).
The espP/pssA gene, which was only recently reported for EHEC strains of serotypes O26:H− (24) and O157:H7 (14), as well as others such as O118:H16/H− (58), was also found in all of our EHEC O156:H25 isolates. The presence of this gene corresponded to the occurrence of a 75-kb virulence-associated plasmid.
In addition to intimin, other LEE-encoded factors such as EspA, EspB, EspC, and Tir were tested by oligonucleotide microarrays. Whereas espA genes were detected in all EHEC O156:H25 isolates, the espB and espC genes were missing. The espB genes were previously detected in bovine isolates of serogroup O156 by Orden et al. (46), but the authors tested enteropathogenic E. coli (EPEC) isolates. The lack of a tir gene in all EHEC O156:H25 isolates is surprising, because this gene might be expected in combination with the intimin gene too. By using PCRs described by China et al. (17), we also detected a tir gene and an espB gene of the α subtype in all EHEC O156:H25 isolates. Perhaps the DNA probes used on the array failed to detect yet unsequenced alleles of both the tir genes and the espB or espC genes, because the variability of these genes seems to be high. Therefore, the divergent results obtained by the PstI restriction digests of the tirα amplicons may point in this direction. It has been shown that additional effector proteins encoded by genes outside the LEE in cryptic or intact prophages are translocated by the LEE-encoded T3SS. This group of non-LEE-encoded effectors also includes the cycle-inhibiting factor Cif (41); the Tir cytoskeleton coupling protein TccP/EspFU (16, 27); the effector proteins NleA/EspI, NleB, and NleC, which are determinants necessary for virulence (31, 35, 40, 44); and EspJ, which may play a role in host survival and pathogen transmission (23). We found some of these factors also in the EHEC O156:H25 isolates. Different nleA gene variants and combinations of the variants were previously reported for members of the O156 serogroup, such as O156:H25 (22). Our study confirms these associations. nleA genes were detected in all of our EHEC O156:H25 isolates. They hybridized in each case with all four tested DNA probes. In our study, the detection of nleB, nleC, tccP, espF, and espJ genes in isolates of serogroup O156 is described for the first time. Interestingly, we found an astA gene, which encodes a heat-stable enterotoxin in all EHEC O156:H25 isolates. This heat-stable enterotoxin is typically found in enterotoxigenic E. coli (ETEC) and enteroaggregative E. coli (EAEC) isolates.
Two non-O156:H25 isolates, such as O103:H43, were identified as typical enteropathogenic E. coli (EPEC) strains. In contrast to the O156:H25 isolates, we detected eae genes of the θ subtype and failed to find stx and hlyAEHEC genes in these strains. Moreover, the hybridization patterns for the genes of LEE-encoded and non-LEE-encoded T3SS varied. O156 isolates with eae-θ and without stx genes were previously reported by Creuzburg and Schmidt (22).
We also analyzed the spatial and temporal behavior of the O156 isolates in the beef herds. In contrast to the O26:H11 isolates that we had analyzed in the same monitoring program (28), the identified EHEC O156:H25 isolates did not represent independent cluster groups on each farm. Despite the relatively large geographic distance between the farms, we found members of the same cluster groups in different farms. However, we isolated members of the dominant cluster 7 repeatedly on several sampling days over a relatively long period of 8 months, although other clusters were only detected on single occasions. Interestingly, we found the same cluster 7 isolates first, in group 1 in farm D and later in group 2. The cattle of group 2 were only born after the animals of group 1 had been slaughtered. This patchy temporal pattern is apparently not a unique property of O156:H25 as we found similar results for cluster groups of the EHEC serotypes O26:H11 and O165:H25 of bovine origin during the same monitoring program, as published elsewhere (28, 29). Transmission of clusters between individual animals was also observed. These results suggest that some EHEC O156:H25 strains as well as some EHEC O26:H11 (28) and EHEC O165:H25 (29) strains had the potential for a longer persistence in the host population, while others did not. The reasons for this difference are not yet clear. The tested genes for adherence factors such as saa, efa, and ifa were missing in the EHEC O156:H25 isolates. On the other hand, the gene for a fimbrial adhesion (lpfA gene) was found in all these isolates.
We distinguished eight different clusters, including four for O156:H25, but complete genetic identity was only found in few isolates. The variations in the O156:H25 clusters may be due to increasing competition between the bacterial populations of the various subtypes in the bovine intestine or to potential interactions between EHEC O156:H25 and the host.
Antimicrobial resistance genes were rarely found, but we detected such genes in an EHEC O156:H25 isolate which was classified as cluster 7. The isolate was found at the end of the detection period for this cluster. This could be an indication of the incorporation of resistance genes by gene transfer, which may be facilitated by the long persistence period of a strain in a cattle herd.
In conclusion, our results showed that the miniaturized E. coli oligonucleotide arrays are an excellent tool for the rapid detection of a large number of virulence markers. In addition, this method provides an overview of the presence of resistance genes. It also allows rapid DNA-based serotyping of important O groups, a technique which can still be improved. Our results further showed that bovine EHEC O156:H25 isolates that can carry virulence factors of EHEC are strongly associated with EHEC-related disease in humans, particularly with severe clinical manifestations such as HC and HUS. Therefore, strains of bovine origin may represent a considerable risk for human infection. Moreover, some clusters of EHEC O156:H25 persisted in cattle and farms over relatively long periods, potentially increasing the risk of transmission to other animals and even to humans.
Supplementary Material
Acknowledgments
We thank H. Tschäpe and A. Fruth for serotyping the E. coli isolates and Ines Engelmann, Jana Sachtschal, Rainer Schuster, and Gisela Rössler for technical assistance.
This study was funded by the German Federal Ministry of Food, Agriculture and Consumer Protection.
Footnotes
Published ahead of print on 25 June 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aktan, I., K. A. Sprigings, R. M. La Ragione, L. M. Faulkner, G. A. Paiba, and M. J. Woodward. 2004. Characterisation of attaching-effacing Escherichia coli isolated from animals at slaughter in England and Wales. Vet. Microbiol. 102:43-53. [DOI] [PubMed] [Google Scholar]
- 2.Anjum, M. F., M. Mafura, P. Slickers, K. Ballmer, P. Kuhnert, M. J. Woodward, and R. Ehricht. 2007. Pathotyping Escherichia coli by using miniaturized DNA microarrays. Appl. Environ. Microbiol. 73:5692-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anjum, M. F., J. D. Tucker, K. A. Springings, M. J. Woodward, and R. Ehricht. 2006. Use of miniaturized protein array for Escherichia coli O serotyping. Clin. Vaccine Immunol. 13:561-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballmer, K., B. M. Korczak, P. Kuhnert, P. Slickers, R. Ehricht, and H. Hächler. 2007. Fast DNA serotyping of Escherichia coli by use of an oligonucleotide microarray. J. Clin. Microbiol. 45:370-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batchelor, M., K. L. Hopkins, E. Liebana, P. Slickers, R. Ehricht, M. Mafura, F. Aarestrup, D. Mevius, F. A. Clifton-Hadley, M. J. Woodward, R. H. Davies, E. J. Threlfall, and M. F. Anjum. 2008. Development of a miniaturized microarray-based assay for the rapid identification of antimicrobial resistance genes in gram-negative bacteria. Int. J. Antimicrob. Agents 31:440-451. [DOI] [PubMed] [Google Scholar]
- 6.Bertin, Y., K. Boukhors, V. Livrelli, and C. Martin. 2004. Localization of the insertion site and pathotype determination of the locus of enterocyte effacement of Shiga toxin-producing Escherichia coli strains. Appl. Environ. Microbiol. 70:61-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bettelheim, K. A. 2007. The non-O157 Shiga-toxigenic (verocytotoxigenic) Escherichia coli; under-rated pathogens. Crit. Rev. Microbiol. 33:67-87. [DOI] [PubMed] [Google Scholar]
- 8.Beutin, L., D. Geier, H. Steinrück, S. Zimmermann, and F. Scheutz. 1993. Prevalence and some properies of verotoxin (Shiga-like toxin)-producing Escherichia coli in seven different species of healthy domestic animals. J. Clin. Microbiol. 31:2483-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bielaszewska, M., B. Middendorf, R. Köck, A. W. Friedrich, A. Fruth, H. Karch, M. A. Schmidt, and A. Mellmann. 2008. Shiga toxin-negative attaching and effacing Escherichia coli: distinct clinical associations with bacterial phylogeny and virulence traits and inferred in-host pathogen evolution. Clin. Infect. Dis. 47:208-217. [DOI] [PubMed] [Google Scholar]
- 10.Bielaszewska, M., R. Köck, A. W. Friedrich, C. von Eiff, L. B. Zimmerhackl, H. Karch, and A. Mellmann. 2007. Shiga toxin-mediated hemolytic uremic syndrome: time to change the diagnostic paradigm? PLoS One 2:e1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanco, J., M. Blanco, J. E. Blanco, A. Mora, E. A. Gonzalez, M. I. Bernardez, M. P. Alonso, A. Coira, A. Rodriguez, M. I. Bernardez, J. Rey, J. M. Alonso, and M. A. Usera. 2003. Verotoxin-producing Escherichia coli in Spain: prevalence, serotypes, and virulence genes of O157:H7 and non-O157 VTEC in ruminants, raw beef products, and humans. Exp. Biol. Med. 228:345-351. [DOI] [PubMed] [Google Scholar]
- 12.Blanco, M., J. E. Blanco, A. Mora, G. Dahbi, M. P. Alonso, E. A. Gonzalez, M. I. Bernardez, and J. Blanco. 2004. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from cattle in Spain and identification of a new intimin variant gene (eae-ξ). J. Clin. Microbiol. 42:645-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blanco, M., J. E. Blanco, A. Mora, J. Rey, J. M. Alonso, M. Hermoso, J. Hermoso, M. P. Alonso, G. Dahbi, E. A. Gonzalez, M. I. Bernardez, and J. Blanco. 2003. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from healthy sheep in Spain. J. Clin. Microbiol. 41:1351-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunder, W., H. Schmidt, and H. Karch. 1997. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol. Microbiol. 24:767-778. [DOI] [PubMed] [Google Scholar]
- 15.Brunder, W., H. Schmidt, and H. Karch. 1996. KatP, a novel catalase-peroxidase encoded by the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology 142:3305-3315. [DOI] [PubMed] [Google Scholar]
- 16.Campellone, K. G., D. Robbins, and J. M. Leong. 2004. EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nick-independent actin assembly. Dev. Cell 7:217-228. [DOI] [PubMed] [Google Scholar]
- 17.China, B., F. Goffaux, V. Pirson, and J. Mainil. 1999. Comparison of eae, tir, espA and espB genes of bovine and human attaching and effacing Escherichia coli by multiplex polymerase chain reaction. FEMS Microbiol. Lett. 178:177-182. [DOI] [PubMed] [Google Scholar]
- 18.China, B., E. Jacquemin, A. C. Devrin, V. Pirson, and J. Mainil. 1999. Heterogeneity of the eae genes in attaching/effacing Escherichia coli from cattle: comparison with human strains. Res. Microbiol. 150:323-332. [DOI] [PubMed] [Google Scholar]
- 19.Clark, C. G., S. T. Johnson, R. H. Easy, J. L. Campell, and F. G. Rodgers. 2002. PCR for detection of cdt-III and the relative frequencies of cytolethal distending toxin variant-producing Escherichia coli isolates from humans and cattle. J. Clin. Microbiol. 40:2671-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coimbra, R. S., F. Grimont, P. Lenormand, P. Burguiere, L. Beutin, and P. A. D. Grimont. 2000. Identification of Escherichia coli O-serogroups by restriction of the amplified O-antigen gene cluster (rfb-RFLP). Res. Microbiol. 151:639-654. [DOI] [PubMed] [Google Scholar]
- 21.Cortes, C., R. de la Fuente, J. Blanco, M. Blanco, J. E. Blanco, G. Dhabi, A. Mora, P. Justel, A. Contreras, A. Sanchez, J. C. Corrales, and J. A. Orden. 2005. Serotypes, virulence genes and intimin types of verotoxin-producing Escherichia coli and enteropathogenic E. coli isolated from healthy dairy goats in Spain. Vet. Microbiol. 110:67-76. [DOI] [PubMed] [Google Scholar]
- 22.Creuzburg, K., and H. Schmidt. 2007. Molecular characterization and distribution of genes encoding members of the type III effector NleA family among pathogenic Escherichia coli strains. J. Clin. Microbiol. 45:2498-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dahan, S., S. Wiles, R. M. La Ragione, A. Best, M. J. Woodward, M. P. Stevens, R. K. Shaw, Y. Chong, S. Knutton, A. Philips, and G. Frankel. 2005. EspJ is a prophage-carried type III effector protein of attaching and effacing pathogens that modulates infection dynamics. Infect. Immun. 73:679-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Djafari, S., F. Ebel, C. Deibel, S. Krämer, M. Hudel, and T. Chakraborty. 1997. Characterization of an exported protease from Shiga toxin-producing Echerichia coli. Mol. Microbiol. 25:771-784. [DOI] [PubMed] [Google Scholar]
- 25.Eklund, M., F. Scheutz, and A. Siitonen. 2001. Clinical isolates of non-O157 Shiga toxin-producing Escherichia coli: serotypes, virulence characteristics, and molecular profiles of strains of the same serotype. J. Clin. Microbiol. 39:2829-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eklund, M., K. Leino, and A. Siitonen. 2002. Clinical Escherichia coli strains carrying stx genes: stx variants and stx-positive virulence profiles. J. Clin. Microbiol. 40:4585-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garmendia, J., A. D. Phillips, M. F. Carlier, Y. Chong, S. Schüller, O. Marches, S Dahan, E. Oswald, R. K. Shaw, S. Knutton, and G. Frankel. 2004. TccP is an enterohaemorrhagic Escherichia coli O157:H7 type III effector protein that couples Tir to the actin-cytoskeleton. Cell. Microbiol. 6:1167-1183. [DOI] [PubMed] [Google Scholar]
- 28.Geue, L., S. Klare, C. Schnick, B. Mintel, K. Meyer, and F. J. Conraths. 2009. Analysis of the clonal relationship of serotype O26:H11 enterohemorrhagic Escherichia coli isolates from cattle. Appl. Environ. Microbiol. 75:6947-6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geue, L., T. Selhorst, C. Schnick, B. Mintel, and F. J. Conraths. 2006. Analysis of the clonal relationship of Shiga toxin-producing Escherichia coli serogroup O165:H25 isolated from cattle. Appl. Environ. Microbiol. 72:2254-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geue, L., M. Segura-Alvarez, F. J. Conraths, T. Kuczius, J. Bockemuhl, H. Karch, and P. Gallien. 2002. A long-term study on the prevalence of Shiga toxin-producing Escherichia coli (STEC) on four German cattle farms. Epidemiol. Infect. 129:173-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gruenheid, S., I. Sekirov, N. A. Thomas, W. Deng, P. O'Donnell, D. Goode, Y. Li, E. A. Frey, N. F. Brown, P. Metalnikov, T. Pawson, K. Ashman, and B. B. Finlay. 2004. Identification and characterization of NleA, a non-LEE-encoded type III translocated virulence factor of enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 51:1233-1249. [DOI] [PubMed] [Google Scholar]
- 32.Johnson, K. E., C. M. Thorpe, and C. L. Sears. 2006. The emerging clinical importance of non-O157 Shiga toxin-producing Escherichia coli. Clin. Infect. Dis. 43:1587-1595. [DOI] [PubMed] [Google Scholar]
- 33.Jores, J., K. Zehmke, J. Eichberg, L. Rumer, and L. H. Wieler. 2003. Description of a novel intimin variant (type zeta) in the bovine O84:NM verotoxin-producing Escherichia coli strain 537/89 and the diagnostic value of intimin typing. Exp. Biol. Med. (Maywood) 228:370-376. [DOI] [PubMed] [Google Scholar]
- 34.Karmali, M. A. 1989. Infection by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2:15-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelly, M., E. Hart, R. Mundy, O. Marches, S. Wiles, L. Badea, S. Luck, M. Tauschek, G. Frankel, R. M. Robins-Browne, and E. L. Hartland. 2006. Essential role of the type III secretion system effector NleB in colonization of mice by Citrobacter rodentium. Infect. Immun. 74:2328-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein, G., and M. Bülte. 2003. Antibiotic susceptibility pattern of Escherichia coli strains with verocytotoxic E. coli-associated virulence factors from food and animal faeces. Food Microbiol. 20:27-33. [Google Scholar]
- 37.Lehmacher, A., H. Meier, S. Aleksic, and J. Bockemühl. 1998. Detection of hemolysin variants of Shiga toxin-producing Escherichia coli by PCR and culture on vancomycin-cefixime-cefsulodin blood agar. Appl. Environ. Microbiol. 64:2449-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liebisch, B., and S. Schwarz. 1996. Evaluation and comparison of molecular techniques for epidemiological typing of Salmonella enterica subsp. enterica serovar Dublin. J. Clin. Microbiol. 34:641-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manna, S. K., C. Manna, K. Batabyal, B. Das, D. Golder, S. Chattopadhyay, and B. K. Biswas. 2010. Serogroup distribution and virulence characteristics of sorbitol-negative Escherichia coli from food and cattle stool. J. Appl. Microbiol. 108:658-665. [DOI] [PubMed] [Google Scholar]
- 40.Marches, O., S. Wiles, F. Dziva, R. M. La Ragione, S. Schüller, A. Best, A. D. Philips, E. L. Hartland, M. J. Woodward, M. P. Stevens, and G. Frankel. 2005. Characterization of two non-locus of enterocyte effacement-encoded type III-translocated effectors, NleC and NleD, in attaching and effacing pathogens. Infect. Immun. 73:8411-8417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marches, O., T. N. Ledger, M. Boury, M. Ohara, X. Tu, F. Goffaux, J. Mainil, I. Rosenshine, M. Sugai, J. De Rycke, and E. Oswald. 2003. Enteropathogenic and eterohaemorrhagic Escherichia coli deliver a novel effector called Cif, which blocks cell cycle G2/M transition. Mol. Microbiol. 50:1553-1567. [DOI] [PubMed] [Google Scholar]
- 42.Mellmann, A., M. Bielaszewska, R. Köck, A. W. Friedrich, A. Fruth, B. Middendorf, D. Harmsen, M. A. Schmidt, and H. Karch. 2008. Analysis of collection of hemolytic uremic syndrome-associated enterohemorrhagic Escherichia coli. Emerg. Infect. Dis. 14:1287-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monecke, S., and R. Ehricht. 2005. Rapid genotyping of methicillin-resistant Staphylococcus aureus (MRSA) isolates using miniaturized microarrays. Clin. Microbiol. Infect. 11:825-833. [DOI] [PubMed] [Google Scholar]
- 44.Mundy, R., L. Petrovska, K. Smollett, N. Simpson, R. K. Wilson, J. Yu, X. Tu, I. Rosenshine, S. Clare, G. Dougan, and G. Frankel. 2004. Identification of a novel Citrobacter rodentium type III secreted protein, EspI, and roles of this and other secreted proteins in infection. Infect. Immun. 72:2288-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orden, J. A., D. Cid, J. A. Ruiz-Santa-Quiteria, S. Garcia, S. Martinez, and R. de la Fuente. 2002. Verotoxin-producing Escherichia coli (VTEC), enteropathogenic E. coli (EPEC) and necrotoxigenic E. coli (NTEC) isolated from healthy cattle in Spain. J. Appl. Microbiol. 93:29-35. [DOI] [PubMed] [Google Scholar]
- 47.Osek, J. 2003. Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1) gene and its relationship with fimbrial and enterotoxin markers in E. coli isolates from pigs with diarrhea. Vet. Microbiol. 91:65-72. [DOI] [PubMed] [Google Scholar]
- 48.Oswald, E., H. Schmidt, S. Morabito, H. Karch, O. Marches, and A. Caprioli. 2000. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect. Immun. 68:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 50.Rey, J., J. E. Blanco, M. Blanco, A. Mora, G. Dahbi, J. M. Alonso, M. Hermoso, J. Hermoso, M. P. Alonso, M. A. Usera, E. A. Gonzalez, M. I. Bernardez, and J. Blanco. 2003. Serotypes, phage types and virulence genes of Shiga-producing Escherichia coli isolated from sheep in Spain. Vet. Microbiol. 94:47-56. [DOI] [PubMed] [Google Scholar]
- 51.Saitou, N., and M. Nei. 1987. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 52.Sandhu, K. S., R. C. Clarke, K. McFadden, A. Brouwer, M. Louie, J. Wilson, H. Lior, and C. L. Gyles. 1996. Prevalence of the eaeA gene in verotoxigenic Escherichia coli strains from dairy cattle in Southwest Ontario. Epidemiol. Infect. 116:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt, H., H. Rüssmann, A. Schwarzkopf, S. Aleksic, J. Heesemann, and H. Karch. 1994. Differentiation in virulence patterns of Escherichia coli possessing eae genes. Med. Microbiol. Immunol. 183:23-31. [DOI] [PubMed] [Google Scholar]
- 54.Segura-Alvarez, M., H. Richter, F. J. Conraths, and L. Geue. 2003. Evaluation of enzyme-linked immunosorbent assays and a PCR test for detection of Shiga toxins for Shiga toxin-producing Escherichia coli in cattle herds. J. Clin. Microbiol. 41:5760-5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharma, V. K. 2002. Detection and quantitation of enterohemorrhagic Escherichia coli O157, O111, and O26 in beef and bovine feces by real-time polymerase chain reaction. J. Food Prot. 65:1371-1380. [DOI] [PubMed] [Google Scholar]
- 56.Vernozy-Rozand, C., M. P. Montet, Y. Bertin, F. Trably, J. P. Girardeau, C. Martin, V. Livrelli, and L. Beutin. 2004. Serotyping, stx2 subtyping, and characterization of the locus of enterocyte effacement island of Shiga toxin-producing Escherichia coli and E. coli O157:H7 strains isolated from the environment in France. Appl. Environ. Microbiol. 70:2556-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, G., C. G. Clark, and F. G. Rodgers. 2002. Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157:H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J. Clin. Microbiol. 40:3613-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wieler, L. H., B. Busse, H. Steinrück, L. Beutin, A. Weber, H. Karch, and G. Baljer. 2000. Enterohemorrhagic Escherichia coli (EHEC) strains of serogroup O118 display three distinctive clonal groups of EHEC pathogens. J. Clin. Microbiol. 38:2162-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson, J. B., R. C. Clarke, S. Renwick, K. Rahn, R. P. Johnson, M. A. Karmali, H. Lior, D. Alves, C. L. Gyles, K. S. Sandhu, S. A. McEwen, and J. S. Spika. 1996. Vero cytotoxigenic Escherichia coli infection in dairy farm families. J. Infect. Dis. 174:1021-1027. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, W. L., B. Kohler, E. Oswald, L. Beutin, H. Karch, S. C. Morabito, A. Caprioli, S. Suerbaum, and H. Schmidt. 2002. Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. J. Clin. Microbiol. 40:4486-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.