Abstract
Dendritic cells (DCs) are of pivotal importance for the initiation of immune responses to control and eliminate viral infections. The molecular mechanisms of hepatitis C virus (HCV) antigen uptake and processing by blood DCs are poorly defined. Here we show that human blood DC subsets acquire HCV independent of the classical HCV entry factors. Following HCV uptake, human plasmacytoid and myeloid DC subsets deliver HCV antigen into distinct endocytotic compartments, which are dedicated to presentation to CD4+ or CD8+ T cells. Our findings support a model of HCV antigen processing and presentation in which DC subsets fulfill distinct functions.
Dendritic cells (DCs) are crucial for the initiation of antiviral immunity. As sentinels of the immune system, DCs capture viral antigens and present them to naive T cells, eliciting immunity. Two distinct pathways are known for the presentation of antigenic peptides on major histocompatibility complex (MHC) molecules. Exogenous antigens are processed and loaded on MHC II molecules to activate CD4+ T cells. In contrast, endogenous antigens are presented by MHC I molecules and activate cytotoxic CD8+ T cells. Additionally, some forms of exogenous antigens can gain access to the MHC I presentation pathway (19). This process, known as cross-presentation, is crucial in the initiation of antiviral immunity when DCs are not targeted directly by viruses or when an infecting virus compromises DC function. Two major DC subsets exist in humans, namely, myeloid DCs (mDCs) and plasmacytoid DCs (pDCs). mDCs are more efficient in priming T-cell responses, while pDCs are the most potent producers of type I interferons (IFN-α/β) upon viral infection (27).
Among the multiple blood-borne viruses, hepatitis C virus (HCV) has created a global health problem of substantial proportions. HCV is a major cause of chronic liver inflammation worldwide and is the leading indication for liver transplantation in developed countries. Therapy for HCV-related chronic hepatitis is limited, and a vaccine protecting against HCV infection is currently unavailable (26). In those individuals who clear the acute infection, a robust and sustained CD4+ and CD8+ T-cell response to multiple HCV epitopes is readily detectable in the blood, suggesting efficient HCV-specific T-cell priming by DCs. Although individuals who develop chronic infection can mount early, vigorous effector T-cell responses, these responses are transient or narrowly focused (17). Recent in vitro studies indicated that HCV is a weak inducer of IFN-α and impairs Toll-like receptor 9 (TLR9)-mediated IFN-α production in plasmacytoid DCs (6, 12, 18), suggesting that HCV attenuates the early innate immune response. Although several ex vivo studies demonstrated impaired DC-mediated cytokine production and maturation in patients with chronic HCV infection (reviewed in reference 11), there is little evidence of global immune dysfunction in these patients. In this context, with a view to mapping the contribution of DCs to initiating an HCV-specific cellular response, we decided to undertake an investigation into a poorly understood aspect of DC biology, namely, the pathways through which human blood DC subsets internalize circulating HCV, and the subsequent intracellular antigen trafficking following HCV uptake by human blood DC subsets.
To elucidate the pathways by which human blood DC subsets acquire HCV, we purified pDCs and mDCs from the blood of healthy subjects and analyzed the expression of putative HCV entry factors, such as the tetraspanin CD81, the scavenger receptor class B type I (SR-BI), and the tight junction protein claudin-1 (CLDN1) (4), on the DC surface. Freshly isolated pDCs (BDCA2+ CD11c− IL-3Rhigh) and mDCs (BDCA2− CD11c+ IL-3Rlow) exhibited an immature phenotype, as demonstrated by the absence of CD80 and the low or moderate expression level of CD86 (Fig. 1A). Using mouse monoclonal anti-CD81 (JS-81; BD Biosciences), polyclonal rat anti-SR-BI (28), and rat anti-CLDN1 (9) antibodies, which target the extracellular domains of the distinct HCV entry factors, along with flow cytometry, we found that different human blood DC subsets express distinct profiles of HCV entry factors. Although both DC subsets expressed low levels of SR-BI, CD81 expression levels were significantly lower in pDCs than in mDCs (P = 0.023). Cell surface expression levels of CLDN1 were lower for pDCs than for mDCs, but this difference did not reach statistical significance (P = 0.144) (Fig. 1B and C). The distinct profiles of HCV entry factors on the DC surface may lead to different interactions of mDCs and pDCs with HCV. To test this hypothesis, we incubated freshly isolated blood DC subsets with iodixanol-purified cell culture-derived infectious HCV (HCVcc) produced in Huh7.5.1 cells by electroporation of the JFH1 genotype 2a genome (24). The produced virus was passaged several times to obtain a high virus titer. Prior to the addition of HCVcc, blood DC subsets were incubated for 45 min with either anti-CD81, anti-SR-BI, or anti-CLDN1 antibody. These antibodies have been shown to inhibit HCVcc infection of Huh7.5.1 cells (9, 28). Following incubation with HCVcc for 2 h at 37°C, mDCs and pDCs were extensively washed, and HCVcc acquisition was quantified by real-time reverse transcription-PCR (RT-PCR) as described previously (16). In brief, total RNA was harvested using an RNeasy Mini kit (Qiagen). HCV RNA was amplified from 100 ng total RNA by use of a SensiMix one-step kit (Labgene Scientific) and was detected using a Rotor-Gene 6000 instrument (Corbett Life Science). The forward and reverse primer sequences were 5′-TCT GCG GAA CCG GTG AGT A-3′ and 5′-GGG CAT AGA GTG GGT TTA TCC-3′, respectively. The probe sequence was 5′-6-carboxyfluorescein-AAA GGA CCC AGT CTT CCC GGC AA-6-carboxytetramethylrhodamine-3′. As shown in Fig. 2A, HCV acquisition by both DC subsets was not altered markedly in the presence of antibodies targeting CD81, SR-BI, or CLDN1 on the DC surface. We were surprised that SR-BI blocking had no effect on HCVcc acquisition by mDCs, since our previous studies indicated a prominent role for SR-BI in HCVcc uptake by monocyte-derived DCs (2). This finding may be explained by the very low expression level of SR-BI on both DC subsets compared to that on in vitro-generated monocyte-derived DCs (2). This observation is supported by the work of Marukian and colleagues (13), who described a >3-log lower expression level of SR-BI RNA in blood DC subsets than in in vitro-generated monocyte-derived DCs. These results suggest that HCV entry factors, which play an important role in mediating HCV entry into hepatocytes (1), are not involved in mediating HCV acquisition by blood DC subsets and that other cell surface molecules, e.g., members of the C-type lectin family, may be involved in HCV acquisition. To address this possibility, we analyzed cell surface expression of DC-SIGN, which has previously been shown to bind recombinant HCV E2 protein (10). DC-SIGN was not detected on the cell surface for either DC subset (Fig. 1B). Our results confirm previously published results by other groups who also did not find DC-SIGN expression on freshly isolated blood DC subsets (15, 21). Additionally, blocking of DC-SIGN by anti-DC-SIGN antibodies (clone 120507; Abcam) did not inhibit HCVcc acquisition by mDCs and pDCs (Fig. 2A), suggesting that HCVcc acquisition by DC subsets does not depend on the C-type lectin DC-SIGN.
FIG. 1.
Phenotypic characterization of mDCs and pDCs. (A) Surface expression of BDCA2, CD11c, interleukin-3 receptor (IL-3R), CD80, and CD86 on freshly isolated pDCs and mDCs was assessed by flow cytometry, using phycoerythrin-conjugated monoclonal antibodies (dark gray histograms) and isotype control antibodies (light gray histograms). (B) Expression of the HCV entry factors CD81, SR-BI, and CLDN1 and the C-type lectin DC-SIGN on pDCs and mDCs was analyzed by flow cytometry, using anti-CD81, anti-SR-BI, anti-claudin-1, and anti-DC-SIGN specific antibodies (open black histograms) and the respective isotype controls (light gray histograms). (C) Expression levels of HCV entry factors on DC subsets are shown as mean fluorescence intensity (MFI) rates. The MFI rate corresponds to the MFI of cells labeled with specific antibody divided by the MFI of cells labeled with the matched isotype control. The means ± standard deviations for independent experiments with pDCs and mDCs from different donors are shown (for CD81, n = 7 donors; for SR-BI, n = 6 donors; and for CLDN1, n = 5 donors). Statistical analysis was carried out using the Mann-Whitney U test. Tests of significance were two-sided, and a P value of ≤0.05 was considered significant (P = 0.023 for comparing CD81 expression levels of mDCs and pDCs). n.s., not significant.
FIG. 2.
HCVcc acquisition by mDCs and pDCs. (A) Role of HCV entry factors in HCVcc acquisition by pDCs and mDCs. DC subsets were preincubated with mouse anti-CD81 (5 μg/ml), rat anti-SR-BI (1:50 dilution), rat anti-claudin 1 (1:50 dilution), anti-DC-SIGN (5 μg/ml), irrelevant mouse IgG (5 μg/ml), or rat IgG (1:50 dilution) for 45 min at 37°C. DC subsets were then incubated with HCVcc (multiplicity of infection [MOI], ∼0.01) for 2 h at 37°C. After extensive washing, the presence of HCVcc RNA was measured by real-time quantitative RT-PCR. As negative controls, DC subsets were incubated with phosphate-buffered saline (PBS) or control iodixanol preparations obtained from culture supernatants of noninfected Huh7.5.1 cells (NC [negative control]). The data represent the means ± standard deviations for triplicate samples from one experiment. The results shown are representative of three independent experiments, using three different donors. (B) Visualization of HCVcc uptake into pDCs and mDCs. For HCVcc internalization, DC subsets were incubated with iodixanol gradient-purified HCVcc (MOI, ∼10) for 3 h at 37°C. Following fixation and permeabilization, cells were stained for HCVcc E2 protein by use of mouse anti-E2 monoclonal antibody (AP33; green), and the nucleus was stained with DAPI (4′,6-diamidino-2-phenylindole; blue). Arrows indicate detectable HCVcc E2 protein. To confirm the specificity of HCV E2 protein labeling, DC subsets were incubated with control iodixanol preparations obtained from culture supernatants of noninfected Huh7.5.1 cells and subsequently labeled with anti-E2 (AP33) antibody (NC [negative control]). (C) For HCVcc double labeling, DC subsets were incubated with mouse anti-E2 (AP33; green) and rabbit anti-core antibody (red). The yellow areas in the overlays indicate colocalization of these two proteins in mDCs and pDCs. (D) HCVcc uptake by pDCs and mDCs was quantified by counting the number of cells that stained positive for E2 protein relative to the total number of cells (n = 300). The results for pDCs and mDCs isolated from five different donors are shown. Statistical analysis was performed using the Mann-Whitney U test. Tests of significance were two-sided, and a P value of ≤0.05 was considered significant. (E) Inhibition of HCVcc uptake in the presence of human anti-HCV serum (1:50 dilution) and antibodies targeting the HCV entry factors (rabbit anti-CD81, 6 μg/ml; rat anti-SR-BI and rat anti-CLDN1, 1:50 dilution). HCVcc uptake by DC subsets was analyzed as described for panel D. The results are shown as the percent inhibition of HCVcc uptake compared to HCVcc uptake in the presence of the respective isotype controls (irrelevant IgG or human control serum) for pDCs and mDCs from four different donors.
Following virus capture, viral antigens are delivered into intracellular compartments which are dedicated to processing and presenting viral antigens to T cells (19). To investigate HCVcc uptake and trafficking in DC subsets, we monitored HCVcc uptake in mDCs and pDCs by HCV E2-specific immunofluorescence and confocal laser scanning microscopy, using a mouse anti-E2 monoclonal antibody (AP33) targeting epitope E2412-423 (22). To confirm the specificity of HCVcc E2 protein staining, DC subsets were incubated with control iodixanol preparations obtained from culture supernatants of noninfected Huh7.5.1 cells and subsequently stained with anti-E2 (AP33) antibody. HCVcc E2 protein was detectable in both DC subsets, while no fluorescence signal was observed in DCs incubated with control iodixanol preparations (Fig. 2B). Furthermore, no fluorescence signal was detected in either DC subset following incubation with HCVcc and subsequent labeling with irrelevant mouse IgG (Invitrogen) (data not shown). Finally, to verify the specificity of HCVcc E2 staining, DC subsets were costained with rabbit anti-core antibody (LifeSpan Biosciences) or irrelevant rabbit IgG (Bethyl Laboratories). Double staining demonstrated colocalization of HCVcc core and E2 protein in both DC subsets, indicating the uptake of specific components of the HCVcc particle (Fig. 2C). Quantification of HCVcc uptake by counting the average number of cells staining positive for HCVcc E2 protein per 300 cells revealed that following 3 h of incubation with HCVcc, mDCs were significantly more efficient at HCVcc uptake than pDCs (P = 0.012) (Fig. 2D). This may be related to the intrinsically lower capability of pDCs than that of conventional DCs for endocytosing antigens (8). To investigate whether HCVcc uptake is mediated by the HCV envelope glycoprotein E2, HCVcc was preincubated with human serum containing high-titer anti-E2 antibodies (20) that efficiently blocked HCVcc infection of Huh7.5.1 cells (24). As shown in Fig. 2E, preincubation of HCVcc with anti-HCV E2-containing serum markedly reduced HCVcc uptake by mDCs (40% ± 14.5% inhibition of HCVcc uptake compared to that with human control serum; P = 0.02), whereas a less potent effect was observed on pDCs (13.4% ± 10.3% inhibition of HCVcc uptake compared to that with human control serum; P = 0.114). These findings suggest that HCVcc uptake by DC subsets is partially mediated by HCVcc E2 protein interaction at the cell surface.
Next, to gain a deeper insight into the mechanisms involved in HCVcc uptake by DC subsets, we studied HCVcc internalization by E2-specific immunofluorescence following blockage of the HCV entry factors. Preincubation of mDCs and pDCs with polyclonal rabbit anti-CD81 (Sigma-Aldrich), rat anti-SR-BI (28), or rat anti-CLDN1 (9) antibody did not markedly inhibit HCVcc uptake (Fig. 2E). These results suggest that HCV cell entry factors which are crucial for viral uptake in hepatocytes do not support HCVcc uptake in DC subsets. One possible explanation may be the fact that DC subsets lack the expression of at least one HCV entry factor or that they do not express sufficient levels of HCV entry factors (Fig. 1B) (13). Thus, future studies are required to identify cell surface molecules on mDCs and pDCs that trigger HCVcc endocytosis. However, we cannot rule out the possibility that HCV enters DC subsets via pinocytosis (14), a constitutive mechanism by which DCs acquire exogenous solute as part of their sentinel function.
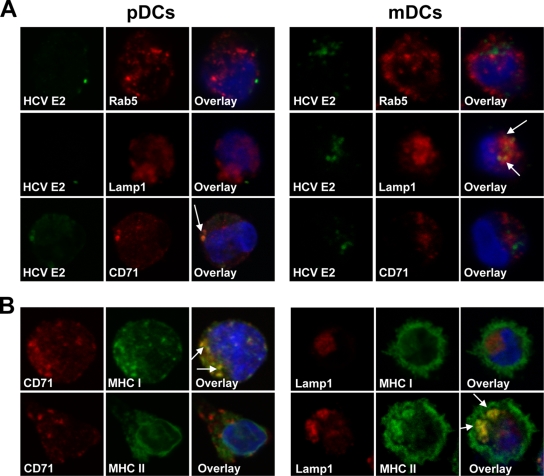
To localize internalized HCVcc particles within DC subsets following HCV uptake, we costained mDCs and pDCs for the early endosome marker Rab5, the lysosome-associated membrane protein 1 (Lamp1), and the transferrin receptor CD71, a marker of recycling endosomes. Within pDCs, HCVcc localized in organelles expressing the transferrin receptor CD71 but not Rab5 or Lamp1, indicating that HCVcc antigen accumulates in recycling endosomes (Fig. 3A). In contrast, HCVcc costained with Lamp1 in mDCs, but not with Rab5 or CD71, suggesting the localization of HCVcc antigen in lysosomal compartments (Fig. 3A). This distinction in HCV antigen localization in the DC subsets was preserved 1 and 3 h after HCV uptake, indicating that differences observed in antigen localization were not due to differences in HCVcc uptake kinetics of mDCs and pDCs. Together, these data suggest that HCV antigens traffic within distinct intracellular compartments in pDCs and mDCs.
FIG. 3.
Distinct intracellular HCVcc trafficking in pDCs and mDCs. (A) DC subsets were incubated for 3 h at 37°C with HCVcc (MOI, ∼10). Cells were then fixed and stained for HCV E2 protein (green) and endocytotic compartments (red), such as early endosomes (Rab5), recycling endosomes (CD71), and lysosomes (Lamp1). Colocalization of HCVcc E2 protein and endocytotic compartments is shown in yellow (indicated by arrows). Cell nuclei were revealed with DAPI staining (blue). In pDCs, HCVcc E2 protein was localized in recycling endosomes, identified by CD71, but not in endocytotic compartments containing Rab5 or Lamp1. In contrast, HCVcc E2 protein colocalized with the lysosome antigen Lamp1 in mDCs, but not with CD71 or Rab5. Representative results from one of three independent experiments are shown. (B) Characterization of the endocytotic compartment containing HCVcc E2 protein in pDCs and mDCs. Plasmacytoid DCs were stained for recycling endosomes (CD71; red) and MHC class I or II molecules (green). The overlay image shows colocalization of CD71 with MHC I but not MHC II molecules (indicated by arrows), indicating that recycling endosomes contain MHC class I molecules. Myeloid DCs were stained for lysosomes (Lamp1; red) and MHC I or II molecules (green). The overlay image shows colocalization of lysosomes with MHC II but not MHC I molecules, indicating that lysosomes contain MHC class II molecules. Data are representative of two independent experiments.
The endosomes and lysosomes of antigen-presenting cells host the processing and assembly reactions that result in the display of peptides on MHC class II molecules. For mDCs, it is known that rapid presentation of exogenous antigens involves loading of peptides onto MHC class II molecules found in late endosomes and lysosomes (25). To further analyze the lysosomal compartment in mDCs harboring HCVcc particles, lysosomal compartments were costained with anti-MHC class I and II antibodies. As shown in Fig. 3B, lysosomal compartments of mDCs colocalized with cytoplasmic pools of MHC class II molecules but not with MHC class I molecules, indicating that HCVcc particles appear to be processed in lysosomes containing MHC class II molecules. Interestingly, in pDCs, the HCVcc-containing, recycling endosomal compartment colocalized with MHC class I molecules but not with MHC class II molecules (Fig. 3B). These findings suggest a mechanism by which HCVcc antigen is delivered to recycling endosomal compartments for processing and loading onto presynthesized pools of MHC class I molecules. Clearly, we cannot exclude the possibility that HCV antigen is also released from the endocytic compartment to the cytosol of the cell, where it gains access to the classical MHC class I pathway. However, Di Pucchio and colleagues (3) demonstrated that pDCs cross-present influenza virus-derived peptides on MHC class I molecules to CD8+ T cells by delivery of internalized virus to recycling endosomes. Thus, it is very likely that HCV-derived antigens are targeted to recycling endosomal compartments to ensure a rapid activation of the HCV-specific CD8+ T-cell response by utilizing a storage pool of MHC class I molecules. However, it should be noted that our in vitro model, which uses freshly isolated blood DC subsets, probably does not fully represent the in vivo scenario. It is likely that DCs that are actively engaged in the immune response to HCV in vivo are phenotypically different from those used in this study. Furthermore, we have to point out that the majority of immunodominant CD4+ and CD8+ T-cell epitopes are derived from HCV nonstructural proteins (5, 23). The mechanism of uptake and intracellular trafficking of HCV nonstructural proteins remains to be determined.
In conclusion, our results demonstrate that HCV antigens traffic within distinct compartments of DC subpopulations. Our data implicate a model of HCV antigen processing and presentation in which mDCs and pDCs perform different functions in HCV-specific CD4+ and CD8+ T-cell stimulation. Since resolution of HCV infection is characterized by a robust CD4+ T-cell response which is essential for HCV-specific CD8+ T-cell function (7), the distinct HCV antigen processing pathways in mDCs and pDCs may ensure the simultaneous initiation of rapid and effective HCV-specific CD4+ and CD8+ T-cell responses. Further investigation is needed to determine whether targeting viral antigens to distinct processing and presentation pathways may present an avenue for improving HCV vaccine delivery and may aid in immunotherapeutic approaches aimed at inducing strong and sustained CD4+ and CD8+ T-cell responses against HCV.
Acknowledgments
We thank J. Barths (INSERM U748, Strasbourg, France) for assistance with flow cytometry and C. Moog, V. Holl (both from INSERM U748, Strasbourg, France), and S. Raghuraman (Liver Diseases Branch, NIH, Bethesda, MD) for helpful discussions. We thank J. Vonesch at the IGBMC (Institut de Génétique et de Biologie Moléculaire et Cellulaire Centre d'Imagerie, Strasbourg, France) for support in confocal microscopy analysis.
This work was supported by the Deutsche Forschungsgemeinschaft, Germany (BA 3643/1-1 to H.B.); the Agence Nationale de la Recherche and the Agence Nationale de Recherche sur le SIDA et les Hépatites Virales, France (2009-183); the Else-Kröner-Fresenius Stiftung, Bad Homburg, Germany (P17//07//A83/06); Dormeur Investment Service, Ltd.; and the European Union (ERC-2008-AdG-233130-HEPCENT).
Footnotes
Published ahead of print on 23 June 2010.
REFERENCES
- 1.Barth, H., T. J. Liang, and T. F. Baumert. 2006. Hepatitis C virus entry: molecular biology and clinical implications. Hepatology 44:527-535. [DOI] [PubMed] [Google Scholar]
- 2.Barth, H., E. K. Schnober, C. Neumann-Haefelin, C. Thumann, M. B. Zeisel, H. M. Diepolder, Z. Hu, T. J. Liang, H. E. Blum, R. Thimme, M. Lambotin, and T. F. Baumert. 2008. Scavenger receptor class B is required for hepatitis C virus uptake and cross-presentation by human dendritic cells. J. Virol. 82:3466-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Pucchio, T., B. Chatterjee, A. Smed-Sorensen, S. Clayton, A. Palazzo, M. Montes, Y. Xue, I. Mellman, J. Banchereau, and J. E. Connolly. 2008. Direct proteasome-independent cross-presentation of viral antigen by plasmacytoid dendritic cells on major histocompatibility complex class I. Nat. Immunol. 9:551-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubuisson, J., F. Helle, and L. Cocquerel. 2008. Early steps of the hepatitis C virus life cycle. Cell. Microbiol. 10:821-827. [DOI] [PubMed] [Google Scholar]
- 5.Gerlach, J. T., A. Ulsenheimer, N. H. Gruner, M. C. Jung, W. Schraut, C. A. Schirren, M. Heeg, S. Scholz, K. Witter, R. Zahn, A. Vogler, R. Zachoval, G. R. Pape, and H. M. Diepolder. 2005. Minimal T-cell-stimulatory sequences and spectrum of HLA restriction of immunodominant CD4+ T-cell epitopes within hepatitis C virus NS3 and NS4 proteins. J. Virol. 79:12425-12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gondois-Rey, F., C. Dental, P. Halfon, T. F. Baumert, D. Olive, and I. Hirsch. 2009. Hepatitis C virus is a weak inducer of interferon alpha in plasmacytoid dendritic cells in comparison with influenza and human herpesvirus type-1. PLoS One 4:e4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grakoui, A., N. H. Shoukry, D. J. Woollard, J. H. Han, H. L. Hanson, J. Ghrayeb, K. K. Murthy, C. M. Rice, and C. M. Walker. 2003. HCV persistence and immune evasion in the absence of memory T cell help. Science 302:659-662. [DOI] [PubMed] [Google Scholar]
- 8.Grouard, G., M. C. Rissoan, L. Filgueira, I. Durand, J. Banchereau, and Y. J. Liu. 1997. The enigmatic plasmacytoid T cells develop into dendritic cells with interleukin (IL)-3 and CD40-ligand. J. Exp. Med. 185:1101-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krieger, S. E., M. B. Zeisel, C. Davis, C. Thumann, H. J. Harris, E. K. Schnober, C. Mee, E. Soulier, C. Royer, M. Lambotin, F. Grunert, V. L. Dao Thi, M. Dreux, F. L. Cosset, J. A. McKeating, C. Schuster, and T. F. Baumert. 2010. Inhibition of hepatitis C virus infection by anti-claudin-1 antibodies is mediated by neutralization of E2-CD81-claudin-1 associations. Hepatology 51:1144-1157. [DOI] [PubMed] [Google Scholar]
- 10.Lai, W. K., P. J. Sun, J. Zhang, A. Jennings, P. F. Lalor, S. Hubscher, J. A. McKeating, and D. H. Adams. 2006. Expression of DC-SIGN and DC-SIGNR on human sinusoidal endothelium: a role for capturing hepatitis C virus particles. Am. J. Pathol. 169:200-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambotin, M., S. Raghuraman, F. Stoll-Keller, T. F. Baumert, and H. Barth. 2010. A look behind closed doors: interaction of persistent viruses with dendritic cells. Nat. Rev. Microbiol. 8:350-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang, H., R. S. Russell, N. L. Yonkers, D. McDonald, B. Rodriguez, C. V. Harding, and D. D. Anthony. 2009. Differential effects of hepatitis C virus JFH1 on human myeloid and plasmacytoid dendritic cells. J. Virol. 83:5693-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marukian, S., C. T. Jones, L. Andrus, M. J. Evans, K. D. Ritola, E. D. Charles, C. M. Rice, and L. B. Dustin. 2008. Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology 48:1843-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mercer, J., and A. Helenius. 2009. Virus entry by macropinocytosis. Nat. Cell Biol. 11:510-520. [DOI] [PubMed] [Google Scholar]
- 15.Meyer-Wentrup, F., D. Benitez-Ribas, P. J. Tacken, C. J. Punt, C. G. Figdor, I. J. de Vries, and G. J. Adema. 2008. Targeting DCIR on human plasmacytoid dendritic cells results in antigen presentation and inhibits IFN-alpha production. Blood 111:4245-4253. [DOI] [PubMed] [Google Scholar]
- 16.Pietschmann, T., M. Zayas, P. Meuleman, G. Long, N. Appel, G. Koutsoudakis, S. Kallis, G. Leroux-Roels, V. Lohmann, and R. Bartenschlager. 2009. Production of infectious genotype 1b virus particles in cell culture and impairment by replication enhancing mutations. PLoS Pathog. 5:e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rehermann, B. 2009. Hepatitis C virus versus innate and adaptive immune responses: a tale of coevolution and coexistence. J. Clin. Invest. 119:1745-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiina, M., and B. Rehermann. 2008. Cell culture-produced hepatitis C virus impairs plasmacytoid dendritic cell function. Hepatology 47:385-395. [DOI] [PubMed] [Google Scholar]
- 19.Steinman, R. M., and H. Hemmi. 2006. Dendritic cells: translating innate to adaptive immunity. Curr. Top. Microbiol. Immunol. 311:17-58. [DOI] [PubMed] [Google Scholar]
- 20.Steinmann, D., H. Barth, B. Gissler, P. Schurmann, M. I. Adah, J. T. Gerlach, G. R. Pape, E. Depla, D. Jacobs, G. Maertens, A. H. Patel, G. Inchauspe, T. J. Liang, H. E. Blum, and T. F. Baumert. 2004. Inhibition of hepatitis C virus-like particle binding to target cells by antiviral antibodies in acute and chronic hepatitis C. J. Virol. 78:9030-9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun, P., S. Fernandez, M. A. Marovich, D. R. Palmer, C. M. Celluzzi, K. Boonnak, Z. Liang, H. Subramanian, K. R. Porter, W. Sun, and T. H. Burgess. 2009. Functional characterization of ex vivo blood myeloid and plasmacytoid dendritic cells after infection with dengue virus. Virology 383:207-215. [DOI] [PubMed] [Google Scholar]
- 22.Tarr, A. W., A. M. Owsianka, J. M. Timms, C. P. McClure, R. J. Brown, T. P. Hickling, T. Pietschmann, R. Bartenschlager, A. H. Patel, and J. K. Ball. 2006. Characterization of the hepatitis C virus E2 epitope defined by the broadly neutralizing monoclonal antibody AP33. Hepatology 43:592-601. [DOI] [PubMed] [Google Scholar]
- 23.Thimme, R., D. Oldach, K. M. Chang, C. Steiger, S. C. Ray, and F. V. Chisari. 2001. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J. Exp. Med. 194:1395-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watts, C. 2004. The exogenous pathway for antigen presentation on major histocompatibility complex class II and CD1 molecules. Nat. Immunol. 5:685-692. [DOI] [PubMed] [Google Scholar]
- 26.Webster, D. P., P. Klenerman, J. Collier, and K. J. Jeffery. 2009. Development of novel treatments for hepatitis C. Lancet Infect. Dis. 9:108-117. [DOI] [PubMed] [Google Scholar]
- 27.Wu, L., and Y. J. Liu. 2007. Development of dendritic-cell lineages. Immunity 26:741-750. [DOI] [PubMed] [Google Scholar]
- 28.Zeisel, M. B., G. Koutsoudakis, E. K. Schnober, A. Haberstroh, H. E. Blum, F. L. Cosset, T. Wakita, D. Jaeck, M. Doffoel, C. Royer, E. Soulier, E. Schvoerer, C. Schuster, F. Stoll-Keller, R. Bartenschlager, T. Pietschmann, H. Barth, and T. F. Baumert. 2007. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology 46:1722-1731. [DOI] [PubMed] [Google Scholar]