Abstract
The involvement of host proteins in the replication and transcription of viral RNA is a poorly understood area for many RNA viruses. For coronaviruses, it was long speculated that replication of the giant RNA genome and transcription of multiple subgenomic mRNA species by a unique discontinuous transcription mechanism may require host cofactors. To search for such cellular proteins, yeast two-hybrid screening was carried out by using the nonstructural protein 14 (nsp14) from the coronavirus infectious bronchitis virus (IBV) as a bait protein, leading to the identification of DDX1, a cellular RNA helicase in the DExD/H helicase family, as a potential interacting partner. This interaction was subsequently confirmed by coimmunoprecipitation assays with cells coexpressing the two proteins and with IBV-infected cells. Furthermore, the endogenous DDX1 protein was found to be relocated from the nucleus to the cytoplasm in IBV-infected cells. In addition to its interaction with IBV nsp14, DDX1 could also interact with the nsp14 protein from severe acute respiratory syndrome coronavirus (SARS-CoV), suggesting that interaction with DDX1 may be a general feature of coronavirus nsp14. The interacting domains were mapped to the C-terminal region of DDX1 containing motifs V and VI and to the N-terminal portion of nsp14. Manipulation of DDX1 expression, either by small interfering RNA-induced knockdown or by overexpression of a mutant DDX1 protein, confirmed that this interaction may enhance IBV replication. This study reveals that DDX1 contributes to efficient coronavirus replication in cell culture.
Coronaviruses cause severe diseases in humans and many other animal species. Severe acute respiratory syndrome coronavirus (SARS-CoV) is the causative agent of SARS (34, 45). Viruses in this family contain a single-stranded, positive-sense RNA genome of 27 to 31 kb. In cells infected with coronaviruses, six to nine mRNA species, including the genome-length mRNA1 and five to eight subgenomic mRNAs (mRNAs 2 to 9), are produced by a discontinuous RNA transcription mechanism (40, 46, 47). Avian infectious bronchitis virus (IBV), a prototype group 3 coronavirus, causes an acute and highly contagious disease in chickens, with a significant impact on the poultry industry worldwide. In IBV-infected cells, six mRNA species are produced (5). Subgenomic mRNAs 2, 3, 4, and 6 encode the four structural proteins, i.e., spike glycoprotein (S), envelope protein (E), membrane protein (M), and nucleocapsid protein (N). The 5′ two-thirds of mRNA1 comprises two large open reading frames (ORFs), 1a and 1b, and encodes polyproteins 1a and 1ab. The two polyproteins are proteolytically cleaved by virus-encoded proteinases into 15 functional nonstructural proteins (nsp2 to nsp16) (18, 25-27, 30-32, 37-39, 54, 59).
The functional roles of coronavirus nonstructural proteins in replication and transcription of viral RNAs are beginning to emerge. For example, nsp14, nsp15, and nsp16 are predicted to possess exonuclease (ExoN), uridylate-specific endoribonuclease (NendoU), and methyltransferase activities, respectively, based on sequence comparison and homology searching (52). The ExoN and NendoU activities of nsp14 and nsp15 were subsequently confirmed by biochemical and structural studies (3, 4, 19, 22, 35, 43). Coronavirus nsp14 contains 3′-to-5′ exonuclease motifs (DE-D-D) (52). SARS-CoV nsp14 substitutions for the DE-D-D residues substantially impaired or abolished this activity (35). This ExoN activity has been shown to be required for efficient RNA synthesis and may be involved in RNA proofreading (1, 8, 9, 35). Deletion of nsp14 or substitution of the ExoN active site residue resulted in a >94% reduction of RNA synthesis in cells transfected with SARS-CoV replicons (1). Alanine substitutions for the ExoN active site residues blocked the recovery of recombinant human coronavirus 229E (HCoV-229E) and resulted in severe defects in viral RNA synthesis in cells electroporated with the mutant full-length RNA (35). In contrast, alanine substitutions for the ExoN active site residues of murine hepatitis virus (MHV) and SARS-CoV resulted in the recovery of viable mutant viruses with growth defects and in 15- and 21-fold decreases in replication fidelity during passage in cells (8, 9). In a more recent study, the SARS-CoV nsp14 protein was identified as a (guanine-N7)-methyltransferase, and the core domain was mapped to the C-terminal half of the protein (6). However, it was less clear if and how cellular proteins were involved in these steps of the coronavirus life cycle. In this study, we present evidence that DDX1, a cellular RNA helicase in the DExD/H helicase family, is associated with coronavirus nsp14 and plays an important enhancement role in coronavirus RNA replication.
DDX1 belongs to the DEAD-box and related DEAH, DExH, and DExD families, commonly referred to as the DExD/H helicase family, in superfamily 2 of helicases. Proteins in the DExD/H helicase family share eight conserved motifs (motifs I, Ia, Ib, II, III, IV, V, and VI) (7). Although only a few proteins have been studied extensively, specific roles and functions may be assigned to the conserved motifs. Most proteins in this family possess ATPase, RNA-binding, unwinding, and annealing activities and are associated with all cellular processes involving RNA metabolism, including pre-mRNA processing, ribosome biogenesis, RNA decay, translation initiation, and transcription regulation (7, 15, 16, 51).
To search for cellular interacting partners, a yeast two-hybrid screen was carried out by using IBV nsp14 as a bait protein, leading to the identification of DDX1 as a potential interacting protein. This interaction was subsequently confirmed in cells coexpressing the two proteins and in IBV-infected cells. In addition, relocation of the endogenous DDX1 protein from the nucleus to the cytoplasm, with a predominant staining pattern in the viral RNA replication site, was observed in IBV-infected cells. Further manipulation of DDX1 expression in virus-infected cells revealed that DDX1 may play an auxiliary role in promotion of coronavirus RNA replication.
MATERIALS AND METHODS
Cells, virus, and antibodies.
Vero and HeLa cells were cultured in complete Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal bovine serum (FBS), penicillin (100 units/ml), and streptomycin (100 μg/ml). All cells were grown at 37°C and supplied with 5% CO2.
Vero cell-adapted IBV strain Beaudette (p65; DQ001339) (13, 29) and the recombinant vaccinia/T7 virus were propagated and titrated on Vero cells (14). Virus stocks were made by 3 repeated freeze-thaw cycles and were kept at −80°C until use. Virus titers were determined as 50% tissue culture infective doses (TCID50) as previously described (60). A recombinant IBV with hemagglutinin (HA)-tagged nsp12 (IBV-HA-RdRp) was obtained by using an established infectious cDNA system (12). The HA tag (YPYDVPDYA) was inserted between amino acids (aa) 3930 and 3940 at the N terminus of nsp12.
Polyclonal antibodies raised in rabbits against IBV N, S, and nsp14 proteins were described previously (28, 59). Anti-actin antibody was purchased from Santa Cruz Biotechnology. Rabbit anti-DDX1 antibodies for indirect immunofluorescence and Western blot assays were from Abcam (ab31963). Mouse anti-HA antibodies for indirect immunofluorescence assays were from ETC (Singapore). Mouse anti-bromodeoxyuridine triphosphate (anti-BrdUTP) antibody was from Invitrogen, and mouse anti-HA, rabbit anti-Flag, and mouse anti-Flag antibodies were from Sigma-Aldrich. Horseradish peroxidase (HRP)-linked goat anti-rabbit secondary antibody and HRP-linked goat anti-mouse secondary antibody were from Dako. HRP-Rec-protein A was from Invitrogen. Alexa Fluor 488-linked anti-rabbit IgG and Alexa Fluor 594-linked anti-mouse IgG were from Molecular Probes.
Plasmid construction.
Plasmid pGBKT7-nsp14, which carries the full-length IBV nsp14 gene, was constructed by cloning an EcoRI/BamHI-digested PCR fragment into EcoRI/BamHI-digested pGBKT7. The same, full-length IBV nsp14 gene was cloned into vector pKT0 to form construct pKT0-nsp14. Plasmid pcDNAHA-Snsp14 (aa 1 to 527) was constructed by cloning the full-length SARS-CoV nsp14 gene from strain sin27744 (nucleotides 17954 to 19534) into vector pcDNAHA under the control of a cytomegalovirus (CMV) promoter. The other two deletion constructs, pcDNAHA-Snsp14N (aa 1 to 285) and pcDNAHA-Snsp14C (aa 286 to 527), were made by cloning the corresponding PCR fragments into the vector pcDNAHA.
Reverse transcription-PCR (RT-PCR) products covering the DDX1 ORF (NM004939) were amplified from H1299 cells by using the forward primer 5′-CATGCCATGGCGGCCTTCTCCGAGAT-3′ and the reverse primer 5′-CGGGATCCTCAGAAGGTTCTGAACAGCT-3′. The PCR fragments were digested with NcoI and BamHI and cloned under the control of a bacteriophage T7 promoter into NcoI- and BamHI-digested pKT0-Flag (with a Flag tag) to form plasmid pFlag-DDX1. The pFlag-DDX1Δ1 and pFlag-DDX1Δ2 constructs were made by cloning the corresponding PCR fragments into pKT0-Flag. Plasmids pACT-DDX1Δ1, pACT-DDX1Δ2, and pACT-DDX1Δ3 were made by cloning the appropriate PCR fragments into the pACT2 vector. The full-length DDX1 gene was also cloned into vectors pKT0 and pXJ40-Flag (with a Flag tag), under the control of a CMV promoter, to form plasmids pKT0-DDX1 and pXJ-F-DDX1, respectively. Plasmids pXJ-F-DDX1m and pKT0-DDX1m, containing three mutations (D370A, E371A, and D373A), were produced by using a QuikChange site-directed mutagenesis kit (Stratagene). To make the construct pXJ-F-DDX1, PCR products containing the wild-type ORF, amplified with the primers 5′-CGGGATCCGCGGCCTTCTCCGAGATG-3′ and 5′-CCGCTCGAGTCAGAAGGTTCTGAA CAGCT-3′, were digested with BamHI and XhoI and ligated into BamHI- and XhoI-digested pXJ40-Flag. All constructs were confirmed by sequencing.
Yeast two-hybrid screening.
The IBV nsp14 gene was cloned into pGBKT7 (pGBKT7-nsp14) as bait to screen a cDNA library prepared from HeLa cells (BD Biosciences), as previously described (58). Briefly, the bait construct pGBKT7-nsp14 was first transformed into the yeast strain AH109, and 100 μg of cDNA library DNA was sequentially transformed into the transformants. The culture was plated on SD/−Leu/−Trp/−His plates, and positive colonies were selected by dotting colonies onto SD/−Leu/−Trp/−His/−X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates. Identification of potential positive genes was carried out by direct PCR amplification of individual yeast colonies followed by automated nucleotide sequencing.
Transient expression in mammalian cells.
Viral or cellular genes cloned into plasmids under the control of the T7 promoter were transiently expressed in mammalian cells by use of a vaccinia virus-T7 system, as previously described (14, 30). Briefly, HeLa cells grown to 90% confluence were infected with a recombinant vaccinia virus (vTF7-3) which expresses the T7 RNA polymerase gene for 2 h at 37°C prior to transfection. The plasmid DNA was transfected into vTF7-3-infected cells by use of Effectene transfection reagent (Qiagen) according to the manufacturer's instructions.
Viral or cellular genes cloned into plasmids under the control of a CMV promoter were transiently expressed in cells by transfection with plasmid DNA, using Lipofectamine 2000 (Invitrogen) according to standard protocols. Cells were harvested at 18 to 24 h posttransfection for coimmunoprecipitation assay or Western blotting.
Coimmunoprecipitation.
Transfected cells were lysed with 250 μl of ice-cold cell lysis buffer (140 mM NaCl, 10 mM Tris-HCl [pH 8.0], 0.5% NP-40). The total cell lysates were clarified by centrifugation at 13,000 rpm for 10 min, and the first antibody was added to the supernatants. After incubation at room temperature for 1 h, protein A-Sepharose beads were added, and incubation was continued for an additional 1 h. In some cases, M2 anti-Flag Sepharose beads (Sigma) were used instead of the primary antibodies and protein A-Sepharose beads. The precipitates were collected by centrifugation, and the beads were washed five times with lysis buffer before being subjected to SDS-PAGE and Western blotting.
For coimmunoprecipitation of DDX1 and nsp14 in IBV-infected cells, Vero cells were mock infected or infected with IBV at a multiplicity of infection (MOI) of approximately 1 PFU/cell and were harvested at 8 and 10 h postinfection. Cells from one 175-cm2 flask were lysed with 2 ml lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.5% NP-40, 0.01% sodium deoxycholate) plus 10 μg/ml of RNase A and 1 protease inhibitor tablet/40 ml (Roche) at 4°C for 1 h. The cytoplasmic extracts were preclarified with protein A/G-agarose beads (Santa Cruz Biotechnology) at 4°C for 30 min. Immunoprecipitation was carried out by mixing the clarified lysates with anti-nsp14 antibodies at 4°C for 3 h, followed by addition of protein A/G-agarose beads and rotation at 4°C for 1 h. The precipitates were washed seven times with the same lysis buffer with rotation for 5 min each time, and polypeptides were eluted with SDS-PAGE loading buffer and then subjected to Western blot analyses. In Western blot assays, HRP-Rec-protein A (Invitrogen) was used instead of goat anti-rabbit-HRP secondary antibody to reduce the IgG background from the antibodies used in the immunoprecipitation step.
Western blot analysis.
The precipitates from coimmunoprecipitation assays or total cell lysates dissolved in 2× SDS loading buffer were subjected to SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad). The membranes then were blocked for 1 h in blocking buffer (5% fat-free milk powder in 1× phosphate-buffered saline with 0.3% Triton X-100 [PBST]) and incubated with 1:2,000-diluted primary antibodies in blocking buffer for 2 h at room temperature. After being washed three times with PBST, the membrane was incubated with 1:2,000-diluted anti-mouse or anti-rabbit IgG antibodies conjugated with horseradish peroxidase (Dako) in blocking buffer for 1 h at room temperature. After membranes were washed three times with PBST, the polypeptides were detected with a chemiluminescence detection kit (ECL; Amersham Biosciences) according to the manufacturer's instructions.
Electroporation of IBV RNA together with siRNA targeting DDX1 or in vitro-transcribed DDX1 RNA.
To check the effect of DDX1 knockdown on IBV infection, small interfering RNA (siRNA) duplexes targeting the DDX1 coding region from nucleotides 888 to 906 (5′-GGAGUUAGCUGAACAAACU-3′) were coelectroporated, together with the in vitro full-length IBV transcripts, into Vero cells. At 24, 36, and 48 h postelectroporation, cells were harvested for Northern blot analysis to check the expression of both IBV and DDX1 RNAs and for Western blot analysis of the levels of DDX1, actin, and IBV S and N proteins.
To check the effects of DDX1 and mutant DDX1 (DDX1m) overexpression on IBV replication, the in vitro-transcribed wild-type and mutant DDX1 RNAs, together with the purified IBV RNA, were electroporated into Vero cells. At 48 h postelectroporation, cells were harvested for Western blot analysis of DDX1 expression and real-time PCR analysis of IBV genomic or subgenomic RNA.
Assembly of the full-length IBV cDNA, in vitro transcription, and electroporation were performed as previously described (12). Sucrose density gradient purification of IBV was performed as previously described (28).
Establishment of stable DDX1 knockdown cells.
To establish stable cell lines, a cDNA fragment (5′-GATCCAGCTGAACTTATCTCTCAAGTTTCAAGAGAACTTGAGATAAGTTCAGCTTTTTTGGAAA-3′) expected to produce a hairpin RNA targeting the DDX1 coding region from nucleotides 1073 to 1091 upon transfection into cells (55) was cloned into the vector pSilencer 2.1-U6 neo (Ambion) and transfected into Vero cells. At 24 h posttransfection, 1 mg/ml of G418 was added to the medium, which was replaced with fresh medium containing the same concentration of G418 at 3-day intervals. G418-resistant clones were picked approximately 30 days later and then amplified. The amplified cell clones were harvested and analyzed by Northern and Western blot analyses, and the DDX1 knockdown cell clones were selected for subsequent studies.
Confocal fluorescence microscopy.
Vero cells were mock infected or infected with a recombinant IBV containing an HA-tagged RdRp protein at an MOI of 1 PFU/cell. At 3 h postinfection, 15 μg/ml of actinomycin D was added to the culture medium, and the cells were fixed at 4, 6.5, and 10 h postinfection. Cells were fixed with 4% formaldehyde, permeabilized with 100% methanol, and blocked with PBST containing 5% normal goat serum. Cells then were incubated with primary antibodies diluted in PBST containing 5% normal goat serum. The DDX1 protein was detected with 1:300-diluted anti-DDX1 rabbit antibody (Abcam), and the HA-tagged RdRp protein was detected with 1:300-diluted anti-HA mouse antibody (ETC, Singapore). After being washed three times, cells were incubated with 1:200-diluted secondary antibodies (Alexa Fluor 488-linked anti-rabbit IgG and Alexa Fluor 594-linked anti-mouse IgG). Images were taken using an Olympus confocal microscope (Fluoview model FV1000).
To examine the colocalization of DDX1 (endogenous DDX1 or exogenous wild-type and mutant DDX1 proteins) with BrdUTP-labeled IBV RNA in IBV-infected cells, Vero cells were either transfected with wild-type and mutant DDX1 proteins or left untransfected and then infected with IBV at an MOI of 1 PFU/cell at 13 h posttransfection. At 3 h postinfection, 15 μg/ml of actinomycin D was added to the culture medium, and the cells were incubated for 4 h to block cellular RNA transcription. BrdUTP was then transfected into the cells by use of SuperFECT (Qiagen) following the manufacturer's instructions to label the newly synthesized IBV RNA. At 3 h post-BrdUTP transfection, cells were washed with 1× PBS, fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and incubated with primary antibodies diluted in 1× PBS containing 5% normal goat serum. The endogenous DDX1 protein or Flag-tagged wild-type and mutant DDX1 proteins were detected with 1:400-diluted rabbit anti-DDX1 antibody or 1:200-diluted rabbit anti-Flag antibody (Sigma-Aldrich), respectively. The BrdUTP-labeled IBV RNA was detected with 1:100-diluted anti-BrdUTP mouse antibodies (Roche). After being washed three times, cells were incubated with 1:200-diluted secondary antibodies (Alexa Fluor 488-linked anti-rabbit IgG and Alexa Fluor 594-linked anti-mouse IgG). Images were taken using an Olympus confocal microscope (Fluoview model FV1000).
Real-time PCR for quantification of viral RNA synthesis.
Total RNA was extracted from IBV-infected cells by use of Trizol reagent (Molecular Research Center, Inc.). RT was performed using Expand reverse transcriptase (Roche). Quantification of RT products was performed by real-time PCR in a LightCycler instrument (Roche), using a LightCycler FastStart DNA master SYBR green I kit (Roche) according to the manufacturer's instructions. The following primers were used for RT: IBV27510R for total positive-strand RNA [RNA(+)], IBV27101F for total negative-strand RNA [RNA(−)], IBV4527F for negative-strand genomic RNA [gRNA(−)], IBV26F for negative-strand subgenomic RNA2 [sgRNA2(−)], IBV20623R for positive-strand subgenomic RNA2 [sgRNA2(+)], and GAPDH reverse primer for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The primer sequences are as follows: for total viral RNA, IBV27101F (forward; 5′-27101GAGTAACATAATGGACCTGT27120-3′) and IBV27510R (reverse; 5′-27510TGCTGTACCCTCGATCGTAC27491-3′); for negative-strand genomic RNA, IBV4527F (forward; 5′-4527TTTAGCAGAACATTTTGACGCAGAT4551-3′) and IBV4829R (reverse; 5′-4829TTAGTAGAACCAACAAACACGACGA4805-3′); for negative- and positive-strand subgenomic RNA2, IBV26F (forward; 5′-26CTATTACACTAGCCTTGCGC TAGA49-3′) and IBV20623R (reverse; 5′-20623GTGCCGTCATAGCTATAGA20605-3′); and for GAPDH, GAPDH forward primer (5′-GACAACTTTGGTATCGTGGAA-3′) and GAPDH reverse primer (5′-CCAGGAAATGAGCTTGACA-3′). For each sample, reactions were performed in triplicate. The amplified DNAs were quantitated by the comparative cycle threshold method for relative quantitation of gene expression. GAPDH was used as an internal control.
RESULTS
Interaction of IBV nsp14 with DDX1.
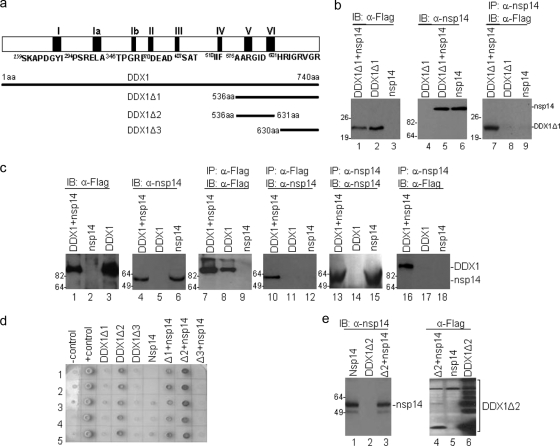
To search for cellular proteins that may be involved in the replication of coronavirus RNA, yeast two-hybrid screening was carried out, using different coronavirus nonstructural proteins as baits. This screening led to the identification of the C-terminal portion of DDX1 (aa 536 to 740; DDX1Δ1) (Fig. 1 a) as a potential interaction partner with IBV nsp14. Coimmunoprecipitation experiments were performed to further test the interaction between DDX1Δ1 and IBV nsp14 in mammalian cells. To facilitate the detection of DDX1Δ1, the protein was tagged with a Flag tag at its N terminus. Analysis of cells expressing the Flag-tagged DDX1Δ1 protein either on its own or together with IBV nsp14, using Western blotting with anti-Flag monoclonal antibody, showed that the Flag-tagged DDX1Δ1 protein was expressed (Fig. 1b). Similarly, analysis of cells expressing nsp14 either on its own or together with the Flag-tagged DDX1Δ1 protein, using anti-nsp14 polyclonal antibodies, showed the presence of nsp14 (Fig. 1b). The same cell lysates were then subjected to immunoprecipitation with anti-nsp14 antibodies. Western blot analysis of the precipitates with anti-Flag antibody detected the Flag-tagged DDX1Δ1 protein only in cells coexpressing the two proteins (Fig. 1b). These results confirmed that the C-terminal portion of the DDX1 protein could indeed interact with IBV nsp14 in mammalian cells.
FIG. 1.
Confirmation of the interaction between DDX1 and IBV nsp14 and mapping of the region in DDX1 responsible for its interaction with nsp14. (a) Diagram showing the regions of DDX1 used in the yeast two-hybrid system to define the interacting domain. (b) Interaction of nsp14 with DDX1Δ1. HeLa cells cotransfected with pFlag-DDX1Δ1+pKT0, pKT0-nsp14+pFlag, and pFlag-DDX1Δ1+pKT0-nsp14 were harvested at 24 h posttransfection and lysed. Total cell lysates were detected directly by Western blotting with anti-Flag and anti-nsp14 antibodies. The precipitates were analyzed by Western blotting with anti-Flag antibody. (c) Interaction of nsp14 with full-length DDX1. HeLa cells cotransfected with pFlag-DDX1+pKT0, pKT0-nsp14+pFlag, and pFlag-DDX1+pKT0-nsp14 were harvested at 24 h posttransfection and lysed. Total cell lysates were either detected directly by Western blotting with anti-Flag and anti-nsp14 antibodies or immunoprecipitated with anti-Flag or anti-nsp14 antibodies. The precipitates were analyzed by Western blotting with anti-Flag or anti-nsp14 antibodies. (d) The full-length DDX1 protein and three fragments, covering the DDX1 sequences from amino acids 536 to 740, 536 to 631, and 630 to 740, were cloned into plasmid pACT2 and cotransformed into yeast strain AH109 with the bait construct pGBKT7-nsp14. The yeast cotransformants, grown on SD/−Trp/−Leu/−His/X-Gal plates, are shown. Yeast strain AH109 was used as a negative control, and strain AH109 carrying pGADT7-T and pGBKT7-53 was used as a positive control. (e) Formation of protein aggregates when DDX1Δ2 was overexpressed in mammalian cells. HeLa cells cotransfected with pFlag-DDX1Δ2+pKT0, pKT0-nsp14+pFlag, and pFlag-DDX1Δ2+pKT0-nsp14 were harvested at 24 h posttransfection and lysed with 2× SDS loading buffer. The total lysates were detected directly by Western blotting with anti-nsp14 or anti-Flag antibodies.
Similar experiments were then performed to check if the full-length DDX1 protein could also interact with IBV nsp14. Efficient detection of the Flag-tagged full-length DDX1 protein was obtained in cells expressing DDX1 either on its own or together with IBV nsp14, using anti-Flag antibody (Fig. 1c, lanes 1 and 3). Analysis of cells expressing nsp14 either on its own or together with DDX1 protein, using anti-nsp14 antibodies, showed the presence of nsp14 (Fig. 1c, lanes 4 and 6). Immunoprecipitation with anti-Flag antibody and subsequent analysis of the precipitates with the anti-Flag antibody detected Flag-tagged DDX1 in cells expressing the protein either on its own or together with nsp14 (Fig. 1c, lanes 7 and 8). Western blot analysis of the same precipitates with anti-nsp14 antibodies detected nsp14 only in cells coexpressing the two proteins (Fig. 1b, lane 10). Similarly, immunoprecipitation with anti-nsp14 antibodies and subsequent analysis of the precipitates by Western blotting with anti-nsp14 antibody showed the presence of nsp14 in cells expressing the protein either on its own or together with DDX1 (Fig. 1c, lanes 13 and 15). Western blot analysis of the same precipitates with anti-Flag antibody detected DDX1 only in cells coexpressing the two proteins (Fig. 1c, lane 16). These results confirmed that the full-length DDX1 protein could also interact with IBV nsp14 in mammalian cells.
To further define the interacting domain in DDX1, we created two more deletion constructs, pACT-DDX1Δ2 (aa 536 to 631) and pACT-DDX1Δ3 (aa 630 to 740), which contain motifs V and VI of DDX1 and the C-terminal domain of DDX1, respectively (Fig. 1a). As shown in Fig. 1d, a positive interaction of DDX1Δ1 with nsp14 and a negative interaction of DDX1Δ3 with nsp14 were observed in yeast cells, implying that the minimal region for interaction with nsp14 may reside in DDX1Δ2 (aa 536 to 631), containing motifs V and VI. However, self-activation was observed in all yeast colonies transformed with DDX1Δ2 (Fig. 1d), suggesting that this portion of DDX1 may be self-associated or may form aggregates when overexpressed. This possibility was tested by overexpression of DDX1Δ2 in mammalian cells. Western blot analysis of HeLa cells expressing DDX1Δ2 either on its own or together with nsp14 showed the formation of massive aggregates in cells overexpressing DDX1Δ2 (Fig. 1e). However, when DDX1Δ2 was coexpressed with nsp14 and a relatively lower level of DDX1Δ2 was expressed, no formation of the protein aggregates was observed (Fig. 1e).
Interaction of DDX1 with SARS-CoV nsp14.
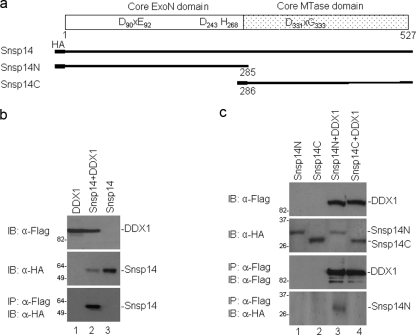
Coimmunoprecipitation assays were performed to check if DDX1 could also interact with nsp14 proteins from other coronaviruses. For this purpose, nsp14 from SARS-CoV, with an HA tag at the N terminus, was cloned and expressed together with the Flag-tagged DDX1 protein (Fig. 2 a). Western blotting of cells expressing the HA-tagged SARS-CoV nsp14 protein either on its own or together with the Flag-tagged DDX1 protein, using anti-HA monoclonal antibody, detected the protein (Fig. 2b). Once again, detection of the Flag-tagged DDX1 protein was obtained in cells expressing the protein either on its own or together with the HA-tagged SARS-CoV nsp14 protein, using Western blotting with anti-Flag monoclonal antibody (Fig. 2b). Immunoprecipitation of the same cell lysates with anti-Flag antibody and subsequent analysis of the precipitates by Western blotting with anti-HA antibodies detected the SARS-CoV nsp14 protein only in cells coexpressing the two proteins (Fig. 2b). These results confirmed that DDX1 could also interact with the SARS-CoV nsp14 protein.
FIG. 2.
Interaction of SARS-CoV nsp14 (Snsp14) with DDX1. (a) Diagram showing the full-length nsp14 protein and two deletion constructs of SARS-CoV nsp14 used in this study. (b) Interaction of SARS-CoV nsp14 with DDX1. HeLa cells expressing Flag-tagged DDX1, HA-tagged Snsp14, or DDX1 plus Snsp14 were harvested at 24 h posttransfection and lysed. The total lysates were either detected directly by Western blotting with anti-Flag and anti-HA antibodies or immunoprecipitated with anti-Flag antibody. The precipitates were analyzed by Western blotting with anti-HA antibody. (c) Interaction of the N-terminal portion of SARS-CoV nsp14 with DDX1. Cells expressing HA-tagged Snsp14N, Snsp14C, Snsp14N plus DDX1, or Snsp14C plus DDX1 were harvested at 24 h posttransfection and lysed. The total lysates were either detected directly by Western blotting with anti-Flag and anti-HA antibodies or immunoprecipitated with anti-Flag antibody. The precipitates were analyzed by Western blotting with anti-Flag and anti-HA antibodies.
To define the interacting domain in SARS-CoV nsp14, we made two more constructs, pcDNAHA-Snsp14N and pcDNAHA-Snsp14C, which covered the SARS-CoV nsp14 sequences from aa 1 to 285 and 286 to 527, respectively (Fig. 2a). Analysis of cells coexpressing the Flag-tagged DDX1 protein with either of the two HA-tagged truncated proteins by Western blotting with anti-Flag monoclonal antibody showed the presence of DDX1 (Fig. 2c). Similarly, Western blot analysis of these cell lysates with anti-HA monoclonal antibody detected Snsp14N and Snsp14C (Fig. 2c). Immunoprecipitation of the same cell lysates with anti-Flag antibody and subsequent analysis of the precipitates by Western blotting with anti-Flag antibody led to the detection of the Flag-tagged DDX1 protein coexpressed with Snsp14N or Snsp14C (Fig. 2c). Western blot analysis of the same precipitates with anti-HA antibody showed the presence of HA-tagged Snsp14N but not HA-tagged Snsp14C (Fig. 2c). These results confirmed that the interacting domain resided in the N-terminal portion of the SARS-CoV nsp14 protein.
Interaction of endogenous DDX1 with nsp14 in IBV-infected Vero cells.
Coimmunoprecipitation assays were then performed to check if the endogenous DDX1 protein could also interact with nsp14 in IBV-infected cells. Since both DDX1 and nsp14 may have RNA-binding activities, binding to a common viral or cellular RNA molecule would result in an indirect association of the two proteins. To rule out this possibility, 10 μg/ml RNase A was added to the lysis buffer and the washing buffer, and total cell lysates from mock- and IBV-infected Vero cells were used at 8 and 10 h postinfection for coimmunoprecipitation with nsp14 antibodies, followed by Western blotting with anti-DDX1 or -nsp14 antibodies. The endogenous DDX1 protein was detected in both mock- and IBV-infected cells by Western blotting with rabbit anti-DDX1 antibodies (Fig. 3). Probing the same membrane with rabbit anti-nsp14 antibodies showed the presence of nsp14 in IBV-infected cells harvested at both time points (Fig. 3). Immunoprecipitation of cell lysates with anti-nsp14 antibodies and subsequent analysis of the precipitates by Western blotting with anti-DDX1 antibodies detected DDX1 only in IBV-infected cells, not in mock-infected cells (Fig. 3). Similarly, probing the same membrane with anti-nsp14 antibodies detected the proteins only in IBV-infected cell lysates (Fig. 3). These results confirmed that the endogenous DDX1 protein could also interact with IBV nsp14 in IBV-infected cells.
FIG. 3.
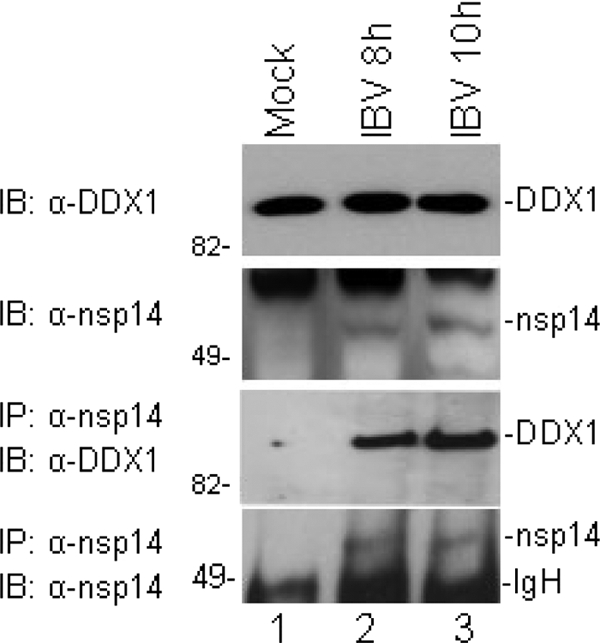
Interaction of endogenous DDX1 with nsp14 in IBV-infected Vero cells. Coimmunoprecipitation of endogenous DDX1 with nsp14 was performed with IBV-infected cells. Vero cells were mock or IBV infected at an MOI of 1 and harvested at 8 and 10 h postinfection. Cells were lysed, and the total lysates were either detected directly by Western blotting with DDX1 (top panel) and anti-nsp14 (second panel) antibodies or immunoprecipitated with anti-nsp14 antibodies. The precipitates were analyzed by Western blotting with anti-DDX1 (third panel) and anti-nsp14 (bottom panel) antibodies.
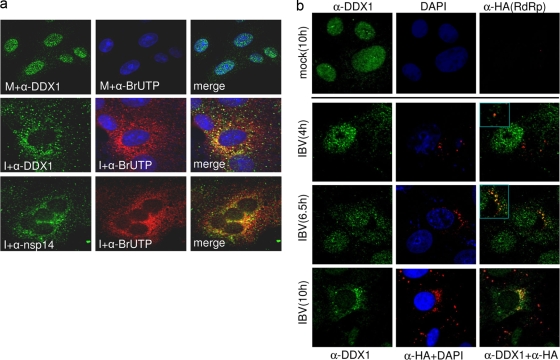
Relocation of endogenous DDX1 from the nucleus to the cytoplasm in IBV-infected Vero cells.
Coronavirus replication occurs in the cytoplasm of infected host cells, and the replication complexes are associated with modified host cell membranes (23, 42). Since DDX1 was located predominantly in the nucleus in normal cells, interaction of nsp14 with DDX1 would probably alter the subcellular localization of DDX1. This possibility was tested by immunofluorescence staining of endogenous DDX1 in IBV-infected cells. As expected, a predominant nuclear localization of DDX1 was observed in mock-infected cells (Fig. 4 a). The majority of DDX1 was found to be relocated to the cytoplasm, with a perinuclear staining pattern, in IBV-infected cells. To test if DDX1 was colocalized with the viral replication/transcription complexes, the subcellular localization of the newly synthesized IBV RNA incorporated with BrdUTP and of the endogenous DDX1 in IBV-infected cells was determined by dual immunofluorescence and confocal microscopy analysis. A perinuclear staining pattern of the DDX1 protein and the newly synthesized IBV RNA was observed at 10 h postinfection (Fig. 4a). The staining pattern of DDX1 was largely, but not perfectly, overlapped with that of the viral RNA in these cells (Fig. 4a). Immunofluorescence staining of nsp14 showed a pattern very similar to the DDX1 staining pattern in the infected cells at the same time point (Fig. 4a). Once again, the staining pattern of nsp14 was largely, but not perfectly, overlapped with that of the viral RNA in these cells (Fig. 4a). Since a suitable anti-DDX1 antibody from another animal species was not available, dual immunofluorescence staining of both DDX1 and nsp14 in IBV-infected cells could not be performed. These data suggest that interaction between DDX1 and nsp14 may be responsible for relocation of DDX1 from the nucleus to the cytoplasm.
FIG. 4.
Relocation of endogenous DDX1 from the nucleus to the cytoplasm in IBV-infected Vero cells. (a) Relocation of endogenous nsp14 from the nucleus to the cytoplasm in IBV-infected cells. Vero cells were mock infected (top row) or infected with IBV at an MOI of 1 (bottom two rows), and the newly synthesized viral RNA was labeled with BrdUTP. Cells were fixed, permeabilized, and double labeled with mouse anti-BrdU antibody and either anti-DDX1 (top two rows) or anti-nsp14 rabbit antibody (bottom row). DDX1 and nsp14 were immunostained with Alexa Fluor 488-linked anti-rabbit IgG, and the BrdUTP-labeled IBV RNA was stained with Alexa Fluor 594-linked anti-mouse IgG. The cellular nuclei were also stained with DAPI (4′,6-diamidino-2-phenylindole). Images were taken using an Olympus confocal microscope. (b) Detailed analysis of the translocation of endogenous nsp14 from the nucleus to the cytoplasm in IBV-infected cells in a time course experiment. Vero cells were infected with a recombinant IBV containing an HA-tagged RdRp protein at an MOI of 1 and were fixed at 4, 6.5, and 10 h postinfection. Cells were stained with rabbit anti-DDX1 and mouse anti-HA antibodies, followed by incubation with a mixture of Alexa Fluor 488-linked anti-rabbit IgG and Alexa Fluor 594-linked anti-mouse IgG. The mock-infected cells, fixed and stained at 10 h posttreatment, were also included as a control.
A detailed time course experiment was then performed to further characterize the translocation of DDX1 from the nucleus to the cytoplasm in IBV-infected cells. To more specifically and sensitively detect and characterize events occurring at early stages of the viral replication cycle, a recombinant IBV with an HA tag at the N terminus of nsp12 (RdRp) was generated and used to infect Vero cells. The recombinant virus displayed very similar growth properties to those of wild-type virus (data not shown). The mock- and IBV-infected cells were fixed at 4, 6.5, and 10 h postinfection and were dually stained with anti-DDX1 and anti-HA antibodies. At 4 h postinfection, the majority of DDX1 remained in the nucleus, with a certain amount of the protein detected in the cytoplasm (Fig. 4b). At 6.5 h postinfection, accumulation of more DDX1 in the cytoplasm was observed (Fig. 4b). At 10 h postinfection, a dramatic increase of DDX1 accumulation in the cytoplasm was detected (Fig. 4b). Staining of the same cells with anti-HA antibody showed gradually increased accumulation of the HA-tagged RdRp protein at the perinuclear region of the infected cells, indicating that viral replication was increased concomitantly (Fig. 4b). Once again, the two staining patterns were largely overlapped (Fig. 4b). In mock-infected cells, DDX1 remained in the nucleus (Fig. 4b).
Effects of DDX1 knockdown by siRNA on replication of IBV RNA.
The effects of the confirmed interaction between DDX1 and coronavirus nsp14 on viral replication were first studied by cointroduction of the in vitro-transcribed full-length RNA derived from a full-length infectious IBV clone (12, 53) with siRNA duplexes targeting either enhanced green fluorescent protein (EGFP) (siGFP; negative control) or DDX1, at nucleotides 888 to 906 (siDDX1), into Vero cells. Typical cytopathic effects (CPE) were observed at 36 h postelectroporation. Analysis of viral RNA replication by Northern blotting showed the presence of five viral mRNA species in cells transfected with both types of siRNA duplexes at 36 h postelectroporation (Fig. 5 a). However, lower levels of the viral RNA species were detected in cells electroporated with siDDX1 than in those electroporated with the siEGFP control at 36 h postelectroporation (Fig. 5a). Analysis of the same RNA preparations by Northern blotting showed a significant reduction of the DDX1 mRNA in cells transfected with siDDX1 compared to that in cells transfected with the control siRNA duplex (Fig. 5a).
FIG. 5.
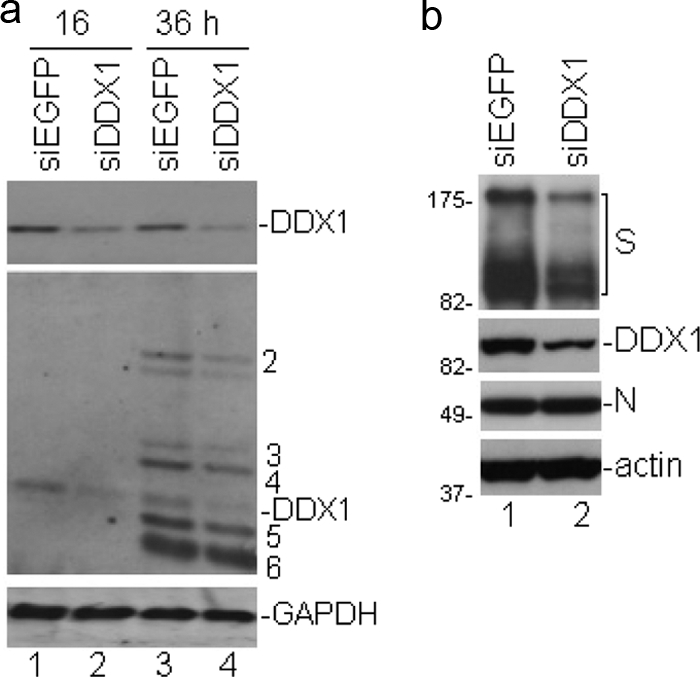
Analysis of effects of DDX1 knockdown by siRNA on the replication of IBV. (a) Northern blot analysis of viral RNA in Vero cells electroporated with either IBV full-length RNA transcripts plus EGFP siRNA (lanes 1 and 3) or IBV full-length RNA transcripts plus DDX1 siRNA (lanes 2 and 4). Cells were harvested at 16 and 36 h postelectroporation, and total RNA was prepared. Ten micrograms of total RNA was separated in a 1% agarose gel and transferred to a Hybond N+ membrane. Viral RNAs were probed with a digoxigenin-labeled DNA probe corresponding to the 3′ 680 nucleotides of the IBV genome. Numbers on the right indicated the subgenomic RNA species of IBV. The same membrane was also probed with DDX1 and GAPDH probes. (b) Western blot analysis of IBV S and N protein expression in Vero cells electroporated with either IBV full-length RNA transcripts plus EGFP siRNA (lane 1) or IBV full-length RNA transcripts plus DDX1 siRNA (lane 2). Cells were harvested at 48 h postelectroporation, and total cell lysates were separated in an SDS-10% polyacrylamide gel. The expression of S and N proteins was analyzed by Western blotting with polyclonal anti-S and anti-N antibodies, respectively. The same membrane was also probed with anti-DDX1 and anti-actin antibodies.
Western blot analysis of cells harvested at 48 h postelectroporation was then carried out to assess viral protein expression. As shown in Fig. 5b, a moderately reduced expression of the S protein was observed in DDX1 knockdown cells compared to that in the control cells (top panel). However, similar levels of N protein were detected in the same lysates (Fig. 5b). These results demonstrated that knockdown of DDX1 inhibited the replication and transcription of coronavirus RNA. Furthermore, it seemed that the inhibitory effect was preferential for the replication of the large subgenomic RNA species of IBV, and this prompted a more detailed study of the replication and growth kinetics of IBV in stable DDX1 knockdown cells.
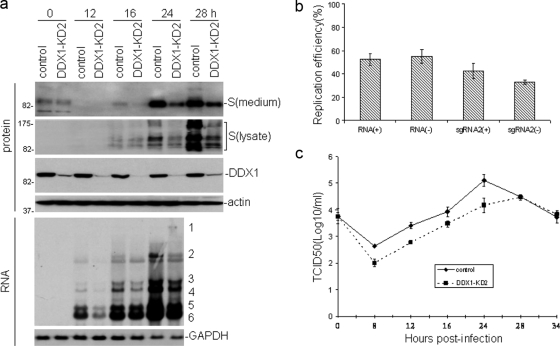
Establishment of stable DDX1 knockdown cell clones by expression of a hairpin RNA targeting DDX1 and detailed characterization of viral replication and growth kinetics in DDX1 knockdown cell clones infected with IBV.
Stable cell clones with DDX1 knockdown were established by expressing a hairpin RNA targeting the DDX1 coding region from nucleotides 1073 to 1091 in Vero cells. Several G418-selected clones were chosen, and the expression of DDX1 at the protein level was analyzed by Western blotting, which showed various knockdown efficiencies in these clones (data not shown). DDX1-KD2, a stable clone with the maximum knockdown efficiency among all clones (Fig. 6 a), was chosen for the subsequent studies. The effect of DDX1 knockdown on IBV replication was characterized by infection of DDX1-KD2 with IBV at an MOI of 1 PFU/cell in time course experiments. Meanwhile, a G418-selected cell clone without detectable DDX1 knockdown was used as a control. As shown in Fig. 6a, infection of DDX1-KD2 cells with IBV consistently showed much lower replication of viral RNA and expression of viral proteins. Western blot analysis of lysates prepared from the IBV-infected cells showed the presence of much less viral S protein in IBV-infected DDX1-KD2 cells than in control cells (Fig. 6a). Analysis of the same cell lysates by Western blotting with anti-DDX1 antibodies confirmed that DDX1 expression at the protein level was significantly reduced in DDX1-KD2 cells (Fig. 6a). The amount of virus released into the culture medium was also analyzed by Western blot analysis of culture medium collected at each time point with anti-S antibodies. At 0 h postinfection, similar amounts of viral S protein were detected, showing that approximately equal amounts of virus were used in the initial infections (Fig. 6a). Very little viral protein was detected at 16 h postinfection (Fig. 6a). At both 24 and 32 h postinfection, a large amount of S protein was detected in the control cells infected with IBV, but much less of the protein was detected in DDX1-KD2 cells infected with the virus (Fig. 6a), confirming that knockdown of DDX1 significantly reduced the replication of IBV and the production of progeny viruses. Consistently, the subgenomic mRNA species were found to be reduced in IBV-infected DDX1-KD2 cells, at all time points, compared to those in the control cells (Fig. 6a). Quantification of the reduction rates by densitometry showed that at 12 h postinfection, the ratios of mRNA species 2, 3, 4, and 5/6 in DDX1-KD2 cells to those in the control cells were 32, 41, 40, and 36%, respectively. The ratios of mRNAs 2, 3, 4, and 5/6 in DDX1-KD2 cells to those in the control cells were 44, 54, 44, and 96%, respectively, at 16 h postinfection, and the ratios were 33, 52, 47, and 65%, respectively, at 24 h postinfection.
FIG. 6.
Analysis of effects of DDX1 knockdown on the replication and growth properties of IBV in a stable DDX1 knockdown cell clone. (a) Northern and Western blot analyses of IBV RNA replication and protein synthesis in IBV-infected DDX1-KD2 cells. Cells were infected in duplicate with IBV at an MOI of 1 and harvested at 0, 12, 16, 24, and 28 h postinfection, and total RNA was prepared from one set of cells. Ten micrograms of total RNA was separated in a 1% agarose gel and transferred to a Hybond N+ membrane. Viral RNAs were probed with a digoxigenin-labeled DNA probe corresponding to the 3′ 680 nucleotides of the IBV genome. Numbers on the right indicated the genomic and subgenomic RNA species of IBV. The same membrane was also probed with DDX1 and GAPDH probes. Total cell lysates were also prepared from the infected cells. Proteins from total cell lysates (second panel) and media (top panel) were separated in an SDS-10% polyacrylamide gel. The expression of S protein, DDX1, and actin was analyzed by Western blotting. (b) Replication efficiency of IBV RNA in virus-infected DDX1-KD2 cells. The relative amounts of RNA(+), RNA(−), sgRNA2(+), and sgRNA2(−) in total RNA extracts from IBV-infected control and DDX1-KD2 cells harvested at 24 h postinfection were determined by real-time RT-PCR. The relative amount of each RNA species detected in IBV-infected control cells was considered 100% replication efficiency for the corresponding RNA species, and the relative amount of each IBV RNA species in virus-infected DDX1-KD2 cells is shown as the percent replication efficiency compared to that in the control cells. The data were generated from three repeat experiments. (c) Growth kinetics of IBV in DDX1-KD2 cells. The control and DDX1-KD2 cells were infected with IBV at an MOI of 1 and harvested at 0, 8, 12, 16, 24, 28, and 34 h postinfection. The virus stocks were prepared by collecting the virus particles released into the cultured medium, and the TCID50 of each viral preparation was determined by infection of five wells of Vero cells in 96-well plates in triplicate with 10-fold serial dilutions of each viral stock.
The IBV RNA(+), RNA(−), sgRNA2(+), and sgRNA2(−) levels in IBV-infected control and DDX1-KD2 cells harvested at 24 h postinfection were quantified by real-time RT-PCR. The relative replication efficiency of each RNA species detected in IBV-infected control cells was considered 100%. Compared to those in IBV-infected control cells, the relative replication efficiencies of RNA(+), RNA(−), sgRNA2(+), and sgRNA2(−) were 52.5, 55, 42.5, and 33%, respectively (Fig. 6b). These results confirmed that knockdown of DDX1 had more profoundly inhibitory effects on large subgenomic RNA species.
The growth kinetics of IBV in DDX1-KD2 cells were then determined by TCID50 assay. As shown in Fig. 6c, IBV reached peak titers at approximately the same time in both the control and DDX1-KD2 cells, but an ∼10-fold-lower peak titer was found in DDX1-KD2 cells infected with IBV.
Inhibition of IBV replication by overexpression of a mutant DDX1 protein.
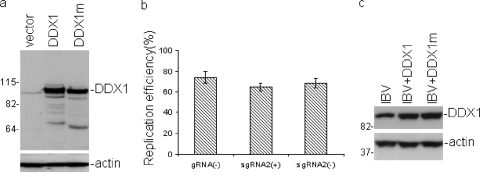
The effects of overexpression of wild-type DDX1 and mutant DDX1 (DDX1m) on IBV replication were then analyzed in the following two ways. Plasmid pXJ-F-DDX1m was constructed by mutation of the DEAD motif to AAAA. These substitutions result in severe reduction/abolishment of the ATPase and helicase activities, but not the RNA-binding activity, of the protein (20, 41). Vero cells were initially transfected with pXJ-F-DDX1 or pXJ-F-DDX1m, infected with IBV at an MOI of 1 PFU/cell at 24 h posttransfection, and harvested at 12 h postinfection. Western blot analysis of the cell lysates with anti-DDX1 antibodies showed efficient expression of DDX1 and DDX1m in the transfected cells (Fig. 7 a). Quantification of viral gRNA(−), sgRNA2(+), and sgRNA2(−) species in IBV-infected cells by real-time RT-PCR showed, unexpectedly, that the relative amounts of the three RNA species detected in cells overexpressing DDX1 were very similar to those in cells transfected with an empty vector, suggesting that overexpression of wild-type DDX1 did not show an enhancement effect on viral replication. In IBV-infected cells transfected with pXJ-F-DDX1m, the relative amounts of the three viral RNA species were 93, 89, and 85%, respectively, compared to those in the IBV-infected cells overexpressing wild-type DDX1 (considered 100% for each of the three RNA species).
FIG. 7.
Analysis of effects of overexpression of wild-type and mutant DDX1 on the replication of IBV. (a) Overexpression of wild-type and mutant DDX1 in Vero cells. Vero cells transfected with pXJ40-Flag (vector), pXJ-F-DDX1 (DDX1), or pXJ-F-DDX1m (DDX1m) were infected with IBV at an MOI of 1 at 24 h posttransfection and were harvested at 12 h postinfection. Proteins from total cell lysates were separated in an SDS-10% polyacrylamide gel. The expression of wild-type and mutant DDX1 was analyzed by Western blotting with polyclonal anti-DDX1 antibodies. The same membrane was also probed with anti-actin antibody. (b) Replication efficiency of IBV in cells transfected with IBV RNA plus either wild-type or mutant DDX1. The purified IBV genomic RNA and a 2-fold excess of in vitro-transcribed wild-type or mutant DDX1 transcript were cointroduced into Vero cells by electroporation. At 48 h postelectroporation, the cells were harvested and total RNA was prepared. The relative amounts of gRNA(−), sgRNA2(+), and sgRNA2(−) were determined by real-time RT-PCR. The relative amount of each RNA species detected in IBV RNA-transfected cells overexpressing wild-type DDX1 was considered 100% replication efficiency for the corresponding RNA species, and the relative amount of each IBV RNA species in virus-infected cells overexpressing mutant DDX1 is shown as the percent replication efficiency compared to that in the control cells. The data were generated from three repeat experiments. (c) Western blot analysis of DDX1 (DDX1m) expression in cells transfected with IBV RNA plus either wild-type or mutant DDX1, using polyclonal anti-DDX1 antibodies. The same membrane was also probed with anti-actin antibody.
Considering the fact that only approximately 30% of cells were usually transfected in a typical transfection experiment with the reagents and conditions used, we speculated that the observed inhibitory effects of DDX1m on IBV replication might be underrepresentative. To address this issue further, IBV was purified by sucrose gradient centrifugation, and the genomic RNA was isolated. The purified genomic RNA and a 2-fold excess of the in vitro-transcribed wild-type or mutant DDX1 transcript were cointroduced into Vero cells by electroporation. This experiment was repeated three times. At 48 h postelectroporation, the viral gRNA(−), sgRNA2(+), and sgRNA2(−) species were quantified by real-time RT-PCR, and the expression level of DDX1 was measured by Western blotting. Compared to those in cells cotransfected with the genomic IBV RNA and the wild-type DDX1 transcript (which were considered 100% for each viral RNA species), the relative replication efficiencies of the three viral RNA species were 74, 64.67, and 68.33%, respectively, in cells cotransfected with the genomic IBV RNA and the mutant DDX1m transcript (Fig. 7b). Analysis of the expression of DDX1 in these cells by Western blotting with anti-DDX1 antibodies showed the presence of higher levels of DDX1 and DDX1m proteins in the transfected cells (Fig. 7c).
Colocalization of mutant DDX1 with nsp14 and newly synthesized viral RNA.
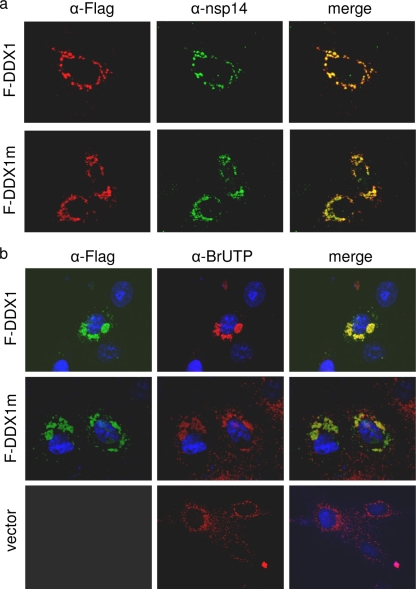
The subcellular localization of wild-type and mutant DDX1 and their colocalization with nsp14 expressed in IBV-infected cells were studied. As shown in Fig. 8 a, the staining patterns of wild-type and mutant DDX1 were very similar. Both patterns were largely overlapped with the staining pattern of nsp14 expressed in IBV-infected cells (Fig. 8a), indicating that DEAD motif mutation did not affect the interaction between nsp14 and DDX1.
FIG. 8.
Colocalization of mutant DDX1 with IBV nsp14 and newly synthesized viral RNA. (a) Colocalization of wild-type and mutant DDX1 with nsp14 expressed in IBV-infected cells. Vero cells overexpressing either Flag-tagged DDX1 or DDX1m were infected with IBV at an MOI of 1. At 10 h post-IBV infection, cells were fixed, permeabilized, and double labeled with mouse anti-Flag antibody and rabbit anti-nsp14 antibody. The Flag-tagged DDX1 and DDX1m proteins were immunostained with Alexa Fluor 594-linked anti-mouse IgG, and nsp14 was stained with Alexa Fluor 488-linked anti-rabbit IgG. Images were taken using an Olympus confocal microscope. (b) Colocalization of newly synthesized IBV RNA with DDX1. Vero cells overexpressing Flag-tagged DDX1 and DDX1m were infected with IBV at an MOI of 1, and the newly synthesized viral RNA was labeled with BrdUTP. Cells were fixed, permeabilized, and double labeled with mouse anti-BrdU antibody and rabbit anti-Flag antibody. The Flag-tagged DDX1 and DDX1m proteins were immunostained with Alexa Fluor 488-linked anti-rabbit IgG, and the BrdUTP-labeled IBV RNA was stained with Alexa Fluor 594-linked anti-mouse IgG. The cellular nuclei were stained with DAPI. Images were taken using an Olympus confocal microscope. Cells transfected with an empty vector and stained as described above were used as a control.
The effect of DEAD motif mutation on the colocalization of DDX1 with the viral RNA replication/transcription compartment was tested. Intracellular localization of the BrdUTP-incorporated, newly synthesized IBV RNA and the overexpressed wild-type or mutant DDX1 protein was determined by dual immunofluorescence and confocal microscopy analysis. At 10 h postinfection, a typical perinuclear staining pattern of the newly synthesized IBV RNA was observed in individual infected cells (Fig. 8b, bottom panels). The staining pattern of DDX1 was largely overlapped with that of IBV RNA in these cells (Fig. 8b, top panels). As shown in the representative images in Fig. 8b, similar but slightly less perfectly merged staining patterns of IBV RNA and DDX1m were observed in IBV-infected cells overexpressing DDX1m (Fig. 8b, middle panels). This observation was supported by examination of more images taken from IBV-infected cells expressing DDX1m and suggests that the DEAD motif might be implicated in the colocalization of DDX1 and viral RNA.
DISCUSSION
It was long speculated that host cell proteins were most likely involved in replication of the giant coronaviral RNA genome and in transcription of the multiple subgenomic mRNA species. However, this was one of the least explored areas in coronavirus research, and controversial data on the involvement of certain host cell proteins in coronavirus replication were available in the literature. For example, several host cell proteins were found by various approaches to bind and interact with coronaviral RNA and may play certain roles in viral RNA replication. One such host protein was the heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1). The protein was shown to form a complex with a mouse hepatitis virus (MHV) polymerase gene product, the N protein, and the viral RNA and to regulate the transcription and replication of MHV RNA (50). However, alteration of the binding affinity of hnRNA A1 by manipulation of the binding site in the RNA genome of MHV did not show much detrimental effect on the replication of MHV RNA, arguing against the possibility that hnRNP A1 was a host factor required for coronavirus mRNA transcription and genome replication (49). In this study, we showed, by yeast-two hybrid screening and biochemical approaches, that nsp14, a replicase gene product from both coronavirus IBV and SARS-CoV, interacts with DDX1, a host cell protein with RNA helicase activity (33). Manipulation of DDX1 expression either by siRNA-induced knockdown or by overexpression of a mutant DDX1 protein confirmed that this interaction played an important enhancement role in the replication of coronavirus RNA.
A large family of cellular proteins with RNA helicase activity was identified in mammalian cells (33). One group of these cellular RNA helicase proteins comprise the DEAD-box and related DEAH, DExH, and DExD protein families. These proteins are involved in multiple steps of RNA metabolism, including RNA transcription, splicing, export, translation, and decay (7, 44). Several members of this family were found to interact with viral proteins and to regulate their functionality. DDX1 was shown to interact with a number of other viral proteins (10, 11, 36). One example is that DDX1 can interact with the Rev protein of human immunodeficiency virus type 1 (HIV-1) and restrict Rev's function in human astrocytes (10, 11). Another DEAD-box protein, DDX3, is involved in the Rev-dependent export of HIV-1 RNA (61) and in regulation of hepatitis C virus (HCV) replication (2). DDX3 was also found to interact with the hepatitis B virus (HBV) polymerase, to incorporate into viral nucleocapsids, and to inhibit reverse transcription (57). In a genome-wide screen with an siRNA library, multiple proteins in the DExD/H helicase family, such as DDX28, DDX42, and DHX15, were shown to promote the replication of West Nile virus and dengue virus (24). It appears that the DExD/H helicase family is actively involved in the replication of diverse viral systems and may play similar enhancement roles in association with different viral components. In this study, we present evidence that DDX1 can interact with nsp14 proteins from both IBV and SARS-CoV. The two coronavirus nsp14 proteins share 51.6% amino acid identity and contain stretches of identical amino acids in the N-terminal region, suggesting that a common interacting domain may exist in this functionally equivalent protein in different coronaviruses.
As an exonuclease, nsp14 may be involved in replication and transcription of the genomic and subgenomic RNAs of coronaviruses (1, 9, 35, 48). Interaction of nsp14 with DDX1 may therefore facilitate this process. It is likely that this interaction enhances the replication and transcription of both genomic and subgenomic RNAs in general. In this regard, DDX1, as an RNA helicase, may provide additional activities facilitating the completion of viral RNA replication and transcription processes, although coronaviruses do encode an RNA helicase (nsp13) with NTPase and helicase activities (12). Recently, DDX1 was shown to interact with RelA (p65), a subunit of NF-κB, and to enhance NF-κB-mediated transcription (21). The intact DEAD domain is required for transcriptional coactivation activity, as trans-dominant-negative mutants of DDX1 lacking the ATP-dependent RNA helicase activity lost transcriptional inducer activity (21). It is likely that DDX1 acts as a transcriptional coactivator, facilitating the recruitment of other viral or cellular factors or stabilization of the viral RNA replication/transcription complexes. However, it would be difficult to pinpoint the precise step(s) in the viral replication cycle in which this interaction is specifically involved.
On the other hand, it was noted that knockdown of DDX1 expression exhibited only a moderate, though significant, effect on viral RNA replication and progeny virus production. The following possibilities were considered. First, DDX1 may function as a cellular cofactor, so the presence of a minute amount of the protein would be adequate. In fact, in both siRNA knockdown cells and the selected DDX1 knockdown stable cell clone, a much reduced but still significant amount of DDX1 protein was detected. Overexpression of DDX1 in Vero cells did not show a detectable enhancement effect on IBV replication, arguing that supplementation with an excess amount of DDX1 does not augment its function in this regard. Alternatively, other members of the DExD/H helicase family may compensate for the enhancement function of DDX1 in IBV replication in these knockdown cells. Continuous propagation of IBV in the DDX1 knockdown cell clone for up to 20 passages did not lead to the detection of aberrant subgenomic RNA species (data not shown), in contrast to that observed in cells transfected with the full-length human coronavirus RNA carrying mutations in the nsp14 protein (35). Furthermore, these serial passages did not lead to increased growth fitness of the virus, and no mutation in nsp14 protein was identified. These observations support the argument that as a cofactor, DDX1 plays an enhancement role in coronavirus replication only. More insights into the essentiality and involvement of DDX1 in coronavirus replication could be provided only by studying coronavirus infection and replication in DDX1 knockout mice and embryonic fibroblast cells derived from these knockout mice. Second, moderate effects on viral replication by knockdown of host genes that were interacting partners of viral replicase gene products were also observed in other viral systems. For instance, the cellular RNA helicase p68 was shown to be an interacting partner of HCV NS5B (17). Knockdown of p68 by siRNA duplexes in a replicon cell line showed no detectable effect on the replication of HCV RNA. However, a reduction in the replication of HCV RNA was observed when the siRNA duplexes and a full-length infectious HCV construct were cotransfected into cells (17).
In summary, our results demonstrate that DDX1 may play an auxiliary function in coronavirus replication by its association with one of the viral replicase proteins, nsp14. It is interesting to speculate that as a general rule, the virus-encoded transcription/replication machinery may function to initiate and maintain a basal level of transcription/replication of viral RNA. Association of viral components with host cell proteins may enhance this process in one way or another. It was recently reported that the in vitro activity of SARS-CoV replication/transcription complexes depends on a cytoplasmic host factor that does not cosediment with the complex (56). Identification and characterization of more cellular factors that are involved in the coronavirus life cycle by systems biology approaches would reveal new insights into the replication mechanisms and pathogenesis of coronaviruses in general.
Acknowledgments
This work was supported by the Agency for Science Technology and Research, Singapore.
Footnotes
Published ahead of print on 23 June 2010.
REFERENCES
- 1.Almazan, F., M. L. Dediego, C. Galan, D. Escors, E. Alvarez, J. Ortego, I. Sola, S. Zuniga, S. Alonso, J. L. Moreno, A. Nogales, C. Capiscol, and L. Enjuanes. 2006. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 80:10900-10906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angus, A. G., D. Dalrymple, S. Boulant, D. R. McGivern, R. F. Clayton, M. J. Scott, R. Adair, S. Graham, A. M. Owsianka, P. Targett-Adams, K. Li, T. Wakita, J. McLauchlan, S. M. Lemon, and A. H. Patel. 2010. Requirement of cellular DDX3 for hepatitis C virus replication is unrelated to its interaction with the viral core protein. J. Gen. Virol. 91:122-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhardwaj, K., L. Guarino, and C. C. Kao. 2004. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 78:12218-12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhardwaj, K., J. Sun, A. Holzenburg, L. A. Guarino, and C. C. Kao. 2006. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J. Mol. Biol. 361:243-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boursnell, M. E., T. D. Brown, I. J. Foulds, P. F. Green, F. M. Tomley, and M. M. Binns. 1987. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 68:57-77. [DOI] [PubMed] [Google Scholar]
- 6.Chen, Y., H. Cai, J. Pan, N. Xiang, P. Tien, T. Ahola, and D. Guo. 2009. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 106:3484-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cordin, O., J. Banroques, N. K. Tanner, and P. Linder. 2006. The DEAD-box protein family of RNA helicases. Gene 367:17-37. [DOI] [PubMed] [Google Scholar]
- 8.Eckerle, L. D., M. M. Becker, R. A. Halpin, K. Li, E. Venter, X. Lu, S. Scherbakova, R. L. Graham, R. S. Baric, T. B. Stockwell, D. J. Spiro, and M. R. Denison. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 6:e1000896. [DOI] [PMC free article] [PubMed]
- 9.Eckerle, L. D., X. Lu, S. M. Sperry, L. Choi, and M. R. Denison. 2007. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 81:12135-12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang, J., E. Acheampong, R. Dave, F. Wang, M. Mukhtar, and R. J. Pomerantz. 2005. The RNA helicase DDX1 is involved in restricted HIV-1 Rev function in human astrocytes. Virology 336:299-307. [DOI] [PubMed] [Google Scholar]
- 11.Fang, J., S. Kubota, B. Yang, N. Zhou, H. Zhang, R. Godbout, and R. J. Pomerantz. 2004. A DEAD box protein facilitates HIV-1 replication as a cellular co-factor of Rev. Virology 330:471-480. [DOI] [PubMed] [Google Scholar]
- 12.Fang, S., B. Chen, F. P. Tay, B. S. Ng, and D. X. Liu. 2007. An arginine-to-proline mutation in a domain with undefined functions within the helicase protein (Nsp13) is lethal to the coronavirus infectious bronchitis virus in cultured cells. Virology 358:136-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang, S. G., S. Shen, F. P. Tay, and D. X. Liu. 2005. Selection of and recombination between minor variants lead to the adaptation of an avian coronavirus to primate cells. Biochem. Biophys. Res. Commun. 336:417-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuller-Pace, F. V. 2006. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 34:4206-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuller-Pace, F. V., and S. Ali. 2008. The DEAD box RNA helicases p68 (Ddx5) and p72 (Ddx17): novel transcriptional co-regulators. Biochem. Soc. Trans. 36:609-612. [DOI] [PubMed] [Google Scholar]
- 17.Goh, P. Y., Y. J. Tan, S. P. Lim, Y. H. Tan, S. G. Lim, F. Fuller-Pace, and W. Hong. 2004. Cellular RNA helicase p68 relocalization and interaction with the hepatitis C virus (HCV) NS5B protein and the potential role of p68 in HCV RNA replication. J. Virol. 78:5288-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 17:4847-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guarino, L. A., K. Bhardwaj, W. Dong, J. Sun, A. Holzenburg, and C. Kao. 2005. Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. J. Mol. Biol. 353:1106-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iost, I., M. Dreyfus, and P. Linder. 1999. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J. Biol. Chem. 274:17677-17683. [DOI] [PubMed] [Google Scholar]
- 21.Ishaq, M., L. Ma, X. Wu, Y. Mu, J. Pan, J. Hu, T. Hu, Q. Fu, and D. Guo. 2009. The DEAD-box RNA helicase DDX1 interacts with RelA and enhances nuclear factor kappaB-mediated transcription. J. Cell. Biochem. 106:296-305. [DOI] [PubMed] [Google Scholar]
- 22.Ivanov, K. A., T. Hertzig, M. Rozanov, S. Bayer, V. Thiel, A. E. Gorbalenya, and J. Ziebuhr. 2004. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. U. S. A. 101:12694-12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knoops, K., M. Kikkert, S. H. Worm, J. C. Zevenhoven-Dobbe, Y. van der Meer, A. J. Koster, A. M. Mommaas, and E. J. Snijder. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krishnan, M. N., A. Ng, B. Sukumaran, F. D. Gilfoy, P. D. Uchil, H. Sultana, A. L. Brass, R. Adametz, M. Tsui, F. Qian, R. R. Montgomery, S. Lev, P. W. Mason, R. A. Koski, S. J. Elledge, R. J. Xavier, H. Agaisse, and E. Fikrig. 2008. RNA interference screen for human genes associated with West Nile virus infection. Nature 455:242-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim, K. P., and D. X. Liu. 1998. Characterisation of a papain-like proteinase domain encoded by ORF1a of the coronavirus IBV and determination of the C-terminal cleavage site of an 87 kDa protein. Adv. Exp. Med. Biol. 440:173-184. [DOI] [PubMed] [Google Scholar]
- 26.Lim, K. P., and D. X. Liu. 1998. Characterization of the two overlapping papain-like proteinase domains encoded in gene 1 of the coronavirus infectious bronchitis virus and determination of the C-terminal cleavage site of an 87-kDa protein. Virology 245:303-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim, K. P., L. F. Ng, and D. X. Liu. 2000. Identification of a novel cleavage activity of the first papain-like proteinase domain encoded by open reading frame 1a of the coronavirus avian infectious bronchitis virus and characterization of the cleavage products. J. Virol. 74:1674-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, D. X., and S. C. Inglis. 1991. Association of the infectious bronchitis virus 3c protein with the virion envelope. Virology 185:911-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, D. X., and S. C. Inglis. 1992. Identification of two new polypeptides encoded by mRNA5 of the coronavirus infectious bronchitis virus. Virology 186:342-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, D. X., S. Shen, H. Y. Xu, and S. F. Wang. 1998. Proteolytic mapping of the coronavirus infectious bronchitis virus 1b polyprotein: evidence for the presence of four cleavage sites of the 3C-like proteinase and identification of two novel cleavage products. Virology 246:288-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, D. X., K. W. Tibbles, D. Cavanagh, T. D. Brown, and I. Brierley. 1995. Identification, expression, and processing of an 87-kDa polypeptide encoded by ORF 1a of the coronavirus infectious bronchitis virus. Virology 208:48-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, D. X., H. Y. Xu, and T. D. Brown. 1997. Proteolytic processing of the coronavirus infectious bronchitis virus 1a polyprotein: identification of a 10-kilodalton polypeptide and determination of its cleavage sites. J. Virol. 71:1814-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luking, A., U. Stahl, and U. Schmidt. 1998. The protein family of RNA helicases. Crit. Rev. Biochem. Mol. Biol. 33:259-296. [DOI] [PubMed] [Google Scholar]
- 34.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 35.Minskaia, E., T. Hertzig, A. E. Gorbalenya, V. Campanacci, C. Cambillau, B. Canard, and J. Ziebuhr. 2006. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 103:5108-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nekhai, S., and K. T. Jeang. 2006. Transcriptional and post-transcriptional regulation of HIV-1 gene expression: role of cellular factors for Tat and Rev. Future Microbiol. 1:417-426. [DOI] [PubMed] [Google Scholar]
- 37.Ng, L. F., and D. X. Liu. 2000. Further characterization of the coronavirus infectious bronchitis virus 3C-like proteinase and determination of a new cleavage site. Virology 272:27-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng, L. F., and D. X. Liu. 1998. Identification of a 24-kDa polypeptide processed from the coronavirus infectious bronchitis virus 1a polyprotein by the 3C-like proteinase and determination of its cleavage sites. Virology 243:388-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng, L. F., and D. X. Liu. 2002. Membrane association and dimerization of a cysteine-rich, 16-kilodalton polypeptide released from the C-terminal region of the coronavirus infectious bronchitis virus 1a polyprotein. J. Virol. 76:6257-6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pasternak, A. O., W. J. Spaan, and E. J. Snijder. 2006. Nidovirus transcription: how to make sense? J. Gen. Virol. 87:1403-1421. [DOI] [PubMed] [Google Scholar]
- 41.Pause, A., and N. Sonenberg. 1992. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 11:2643-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perlman, S., and J. Netland. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 7:439-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ricagno, S., M. P. Egloff, R. Ulferts, B. Coutard, D. Nurizzo, V. Campanacci, C. Cambillau, J. Ziebuhr, and B. Canard. 2006. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc. Natl. Acad. Sci. U. S. A. 103:11892-11897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rocak, S., and P. Linder. 2004. DEAD-box proteins: the driving forces behind RNA metabolism. Nat. Rev. Mol. Cell. Biol. 5:232-241. [DOI] [PubMed] [Google Scholar]
- 45.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 46.Sawicki, S. G., and D. L. Sawicki. 2005. Coronavirus transcription: a perspective. Curr. Top. Microbiol. Immunol. 287:31-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sawicki, S. G., D. L. Sawicki, and S. G. Siddell. 2007. A contemporary view of coronavirus transcription. J. Virol. 81:20-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sawicki, S. G., D. L. Sawicki, D. Younker, Y. Meyer, V. Thiel, H. Stokes, and S. G. Siddell. 2005. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 1:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen, X., and P. S. Masters. 2001. Evaluation of the role of heterogeneous nuclear ribonucleoprotein A1 as a host factor in murine coronavirus discontinuous transcription and genome replication. Proc. Natl. Acad. Sci. U. S. A. 98:2717-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi, S. T., P. Huang, H. P. Li, and M. M. Lai. 2000. Heterogeneous nuclear ribonucleoprotein A1 regulates RNA synthesis of a cytoplasmic virus. EMBO J. 19:4701-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silverman, E., G. Edwalds-Gilbert, and R. J. Lin. 2003. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene 312:1-16. [DOI] [PubMed] [Google Scholar]
- 52.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan, Y. W., S. Fang, H. Fan, J. Lescar, and D. X. Liu. 2006. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 34:4816-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tibbles, K. W., I. Brierley, D. Cavanagh, and T. D. Brown. 1996. Characterization in vitro of an autocatalytic processing activity associated with the predicted 3C-like proteinase domain of the coronavirus avian infectious bronchitis virus. J. Virol. 70:1923-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tingting, P., F. Caiyun, Y. Zhigang, Y. Pengyuan, and Y. Zhenghong. 2006. Subproteomic analysis of the cellular proteins associated with the 3′ untranslated region of the hepatitis C virus genome in human liver cells. Biochem. Biophys. Res. Commun. 347:683-691. [DOI] [PubMed] [Google Scholar]
- 56.van Hemert, M. J., S. H. van den Worm, K. Knoops, A. M. Mommaas, A. E. Gorbalenya, and E. J. Snijder. 2008. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 4:e1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, H., S. Kim, and W. S. Ryu. 2009. DDX3 DEAD-box RNA helicase inhibits hepatitis B virus reverse transcription by incorporation into nucleocapsids. J. Virol. 83:5815-5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, L., J. P. Tam, and D. X. Liu. 2006. Biochemical and functional characterization of Epstein-Barr virus-encoded BARF1 protein: interaction with human hTid1 protein facilitates its maturation and secretion. Oncogene 25:4320-4331. [DOI] [PubMed] [Google Scholar]
- 59.Xu, H. Y., K. P. Lim, S. Shen, and D. X. Liu. 2001. Further identification and characterization of novel intermediate and mature cleavage products released from the ORF 1b region of the avian coronavirus infectious bronchitis virus 1a/1b polyprotein. Virology 288:212-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamada, Y., and D. X. Liu. 2009. Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J. Virol. 83:8744-8758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yedavalli, V. S., C. Neuveut, Y. H. Chi, L. Kleiman, and K. T. Jeang. 2004. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119:381-392. [DOI] [PubMed] [Google Scholar]