Abstract
The pseudorabies virus (PRV) Us2 protein binds to the extracellular-regulated kinase (ERK) and inhibits the activation of ERK nuclear targets by sequestering cytoplasmic ERK on cellular membranes. Utilizing a series of Us2 truncations, we determined that the minimal portion of Us2 required for interaction with ERK is contained within its amino-terminal 214 amino acids. The loss of the ability of Us2 to bind to ERK in coimmunoprecipitation experiments was accompanied by a failure of Us2 to form oligomers, raising the possibility that higher-order Us2 structures are required for ERK interaction. To map the Us2 interaction site on ERK, we introduced mutations into the region of ERK that interacts with the ERK kinase, MEK, or into the common docking (CD) domain that mediates interactions with many ERK substrates. ERK carrying mutations within the MEK binding region maintained the ability to bind Us2, whereas ERK carrying mutations within the CD domain did not. Furthermore, the ERK CD domain was required for the Us2-mediated recruitment of ERK to membranes. Taken together, these findings suggest that Us2 regulates ERK activity by spatially restricting ERK localization and also by interfering with select ERK-substrate interactions.
Alphaherpesviruses are an important group of human and animal pathogens, the majority of which establish life-long latent infections in the peripheral nervous systems of their hosts. The swine pathogen pseudorabies virus (PRV) is a well-studied alphaherpesvirus whose analysis has contributed greatly to our understanding of the molecular and cellular biology of virion assembly, as well as how viruses of this group spread and cause disease in the nervous system (25). Herpesviruses are large enveloped viruses that contain a linear double-stranded DNA genome that, in the case of PRV, is 143 kbp in length and is predicted to encode in excess of 70 proteins (12). Between the icosahedral nucleocapsid, which contains the virus genome, and the virion envelope is a structure called the tegument. The tegument is the most complex subvirion compartment, housing 20 or so different virus-encoded proteins, a number of host proteins, and messenger RNAs. In recent years, significant insight has been gained into the complex series of protein-protein interactions required for tegument assembly (20). Despite these advances, little is known about the functions of most tegument proteins. Because tegument proteins are delivered to the cytoplasm of cells upon virus infection, they have an opportunity to function prior to the expression of virus genes. A widely held view is that one role of the tegument is to establish a cellular environment conducive to virus infection (11). In addition to these functions that take place early in infection, it is clear that a number of tegument proteins function later in infection, during virion maturation and assembly (21).
The Us2 gene encodes a tegument component that is conserved among most of the alphaherpesviruses, with one notable exception being varicella-zoster virus. While nonessential for the growth of virus in cell culture, the Us2 gene is deleted from an attenuated PRV vaccine strain, Bartha, as well as from the attenuated equine herpesvirus 1 (EHV-1) vaccine strain, RacH, strongly suggesting that Us2 is an important virulence determinant during natural infections (8, 9, 16). Studies of the EHV-1 Us2 protein have shown that it associates with cellular and viral membranes and enhances virus entry and the cell-to-cell spread of infection (19).
We previously demonstrated that PRV Us2 binds to the extracellular-regulated kinase (ERK), an important effector serine/threonine kinase in the Raf/MEK/ERK signaling module (18). The ERK mitogen-activated protein kinase (MAPK) pathway facilitates cellular responses to extracellular stimuli. It is a key signaling pathway that is important for the regulation of cell growth, division, apoptosis, and differentiation, and it is activated upon the binding of numerous growth factors, hormones, and cytokines to their cognate receptors (4, 14, 24, 26, 31). Upwards of 70 different substrates have been identified for ERK (13, 15). While the majority of ERK substrates are transcription factors that reside in the nucleus of the cell, others are found in the cytosol, on the cytoskeleton, and on membranes. Importantly, after ERK activation by a given stimulus, only a subset of ERK substrates are phosphorylated. The restriction of ERK activity toward select substrates is thought to be mediated in part by ERK scaffolding molecules that spatially regulate ERK enzymatic activity and direct it toward the appropriate, stimulus-specific substrates (13, 17, 22, 29). We have demonstrated that Us2 binds to and localizes ERK to the plasma membrane as well as to a perinuclear vesicular compartment, thereby preventing the translocation of ERK into the nucleus (18). However, Us2 does not prevent ERK activation, nor does it inhibit ERK enzymatic activity (18). Furthermore, both ERK and Us2 activities are required for the efficient release of infectious virus from cells (18), and enveloped viruses accumulate in cytoplasmic vesicles in cells infected with Us2 null mutants (30). Thus, it appears that Us2 acts as an ERK scaffold used by the virus to divert ERK activity to cellular membranes during a late stage of virus egress. In this study, we sought to define the mechanism by which Us2 and ERK interact.
MATERIALS AND METHODS
Cell lines.
Vero cells, PK15 cells, and HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) in a 5% CO2 environment. NIH 3T3 cells, obtained from the ATCC, were maintained in DMEM containing 10% newborn calf serum (NCS).
Expression plasmids.
The Us2 expression plasmids pCC34 (wild-type Us2), pCC35 (Us2GAAX), pJR73 (3XFLAG-Us2), and pJR47 (GAL4DBD-Us2) have been described previously (3, 18). The ERK2-enhanced green fluorescent protein (EGFP) expression construct was kindly provided by N. W. Bunnet, UCSF (5). The pCMV-myc vector was a kind gift from P. Sadowski, University of Toronto. Plasmids encoding FLAG-tagged Us2 C-terminal truncation mutants were constructed by amplifying Us2 from pCC34 using Pfx polymerase (Invitrogen, Burlington, Ontario, Canada)-mediated PCR using forward primer PRVUs2 F EcoRV and reverse primers PRVUs2N37, PRVUs2N71, PRVUs2N84, PRVUs2N146, PRVUs2N157, and PRVUs2N214 (see Table S1 in the supplemental material). The PCR products were digested with EcoRV and SalI and cloned into pFLAG-CMV-2 (Sigma, St. Louis, MO) and pGBKT7 (Clontech, Palo Alto, CA) that had been digested with EcoRV-SalI and SmaI-SalI, respectively. Plasmids encoding Us2 LxxRR mutants were constructed by site-directed mutagenesis using primers PRVUs2 F EcoRI, R183K/R184K F/R, R183A/R184A F/R, R183-184A F/R (LAAAA), and PRVUs2 R SalI to amplify Us2 from pCC34. The PCR products were digested with EcoRI and SalI and cloned in frame into pCIneo and pGBKT7 that had been digested with EcoRI and SalI. The PRV Us2 AAAAA (LTRRR/5A) mutant was constructed by amplifying Us2 from R183-185A-pCIneo plasmid using primers PRVUs2 F EcoRI, PRVUs2LxxRR5A F/R, and PRVUs2 R SalI. The PCR product was digested with EcoRI and SalI and cloned in frame into pCIneo and pGBKT7. Myc-tagged Us2 was constructed by amplifying Us2 from pCC34 using primers PRVUs2 F EcoRI and PRVUs2 R BglII. The PCR product was digested with EcoRI and BglII and cloned in frame into pCMV-myc that had been digested with EcoRI and BamHI. To subclone murine ERK2 into pACT2 (Clontech, Palo Alto, CA), pML32 (18) was digested with BamHI and the purified insert treated with the Klenow fragment to produce a blunt-ended product. The insert was ligated to pACT2 that was digested with NcoI and treated with the Klenow fragment. Plasmids encoding ERK2 mutants were constructed by site-directed mutagenesis using primers pACT2 MUT F2, H229R F/R, N235K F/R, Y260N F/R, D315A/D318A F/R, and ERK2 R XhoI to amplify the gene from ERK2-pACT2. The PCR products were cloned into pCR-BluntII-TOPO vector using a Zero Blunt TOPO PCR cloning kit (Invitrogen, Burlington, Ontario, Canada) according to the manufacturer's instructions. Positive clones were digested with NcoI and XhoI, and the inserts were cloned in-frame into pACT2 that had been similarly digested. ERK2 D315A/D318A mutant was cloned into pEGFP-C1 (Clontech, Palo Alto, CA) by amplifying the gene from ERK2 D315A/D318A-pACT2 using forward primer ERK2 F HindIII and reverse primer ERK2 R SalI. The PCR product was cloned into pCR-BluntII-TOPO vector using a Zero Blunt TOPO PCR cloning kit. A clone containing the PCR product was digested with HindIII and SalI, and the insert was cloned in-frame into pEGFP-C1 that had been similarly digested.
Antibodies.
The production of Us2 antiserum (used at a 1:10,000 dilution for immunoblotting and a 1:500 dilution for immunostaining) was described previously (3). Monoclonal antisera against total ERK1/2 (1:1,000 dilution for immunoblotting) and phospho-ERK1/2 (1:1,000 dilution for immunoblotting) were purchased from Cell Signaling Technology (Beverly, MA). Mouse monoclonal antiserum against GFP (1:1,000 dilution for immunoblotting) was purchased from Clontech (Palo Alto, CA). Mouse monoclonal antiserum against c-myc (1:2,000 dilution for immunoblotting) was purchased from Roche Diagnostics (Laval, Canada). Anti-FLAG polyclonal antibody was purchased from Sigma (St. Louis, MO).
Yeast two-hybrid analysis.
The PRV Us2 C-terminal and N-terminal truncations were cloned into pGBKT7 (Trp) (Clontech, Palo Alto, CA). Wild-type ERK1, ERK2, and four ERK2 mutants, H229R, N235K, Y260N, and D315A/D318A, were cloned into pACT2 (Leu) (Clontech, Palo Alto, CA). Plasmids were cotransformed into Saccharomyces cerevisiae strain YCH1. The absence of tryptophan from the growth medium selected for the Us2 plasmids, and the absence of leucine from the medium selected for the ERK2 plasmids. In these assays, interaction between Us2 and ERK brings the GAL4 DNA binding domain (DBD) and activation domain (AD) in proximity, leading to the transcription of the reporter genes HIS3 and ADE2, which allows yeast to grow on medium lacking histidine and adenine, respectively. Selected clones were grown on −Trp/−Leu/−His dropout medium (medium stringency) or −Trp/−Leu/−Ade/−His (high stringency) to select for two-hybrid interactions.
Immunoprecipitation (IP).
HEK293T cells were grown to 80% confluence in 150-mm dishes and transfected with FLAG-tagged wild-type Us2 (pJR73) or C-terminal Us2 truncation expression plasmids and ERK2-EGFP, EGFP-ERK2 D315A/D318A, or pEGFP-C1 (Clontech, Palo Alto, CA) expression construct. For Us2 oligomerization experiments, cells were cotransfected with pJR73 or pFLAG-CMV-2 and Myc-Us2 expression constructs. Twenty-four hours posttransfection, cells were washed with phosphate-buffered saline (PBS) and scraped into 1 ml of lysis buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) containing protease inhibitors (Roche Diagnostics, Laval, Canada) and phosphatase inhibitors (50 mM NaF, 10 mM Na3VO4) and centrifuged at 13,000 rpm for 10 min. Supernatants were transferred to a fresh microcentrifuge tube containing 40 μl of anti-FLAG M2 affinity gel (Sigma, St. Louis, MO) and rocked overnight at 4°C. Anti-FLAG beads were pelleted by brief centrifugation at 9,000 rpm. Supernatants were discarded, and the beads were washed three times successively with 1 ml Tris-buffered saline (TBS). Immune complexes were resuspended in 50 μl of SDS-PAGE sample buffer and boiled for 5 min. Samples were analyzed by SDS-PAGE on 12% gels, followed by Western blotting.
To determine if Us2 oligomers were capable of interacting with ERK, cells were transfected with pJR73 or pFLAG-CMV-2, Myc-Us2 or pCMV-myc, and ERK2-EGFP expression constructs. Coimmunoprecipitations were performed as described above to isolate FLAG-Us2. The immune complexes were washed three times with TBS and then subjected to competitive elution by using 3× FLAG peptide (Sigma, St. Louis, MO) according to the manufacturer's instructions. Eluates then were incubated with anti-myc antibody (Roche Diagnostics, Laval, Canada) overnight at 4°C. The antigen-antibody complexes were recovered using protein G agarose (Pierce Biotechnology, Rockford, IL). Protein G agarose was washed three times successively with 1 ml TBS. Immune complexes were resuspended in SDS-PAGE sample buffer and boiled for 5 min prior to Western blot analysis.
Immunofluorescence microscopy.
Vero cells were seeded onto glass coverslips in 6-well plates and grown to 30 to 40% confluence. Cells were transfected with 1 μg of plasmid DNA using FuGENE 6 (Roche, Laval, Canada) according to the manufacturer's instructions. At 24 h posttransfection, cells were rinsed with PBS and then fixed in 4% paraformaldehyde-PBS for 10 min at room temperature. Cells were rinsed with PBS and permeabilized in 1% bovine serum albumin (BSA)-PBS containing 0.1% Triton X-100 at room temperature for 5 min. Cells were rinsed three times with PBS and incubated with Us2 goat polyclonal antiserum diluted in 1% BSA-PBS (1:500) for 1 h. Cells were rinsed three times with TBST (TBS containing 0.05% Tween 20) and incubated with Alexa 568-conjugated secondary antibodies (Invitrogen, Burlington, Ontario, Canada) and diluted in 1% BSA-PBS (1:500) for 1 h. To visualize nuclei, cells were stained with Hoechst 33342 (Sigma, St. Louis, MO) diluted to 0.5 μg/ml in PBS for 7 min at room temperature, followed by three rinses with PBS. Coverslips were mounted onto glass slides, and digital images were captured using an Olympus FV1000 laser-scanning confocal microscope.
Fluorescence recovery after photobleaching (FRAP) analyses.
PK15 cells seeded onto 35-mm glass-bottom dishes (MatTek, Ashland, MA) were cotransfected with the indicated plasmids using the FuGENE 6 transfection reagent. Twenty-four hours after transfection, the medium was replaced with warm DMEM (lacking phenol red)-10% FCS, and the cells were mounted onto an Olympus FV1000 confocal microscope and maintained at 37°C in a humidified 5% CO2 environment. All image acquisition parameters were controlled using Olympus Fluoview software version 1.7.3.0. EGFP was excited using a 488-nm laser line set at 5% power. Images (512 by 512 pixels) were obtained using a 60× (1.42-numerical aperture) oil immersion objective and a digital zoom factor of 4. Images were collected at a rate of 0.2 frames per second. Three frames were collected before the regions bounded by the red rectangles were photobleached by repeated scanning with a 488-nm laser set at 80% power for a total of 200 ms. Before, during, and after photobleaching, fluorescence intensity in bleached and unbleached control regions was measured using Fluoview 1.7.3.0, and the data were exported into Microsoft Excel for graphical presentation.
Elk-1 dual-reporter assay.
Elk-1 activation was measured using the dual-luciferase reporter system (Promega, Madison, WI). NIH 3T3 cells were grown to 40 to 60% confluence in 24-well plates, and each well was cotransfected with 30 ng of GAL4-fused Elk-1 (pFA2-Elk-1) (Stratagene, La Jolla, CA), 200 ng of luciferase reporter linked to a GAL4 binding element (pFR-Luc) (Stratagene, La Jolla, CA), 50 ng Renilla-luciferase (pRL-SV40) (Promega, Madison WI), 30 ng of constitutively active MEK (pFC-MEK1) (Stratagene, La Jolla, CA), and 200 ng of wild-type Us2, mutant Us2, or pCIneo (empty vector control) expression plasmid using the TransIT-3T3 transfection kit (Mirus, Madison, WI) according to the manufacturer's instructions. Transfections with each plasmid combination were performed in triplicate. Cells were incubated in medium containing 10% NCS for the first 24 h and in medium containing 0.5% NCS for the final 24 h to minimize background ERK activity. Elk-1-dependent transcription (i.e., luciferase activity) was measured using a Promega dual-luciferase reporter system. In some experiments, the MEK inhibitor U0126 (Promega, Madison WI) was included at a concentration of 30 μM.
RESULTS AND DISCUSSION
Us2 confers the properties of a membrane protein upon ERK.
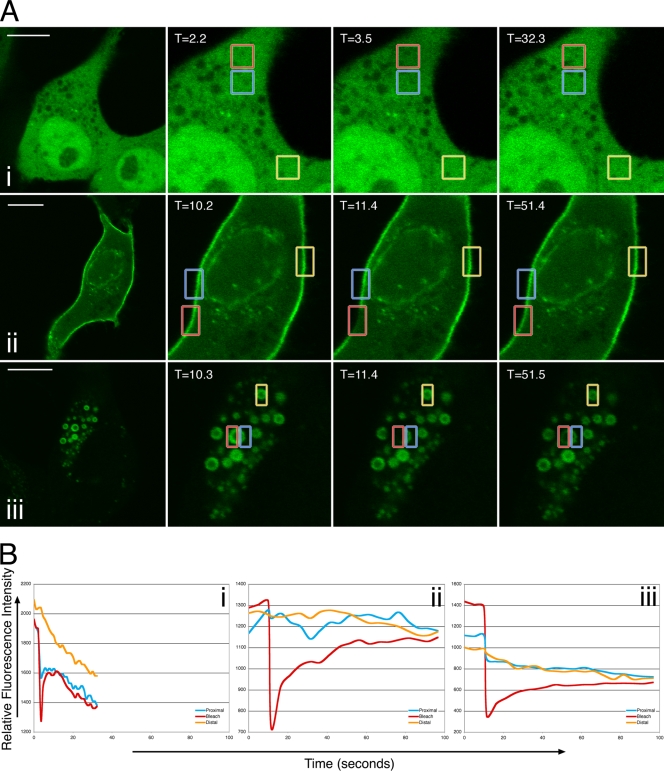
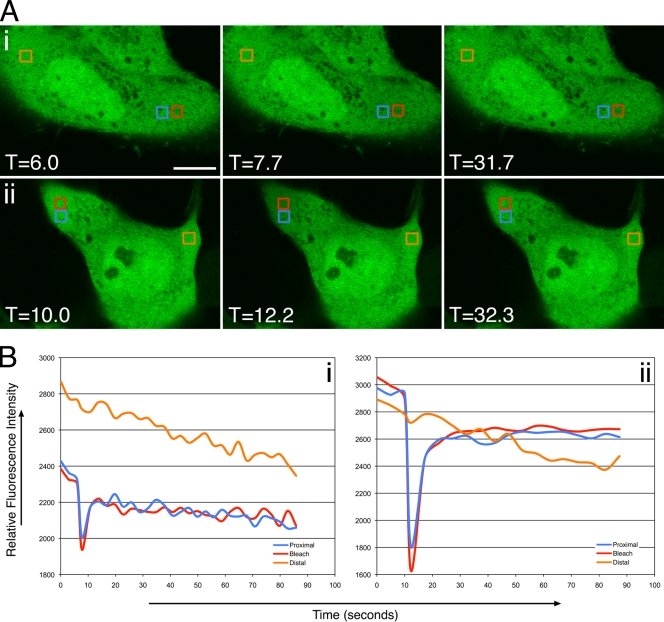
To examine the dynamics of ERK localization in Us2-expressing cells, we performed FRAP experiments (Fig. 1). Cells were cotransfected with either the empty vector pCIneo (Fig. 1A, row i) or a Us2 expression plasmid (Fig. 1A, row ii) along with an ERK2-EGFP expression construct. In the absence of Us2, ERK was localized diffusely throughout the nucleus and cytoplasm of transfected cells. In contrast, the majority of ERK localized to the plasma membrane, to the nuclear envelope, and to punctate vesicular structures in cells that were cotransfected with Us2. The photobleaching of ERK2-EGFP in pCIneo-cotransfected cells resulted in the rapid recovery of the fluorescent signal into the photobleached region within 4 s (Fig. 1B, graph i; also see Movie S1 in the supplemental material). These data are consistent with ERK2-EGFP behaving as a soluble cytoplasmic protein. The photobleaching of ERK2-EGFP that was localized to the plasma membrane in Us2-expressing cells resulted in a much lower rate of recovery of fluorescence into the bleached region, with the maximal recovery observed by 35 s after bleaching (Fig. 1B, graph ii; also see Movie S2 in the supplemental material). These findings suggest that the interaction between Us2 and ERK changes the diffusion properties of ERK such that, in the presence of Us2, ERK behaves like a membrane protein. PRV Us2 associates with membranes through the posttranslational addition of a farnesyl group to its C-terminal CAAX motif (3). The ability of ERK to bind to membrane-tethered Us2 likely explains these findings.
FIG. 1.
FRAP analysis of ERK2-EGFP in the absence and presence of Us2. (A) Images of live PK15 cells cotransfected with pCIneo and ERK2-EGFP (i), Us2 and ERK2-EGFP (ii), or Us2GAAX and ERK2-EGFP expression plasmids (iii) immediately before, immediately after, or 28.8 s (i), 40 s (ii), and 40.1 s (iii) after the area outlined in the red box were photobleached using an Olympus FV1000 laser-scanning confocal microscope. The boxed blue area is proximal to the photobleached region, and the boxed orange area is distal to the photobleached region. The fluorescence signal is ERK2-EGFP. Scale bars are 10 μm. (B) Quantitation of fluorescence intensity of the photobleached area, as well as control proximal and distal areas, over time. Shown are data for PK15 cells cotransfected with pCIneo and ERK2-EGFP (i), cells cotransfected with Us2 and ERK2-EGFP expression plasmids (ii), and cells cotransfected with Us2GAAX and ERK2-EGFP expression plasmids (iii).
To determine the role of Us2 prenylation in ERK membrane localization, we mutated the cysteine of the Us2 CAAX motif to a glycine, thereby abrogating the ability of Us2 to be prenylated inside cells (3, 7). This mutant Us2 protein was designated Us2GAAX. When Us2GAAX and ERK2-EGFP were coexpressed, ERK2-EGFP localized to large cytoplasmic vesicles but was not present at the plasma membrane, indicating that the prenylation of Us2 is necessary for plasma membrane localization (Fig. 1A, row iii). When the ERK2-EGFP in one half of a large cytoplasmic vesicle was photobleached, the migration of ERK2-EGFP from the nonbleached portion of the vesicle into the bleached region occurred with maximal recovery observed by 40 s after bleaching (Fig. 1A, row iii, and B, graph iii; also see Movie S3 in the supplemental material). These findings suggest that, like wild-type Us2, Us2GAAX imparts the properties of a membrane protein upon ERK2-EGFP.
The finding that nonprenylated Us2 maintained the ability to associate with membranes was unexpected, because the loss of this lipid modification was predicted to result in the loss of Us2-membrane association. The majority of alphaherpesvirus Us2 proteins do not contain signals for prenylation or other lipid modifications, nor do they contain putative membrane-spanning domains. Curiously, Meindl and Osterrieder have demonstrated that EHV-1 Us2, a nonprenylated protein, is indeed membrane associated (19). It may be that nonprenylated Us2 isoforms associate with membranes through interaction with other membrane components.
Mapping Us2 determinants required for interaction with ERK.
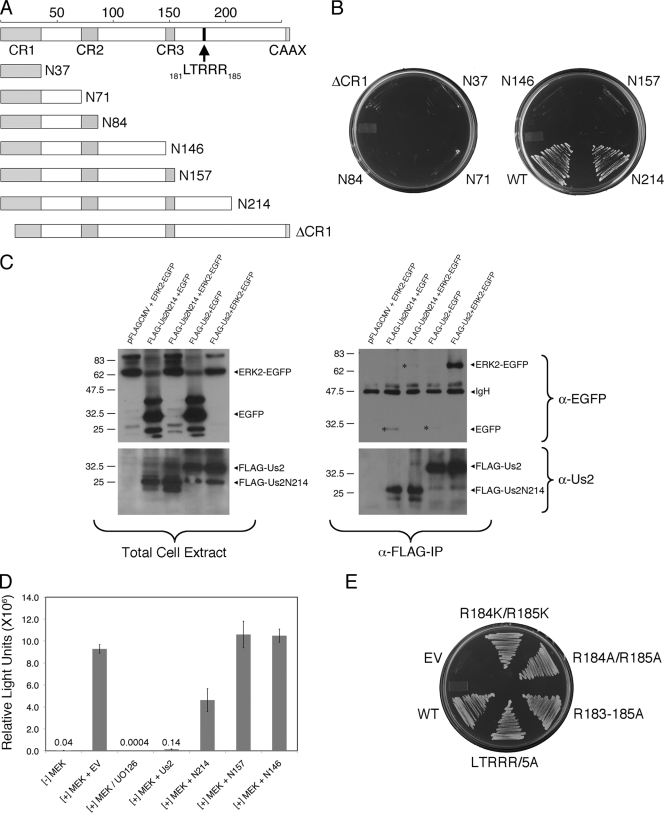
To identify the minimal portion of Us2 required for interaction with ERK, we began by constructing a series of C-terminal truncation mutants (Fig. 2A). Each truncation was cloned into a yeast expression vector, pGBKT7, for yeast two-hybrid assays and a mammalian expression vector, pFLAG-CMV-2, for coimmunoprecipitation (co-IP) studies and Elk-1 trans-reporter assays. We first examined protein-protein interactions in yeast by coexpressing Us2 truncation mutants fused to the GAL4 DNA binding domain (DBD) and ERK2 fused to the GAL4 activation domain (AD). Only yeast strains cotransformed with ERK2 and wild-type Us2 or Us2N214 expression plasmids were able to grow on medium lacking histidine, indicating a two-hybrid interaction (Fig. 2B). We also found that the deletion of the first 14 N-terminal residues (ΔCR1) of Us2 disrupted the ERK interaction (Fig. 2A and B). We then performed IP experiments to determine if N214 was able to interact with ERK in mammalian cells. Immunoprecipitates and total cell extracts were analyzed by Western blotting (Fig. 2C). The results show that while both FLAG-tagged Us2 and EGFP-fused ERK2 were expressed efficiently, only full-length Us2 could pull down ERK2-EGFP.
FIG. 2.
Mapping Us2 determinants required for interaction with ERK. (A) Cartoon of the PRV Us2 protein. The top line shows full-length Us2 protein and highlights the conserved regions CR1, CR2, and CR3, the C-terminal CAAX motif, which serves as a signal for Us2 prenylation, and an LTRRR motif located at residues 181 to 185. The structure of six C-terminal truncation mutants and one N-terminal truncation mutant analyzed in this study are shown. (B) Truncation mutant N214 interacts with ERK in yeast. Expression plasmids for the Us2 truncations shown in panel A fused to the Gal4 DBD were cotransformed with a Gal4 AD ERK2 expression plasmid into yeast. Transformants were plated on medium lacking leucine, tryptophan, and histidine (medium stringency). (C) Coimmunoprecipitation of Us2 and ERK. FLAG-tagged Us2 and ERK2-EGFP fusion proteins were coexpressed in HEK293T cells and immunoprecipitated with anti-FLAG agarose. Total cell extracts (left panels) and immunoprecipitates (right panels) were analyzed by Western blotting using antibodies reactive with Us2 (α-Us2; bottom blots) and EGFP (α-EGFP; top blots). Asterisks shown in the top right blot indicate the position of background levels of EGFP and ERK2-EGFP that bind nonspecifically to FLAG-Us2 fusion proteins. (D) Elk-1 luciferase reporter assays of Us2 truncation mutants. NIH 3T3 cells were transfected with constitutively active MEK, empty vector (EV) or Us2, GAL4-fused Elk-1, and luciferase reporter linked to GAL4 binding sequences. Luciferase activity is expressed as relative light units. Combinations of plasmids are indicated under the x axis. Each assay was performed in triplicate, and the error bars represent the standard deviations between replicate assays. (E) PRV Us2 LTRRR mutants interact with ERK in yeast. Expression plasmids for the indicated Us2 mutants and ERK2 were cotransformed into yeast and plated onto medium lacking leucine, tryptophan, histidine, and adenine (high stringency). Growth is indicative of a two-hybrid interaction.
Elk-1 trans-reporter assays that utilize an ERK-dependent Elk-1-responsive luciferase expression construct were performed to determine the effects of Us2 truncations on the ERK signaling pathway. Upon the activation of ERK by the MAPK kinase, MEK, ERK dimerizes and enters the nucleus, where it phosphorylates many transcription factors, including Elk-1. Previous studies indicated that wild-type Us2 sequestered ERK in the cytoplasm, preventing it from entering the nucleus to activate Elk-1 and resulting in a dramatic decrease in luciferase activity from the reporter construct (18). In the absence of constitutively active MEK, a basal level of luciferase activity was observed (Fig. 2D). As expected, the inclusion of a constitutively active MEK expression construct in the transfection cocktail led to a 240-fold activation of luciferase activity. The incubation of these cells with the MEK inhibitor, UO126, dramatically decreased luciferase activity to basal levels. These control experiments confirmed that the assay was dependent upon ERK activity for the production of luciferase. Similarly to what has been reported previously, the inclusion of wild-type Us2 led to a 67-fold reduction in luciferase activity (18). Consistent with the yeast two-hybrid assays (Fig. 2B), the N157 and N146 truncation mutants did not inhibit luciferase activity, whereas the N214 truncation showed a 2-fold inhibition of ERK-dependent Elk-1 activity. In summary, the N214 mutant demonstrated a strong interaction with ERK in yeast cells and demonstrated the modest inhibition of ERK signaling in NIH 3T3 cells. However, FLAG-N214 was unable to coprecipitate ERK2-EGFP from HEK293T cell lysates, which may indicate that the N214-ERK interaction is not strong enough to withstand the presence of detergents in the co-IP assays. The coexpression of the N214 truncation with ERK2-EGFP and subsequent analysis by immunofluorescence microscopy revealed the substantial colocalization of the two proteins; however, as both proteins localized throughout the cytoplasm and to cytoplasmic aggregates, these data could not reliably report on the interaction between these molecules (data not shown).
Between residues 157 and 214 of Us2, there is a putative LxxRR motif that we hypothesized interacted with ERK. The LxxRR motif, found in the ERK substrates MNK1 and MNK2 (23) and all Rsk isoforms (6), is required for binding to the ERK common docking (CD) domain. MNK1 contains an LARRR motif, whereas MNK2 and the Rsk isoforms contain an LAQRR motif. The LTRRR motif of PRV Us2 is located from residues 181 to 185 (Fig. 2A). To determine whether the LTRRR motif is required for Us2 binding to ERK, we first constructed LTRKK (R184K/R185K) and LTRAA (R184A/R185A) mutants that replaced the last two arginine residues with either lysine or alanine (28). Both mutants maintained their ability to interact with ERK in yeast (Fig. 2E). In addition to the R184K/R185K and R184A/R185A mutants, we also constructed an LAAAA (R183 to 185A) mutant to abolish positive charges in this motif and an AAAAA (LTRRR/5A) mutant to additionally replace the leucine, which might be critical in ERK docking (23), with alanine. Both the R183 to 185A and LTRRR/5A mutants were able to interact with ERK (Fig. 2E), indicating that LTRRR is not a functional ERK binding motif.
Taken together, these data suggest that the N-terminal 214 amino acids of Us2 are necessary and sufficient for an interaction with ERK in yeast cells but are not sufficient for detectible interactions in coimmunoprecipitation experiments. These findings suggest that Us2 secondary or tertiary structure, rather than a linear amino acid sequence, is required for the ERK interaction. To analyze this further, we examined the ability of Us2 to form higher-order structures.
Us2 forms oligomeric structures.
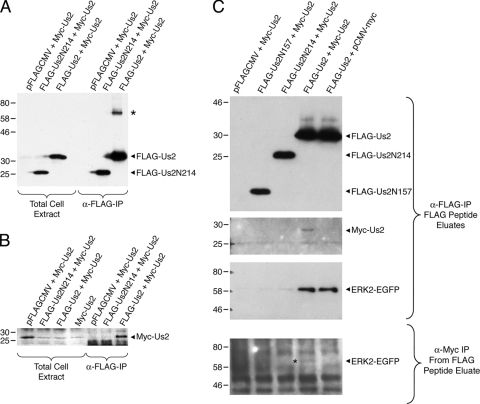
In the course of our studies, we noted an additional band that reacted with the anti-FLAG antiserum on Western blots of Us2 immunoprecipitates (Fig. 3A, asterisk). This species had a relative mobility of roughly twice that of Us2, which is consistent with the idea that Us2 can form dimers. To test this idea, we constructed a Myc-tagged Us2 expression plasmid and coexpressed it with FLAG-Us2 and FLAG-Us2N214 expression constructs. If Us2 forms oligomers, then one would predict that Myc-Us2 would be immunoprecipitated by FLAG-Us2. FLAG-tagged Us2s were immunoprecipitated, and the precipitated material was analyzed for the presence of Myc-Us2 (Fig. 3B). Whereas FLAG-Us2 and FLAG-Us2N214 were efficiently expressed and recovered in the immunoprecipitates (Fig. 3A), only FLAG-Us2 pulled down Myc-Us2 (Fig. 3B). The data show that Myc-Us2 formed oligomers with full-length Us2 but not with the Us2N214 truncation mutant. To investigate whether oligomeric Us2 was capable of interacting with ERK, we performed sequential coimmunoprecipitation experiments. Protein complexes from cell lysates containing FLAG-Us2, Myc-Us2, and ERK2-EGFP first were immunoprecipitated by using anti-FLAG agarose, and the protein complexes were competitively eluted using a FLAG peptide. The eluted complexes then were immunoprecipitated by using anti-myc antibody, and these immune complexes were analyzed for the presence of ERK2-EGFP by Western blotting. The data demonstrate that ERK2-EGFP was immunoprecipitated specifically in the complexes containing oligomeric Us2 (Fig. 3C). Taken together, these findings raise the possibility that Us2 oligomerization is required for robust interaction with ERK, and that the inability of Us2N214 to oligomerize might explain why it fails to interact with ERK efficiently.
FIG. 3.
Us2 forms oligomers. FLAG-tagged and Myc-tagged Us2 expression constructs were cotransfected into HEK293T cells. Proteins were immunoprecipitated using anti-FLAG agarose and analyzed by Western blotting. Total cell extracts and IPs were probed with anti-FLAG (A) and anti-myc (B) antibodies to detect the presence of Us2. An asterisk indicates a protein species with a relative mobility roughly twice that of Us2. (C) FLAG-tagged Us2, Myc-Us2, and ERK2-EGFP expression constructs were cotransfected into 293T cells, and protein complexes were immunoprecipitated using anti-FLAG agarose and subsequently eluted using a FLAG peptide. The top three blots show the analysis of FLAG-Us2, Myc-Us2, and ERK2-EGFP present in the eluted fraction. Protein complexes were immunoprecipitated from the FLAG elutes using anti-myc antibody and detected for the presence of ERK2-EGFP (asterisk) by Western blotting (bottom blot).
Mapping ERK determinants required for interaction with Us2.
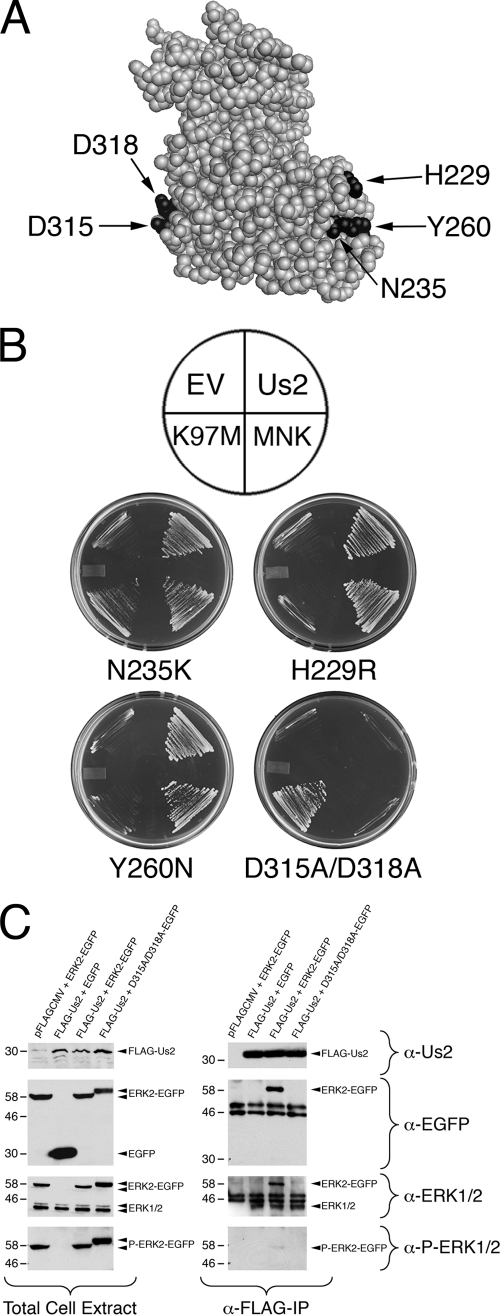
The ERK structure has three notable features: an activation loop, a substrate docking site, called the CD domain, and a MEK binding site, termed the MAPK insert. To identify the Us2 binding site on ERK, we constructed four ERK mutants, H229R, N235K, Y260N, and D315A/D318A (Fig. 4 A). Mutants H229R and N235K locate close to the MAPK insert, whereas Y260N locates in the MAPK insert. Based on previous studies, the H229R and Y260N mutations prevent MEK binding and the N235K mutation inhibits MEK binding (27). The D315A/D318A mutations are in the CD domain and have been shown to prevent the binding of the ERK substrate, MNK, to ERK (27). A yeast two-hybrid assay was used to determine the ability of these mutants to interact with Us2, MNK, and MEK K97M, a kinase-dead mutant of MEK that shows stronger interaction with ERK than wild-type MEK (27). As expected, the H229R and Y260N mutants lost their interaction with MEK K97M, whereas N235K maintained a weak interaction with MEK K97M (Fig. 4B). All three of these mutants interacted with MNK, which was expected because the CD domain remained intact. Furthermore, these mutants maintained the ability to bind to Us2. Whereas the D315A/D318A mutant interacted with MEK K97M, it failed to bind MNK, as shown previously (Fig. 4B) (27). Significantly, the D315A/D318A mutant also failed to interact with Us2, indicating that Us2 binds to ERK via its CD domain.
FIG. 4.
PRV Us2 interacts with ERK at its common docking domain. (A) Structure of ERK2 (1). Four mutations, H229R, N235K, Y260N, and D315A/D318A, were introduced into ERK2, and the positions of these key residues are indicated by the arrows. (B) Yeast two-hybrid analysis. Expression plasmids for the ERK2 mutants were cotransformed into yeast strain YCH1 along with empty vector (EV), wild-type Us2, the ERK substrate MNK, or the MEK kinase-dead mutant K97M to determine protein-protein interactions as described for Fig. 2. The top image indicates the position of the ERK-interacting partners on the plates shown underneath. Growth is indicative of a two-hybrid interaction. (C) Coimmunoprecipitation of Us2 and ERK. FLAG-tagged Us2 was cotransfected with either wild-type ERK2-EGFP fusion or EGFP-D315A/D318A for 24 h. Proteins were immunoprecipitated using anti-FLAG agarose and subjected to Western blot analysis. Cell lysates (left panel) and immunoprecipitates (right panel) were probed using Us2 (α-Us2) antiserum, anti-EGFP (α-EGFP), anti-ERK1/2 (α-ERK1/2), and anti-phospho-ERK1/2 (α-P-ERK1/2) antibodies.
IP experiments were performed to confirm the interaction of Us2 with the ERK CD domain in mammalian cells. FLAG-tagged Us2 and ERK2 or D315A/D318A-EGFP expression plasmids were cotransfected into HEK293T cells for 24 h. Us2 was immunoprecipitated with anti-FLAG agarose. Immunoprecipitates and total cell extracts were analyzed by Western blotting using antiserum against Us2, EGFP, ERK1/2, and phospho-ERK1/2. Both Us2 and wild-type ERK2-EGFP or EGFP-D315A/D318A, or EGFP alone, were efficiently coexpressed in cells (Fig. 4C, left). Moreover, the ERK and D315A/D318A-EGFP fusion proteins were readily detected with anti-phospho-ERK1/2 antibody, indicating that these proteins maintained their ability to interact with, and be phosphorylated by, MEK (Fig. 4C, left). Consistent with the yeast two-hybrid results (Fig. 4B), only wild-type ERK2-EGFP and endogenous ERK could be pulled down by Us2. These findings indicate that ERK D315 and D318 residues are critical for Us2 interaction in animal cells (Fig. 4C, right).
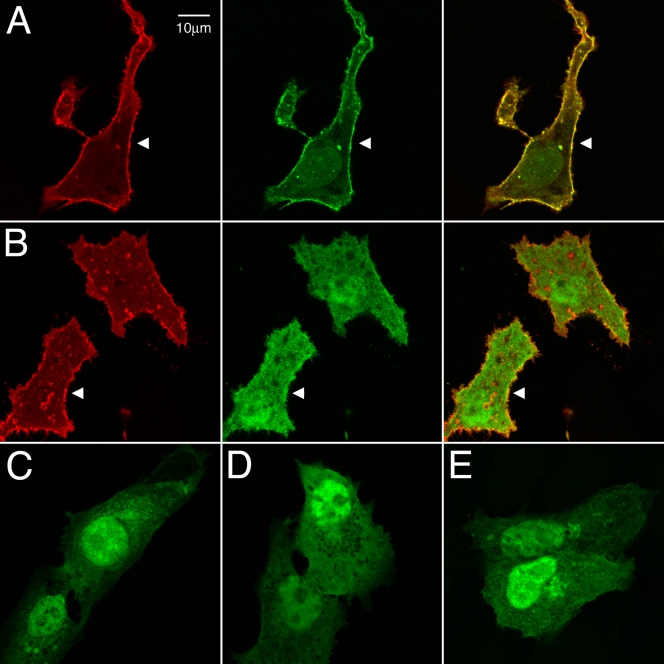
Us2 and the ERK EGFP expression plasmids were cotransfected into Vero cells to determine their subcellular localization (Fig. 5). In the absence of Us2, EGFP, ERK2-EGFP, and EGFP-D315A/D318A, all showed a diffuse nuclear and cytoplasmic distribution (Fig. 5C to E). When coexpressed with PRV Us2, wild-type ERK2-EGFP was relocalized to the plasma membrane, where it colocalized with Us2 (Fig. 5A, arrowhead). In sharp contrast, the localization of the EGFP-D315A/D318A mutant was unaffected in the presence of Us2 (Fig. 5B, arrowhead).
FIG. 5.
PRV Us2 does not alter subcellular localization of EGFP-D315A/D318A. Vero cells were cotransfected with Us2 and EGFP-ERK2 expression plasmids. Twenty-four hours after transfection, cells were stained for Us2 using Us2 antiserum. Arrowheads indicate points of interest. Scale bars are 10 μm. (A) Localization of Us2 (red) and wild-type ERK2-EGFP (green). (B) Localization of Us2 and EGFP-D315A/D318A. (C) Localization of EGFP-ERK2 in the absence of Us2. (D) Localization of EGFP-D315A/D318A in the absence of Us2. (E) Localization of EGFP in the absence of Us2.
Finally, FRAP experiments were performed to determine the influence of wild-type Us2 expression on the diffusion properties of EGFP-D315A/D318A (Fig. 6). In the absence of Us2, EGFP-D315A/D318A (Fig. 6A, row i, and B, graph i) behaved very similarly to ERK2-EGFP (Fig. 1A, row i, and B, graph i), with the rapid recovery of the fluorescent signal into the photobleached region occurring within 5 s. The diffusion of EGFP-D315A/D318A in the presence of Us2 (Fig. 6A, row ii, and B, graph ii) was indistinguishable from its behavior in the absence of Us2. Collectively, these data indicate that Us2 interacts with ERK via the CD domain, and that this interaction facilitates the tethering of ERK to the plasma membrane.
FIG. 6.
FRAP analysis of EGFP-D315A/D318A in the absence and presence of Us2. (A) Images of live PK15 cells cotransfected with pCIneo and EGFP-D315A/D318A (i) or Us2 and EGFP-D315A/D318A expression plasmids (ii) immediately before, immediately after, or 24 (i) and 20.1 (ii) s after the area outlined in the red box was photobleached using an Olympus FV1000 laser-scanning confocal microscope. The boxed blue area is proximal to the photobleached region, and the boxed orange area is distal to the photobleached region. The fluorescence signal is ERK2-EGFP. Scale bars, 10 μm. (B) Quantitation of the fluorescence intensity of the photobleached area, as well as control proximal and distal areas, over time. Data for PK15 cells cotransfected with pCIneo and EGFP-D315A/D318A (i) and cells cotransfected with Us2 and EGFP-D315A/D318A expression plasmids (ii) are shown.
Previous studies showed that ERK isolated in the presence of Us2 maintained the ability to phosphorylate Elk-1 in vitro, suggesting that Us2 did not inhibit ERK enzymatic activity (18). Interestingly, the ERK-mediated activation of Elk-1 was inhibited by Us2 in transfected cells (Fig. 2D) (18). Because Elk-1 normally resides in the nucleus and Us2 restricts ERK localization to the cytoplasm, these observations suggest that Us2 spatially regulates ERK activity. The data presented in this study indicate that, like many ERK substrates, Us2 interacts with ERK via the CD domain (Fig. 4 and 5). However, our experiments suggest that Us2 is not modified by phosphorylation (data not shown).
It may be that Us2 also regulates ERK activity by competing for ERK substrate binding. How might ERK bound to Us2 interact with its substrates? While many ERK substrates bind to the CD domain, ERK substrates that contain an FXFP motif bind to the ERK MAP kinase insert, which is distal to the CD domain (10). It may be that ERK bound to Us2 has access only to substrates that bind the MAP kinase insert. Experiments to identify proteins phosphorylated by ERK in the presence of Us2 should help sort this out. It also is possible that ERK bound to Us2 is capable of activating all of its potential substrates, and that Us2 spatially regulates ERK activity simply by restricting its cellular localization. Previous studies have shown that the cytoplasmic ERK scaffold, KSR, binds to the ERK MAP kinase insert via its FXFP motif (10). However, KSR binding to ERK does not interfere with its interaction with other substrates that also use the MAP kinase insert to bind ERK (2). The dimerization of ERK allows it to form simultaneous interactions with both KSR and cytoplasmic substrates (2). At this time, we do not know if Us2 interacts with ERK monomers, dimers, or both. Determining the stoichiometry of the Us2-ERK interaction will provide important clues to further our understanding of the regulation of ERK activity by Us2 and help define the role of this complex in alphaherpesvirus egress.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grant AI48626, CFI award 16389, and CIHR operating grant 93804 to B.W.B. M.-H.K. was supported in part by R. Samuel McLaughlin and Robert John Wilson fellowships.
We thank M. Cobb (University of Texas Southwestern Medical Center) for providing MNK1 and MEK K97M cDNAs, N. Bunnet (University of California, San Francisco) for the ERK2-EGFP expression plasmid, and P. Sadowski (University of Toronto) for providing pCMV-myc. We are grateful to C. Calton and J. Randall for excellent technical assistance. We acknowledge members of the Banfield laboratory for helpful discussions.
Footnotes
Published ahead of print on 16 June 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Canagarajah, B. J., A. Khokhlatchev, M. H. Cobb, and E. J. Goldsmith. 1997. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell 90:859-869. [DOI] [PubMed] [Google Scholar]
- 2.Casar, B., A. Pinto, and P. Crespo. 2008. Essential role of ERK dimers in the activation of cytoplasmic but not nuclear substrates by ERK-scaffold complexes. Mol. Cell 31:708-721. [DOI] [PubMed] [Google Scholar]
- 3.Clase, A. C., M. G. Lyman, T. del Rio, J. A. Randall, C. M. Calton, L. W. Enquist, and B. W. Banfield. 2003. The pseudorabies virus Us2 protein, a virion tegument component, is prenylated in infected cells. J. Virol. 77:12285-12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cobb, M. H., and E. J. Goldsmith. 1995. How MAP kinases are regulated. J. Biol. Chem. 270:14843-14846. [DOI] [PubMed] [Google Scholar]
- 5.DeFea, K. A., J. Zalevsky, M. S. Thoma, O. Dery, R. D. Mullins, and N. W. Bunnett. 2000. Beta-arrestin-dependent endocytosis of proteinase-activated receptor 2 is required for intracellular targeting of activated ERK1/2. J. Cell Biol. 148:1267-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gavin, A. C., and A. R. Nebreda. 1999. A MAP kinase docking site is required for phosphorylation and activation of p90(rsk)/MAPKAP kinase-1. Curr. Biol. 9:281-284. [DOI] [PubMed] [Google Scholar]
- 7.Hancock, J. F., A. I. Magee, J. E. Childs, and C. J. Marshall. 1989. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell 57:1167-1177. [DOI] [PubMed] [Google Scholar]
- 8.Hermann, S., B. Heppner, and H. Ludwig. 1984. Pseudorabies viruses from clinical outbreaks and latent infections grouped into four major genome types. Curr. Top. Vet. Med. Anim. Sci. 27:387-401. [Google Scholar]
- 9.Hübert, P. H., S. Birkenmaier, H. J. Rziha, and N. Osterrieder. 1996. Alterations in the equine herpesvirus type-1 (EHV-1) strain RacH during attenuation. Zentralbl. Veterinarmed. B 43:1-14. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs, D., D. Glossip, H. Xing, A. J. Muslin, and K. Kornfeld. 1999. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 13:163-175. [PMC free article] [PubMed] [Google Scholar]
- 11.Kalejta, R. F. 2008. Tegument proteins of human cytomegalovirus. Microbiol. Mol. Biol. Rev. 72:249-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klupp, B. G., C. J. Hengartner, T. C. Mettenleiter, and L. W. Enquist. 2004. Complete, annotated sequence of the pseudorabies virus genome. J. Virol. 78:424-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolch, W. 2005. Coordinating ERK/MAPK signaling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 6:827-837. [DOI] [PubMed] [Google Scholar]
- 14.Kolch, W. 2000. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. J. 351:289-305. [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis, T. S., P. S. Shapiro, and N. G. Ahn. 1998. Signal transduction through MAP kinase cascades. Adv. Cancer Res. 74:49-139. [DOI] [PubMed] [Google Scholar]
- 16.Lomniczi, B., S. Watanabe, T. Ben-Porat, and A. S. Kaplan. 1984. Genetic basis of the neurovirulence of pseudorabies virus. J. Virol. 52:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luttrell, L. M. 2003. ‘Location, location, location’: activation and targeting of MAP kinases by G protein-coupled receptors. J. Mol. Endocrinol. 30:117-126. [DOI] [PubMed] [Google Scholar]
- 18.Lyman, M. G., J. A. Randall, C. M. Calton, and B. W. Banfield. 2006. Localization of ERK/MAP kinase is regulated by the alphaherpesvirus tegument protein Us2. J. Virol. 80:7159-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meindl, A., and N. Osterrieder. 1999. The equine herpesvirus 1 Us2 homolog encodes a nonessential membrane-associated virion component. J. Virol. 73:3430-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mettenleiter, T. C. 2006. Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet. Microbiol. 113:163-169. [DOI] [PubMed] [Google Scholar]
- 21.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2009. Herpesvirus assembly: an update. Virus Res. 143:222-234. [DOI] [PubMed] [Google Scholar]
- 22.Murphy, L. O., and J. Blenis. 2006. MAPK signal specificity: the right place at the right time. Trends Biochem. Sci. 31:268-275. [DOI] [PubMed] [Google Scholar]
- 23.Parra, J. L., M. Buxade, and C. G. Proud. 2005. Features of the catalytic domains and C termini of the MAPK signal-integrating kinases Mnk1 and Mnk2 determine their differing activities and regulatory properties. J. Biol. Chem. 280:37623-37633. [DOI] [PubMed] [Google Scholar]
- 24.Peyssonnaux, C., and A. Eychene. 2001. The Raf/MEK/ERK pathway: new concepts of activation. Biol. Cell 93:53-62. [DOI] [PubMed] [Google Scholar]
- 25.Pomeranz, L. E., A. E. Reynolds, and C. J. Hengartner. 2005. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. 69:462-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robbins, D. J., M. Cheng, E. Zhen, C. A. Vanderbilt, L. A. Feig, and M. H. Cobb. 1992. Evidence for a Ras-dependent extracellular signal-regulated protein kinase (ERK) cascade. Proc. Natl. Acad. Sci. U. S. A. 89:6924-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson, F. L., A. W. Whitehurst, M. Raman, and M. H. Cobb. 2002. Identification of novel point mutations in ERK2 that selectively disrupt binding to MEK1. J. Biol. Chem. 277:14844-14852. [DOI] [PubMed] [Google Scholar]
- 28.Tanoue, T., M. Adachi, T. Moriguchi, and E. Nishida. 2000. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2:110-116. [DOI] [PubMed] [Google Scholar]
- 29.Torii, S., K. Nakayama, T. Yamamoto, and E. Nishida. 2004. Regulatory mechanisms and function of ERK MAP kinases. J. Biochem. 136:557-561. [DOI] [PubMed] [Google Scholar]
- 30.Wagenaar, F., J. M. Pol, B. Peeters, A. L. Gielkens, N. de Wind, and T. G. Kimman. 1995. The US3-encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus. J. Gen. Virol. 76:1851-1859. [DOI] [PubMed] [Google Scholar]
- 31.Widmann, C., S. Gibson, M. B. Jarpe, and G. L. Johnson. 1999. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 79:143-180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.