Abstract
The N termini of the capsid proteins VP1 and VP2 of adeno-associated virus (AAV) play important roles in subcellular steps of infection and contain motifs that are highly homologous to a phospholipase A2 (PLA2) domain and nuclear localization signals (NLSs). To more clearly understand how virion components influence infection, we have generated mutations in these regions and examined their effects on subcellular trafficking, capsid stability, transduction, and sensitivity to pharmacological enhancement. All mutants tested assembled into capsids; retained the correct ratio of VP1, VP2, and VP3; packaged DNA similarly to recombinant AAV2 (rAAV2); and displayed similar stability profiles when heat denatured. Confocal microscopy demonstrated that these mutants trafficked through a perinuclear region in the vicinity of the Golgi apparatus, with a subset of mutants displaying more-diffuse localization consistent with an NLS-deficient phenotype. When tested for viral transduction, two mutant classes emerged. Class I (BR1−, BR2−, and BR2+K) displayed partial transduction, whereas class II (VP3only, 75HD/AN, BR3−, and BR3+K) were severely defective. Surprisingly, one class II mutant (BR3+K) trafficked identically to rAAV2 and accumulated in the nucleolus, a step recently described by our laboratory that occurs with wild-type infection. The BR3+K mutant, containing an alanine-to-lysine substitution in the third basic region of VP1, was 10- to 100-fold-less infectious than rAAV2 in transformed cell lines (such as HEK-293, HeLa, and CV1-T cells), but in contrast, it was indistinguishable from rAAV2 in several nontransformed cell lines, as well as in tissues (liver, brain, and muscle) in vivo. Complementation studies with pharmacological adjuvants or adenovirus coinfection suggested that additional positive charges in NLS regions restrict mobilization in the nucleus and limit transduction in a transformed-cell-specific fashion. Remarkably, besides displaying cell-type-specific transduction, this is the first description of a capsid mutant indicating that nuclear entry is not sufficient for AAV-mediated transduction and suggests that additional steps (i.e., subnuclear mobilization or uncoating) limit successful AAV infection.
Interest in adeno-associated virus (AAV) has been fueled by its enormous potential as a gene therapy tool, given that it has never been associated with disease, it can infect a wide variety of mitotic and postmitotic cells, and therapeutic transgenes can easily be substituted in place of its coding elements (4). As a member of the parvovirus family, AAV is one of the smallest and simplest viruses. It has a diameter of only 25 nm, is nonenveloped, and cannot replicate on its own. The single-stranded wild-type viral genome is flanked by two inverted terminal repeats (TR) that are the only elements necessary for packaging. Three capsid proteins (VP1, VP2, and VP3) are encoded in an overlapping reading frame, each beginning with a different start codon but sharing a common C terminus and stop codon. Capsids are comprised of 60 proteins, with VP1, VP2, and VP3 in a ratio of approximately 1:1:10, respectively (6, 44).
Although AAV has a several properties that make it an attractive vehicle for gene therapy, transduction is difficult to achieve in many cell types due to inefficient subcellular trafficking (12, 29, 54). At the cellular level, infection commences with receptor attachment, which is mediated by glycoproteins such as heparan sulfate (49, 58, 59). After coreceptor activation, AAV2 is endocytosed from the cell surface in a clathrin- and dynamin-dependent process (2, 3, 17). Subsequently, virions have been reported to traffic through early endosomes (3), late endosomes, recycling endosomes, and lysosomes (14), and AAV2 and AAV5 have been visualized in the Golgi apparatus (2, 47).
As the AAV virion traffics toward the nucleus, amino acid residues exposed on the capsid surface interact with the subcellular environment and influence its navigation, either directly or indirectly. Critical components in the capsid that are necessary for subcellular navigation are harbored near the N termini of VP1 and VP2 (7, 23, 71). These regions of VP1 and VP2 are normally inaccessible to antibodies and are thought to be tucked away in the capsid interior during assembly, but at some point during infection (prior to endosomal escape), a conformational change promotes their translocation to the capsid exterior (5, 36, 56). Two functional elements have been identified in these N-terminal regions of VP1/2, a phospholipase A2 (PLA2) domain and putative nuclear localization signals (NLS). These elements are conserved in almost all parvoviruses (7), with a sequence of about 70 amino acids in VP1 being highly homologous to the catalytic domain of secretory PLA2. Mutating the histidine and aspartic acid catalytic dyad of PLA2 to alanine and asparagine (75HD/AN) does not affect cell surface attachment or endocytosis of the virus but strongly inhibits PLA2 activity and renders the virus severely defective, which has been shown to result in a decrease in transduction of roughly 3 orders of magnitude (23, 56, 71). Phospholipase action is surmised to mediate the membrane disruption of a vesicular compartment, which would permit the escape of the virion into the cytosol, although this has yet to be directly demonstrated for AAV.
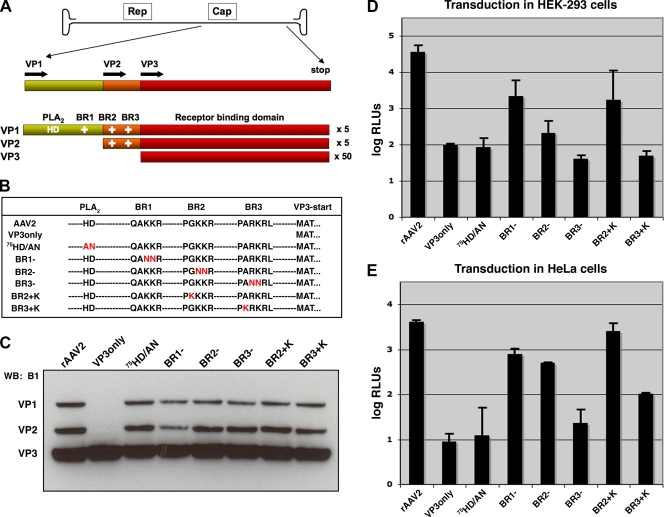
The vast majority of genomes remain outside the nuclear membrane (12, 29), but ultimately, the transgene must be delivered to the nucleus, transition from a single-stranded genome to a double-stranded intermediate, and convert to a stable double-stranded form before transduction is considered complete (62). We have previously shown that capsids are capable of entering the nucleus intact (33), but the precise location for uncoating is still debated. The transport of macromolecules such as parvoviruses across the nuclear membrane is an ATP-dependent process and is mediated by soluble cytosolic factors, like Ran-1, that interact with specific NLSs on target proteins (25, 46). NLSs have been identified on VP1s from various parvoviruses that are integral to nuclear targeting (40, 41, 60). The first 10 residues of VP1 in canine parvovirus has been identified as an NLS (60), and minute virus of mice is known to have a bipartite NLS (41), yet these residues are not conserved in AAV. Several patches of hydrophilic, basic regions (also considered basic clusters) have been identified in AAV2 capsid proteins (Fig. 1 A), termed BR1 (only in VP1), BR2, and BR3 (in VP1 and VP2) (27, 56). Mutating these regions results in a graduated loss of infectivity compared to recombinant AAV2 (rAAV2): roughly 1 log order for BR1 (120QAKKR/QANNR), 2 log orders for BR2 (140PGKKR/PGNNR), and 3 log orders for BR3 (168PARKR/PANNR) (27, 67). While it is known that these mutations negatively impact the infectivity of AAV2, their influence on subcellular trafficking in the context of the virion has not been studied in depth.
FIG. 1.
Mutations in VP1/2 N termini affect transduction of rAAV. (A) The AAV genome contains genes that code for Rep proteins involved in replication and Cap proteins that comprise the capsid. Specific Cap proteins harbor a phospholipase domain (PLA2) (HD) and basic regions (BR) (+) that are putative nuclear localization signals. (B) Capsid mutants used in this study are listed by sequence, and amino acid changes are shown in red letters. (C) Western blot (WB) analysis of purified virus (5 × 109 vg) blotted with antibody B1, which detects VP1, VP2, and VP3. (D and E) Luciferase assay of transduction in HEK-293 cells (D) and HeLa cells (E). Cells were inoculated with rAAV2 or mutants that package TR-CMV-Luc (10,000 vg/cell), and luciferase activity was quantified 24 h later. Error bars represent standard deviations from three independent samples.
Evidence from one of the most robust and well-studied NLSs, found in the large T antigen of simian virus 40 (SV40), suggests that the number of positive charges in an NLS correlates to the strength of its function (34, 35). The putative NLS regions in AAV are not as extensive as those in other viruses, so we reasoned that this might at least partially explain why the nuclear targeting of AAV is inefficient. Thus, we attempted to see if the BR2 and BR3 regions could be manipulated to (i) tolerate an additional positive charge and (ii) mediate enhanced nuclear targeting and transduction. Thus, for these studies we have engineered a collection of mutant capsids containing amino acid substitutions or deletions in the N termini of VP1 and VP2 and have explored how these mutations influence subcellular trafficking and transduction. Here we report that these mutants display subtle differences in their trafficking profiles: in contrast to rAAV2, mutants that lack a PLA2 domain or positive charge in NLS regions cannot be detected in the nucleus. The BR1− mutant targets mainly to the Golgi apparatus, whereas BR2− and BR3− display a slightly more diffuse localization, distal from the nucleus. We have generated one mutant, containing a lysine-to-alanine substitution in BR3 (168PARKR/PKRKR [BR3+K]), that was 10- to 100-fold-less infectious than rAAV2 in certain cell types but was able to transduce as well as rAAV2 in mouse liver, muscle, and rat brain in vivo and other cultured cells in vitro. Surprisingly, despite BR3+K being defective in transducing HeLa cells, the capsid was still able to enter the nucleus with the transgene. In addition to suggesting that nuclear entry is not sufficient for transduction, our results imply that additional positive charges in NLS regions may restrict mobilization in the nucleus and limit transduction in a cell-specific manner that appears to be linked to the transformation state of the cell. The availability of these mutant capsids and permissive and nonpermissive cell lines should aid in dissecting specific steps in subcellular trafficking required for successful AAV infection, which might eventually advance the design of AAV-based vectors.
MATERIALS AND METHODS
Cell culture.
HeLa, HEK-293, C2C12, and HepG2 cells were maintained at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) that was supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 U/ml penicillin, and 100 g/ml streptomycin. CV1 and CV1-T cells (also known as Cos-7 cells) were maintained under the same conditions but with Eagle's minimum essential medium (MEM) supplemented with 10% FBS and 1% penicillin-streptomycin.
PCR site-directed mutagenesis.
Capsid mutations were generated in the pXR2 backbone as previously described (27), with minor modifications. PCR was performed with the Expand Long Template PCR kit (Roche) according to cycle guidelines listed in the manual for QuikChange site-directed mutagenesis (Stratagene). PCR products were digested with DpnI and transformed into DH10B bacteria, which were selected on ampicillin-containing agar plates. Single colonies were picked and grown in liquid cultures overnight, and plasmids were isolated by column purification (Qiagen). Mutations were verified by sequencing of the plasmids utilizing the University of North Carolina—Chapel Hill (UNC-CH) Automated DNA Sequencing Facility with a 3100 Genetic Analyzer (Applied Biosystems).
Virus production.
Virus was produced in HEK-293 cells as previously described (69). Briefly, polyethylenimine (linear molecular weight, ∼25,000) was used to for the triple transfection of the pXR2 cap and rep plasmid, the pXX6-80 helper plasmid, and a TR-Luciferase or TR-eGFP reporter plasmid containing either firefly luciferase (Luc) or enhanced green fluorescent protein (GFP) transgenes flanked by inverted terminal repeats. Cells were harvested at between 48 and 72 h posttransfection, and virus was purified by cesium chloride gradient density centrifugation. After identifying peak fractions by dot blot hybridization, virus was dialyzed into 1× phosphate-buffered saline (PBS) containing 5% sorbitol. Titers were calculated by quantitative PCR (qPCR) according to established procedures (33) by using a LightCycler 480 instrument with Sybr green (Roche) and primers designed against the Luc transgene, 5′ AAA AGC ACT CTG ATT GAC AAA TAC 3′ (forward) and 5′ CCT TCG CTT CAA AAA ATG GAA C 3′ (reverse), or the GFP transgene, 5′ AGC AGC ACG ACT TCT TCA AGT CC 3′ (forward) and 5′ TGT AGT TGT ACT CCA GCT TGT GCC 3′ (reverse). Conditions used for the reaction were as follows: 1 cycle at 95°C for 10 min; 45 cycles at 95°C for 10 s, 62°C for 10 s, and 72°C for 10 s; and 1 cycle at 95°C for 30 s, 65°C for 1 min, and 99°C for acquisition. Adenovirus (Ad) mutant stocks were generously provided by Pat Hearing or generated in the laboratory, produced and purified as previously described (20, 30).
Confocal immunofluorescence microscopy.
Similar to what we have previously described (26), HeLa cells (5 × 104 cells/well) were plated onto poly-l-lysine-coated 12-mm glass coverslips (thickness, 1.5) between 4 and 16 h before inoculation. Recombinant virions were added to cell media at the indicated particle numbers. No virus was added to control wells. Where indicated, hydroxyurea (HU; Sigma) was added at 10 mM for 12 h and washed off extensively prior to virus administration. A proteasome inhibitor (MG132; Calbiochem) was present for the duration of virus inoculation at 2 μM, when used. At the indicated time points, cells were washed three times with PBS and then fixed with 2% paraformaldehyde for 15 min at room temperature. The cells were then permeabilized with 0.1% Triton X-100 in PBS for 5 min at room temperature. Following four washes with PBS, the permeabilized cells were blocked with immunofluorescence buffer (IFB) (20 mM Tris [pH 7.5], 137 mM NaCl, 3 mM KCl, 1.5 mM MgCl2, 5 mg/ml bovine serum albumin, 0.05% Tween) for 30 min at room temperature. The cells were incubated with primary antibody to detect intact capsids (monoclonal antibody [MAb] A20 [1:10], nucleolin (NCL) [Ab22758, 1:1,000; Abcam], lamin B1 [Ab16048, 1:750; Abcam], giantin [Ab24586, 1:1,000; Abcam], and polyclonal anti-VP1 (αVP1)-147/148 [1:1,000]) diluted in IFB for 1 h at 37°C or overnight at 4°C. The cells were then incubated in secondary antibody diluted 1:5,000 in IFB (anti-mouse Alexa Fluor 488 or anti-rabbit Alexa Fluor 568; Molecular Probes) for 1 h at 37°C. After six washes in PBS, coverslips were mounted cell side down onto glass slides with mounting medium (Prolong Antifade gold with DAPI [4′,6′-diamidino-2-phenylindole]; Molecular Probes). Images were captured on a Leica SP2 AOBS upright laser scanning confocal microscope and processed by using Adobe Photoshop.
Three-dimensional (3D) rendering of infection.
Confocal z-stack sections of HeLa cells fixed 16 h after infection with either rAAV2 or mutant virions (10,000 vector genomes [vg]/cell) were processed and rendered in three dimensions by using Volocity software (Improvision) in the Michael Hooker Microscopy Facility.
Cell fractionation and nucleolar isolation.
Nucleoli were isolated from cell fractionations as previously described (43), with minor modifications allowing for viral infection (33). Briefly, five 15-cm plates of HeLa cells at 90% confluence were used for each preparation. rAAV2 or mutant virions were incubated with cells for 16 h at 37°C (2,000 vg/cell). Cells were washed three times with ice-cold PBS, trypsinized, and harvested by centrifugation at 218 × g for 5 min. The cell pellet was resuspended in hypotonic buffer and homogenized on ice using a tight pestle to the point where >90% of cells were burst (approximately 15 strokes) and leaving nuclei intact. The homogenate was spun at 218 × g for 5 min to separate the crude nuclear pellet from the postnuclear supernatant (∼5 ml). Nuclei were further purified from cytoplasmic contaminants by spinning through a 0.35 M sucrose cushion to give a nuclear-associated pellet. Nuclei were lysed by limited sonication on ice, and the nuclear suspension in 0.35 M sucrose was layered over a 0.88 M sucrose cushion to separate nucleoli (200 μl) from bulk nuclear fractions (∼5 ml). Nucleoli were further purified by another spin through sucrose, and their integrity and purity were verified by phase-contrast microscopy and Western blotting.
Transduction assays.
At least 4 h prior to infection or drug treatment, cells were plated in 24-well plates at densities to approximate 70% confluence (2 × 105 cells/well for HEK-293 cells and 7 × 104 cells/well for other lines). For drug studies, cells were handled as stated above. Cells were infected with purified rAAV2 at the designated number of vector genomes per cell and typically harvested after 24 h unless otherwise noted. Luciferase activity was measured in accordance with the manufacturer's instructions (Promega) using a Wallac1420 Victor2 automated plate reader. Error bars represent standard deviations from samples scored in triplicate. Graphs are representative of data sets from at least three independent assays.
Western blotting.
As reported previously (33), samples from purified virus or cell fractionations were loaded onto NuPage 10% Bis-Tris gels and typically run for 3 h at 150 V in 1× NuPage MOPS (morpholinepropanesulfonic acid) buffer. Electrophoresis was performed with the XCell SureLock minicell (Invitrogen). Proteins were transferred onto a Hybond ECL membrane utilizing the XCell II blot module (Invitrogen) according to the manufacturer's protocol. Membranes were blocked in 5% nonfat dry milk (NFDM) dissolved in 1× PBS-Tween (0.1%) (PBST). For the detection of capsid proteins, primary antibody (MAb B1) was diluted 1:20 with 2.5% NFDM in PBST and incubated for 1 h at room temperature or overnight at 4°C. After washing with PBST, blots were incubated for 1 h at room temperature with anti-mouse horseradish peroxidase (HRP) secondary antibody diluted 1:5,000 in 2.5% NFDM with PBST. Following multiple washes with PBST, SuperSignal West Femto maximum-sensitivity substrate (Pierce) was added to each membrane according to the manufacturer's protocol for development. Each membrane was then exposed to Hyperfilm ECL (Amersham).
Immunoblot after limited heat treatment.
To examine capsid stability and VP1 exposure, purified virions (1 × 109 vg/sample) were heat treated over a range of temperatures between 37°C and 75°C for 10 min in PBS, and samples were then split for detection with separate antibodies (2.5 × 109 vg/well) blotted onto a nitrocellulose membrane through a dot blotting apparatus. The membrane was processed as described above for Western blotting by using antibodies to detect intact capsids (monoclonal A20 [1:10]), exposed VP1 N termini (monoclonal A1 [1:20] and polyclonal αVP1-147/148 [1:1,000]), disassembled subunits (monoclonal B1 [1:2,000]), and appropriate secondary antibodies conjugated to HRP. Data are from representative blots from experiments performed in triplicate.
EM.
As previously described (15), purified and dialyzed full rAAV2 or BR3+K virus particles (109 vg) were pipetted onto a glow-discharged copper grid. The grid was washed twice with water and then stained with 2% uranyl acetate. Electron microscopy (EM) images were taken with a LEO EM 910 transmission electron microscope at several magnifications at the University of North Carolina Microscopy Services Laboratories.
FISH to detect viral genomes.
HeLa cells were plated onto chamber slides (Nunc) coated with poly-l-lysine (25 × 103 cells/well) 24 h prior to infection. rAAV2 or BR3+K virions were administered (10,000 vg/cell) for 16 h before the cells were washed four times with PBS and fixed with 4% paraformaldehyde for 15 min at room temperature. Cells were then treated with protease K (0.5 μg/ml) for 5 min, washed with PBS, and fixed again in paraformaldehyde. The samples were then treated with triethanolamine-HCl with acetic anhydride for 10 min at room temperature, washed with PBS, and prehybridized for 2 h at 65°C in prewarmed hybridization buffer. A digoxigenin-labeled sense-strand RNA probe complementary to the vector genome was synthesized by using T7 polymerase and spanned a 1-kb fragment of the firefly luciferase transgene. The probe was added to samples in hybridization buffer overnight at 65°C, and after washing steps, the digoxigenin label was detected by using the Roche Fast Red kit for fluorescent in situ hybridization (FISH), and the slides were then covered with coverslips and mounted with DAPI to detect cell nuclei.
Vector administration and animal studies.
Housing and handling of BALB/c mice and Sprague-Dawley (Harlan) rats used in the current study were carried out in compliance with National Institutes of Health guidelines and approved by the IACUC at the University of North Carolina—Chapel Hill. Recombinant AAV2 vectors or mutants packaging Luc transgenes were administered through the intramuscular (1 × 1010 vg into the hind limb; gastrocnemius muscle) in a volume of 50 μl PBS or through the intravenous route (tail vein) in a volume of 200 μl PBS. Bioluminescence of Luc expression was visualized by using a Xenogen IVIS100 imaging system (Caliper Lifesciences) after intraperitoneal injection of luciferin substrate (120 mg/kg of body weight; Nanolight). Image acquisition and analysis were carried out by using Living Image software. Data are representative images from only one mouse, but results were reproduced consistently using three or four mice for each vector group, and average values are included in the supplemental material.
For vector infusions into rat brain, animals were anesthetized with isoflurane and placed into a stereotaxic frame. Using a 32-gauge stainless steel injector and a Fisher Scientific infusion pump, rats (n = 2) received 3 μl per hemisphere (total of 3.8 × 108 vg) AAV2-CBA-GFP (left hemisphere) and BR3+K-CBA-GFP (right hemisphere) into the striatum (bregma, +1.0 mm; lateral, ±3.0 mm; vertical, −5.0 mm [based on a brain atlas {48}]) over 15 min. The injector was left in place for 3 min after infusion to allow diffusion from the injector. Two weeks after intracerebral injection, animals received an overdose of pentobarbital (100 mg/kg intraperitoneally) and were perfused transcardially with ice-cold 100 mM sodium PBS (pH 7.4), followed by 4% paraformaldehyde in phosphate buffer (PB) (pH 7.4). After brains were postfixed for 48 h at 4°C in paraformaldehyde-PB, 40-μm coronal sections were cut using a vibrating-blade microtome and subjected to immunohistochemical processing for GFP. Colorimetric GFP detection was performed by incubating free-floating sections with a rabbit polyclonal antibody to GFP (1:1,000; Millipore). Briefly, sections were blocked in 10% normal goat serum and 0.1% Triton X-100 in PBS for 1 h. The sections were then incubated with anti-GFP primary antibody (prepared in 3% normal serum-0.2% Triton X-100) for 48 to 72 h at 4°C with gentle agitation. Immunoperoxidase staining was then performed by using the anti-rabbit Vectastain Elite ABC kit (Vector Laboratories) according to the manufacturer's instructions, with 3,3′-diaminobenzidine (DAB) substrate and nickel-cobalt intensification of the reaction product. Brain sections were mounted onto slides, covered with a coverslip, and digitized by using a Scan-Scope slide scanner (Aperio Technologies). Virtual slides were viewed and images were captured by using the ImageScope software package (v. 10.0; Aperio Technologies).
Statistical analyses.
When luciferase activity after the transduction of rAAV2 and the BR3+K mutant was evaluated (see Fig. 7A and B), a two-tailed Student's t test was used to compare means. A P value of ≤0.01 was considered significant and is marked in the figures with an asterisk. All data were analyzed by using Excel software (Microsoft).
FIG. 7.
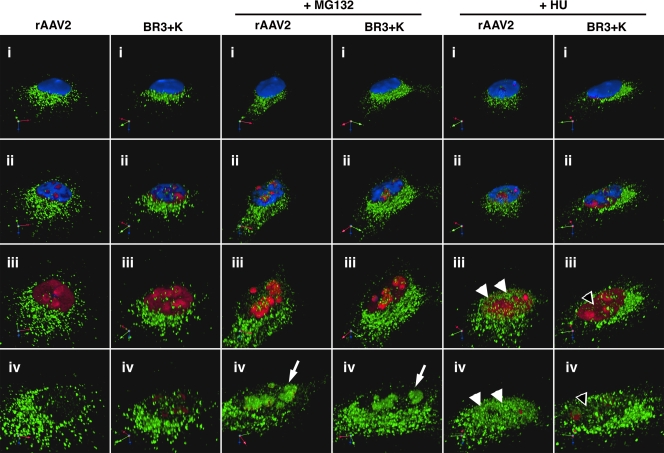
HU treatment does not mobilize BR3+K capsids to the nucleoplasm. To visualize whether BR3+K mutant capsids could be mobilized into the nucleoplasm in response to HU, we fixed and stained infected HeLa cells for indirect immunofluorescence of capsids (green), nucleolin (red), and nuclei (blue). Roughly 40 confocal sections that span 5 μm were stacked and rendered in three dimensions by using Volocity software (i). A movie was generated to explore capsid localization in detail by digitally gating first the blue channel (DAPI, representing the nucleus) and then the red channel (nucleolin, representing the nucleolus), along with increasing the zoom. Images were captured at successive stages in the movie to reveal the nuclear interior (ii and iii) and the interior of the nucleolus (iv). Both rAAV2 and the BR3+K mutant accumulate in the nucleolus in the presence of proteasome inhibitor (white arrows). Only rAAV2 is localized to nucleoplasmic sites following HU treatment (white arrowheads, rAAV2; empty arrowheads, BR3+K mutant).
RESULTS
Mutations in VP1/2 N termini affect transduction of rAAV.
Through PCR mutagenesis we have generated capsid mutants that lack VP1 and VP2 (VP3only), lack PLA2 activity (75HD/AN) (23), lack positive charges in VP1 and VP2 (BR1−, BR2−, and BR3−), or have additional positive charges in the latter basic regions (BR2+K and BR3+K) (Fig. 1B). Each of these mutants was found to assemble into capsids and produce titers of magnitudes similar to that of rAAV2 (data not shown). Western blot analysis of 5 × 109 vector genomes shows that VP3only capsids carry no detectable levels of VP1 or VP2, whereas the remaining mutants contain appropriate stoichiometric ratios of VP1, VP2, and VP3 at roughly 1:1:10, respectively (Fig. 1C). After assessing several different preparations of virus, no significant variation in VP1 or VP2 incorporation was found to be different from rAAV2 in any of the mutants, as depicted by the representative blot shown in Fig. 1C. These results corroborate previous reports that VP1 and VP2 do not play an obvious role in packaging (63). However, mutations in these capsid proteins have striking effects on transduction, with similar profiles for HEK-293 and HeLa cells (Fig. 1D and E). Previously, we identified through prediction mapping software that there are four potential NLS regions in AAV2 (27). The first three basic regions, BR1, BR2, and BR3, were found to be important for transduction, but the fourth was required for capsid assembly. Mutation of BR2 and BR3 to include neutral amino acids was shown to have the most negative impact on transduction, similar to what Sonntag et al. observed previously after mutating these basic regions to negative amino acids (56). Since these regions appeared to be most important for transduction, we wanted to assess whether these regions would tolerate an additional positive charge in an attempt to increase the strength of their NLS activities, similar to what was demonstrated previously for the large T antigen of SV40 (34, 35, 64). The VP3only, 75HD/AN, BR3−, and BR3+K mutants seemed to be the most defective of the group, as it was difficult to distinguish any difference from background light units (log relative light units [RLUs], ≤2). Thus, these mutants could be divided into two classes: class I mutants (BR1−, BR2−, and BR2+K), which were partially deficient, and class II mutants (VP3only, 75HD/AN, BR3−, and BR3+K), which were markedly deficient. It is noteworthy that these results were consistent when using different reporters (Luc or GFP), different promoters (cytomegalovirus [CMV] or CBA), and a wide range of particle numbers (from 100 to 20,000 vg/cell) (data not shown). In each case the BR3+K mutant was severely compromised in HEK-293 and HeLa cells, transducing at least an order of magnitude less efficiently than rAAV2.
Subcellular localization of capsid mutants.
Previous studies demonstrated that a significant percentage of AAV capsids colocalize with the Golgi apparatus during subcellular trafficking (2, 47); therefore, we used this area as a point of reference for analyzing whether mutants traffic differently than rAAV2 in HeLa cells, in an attempt to link capsid localization with a corresponding defect in transduction. HeLa cells were inoculated with rAAV2 or mutant virions for 16 h and prepped samples for immunofluorescence microscopy using antibodies to detect intact capsids (A20) or a Golgi marker (giantin). A variety of localization patterns was observed for rAAV2 and mutant capsids; however, cells that were most representative of the population were selected for analysis. Using z-stacked confocal slices of inoculated cell samples, we were able to generate three-dimensional renderings of infection with volume imaging software. We found that all mutants were able to bind and efficiently internalize into HeLa cells to the same extent. These vectors displayed either partial or significant colocalization with the Golgi apparatus (Fig. 2). We observed pronounced Golgi localization with the BR1− mutant, more so than with any other mutant, although the significance of this is not yet clear (see Discussion). The BR2− and BR3− capsid mutants appear to have diffuse localization throughout the cell in addition to localizing to the Golgi. This finding supports the idea that BR2 and BR3 are important for nuclear localization, but it is unknown if these mutant capsids remain in endosomal compartments at distal regions or have escaped into the cytoplasm. Considering the observation that a population of even the most defective class II mutants traffic to the Golgi apparatus during infection, namely, VP3only and 75HD/AN particles, one could speculate that accumulation in Golgi-associated vesicles is a default sorting pathway taken by the virus, which does not require PLA2 or NLS function.
FIG. 2.
Subcellular localization of capsid mutants. Volume imaging software was used to process z-stacked confocal images of HeLa cells infected with capsid mutants (10,000 vg/cell) for 16 h. Capsids (green) are shown in relation to the Golgi marker giantin (red) and nuclei (blue). All mutants, along with rAAV2, display some level of colocalization with the Golgi apparatus, with the BR1− mutant being the most intense. The BR2− and BR3− capsid mutants appear to have diffuse localization throughout the cell in addition to localizing to the Golgi apparatus compared to the localization of rAAV2 and other mutants.
Exposure of VP1 N termini.
Many viruses undergo a maturation process during entry into cells that is necessary for infection to progress. Parvoviruses follow this paradigm, with late subcellular events of infection hinging on the exposure of VP1/2 N termini (36, 56). To establish whether VP1/2 N termini were appropriately exposed in mutant capsids during infection, we employed antibodies that have been generated to specifically detect either intact capsids (A20), VP1 N termini (A1/anti-VP1) with the exposed phospholipase domain, or disassembled subunits (B1) (Fig. 3 A) (66). The extrusion of VP1/2 N termini can be induced in vitro by limited heat treatment or urea. Kronenberg et al. reported previously that empty capsids fail to expose VP1 at temperatures that expose VP1 in rAAV2 (37). Here, we have also demonstrated this result by treating purified rAAV2 and empty capsids with a range of temperatures and subsequently immunoblotting samples onto nitrocellulose with the above-mentioned antibodies (Fig. 3B). In contrast to empty capsids, virions with mutations in VP1 N termini are able to expose VP1 similarly to rAAV2 after limited heat treatment. Epitopes on VP1 become exposed after incubation at 60°C (reactive to anti-VP1), with capsids remaining intact, as indicated by A20 reactivity and the lack of B1 reactivity (Fig. 3C).
FIG. 3.
Exposure of VP1 N termini. (A) Antibodies that specifically detect either intact capsids (A20), VP1 N termini (A1/αVP1), or disassembled subunits (B1) permit analysis of conformational changes in vitro in response to limited heat treatment or in cells during infection (i.e., exposure of the phospholipase and NLS motifs). (B) Immunoblot showing exposure of VP1 N termini. rAAV2 virions or empty capsids were exposed to limited heat treatment after limited heat treatment at designated temperatures for 10 min, applied onto nitrocellulose membranes, incubated with the antibodies shown, and developed after processing for chemiluminescence. Empty capsids are more resistant to exposing VP1. (C) Immunoblot comparing VP1 exposure among capsid mutants (1 × 109 vg/well). Capsids disassemble prior to 75°C, as indicated by the loss of A20 reactivity and gain of B1 reactivity. An intermediate conformational change exists after heating capsids to 60°C, where VP1 epitopes are detected but capsids remain intact. (D) Immunofluorescence microscopy demonstrating VP1 exposure during infection. Capsids (green) display VP1 exposure (red) in a perinuclear region. Empty capsids, which are deficient in VP1 exposure, and VP3only particles, which lack VP1, show no significant signal from αVP1 (amino acids [aa] 2 to 16). The two mutants that are least infectious, 75HD/AN and BR3−, have not lost the ability to expose VP1 during infection, implicating dysfunction in later steps during infection as the cause of the lost transduction.
We also visualized VP1 exposure during infection in HeLa cells using indirect immunofluorescence confocal microscopy (Fig. 3D). The signal for exposed VP1 (red) (anti-VP1) overlaps with the signal for capsids (green) (A20) in a perinuclear region that corresponds mainly to the Golgi apparatus and associated COPI vesicles (data not shown). Supporting our findings from immunoblotting capsids after limited heat treatment, we demonstrate here that we cannot detect a significant exposure of VP1 N termini on empty capsids, which provides an explanation of why empty capsids do not enter the nucleus and fail to accumulate in the nucleolus (33). As a negative control, VP3only particles, which lack VP1, show no significant signal from αVP1. Additionally, we found that other mutants (75HD/AN and BR3−) are still capable of exposing VP1 during entry. Similar results were obtained with antibody A69, which detects the N termini of both VP1 and VP2 (data not shown). Thus, we conclude that these capsid mutants are deficient after the conformational change, at steps in infection such as endosomal escape or nuclear targeting.
Effect of proteasome inhibition on nucleolar accumulation and transduction of capsid mutants.
Since all mutant capsids except VP3only were able to expose VP1/2 N termini, we next tested whether any capsids could be detected in the nucleus and if their deficient transduction profiles could be rescued with a pharmacological adjuvant. Proteasome inhibitors are known to be one of the most potent agents for enhancing the transduction of AAV in certain cell types and have been shown to increase the nuclear accumulation of virions (13). The specific mechanisms by which proteasome inhibitors act to improve transduction remain unclear, but several hypotheses have been put forth: they increase the nuclear delivery of AAV (18), increase the ubiquitination of capsids (70), inhibit the clearance of viral capsids, inhibit the clearance of viral genomes (16), and potentiate the accumulation of virions in the nucleolus (33). We explored the effects of the proteasome inhibitor MG132 on capsid localization using indirect immunofluorescence microscopy (Fig. 4 A). In the presence of MG132, rAAV2, BR2+K, and BR3+K capsids (green) were found to accumulate in the nucleus. We found that the VP3only, 75HD/AN, BR1−, BR2−, and BR3− mutants were restricted mostly to extranuclear regions and were not observed within the nuclear membrane (dotted line traced from DAPI staining). We corroborated these findings by subcellular fractionation and detected the most abundant capsid protein, VP3, from rAAV2, BR2+K, and BR3+K capsids in nucleolar isolations (Fig. 4B). To a much lesser extent, capsid proteins from the BR1− and BR2− mutants were detectable at faint levels in these fractions.
FIG. 4.
Effect of proteasome inhibition on nucleolar accumulation and transduction of capsid mutants. MG132 (2 μM) was administered to HeLa cells simultaneously with virions (10,000 vg/cell), and after 16 h, samples were prepped for immunofluorescence microscopy. rAAV2, BR2+K, and BR3+K capsids (green) are found to accumulate in the nucleus (white arrows), and VP3only, 75HD/AN, BR1−, BR2−, and BR3− capsids are restricted mostly to extranuclear regions. (B) Subcellular fractionation of nucleoli after infection. HeLa cells were infected with mutant virions (1,000 vg/cell) for 16 h and subjected to subcellular fractionation. Capsid proteins from rAAV2 and the BR2+K and BR3+K mutants were detected in nucleolar fractions by Western blotting. (C) Luciferase assay of transduction in the presence of proteasome inhibitor. HEK-293 cells were infected (10,000 vg/cell) for 24 h in the presence of MG132 (2 μM) and then scored for transduction efficiency. Error bars represent standard deviations from three independent samples.
Interestingly, the pattern for transduction enhancement with proteasome inhibitors did not match the observed effect on capsid localization. Only rAAV2, BR1−, and BR2+K transduction profiles were augmented by MG132 treatment, as determined by a luciferase assay (Fig. 4C). The most perplexing discrepancy was seen with the BR3+K mutant, a mutant that was found to accumulate in nucleoli in response to MG132, as shown by indirect immunofluorescence and subcellular fractionation. Surprisingly, the transduction of this mutant was not enhanced in the presence of MG132. This finding raised several questions. Do BR3+K capsids release their genome prematurely, prior to nuclear entry, or are BR3+K capsids unable to release their genome once within the nucleus? Furthermore, will the transduction of BR3+K be rescued under certain conditions? We have attempted to address these questions with the remainder of the study.
Stability of BR3+K capsids compared to that of rAAV.
It is possible that BR3+K capsids are either less or more structurally stable than rAAV2, which could promote an early loss of genomes before capsids have entered the nucleus or, conversely, prevent genome ejection/uncoating after the capsid has entered the nucleus. Thus, we set out to determine if BR3+K capsids were physically different from rAAV2 in terms of their ultrastructural appearance as judged by electron microscopy, capsid sensitivity to limited heat treatment, and genome sensitivity to DNase. No obvious structural differences were apparent between rAAV2 and BR3+K capsids when examined by electron microscopy (Fig. 5 A). We also submitted these capsids to a refined heat gradient using eight temperatures between 37°C and 75°C. In this assay rAAV2 and BR3+K capsids were comparable with respect to thermostability, as VP1 N termini become exposed after 57°C, and disassembled subunits could be detected prominently for both capsids after samples reached 65°C (Fig. 5B). A subtle difference in the B1 signal was seen at lower temperatures, with BR3+K samples being slightly more reactive, but this could not be reliably reproduced (data not shown). Next, we tested if BR3+K genomes were more sensitive to DNase treatment, which would indicate that genomes in BR3+K capsids might be less protected during infection. After heat treatment, virions were treated with DNase for 30 min, the DNase was then inactivated with EDTA, and the remaining genomes were quantified by qPCR. For both rAAV2 and the BR3+K mutant, genomes first became susceptible to DNase once temperatures that extruded VP1 N termini were reached (Fig. 5C), similar to what was observed previously for other parvoviruses (10, 11, 42, 50). Again, no difference in sensitivity to DNase between rAAV2 and BR3+K virions was reliably reproduced. From these assays, we conclude that the BR3+K mutant has no observable effects on capsid ultrastructure, thermostability, or DNase sensitivity.
FIG. 5.
Stability of BR3+K capsids compared to rAAV. (A) Purified virus preparations were subjected to uranyl-acetate negative staining and applied onto discharged copper grids for electron microscopy. The scale bar represents 50 nm. (B) Immunoblotting of antibody reactivity after limited heat treatment (10 min at designated temperatures) indicates that rAAV2 and BR3+K capsids have similar macromolecular stabilities. Capsids are no longer intact at 75°C (A20), VP1 N termini become exposed after 57°C, and disassembled subunits can be detected prominently for both capsids at 65°C. (C) After heat treatment, virions were treated with DNase and submitted to qPCR to compare genome protections. rAAV2 and the BR3+K mutant have similar patterns of DNase sensitivity after heat treatment. The line graph is representative of data from one of three experiments.
Fluorescent in situ hybridization depicting that DNA from the BR3+K mutant is delivered to the nucleus during infection.
Because the above-described in vitro assays of stability did not uncover any differences between rAAV2 and BR3+K capsids, we hypothesized that an uncoating mutant phenotype may be more striking in the context of infection. Therefore, to determine if BR3+K capsids jettison their genomes into the cytoplasm or, in contrast, carry their genomes into the nucleus where the capsid may fail to uncoat, we used fluorescent in situ hybridization (FISH) to visualize the localization of viral genomes. HeLa cells were inoculated with rAAV2 or the BR3+K mutant for 16 h and prepped for FISH by using an RNA probe that detects single-stranded TR-GFP genomes of minus-strand polarity, to avoid the detection of transcribed mRNA. Similar to what was found with capsid localization, the majority of viral genomes (red) accumulated in a perinuclear region (Fig. 6 ii and iii). When the proteasome inhibitor MG132 was administered concurrently with virus, we found that the genomes were delivered to the nucleus and accumulated in the nucleolus (Fig. 6v and vi). When uninfected cells were processed for FISH, no signal could be detected, indicating that the probe is specific to the viral genome (Fig. 6i and iv). Since the BR3+K mutant fails to transduce HeLa cells, yet BR3+K capsids and genomes are found to enter the nucleus, this strongly suggests that the mutant is impeded from transitioning to a nuclear location in this cell type where uncoating can occur. Perhaps the most salient conclusion from this finding is that the nuclear entry of AAV is not sufficient for transduction.
FIG. 6.
Fluorescent in situ hybridization depicting that DNA from the BR3+K mutant is delivered to the nucleus during infection. HeLa cells were infected with rAAV2 or the BR3+K mutant (10,000 vg/cell) for 16 h in the absence (left) or presence (right) of MG132. Cells were prepped for FISH using an RNA probe complementary to the antisense strand of the TR-Luc genome. No staining in control samples (i and iv) indicates a lack of nonspecific binding from the probe. Both rAAV2 (v) and BR3+K (vi) genomes (red) are found to accumulate in the nucleus, specifically in nucleolar regions during proteasome inhibition.
BR3+K capsids cannot be efficiently mobilized to the nucleoplasm.
Previous work identified that at least two separate trafficking pathways in the nucleus can be taken by rAAV2 capsids: a pathway that results in nucleolar accumulation and a second pathway that mobilizes capsids to the nucleoplasm (33). In a prior work we have demonstrated that hydroxyurea (HU) disrupts the nucleolus, which is consistent with data from other reports (51). Treatment with HU results in a reorganization of nucleolar proteins, and concomitantly, virus capsids localize to the nucleoplasm (33). As we have shown that the BR3+K mutant can accumulate in the nucleolus in the presence of proteasome inhibitors, we next asked if this mutant could be mobilized to the nucleoplasm in response to HU treatment. To build on our earlier findings, we inoculated HeLa cells with rAAV2 or the BR3+K mutant for 16 h after pretreatment with HU (10 mM for 12 h) and prepped samples for confocal immunofluorescence. After rendering z-stacked sections into 3D projections (described in Materials and Methods), a movie was generated to visualize capsids that had been internalized into the cell (Fig. 7, and see Movie S1 in the supplemental material). The channels for the nucleus (blue) and nucleoli (red) were digitally subtracted (Fig. 7ii to iv and Movie S1) to reveal that rAAV2 and BR3+K virions (green) accumulate in HeLa cell nucleoli when infection is performed in the presence of MG132 (Fig. 7 and Movie S1). However, only rAAV2 capsids were significantly affected by HU and mobilized to the nucleoplasm (Fig. 7 and Movie S1). BR3+K capsids were not efficiently mobilized to the nucleoplasm following HU treatment, which further implicates the nucleoplasm, and not the nucleolus, as a site for uncoating.
Adenovirus potentiates transgene expression from the BR3+K mutant.
Since the possibility remains that a small percentage of BR3+K virions are able to infect HEK-293 and HeLa cells, we tested whether the classical helper virus of AAV, adenovirus (Ad), could potentiate the transduction of this mutant (see Fig. S2 in the supplemental material). Ad was proposed previously to have multiple effects on AAV transduction, from early effects on AAV internalization (3) to the nuclear delivery of AAV capsids (68), subnuclear effects on the Mre11-Rad50-Nbs1 DNA repair complex (55), effects that increase the efficiency of second-strand synthesis of AAV genomes (20, 21), and effects that increase AAV mRNA stability (52). The coadministration of Ad with rAAV2 and the BR3+K mutant in HEK-293 and HeLa cells dramatically potentiated transduction (Fig. S2C and S2D). However, Ad did not rescue the transduction of the BR3+K mutant to the level of rAAV2; it increased the transduction of the mutant and rAAV2 only by the same order of magnitude. By testing the BR3+K mutant against a battery of Ad mutants, we found that Ad E4 ORF3 and ORF6 are necessary for the Ad-mediated transduction enhancement of the BR3+K mutant (Fig. S2E and S2F). This finding agrees with what Ferrari et al. reported previously for rAAV2, which is that the enhancement is through E4 ORF3- and ORF6-mediated effects on AAV genome processing (20). Importantly, the observed enhancement that Ad has on the BR3+K mutant indicates that a small number of BR3+K virions are capable of transducing HEK-293 and HeLa cells despite most capsids being unable to transition to the nucleoplasm. Taking this finding into account together with the fact that measurement of transduction levels in HEK-293 and HeLa cell lines is not always an accurate predictor of how an AAV vector will perform in other targets (see Discussion), we therefore sought to explore whether the restricted transduction profile of the BR3+K mutant was universal in other cells and tissues.
BR3+K transduction efficiency is lower than that of rAAV2 in transformed cells.
We began to amass a collection of cell lines in which the BR3+K mutant appeared restricted compared to rAAV2 (HEK-293 cells, HeLa cells, and W162 cells) (Fig. 8 A and data not shown). Surprisingly, we identified several cell lines where there were no obvious differences in transduction between the BR3+K mutant and rAAV2 (Fig. 8B). The most striking difference between these groups is that the former group is transformed (HEK-293 [transformed with the Ad E1 gene], HeLa [cervical cancer cells associated with human papillomavirus], and W162 [transformed with the Ad E4 gene] cells) compared to the latter group (HepG2 cells, C2C12 cells, and CV1 cells), which are mostly normal cell lines. Although HepG2 cells may appear to be an exception in the second group, this immortalized line, derived from a hepatic adenocarcinoma, does not display viral antigens and retains many characteristics of normal hepatocytes (31).
FIG. 8.
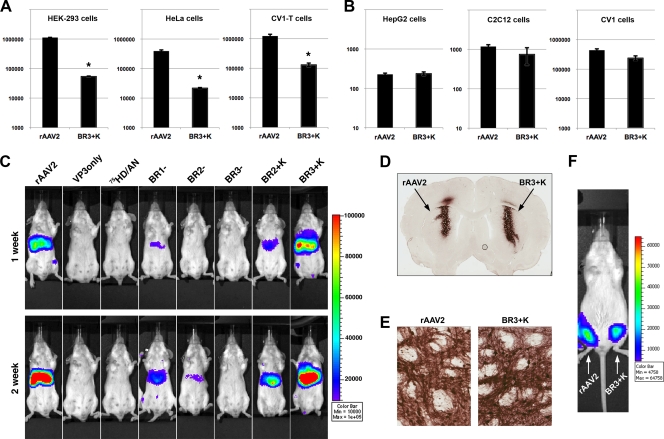
The BR3+K mutant is significantly less efficient than rAAV2 in transformed cells but is indistinguishable from rAAV2 in liver, brain, and muscle tissue in vivo. (A) Transformed cell lines such as HEK-293, HeLa, or CV1-T cells were inoculated with rAAV2 or the BR3+K mutant (1,000 vg/cell) (TR-CBA-Luc transgene) and assayed for transduction efficiency after 24 h. rAAV2 transduction was at least 10-fold-more efficient than that of the BR3+K mutant in these cell lines. (B) The normal cell lines HepG2 (human liver cells), C2C12 (mouse myoblasts), and CV1 (green monkey kidney cells) were assayed for transduction efficiency of rAAV2 or the BR3+K mutant under the same conditions as those described above (A). The transduction efficiencies of rAAV2 and the BR3+K mutant in nontransformed cells were similar (within 2-fold) (*, P value of ≤0.01 for A and B). Note that HepG2 cells are derived from an adenocarcinoma but contain no viral antigens and display many characteristics of normal hepatocytes. (C) rAAV2 or mutant virions (1010 vg) were administered by tail vein injection to female BALB/c mice, and bioluminescent luciferase activity was visualized 1 and 2 weeks later. The color scale bar represents photons/s/cm2, and data shown are representative images of a single mouse from three to four mice per vector group. (D) rAAV2 or BR3+K packaging GFP transgenes (3.8 × 108 vg) was infused into rat striatum by intracerebral injection. Two weeks postinjection, brains were fixed and coronal sections were processed to detect GFP using DAB as a substrate and nickel-cobalt intensification. (E) Higher magnification of images from coronal sections similar to those in D depicting the transduction of neurons and associated processes (dark staining). (F) Bioluminescence images of transduction following intramuscular injections of rAAV2 (right hind limb) and BR3+K (left hind limb) packaging Luc transgenes acquired 2 weeks postinjection.
The availability of CV1 cells and transformed CV1-T cells allowed us to assess whether effects related to cellular transformation were a primary factor in prohibiting BR3+K gene delivery compared to rAAV2. CV-1 cells were derived from the kidney of a male African green monkey in 1964 (32) and transformed to CV1-T cells by the genetic transfer of large T antigen from SV40 (24). We found that the transduction efficiency of the BR3+K mutant was at least 10-fold lower than that of rAAV2 in CV1-T cells (Fig. 8A). However, in nontransformed CV1 cells, the difference between rAAV2 and BR3+K transduction was found to be less than 2-fold (Fig. 8B). These results were on a scale similar to what we have seen for other transformed and nontransformed cell lines (compare Fig. 8A and B), supporting the hypothesis that the cell-type-specific restriction of the BR3+K mutant is correlated with the transformed status of the target.
The BR3+K mutant is able to transduce liver, brain, and muscle tissue in vivo.
To determine if mutant virions performed similarly in nontransformed cells in vivo as they did in vitro, we administered 1010 vg by tail vein injection to female BALB/c mice. At 1 week and 2 weeks postinjection, we visualized the bioluminescence of the luciferase reporter in the liver of these mice (Fig. 8C). Transduction profiles closely matched those observed for HEK-293 and HeLa cells except for the BR3+K mutant, which produced transgene expression levels as high as those of rAAV2 (compare Fig. S3A in the supplemental material to Fig. 1D and E). In addition to examining BR3+K transduction in liver, we also explored whether this mutant would transduce brain and muscle tissue. We infused rAAV2 or the BR3+K mutant into rat striatum by intracerebral injection, and after 2 weeks, coronal sections were fixed and processed for the immunohistochemical detection of GFP transgenes (Fig. 8D). No obvious differences were apparent between rAAV2 and BR3+K transductions in terms of intensity or cellular targets viewed at higher magnifications (Fig. 8E). We have also compared rAAV2 and BR3+K transductions following intramuscular injections into the hind limb of female BALB/c mice, and again, the transduction patterns of the two vectors were indistinguishable in vivo (Fig. 8F and Fig. S3B). Furthermore, the nonrestricted transduction pattern of the BR3+K mutant in vivo in liver and muscle tissue closely matched what was observed for cultured cell lines in vitro for HepG2 cells (human liver cells) and C2C12 cells (mouse myoblasts). These results demonstrate that the transduction of the BR3+K mutant is not species specific and that BR3+K infection is not restricted compared to that of rAAV2 in normal cells and tissues, in contrast to transformed cells, emphasizing the importance of testing AAV-based vector transduction with a variety of targets.
DISCUSSION
Several adeno-associated virus (AAV) serotypes are emerging as popular tools for gene therapy. While research efforts on the AAV capsid have focused largely on how residues can be manipulated to avoid neutralizing antibodies or direct targeting to specific tissues, how capsid residues modulate subcellular trafficking and influence transduction is less understood. Research on subcellular events in AAV infection has garnered increasing interest, as several groups are now demonstrating that the nuclear delivery of the genome is a major barrier to transduction (16, 18, 28, 56, 68, 72). To more clearly understand the trafficking profile of the prototypical serotype, AAV2, and to explore how virion components modulate subcellular trafficking, we have studied a panel of capsid mutants with deletions or substitutions in the unique N-terminal regions of the capsid proteins VP1 and VP2 (Fig. 1). Here we report that these mutants are able to assemble into capsids, package DNA similarly to the wild type, internalize into cells, and traffic through a perinuclear region in the vicinity of the Golgi apparatus. These mutants could be grouped into two classes: class I mutants (BR1−, BR2−, and BR2+K), which were partially deficient, and class II mutants (VP3only, 75HD/AN, BR3−, and BR3+K), which were markedly deficient. Mutations that introduced neutral residues into the positively charged NLS-like regions (BR2− and BR3−) yielded capsids that had a more diffuse localization pattern consistent with a loss of NLS activity. One particularly striking class II mutant, BR3+K, was found to carry its genome to the nucleus but was 10- to 100-fold-less infectious than rAAV2 in transformed cell lines such as HEK-293, HeLa, W162, and CV1-T cells. We speculate that additional positive charges in VP1 or VP2 may restrict the nuclear mobilization or uncoating of capsids and limit transduction in a transformed-cell-specific fashion.
For several years now, the subcellular trafficking of AAV upstream of nuclear entry has been studied. Although the details are not yet in focus, a general route of trafficking has been established, from virus binding, to endocytosis, to perinuclear accumulation, which depends on the activation of Rac1 and phosphatidylinositol 3-kinase (53). In our studies, a significant fraction of mutant capsids appear to traffic through the Golgi apparatus (Fig. 2), as was previously shown for AAV2 and AAV5 (2, 47). It is interesting that classical retrograde trafficking from the Golgi apparatus depends on COPI interactions with cytoplasmically exposed dilysine (KKXX) sequences on target proteins (9, 38). Transmembrane proteins that present this sequence to the cytoplasm are recognized by components of the coatomer protein complex, which then send these targets in vesicles from the Golgi apparatus toward the endoplasmic reticulum (ER). It seems plausible that the cell could process rAAV2 capsids through this sorting pathway, since two dilysine sequences are found in the N terminus of VP1, and mutating one of them (BR1− [120QAKKR/QANNR]) appears to increase localization in Golgi-associated regions (Fig. 2 and 4).
It is important to consider that the cellular environment could play a role in regulating the function of VP1. Specifically, concerning the viral phospholipase domain in VP1, it should be mentioned that Golgi membranes are known to be a primary target for endogenous cytosolic PLA2 activity (22). Our observation that Golgi-associated capsids display exposed VP1 N termini (Fig. 3 and data not shown) could be interpreted as the virus attempting to target an area where the chance of PLA2-mediated membrane disruption is high. The propensity of cellular PLA2 to cleave phospholipids into arachadonic acid is typically regulated by both intracellular Ca2+ levels and the phosphorylation of residues near the catalytic domain of PLA2. Not surprisingly, the N terminus of VP1 contains several potential phosphorylation sites and a GXG calcium-binding site. Structural modeling of VP1 predicts that the N terminus can translocate through the 5-fold axis of symmetry in the capsid and permit the exposure of the first 185 residues in VP1 (36). Since it requires only 18 to 20 amino acids to span a phospholipid bilayer, this would be of sufficient length to allow the presentation of the NLS regions and dilysine sequences to the cytosol if the PLA2 domain had penetrated through a Golgi or endosomal membrane. Perhaps future work will uncover whether the PLA2 domain in AAV operates optimally in a specific vesicular compartment, prefers a specific phospholipid substrate, or functions at multiple cellular membranes such as the endosome and the nuclear envelope or if its activity is regulated by components of the cellular environment.
Clearly, infection by AAV and other parvoviruses requires positive charges near the N terminus of VP1 (27, 41, 56, 61) that putatively direct nuclear targeting. As stated above, the nuclear accumulation of AAV virions is potentiated through an unknown mechanism when the ubiquitin/proteasome system is compromised. Rather strikingly, proteasome inhibitors drove the accumulation of BR3+K mutant virions into the nucleolus, as we have previously described for rAAV2 (33), but did not increase the transduction of this mutant (Fig. 4 and 6). This result yielded a clear separation of proteasome inhibitor-mediated effects on the capsid from effects on gene expression and suggested that BR3+K transduction is blocked at a point after nuclear entry. Apparently, a few BR3+K genomes are able to serve as templates for transgene expression, since transduction is enhanced by Ad E4 genes (see Fig. S2 in the supplemental material), which increase the efficiency of genome or mRNA processing and are not thought to affect the AAV capsid. Taken together, our results provide evidence that the primary mechanism by which proteasome inhibitors increase AAV transduction occurs upstream of nucleoplasmic mobilization or uncoating. We attempted to see if proteasome inhibitor-mediated effects on the capsid could be combined with Ad-mediated effects on the genome to rescue BR3+K transduction, but cooperation or synergy could not be obtained (data not shown), most likely due Ad E4 ORF3 and ORF6 requiring active proteasomes to enhance viral gene expression (8). Nonetheless, a major conclusion from our experiments demonstrating that the BR3+K mutant enters the nucleus in HeLa cells (without being infectious) is that nuclear entry is not sufficient for the transduction of AAV, at least in transformed cells.
Our knowledge of how cellular parameters govern AAV transduction is still in its infancy, and many serotypes behave differently in vitro compared to in vivo. AAV8 is a prototypical example; it is heralded as one of the better capsids for targeting mouse liver in vivo and is capable of transducing ∼100% of mouse hepatocytes after injection through the portal or tail vein (45). In contrast, AAV8 does not efficiently transduce cells in tissue culture. This pattern is somewhat similar to what we have observed for the BR3+K mutant, where it was unable to efficiently transduce several transformed cell lines in vitro (Fig. 1 and 8) but was indistinguishable from rAAV2 in untransformed cell lines and tissues in vivo (Fig. 8 and 9). Cellular transformation from small DNA tumor viruses, such as Ad, SV40, and papillomavirus, centers on the inactivation of the tumor suppressor p53 (39). Studies of the large T antigen from SV40 have shed light on the multitude of pathways that are affected in transformed cells, including molecular interactions that interfere with the heat shock chaperone hsc70, the retinoblastoma family (Rb family) of tumor suppressors, and p53 (1). Future work may uncover how the perturbation of these factors may specifically influence and restrict BR3+K transduction.
FIG. 9.
Model to illustrate the transformed-cell-type-specific defect of the BR3+K mutant. (A) The entry pathway of rAAV2 proceeds from binding glycoprotein receptors at the cell surface to endocytosis, trafficking to a perinuclear compartment, escape into the cytoplasm, nuclear entry, and mobilization to an unknown region of the nucleoplasm. (B) The trafficking of the BR3+K mutant is indistinguishable from that of rAAV2 except that in transformed cells, it does not mobilize to the nucleoplasm as efficiently as rAAV2 (Fig. 7, and see Movie S1 in the supplemental material). Cellular transformation modifies several nuclear processes that could influence the trafficking and transduction of the BR3+K mutant (see Discussion). These changes likely affect the processing of the BR3+K mutant after nuclear entry but prior to gene expression (subnuclear localization or uncoating).
Although no consensus sequence has been identified for nucleolar targeting, proteins that are trafficked to the nucleolus contain sequences that resemble NLS motifs (19). It is possible that the additional lysine residue in BR3 could essentially “target and trap” the virus in the nucleolus, preventing it from uncoating in the nucleoplasm (Fig. 9). If the lysine substitution in the BR3+K mutant generates a stronger nucleolar localization signal, this may explain why the BR3+K mutant is less infectious than rAAV2 in transformed cells, given that high levels of nucleolar proteins are found in tumor cells (57) to the degree that in certain cases, levels of nucleolar proteins are used as a diagnostic marker for cancer. We know from our previous studies that infectious virions can accumulate in the nucleolus (33). Treatments that disrupt the nucleolus (such as HU or small interfering RNA [siRNA]-mediated knockdown of nucleolar proteins) result in the mobilization of capsids into the nucleoplasm and correlate with increases in transduction. The BR3+K mutant might be terminally targeted to the nucleolus in transformed cells and unable to escape to an environment favorable for uncoating. In contrast, rAAV2 might contain weaker nucleolar localization properties, which could function to target capsids to the stable environment of the nucleolus during assembly (65) or infectious entry (33), yet be flexible enough to transition to the nucleoplasm if favorable conditions arose in the cell that were permissive for infection. With future work it will be interesting to determine if other serotypes besides AAV2 employ this subcellular strategy of maneuvering to nucleolar reservoirs or mobilizing to the nucleoplasm, since a high degree of sequence conservation is found in the NLS-like regions of VP1 and VP2. Ultimately, the BR3+K mutant and other vectors that we have generated in this study should prove valuable as we continue to dissect the trafficking and transduction profiles of AAV variants with new molecular tools.
Supplementary Material
Acknowledgments
This research was funded from NIH fellowship F31NS060688 (NINDS to J.S.J.) and from grants 5P01HL051818-15 (NHLBI to R.J.S.), 1R01AI080726-01A2 and 1R01DK084033-01 (NIH to C.L. and R.U.S.), and 5R01NS035633-12 (NINDS to T.J.M.).
We thank members of the UNC Gene Therapy Center for productive discussions, specifically those in the laboratories of R. Jude Samulski, Aravind Asokan, and Tal Kafri. We greatly appreciate Swati Yadav for calculating virus titers by qPCR, and we thank the Michael Hooker Microscopy Facility and the UNC In Situ Hybridization Core for providing resources.
Footnotes
Published ahead of print on 23 June 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ahuja, D., M. T. Saenz-Robles, and J. M. Pipas. 2005. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene 24:7729-7745. [DOI] [PubMed] [Google Scholar]
- 2.Bantel-Schaal, U., B. Hub, and J. Kartenbeck. 2002. Endocytosis of adeno-associated virus type 5 leads to accumulation of virus particles in the Golgi compartment. J. Virol. 76:2340-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartlett, J. S., R. Wilcher, and R. J. Samulski. 2000. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J. Virol. 74:2777-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berns, K. I., and C. Giraud. 1996. Biology of adeno-associated virus. Curr. Top. Microbiol. Immunol. 218:1-23. [DOI] [PubMed] [Google Scholar]
- 5.Bleker, S., F. Sonntag, and J. A. Kleinschmidt. 2005. Mutational analysis of narrow pores at the fivefold symmetry axes of adeno-associated virus type 2 capsids reveals a dual role in genome packaging and activation of phospholipase A2 activity. J. Virol. 79:2528-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buller, R. M., and J. A. Rose. 1978. Characterization of adenovirus-associated virus-induced polypeptides in KB cells. J. Virol. 25:331-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canaan, S., Z. Zadori, F. Ghomashchi, J. Bollinger, M. Sadilek, M. E. Moreau, P. Tijssen, and M. H. Gelb. 2004. Interfacial enzymology of parvovirus phospholipases A2. J. Biol. Chem. 279:14502-14508. [DOI] [PubMed] [Google Scholar]
- 8.Corbin-Lickfett, K. A., and E. Bridge. 2003. Adenovirus E4-34kDa requires active proteasomes to promote late gene expression. Virology 315:234-244. [DOI] [PubMed] [Google Scholar]
- 9.Cosson, P., and F. Letourneur. 1994. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science 263:1629-1631. [DOI] [PubMed] [Google Scholar]
- 10.Cotmore, S. F., A. M. D'Abramo, Jr., C. M. Ticknor, and P. Tattersall. 1999. Controlled conformational transitions in the MVM virion expose the VP1 N-terminus and viral genome without particle disassembly. Virology 254:169-181. [DOI] [PubMed] [Google Scholar]
- 11.Cotmore, S. F., S. Hafenstein, and P. Tattersall. 2010. Depletion of virion-associated divalent cations induces parvovirus minute virus of mice to eject its genome in a 3′-to-5′ direction from an otherwise intact viral particle. J. Virol. 84:1945-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding, W., Z. Yan, R. Zak, M. Saavedra, D. M. Rodman, and J. F. Engelhardt. 2003. Second-strand genome conversion of adeno-associated virus type 2 (AAV-2) and AAV-5 is not rate limiting following apical infection of polarized human airway epithelia. J. Virol. 77:7361-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding, W., L. Zhang, Z. Yan, and J. F. Engelhardt. 2005. Intracellular trafficking of adeno-associated viral vectors. Gene Ther. 12:873-880. [DOI] [PubMed] [Google Scholar]
- 14.Ding, W., L. N. Zhang, C. Yeaman, and J. F. Engelhardt. 2006. rAAV2 traffics through both the late and the recycling endosomes in a dose-dependent fashion. Mol. Ther. 13:671-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiPrimio, N., A. Asokan, L. Govindasamy, M. Agbandje-McKenna, and R. J. Samulski. 2008. Surface loop dynamics in adeno-associated virus capsid assembly. J. Virol. 82:5178-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Douar, A. M., K. Poulard, D. Stockholm, and O. Danos. 2001. Intracellular trafficking of adeno-associated virus vectors: routing to the late endosomal compartment and proteasome degradation. J. Virol. 75:1824-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duan, D., Q. Li, A. W. Kao, Y. Yue, J. E. Pessin, and J. F. Engelhardt. 1999. Dynamin is required for recombinant adeno-associated virus type 2 infection. J. Virol. 73:10371-10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duan, D., Y. Yue, Z. Yan, J. Yang, and J. F. Engelhardt. 2000. Endosomal processing limits gene transfer to polarized airway epithelia by adeno-associated virus. J. Clin. Invest. 105:1573-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emmott, E., and J. A. Hiscox. 2009. Nucleolar targeting: the hub of the matter. EMBO Rep. 10:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrari, F. K., T. Samulski, T. Shenk, and R. J. Samulski. 1996. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J. Virol. 70:3227-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisher, K. J., G. P. Gao, M. D. Weitzman, R. DeMatteo, J. F. Burda, and J. M. Wilson. 1996. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J. Virol. 70:520-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh, M., D. E. Tucker, S. A. Burchett, and C. C. Leslie. 2006. Properties of the group IV phospholipase A2 family. Prog. Lipid Res. 45:487-510. [DOI] [PubMed] [Google Scholar]
- 23.Girod, A., C. E. Wobus, Z. Zadori, M. Ried, K. Leike, P. Tijssen, J. A. Kleinschmidt, and M. Hallek. 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 83:973-978. [DOI] [PubMed] [Google Scholar]
- 24.Gluzman, Y. 1981. SV40-transformed simian cells support the replication of early SV40 mutants. Cell 23:175-182. [DOI] [PubMed] [Google Scholar]
- 25.Gorlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 26.Grieger, J. C., J. S. Johnson, B. Gurda-Whitaker, M. Agbandje-McKenna, and R. J. Samulski. 2007. Surface-exposed adeno-associated virus Vp1-NLS capsid fusion protein rescues infectivity of noninfectious wild-type Vp2/Vp3 and Vp3-only capsids but not that of fivefold pore mutant virions. J. Virol. 81:7833-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grieger, J. C., S. Snowdy, and R. J. Samulski. 2006. Separate basic region motifs within the adeno-associated virus capsid proteins are essential for infectivity and assembly. J. Virol. 80:5199-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen, J., K. Qing, H. J. Kwon, C. Mah, and A. Srivastava. 2000. Impaired intracellular trafficking of adeno-associated virus type 2 vectors limits efficient transduction of murine fibroblasts. J. Virol. 74:992-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hauck, B., W. Zhao, K. High, and W. Xiao. 2004. Intracellular viral processing, not single-stranded DNA accumulation, is crucial for recombinant adeno-associated virus transduction. J. Virol. 78:13678-13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang, M. M., and P. Hearing. 1989. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J. Virol. 63:2605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Javitt, N. B. 1990. Hep G2 cells as a resource for metabolic studies: lipoprotein, cholesterol, and bile acids. FASEB J. 4:161-168. [DOI] [PubMed] [Google Scholar]
- 32.Jensen, F. C., A. J. Girardi, R. V. Gilden, and H. Koprowski. 1964. Infection of human and simian tissue cultures with Rous sarcoma virus. Proc. Natl. Acad. Sci. U. S. A. 52:53-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson, J. S., and R. J. Samulski. 2009. Enhancement of adeno-associated virus infection by mobilizing capsids into and out of the nucleolus. J. Virol. 83:2632-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalderon, D., B. L. Roberts, W. D. Richardson, and A. E. Smith. 1984. A short amino acid sequence able to specify nuclear location. Cell 39:499-509. [DOI] [PubMed] [Google Scholar]
- 35.Kalderon, D., and A. E. Smith. 1984. In vitro mutagenesis of a putative DNA binding domain of SV40 large-T. Virology 139:109-137. [DOI] [PubMed] [Google Scholar]
- 36.Kronenberg, S., B. Bottcher, C. W. von der Lieth, S. Bleker, and J. A. Kleinschmidt. 2005. A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden VP1 N termini. J. Virol. 79:5296-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kronenberg, S., J. A. Kleinschmidt, and B. Bottcher. 2001. Electron cryo-microscopy and image reconstruction of adeno-associated virus type 2 empty capsids. EMBO Rep. 2:997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Letourneur, F., E. C. Gaynor, S. Hennecke, C. Demolliere, R. Duden, S. D. Emr, H. Riezman, and P. Cosson. 1994. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell 79:1199-1207. [DOI] [PubMed] [Google Scholar]
- 39.Levine, A. J. 2009. The common mechanisms of transformation by the small DNA tumor viruses. The inactivation of tumor suppressor gene products: p53. Virology 384:285-293. [DOI] [PubMed] [Google Scholar]
- 40.Lombardo, E., J. C. Ramirez, M. Agbandje-McKenna, and J. M. Almendral. 2000. A beta-stranded motif drives capsid protein oligomers of the parvovirus minute virus of mice into the nucleus for viral assembly. J. Virol. 74:3804-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lombardo, E., J. C. Ramirez, J. Garcia, and J. M. Almendral. 2002. Complementary roles of multiple nuclear targeting signals in the capsid proteins of the parvovirus minute virus of mice during assembly and onset of infection. J. Virol. 76:7049-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mani, B., C. Baltzer, N. Valle, J. M. Almendral, C. Kempf, and C. Ros. 2006. Low pH-dependent endosomal processing of the incoming parvovirus minute virus of mice virion leads to externalization of the VP1 N-terminal sequence (N-VP1), N-VP2 cleavage, and uncoating of the full-length genome. J. Virol. 80:1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mauramatsu, M., K. Smetana, and H. Busch. 1963. Quantitative aspects of isolation of nucleoli of the Walker carcinosarcoma and liver of the rat. Cancer Res. 23:693-697. [PubMed] [Google Scholar]
- 44.Muzyczka, N. 1992. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr. Top. Microbiol. Immunol. 158:97-129. [DOI] [PubMed] [Google Scholar]
- 45.Nakai, H., S. Fuess, T. A. Storm, S. Muramatsu, Y. Nara, and M. A. Kay. 2005. Unrestricted hepatocyte transduction with adeno-associated virus serotype 8 vectors in mice. J. Virol. 79:214-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nigg, E. A. 1997. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature 386:779-787. [DOI] [PubMed] [Google Scholar]
- 47.Pajusola, K., M. Gruchala, H. Joch, T. F. Luscher, S. Yla-Herttuala, and H. Bueler. 2002. Cell-type-specific characteristics modulate the transduction efficiency of adeno-associated virus type 2 and restrain infection of endothelial cells. J. Virol. 76:11530-11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paxinos, G., and C. Watson. 1998. The rat brain in stereotaxic coordinates, 4th ed. Academic Press, San Diego, CA.
- 49.Qing, K., C. Mah, J. Hansen, S. Zhou, V. Dwarki, and A. Srivastava. 1999. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat. Med. 5:71-77. [DOI] [PubMed] [Google Scholar]
- 50.Ros, C., C. Baltzer, B. Mani, and C. Kempf. 2006. Parvovirus uncoating in vitro reveals a mechanism of DNA release without capsid disassembly and striking differences in encapsidated DNA stability. Virology 345:137-147. [DOI] [PubMed] [Google Scholar]
- 51.Rubbi, C. P., and J. Milner. 2003. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 22:6068-6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Samulski, R. J., and T. Shenk. 1988. Adenovirus E1B 55-Mr polypeptide facilitates timely cytoplasmic accumulation of adeno-associated virus mRNAs. J. Virol. 62:206-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanlioglu, S., P. K. Benson, J. Yang, E. M. Atkinson, T. Reynolds, and J. F. Engelhardt. 2000. Endocytosis and nuclear trafficking of adeno-associated virus type 2 are controlled by rac1 and phosphatidylinositol-3 kinase activation. J. Virol. 74:9184-9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanlioglu, S., M. M. Monick, G. Luleci, G. W. Hunninghake, and J. F. Engelhardt. 2001. Rate limiting steps of AAV transduction and implications for human gene therapy. Curr. Gene Ther. 1:137-147. [DOI] [PubMed] [Google Scholar]
- 55.Schwartz, R. A., J. A. Palacios, G. D. Cassell, S. Adam, M. Giacca, and M. D. Weitzman. 2007. The Mre11/Rad50/Nbs1 complex limits adeno-associated virus transduction and replication. J. Virol. 81:12936-12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sonntag, F., S. Bleker, B. Leuchs, R. Fischer, and J. A. Kleinschmidt. 2006. Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. J. Virol. 80:11040-11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Storck, S., M. Shukla, S. Dimitrov, and P. Bouvet. 2007. Functions of the histone chaperone nucleolin in diseases. Subcell. Biochem. 41:125-144. [DOI] [PubMed] [Google Scholar]
- 58.Summerford, C., J. S. Bartlett, and R. J. Samulski. 1999. AlphaVbeta5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat. Med. 5:78-82. [DOI] [PubMed] [Google Scholar]
- 59.Summerford, C., and R. J. Samulski. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 72:1438-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vihinen-Ranta, M., L. Kakkola, A. Kalela, P. Vilja, and M. Vuento. 1997. Characterization of a nuclear localization signal of canine parvovirus capsid proteins. Eur. J. Biochem. 250:389-394. [DOI] [PubMed] [Google Scholar]
- 61.Vihinen-Ranta, M., D. Wang, W. S. Weichert, and C. R. Parrish. 2002. The VP1 N-terminal sequence of canine parvovirus affects nuclear transport of capsids and efficient cell infection. J. Virol. 76:1884-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, J., J. Xie, H. Lu, L. Chen, B. Hauck, R. J. Samulski, and W. Xiao. 2007. Existence of transient functional double-stranded DNA intermediates during recombinant AAV transduction. Proc. Natl. Acad. Sci. U. S. A. 104:13104-13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Warrington, K. H., Jr., O. S. Gorbatyuk, J. K. Harrison, S. R. Opie, S. Zolotukhin, and N. Muzyczka. 2004. Adeno-associated virus type 2 VP2 capsid protein is nonessential and can tolerate large peptide insertions at its N terminus. J. Virol. 78:6595-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Welsh, J. D., C. Swimmer, T. Cocke, and T. Shenk. 1986. A second domain of simian virus 40 T antigen in which mutations can alter the cellular localization of the antigen. Mol. Cell. Biol. 6:2207-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wistuba, A., A. Kern, S. Weger, D. Grimm, and J. A. Kleinschmidt. 1997. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J. Virol. 71:1341-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wobus, C. E., B. Hugle-Dorr, A. Girod, G. Petersen, M. Hallek, and J. A. Kleinschmidt. 2000. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J. Virol. 74:9281-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu, P., W. Xiao, T. Conlon, J. Hughes, M. Agbandje-McKenna, T. Ferkol, T. Flotte, and N. Muzyczka. 2000. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 74:8635-8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiao, W., K. H. Warrington, Jr., P. Hearing, J. Hughes, and N. Muzyczka. 2002. Adenovirus-facilitated nuclear translocation of adeno-associated virus type 2. J. Virol. 76:11505-11517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiao, X., J. Li, and R. J. Samulski. 1998. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 72:2224-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yan, Z., R. Zak, G. W. Luxton, T. C. Ritchie, U. Bantel-Schaal, and J. F. Engelhardt. 2002. Ubiquitination of both adeno-associated virus type 2 and 5 capsid proteins affects the transduction efficiency of recombinant vectors. J. Virol. 76:2043-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zadori, Z., J. Szelei, M. C. Lacoste, Y. Li, S. Gariepy, P. Raymond, M. Allaire, I. R. Nabi, and P. Tijssen. 2001. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 1:291-302. [DOI] [PubMed] [Google Scholar]
- 72.Zhong, L., W. Li, Z. Yang, K. Qing, M. Tan, J. Hansen, Y. Li, L. Chen, R. J. Chan, D. Bischof, N. Maina, K. A. Weigel-Kelley, W. Zhao, S. H. Larsen, M. C. Yoder, W. Shou, and A. Srivastava. 2004. Impaired nuclear transport and uncoating limit recombinant adeno-associated virus 2 vector-mediated transduction of primary murine hematopoietic cells. Hum. Gene Ther. 15:1207-1218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.