Abstract
Hantavirus nucleocapsid protein (N) can replace the cellular cap-binding complex, eukaryotic initiation factor 4F (eIF4F), to mediate translation initiation. Although N can augment translation initiation of nonviral mRNA, initiation of viral mRNA by N is superior. All members of the Bunyaviridae family, including the species of the hantavirus genus, express either three or four primary mRNAs from their tripartite negative-sense genomes. The 5′ ends of the mRNAs contain nonviral heterologous oligonucleotides that originate from endonucleolytic cleavage of cellular mRNA during the process of cap snatching. In the hantaviruses these caps terminate with a 3′ G residue followed by nucleotides arising from the viral template. Further, the 5′ untranslated region (UTR) of viral mRNA uniformly contains, near the 5′ end, either two or three copies of the triplet repeat sequence, UAGUAG or UAGUAGUAG. Through analysis of a panel of mutants with mutations in the viral UTR, we found that the sequence GUAGUAG is sufficient for preferential N-mediated translation initiation and for high-affinity binding of N to the UTR. This heptanucleotide sequence is present in viral mRNA containing either two or three copies of the triplet repeat.
Eukaryotic translation initiation involves the recognition of capped mRNA by the heterotrimeric eukaryotic initiation factor 4F (eIF4F) cap-binding complex (3, 6, 28). The components of eIF4F are the cap-binding peptide eIF4E (28, 33), the prototypic DEAD box RNA helicase eIF4A (9, 29), and the bridging peptide eIF4G (3, 8, 15). During translation initiation, eIF4F recruits the 43S preinitiation complex to the mRNA cap via eIF4G (10, 11). The preinitiation complex is composed of the small ribosomal subunit, additional initiation factors (eIF3, -1, -1A, and -5), and a ternary complex containing methionine-loaded initiator tRNA and eIF2 coupled with GTP (10). This large set of proteins scans mRNA from the 5′ cap, and when a start codon is encountered, the large ribosomal subunit and additional initiation factors are recruited and translation begins (13, 14). Hantavirus nucleocapsid protein (N) is a surrogate for the eIF4F cap-binding complex (18). Consequently, N mediates translation initiation in trans, whereas viral internal ribosomal entry sites (IRESs) facilitate initiation in cis using components of eIF4F (7, 25). N augments translation of capped nonviral RNA. However, N-mediated translation of capped RNAs containing the hantaviral mRNA 5′ untranslated region (UTR) is more robust (18). This indicates that the viral UTR likely contains a cis-acting signal necessary for preferential initiation of viral mRNA.
The genomes of all members of the Bunyaviridae family, including the hantaviruses, consist of three negative-stranded RNA molecules, designated S, M, and L. These genome segments express N, the envelope glycoproteins (Gn and Gc), and viral RNA-dependent RNA polymerase (RdRp), respectively (30, 31). A hallmark of the viral genome is that each segment contains nucleotides with terminal complementarity that undergo base pairing to form a panhandle structure (24, 26, 27). For hantaviruses this includes a nucleotide triplet repeated three times in tandem at both the 5′ end (5′-UAGUAGUAG [the third triplet in the minus strands of some species is UAU]) and the 3′ end (3′-AUCAUCAUC).
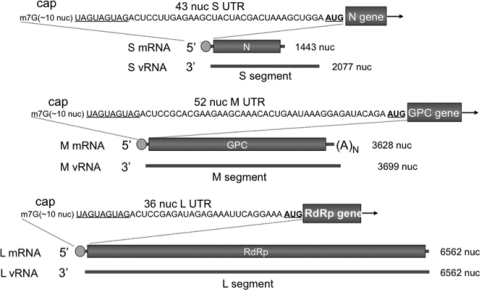
Hantavirus replication is exclusively cytoplasmic. mRNA synthesis involves an influenza virus-like cap snatching mechanism, resulting in the generation of viral mRNAs with 5′ caps derived from cellular mRNAs (4, 5, 12). Sequence analysis of the 5′ ends of viral mRNAs is consistent with a “prime-and-realign” model for transcription initiation (5). In this model, capped cellular oligonucleotides with a 3′-terminal G are generated and base pair with one of three possible C residues resident in the tandem triplet repeat at the 3′ terminus of the RNA template. After synthesis of a few nucleotides complementary to the triplet repeat, the nascent mRNA may sometimes realign to a position upstream on the viral RNA (vRNA) template. Thus, viral mRNA harbors 5′ nonviral caps ranging from 8 to 17 nucleotides in length, followed by viral sequences containing three or two triplet repeats depending on whether or not complete realignment has occurred. For the mRNA copied from the S and M segments, transcription terminates before the end of the template (12). In contrast, mRNA from the L segment is apparently copied from the entire minus-strand template. Figure 1 provides a summary of some of the salient features of hantaviral mRNA along with the 5′ UTR sequences of the Sin nombre hantavirus (SNV) mRNAs from the S, M, and L segments.
FIG. 1.
Organization of hantaviral mRNAs. The S, M, and L genome segments of SNV are displayed in 3′-to-5′ orientation. The corresponding mRNA is shown above the vRNA from which it is derived. Green circles represent cellular 5′ oligomeric caps acquired from cap snatching. The 5′ UTR sequence of each mRNA is also shown. For the sake of simplicity, three copies of the triplet repeat are shown, although some mRNAs contain two copies of the repeat.
The nucleocapsid protein (N) of SNV is a multifunctional viral component. As previously adumbrated, N is a surrogate for the eIF4F cap-binding complex and is actively involved in translation initiation. Additionally, N binds the viral RNA panhandle with specificity and is likely involved in encapsidation and packaging of the viral genome (22). N is also an RNA chaperone that is capable of unwinding RNA duplexes. This activity of N both facilitates the formation of the panhandle and then unwinds the panhandle remaining in association with the 5′ end of the viral genome, which is likely a prelude to transcription initiation (19, 20). N also binds 5′ caps of cellular mRNA and sequesters these caps in cytoplasmic processing bodies (P bodies) for use in cap snatching and transcription initiation (17). Ns from other hantavirus species likely have the same or overlapping activities. Hantaan virus N inhibits the translocation of the transcription factor NF-κB to the nucleus, which is consistent with a role in blocking induction of innate and acquired immune responses by the host (32).
Here we show that the SNV UTR contains a cis-acting signal necessary for preferential translation initiation of viral mRNA by N. Our data suggest a model in which interaction of N with both the 5′ cap and the triplet repeats mediates efficient translation of viral mRNA.
MATERIALS AND METHODS
Oligonucleotides, enzymes, and other reagents.
PCR primers were from Sigma Genosys, restriction enzymes were from New England Biolabs, “Proof-Pro” DNA polymerase was from Gene Choice, DNase I was from Invitrogen, and T7 transcription reagents were from Fermentase or Promega. Rabbit reticulocyte lysates and 5′ mRNA cap analog were from Promega. Radioactive reagents, including [35S]methionine and [32P]CTP were from Perkin-Elmer. All other chemicals were purchased from Sigma.
SNV nucleocapsid protein expression and purification.
As reported previously, the SNV nucleocapsid gene was expressed in Escherichia coli with a C-terminal His tag, and N was purified using Ni-nitrilotriacetic acid (NTA) beads (16).
Preparation of mRNA substrates by in vitro T7 transcription reaction.
We synthesized mRNA molecules for translational expression of green fluorescent protein (GFP) and N in rabbit reticulocyte lysates, using a Ribomax T7 kit (Promega). All mRNA molecules contained a 5′ UTR sequence followed by the appropriate coding region and a 30-nucleotide-long 3′ poly(A) tail. The 5′ UTR sequences of all mRNAs were either a nonviral sequence; a native 5′ viral UTR corresponding to the SNV S, L, or M segment; or a derivative of the S segment UTR. The GFP gene was PCR amplified from pEGFP-C1 (22) using the forward primer 5′-CTAGCTAATACGACTCACTATAGGTAGTAGTAGACTCCTTGAGAAGCTACTACGACTAAAGCTGGA ATGGTGAGCAAGGGCGAGGAGCTGTTCACCGGG3′ and the reverse primer 5′-(T)30AATAAACAAGTTAACAACAACAATTGC. The forward primer contained a proximal T7 RNA polymerase promoter, followed by a 42-nucleotide-long SNV S mRNA UTR sequence and then a sequence complementary to beginning of the GFP gene. The reverse primer contained a sequence to generate a poly(A) tail upon transcription. The PCR product was gel purified and used as a template in T7 transcription reactions. A parallel strategy was used to synthesize capped RNAs with nonviral, M segment RNA, L segment RNA, or S mRNA UTR derivatives preceding the GFP gene, using forward primers of appropriate sequence and the reverse primer described above. A similar strategy was used for the PCR amplification template DNAs with the SNV N reporter gene rather than the GFP gene. pGEX-SNV N, containing the N gene (22), was used as the template in these amplification reactions.
T7 transcription reactions were carried out at 37°C for 3 h. The DNA template was removed with DNase I, and the mRNA purified with RNeasy (Qiagen) and stored in 10-μl aliquots at −70°C. All mRNA molecules were capped by the incorporation of m7G cap analog in the transcription reaction mixtures according to the manufacturer's protocol (Promega).
Capped oligoribonucleotides corresponding to the 5′ mRNA UTRs were synthesized by T7 transcription reactions and used in filter binding studies with SNV N. A single-stranded DNA template containing a terminal T7 promoter juxtaposed to the sequence of interest was PCR amplified using two opposing primers, gel purified, and used as the template in the T7 transcription reaction. T7 transcription reactions were carried out as described above. [α-32P]CTP RNAs were fractionated on denaturing 14% polyacrylamide gels. Gel slices containing RNA of the correct size were excised from the gel, crushed, and incubated with 500 μl of probe elution buffer (0.5 M NH4 acetate, 1 mM EDTA, 0.2% SDS) overnight, followed by centrifugation at 13,000 rpm for 10 min. RNA was precipitated from the supernatant by the addition of 0.5 M NH4 acetate and 2.5 volumes of ethanol at −20°C for 30 min. Samples were centrifuged at 13,000 rpm for 30 min, and the pellet was air dried, resuspended in 100 μl of RNase-free water, and stored in 10-μl aliquots at −70°C.
In vitro translation in rabbit reticulocyte lysates.
Nuclease-treated rabbit reticulocyte lysates (Promega) were used for the translation of mRNA in the presence and absence of supplemented, bacterially expressed SNV N. Translation reactions were carried out in 50 μl containing 35 μl of rabbit reticulocyte lysate, 1 μl amino acid mixture minus methionine (1 mM), 1 μl [35S]methionine (1,175 Ci/mmol), 2 μl RNase inhibitor (40 U/μl), 4 μl mRNA substrate in water (containing 2 μl of GFP mRNA [250 ng/μl] and 2 μl of SNV N mRNA [250 ng/μl]), and 7 μl of RNase-free water. Reaction mixtures were incubated at 30°C for 90 min. Samples were electrophoresed on 15% SDS gels, and the expression of N and GFP was quantified by phosphorimage analysis.
Filter binding.
Interaction of SNV N with labeled, 5′-capped RNAs was examined by filter binding. All binding reactions were carried out in RNA binding buffer at a constant concentration of RNA and with increasing concentrations of N protein as previously described (22). Reaction mixtures were incubated at room temperature for 30 to 45 min and filtered through nitrocellulose membranes under vacuum. Filters were washed with 10 ml of RNA binding buffer and dried. The amount of RNA retained on the filter at different input concentrations of N was measured using a scintillation counter. Data points were fit to a hyperbolic equation using the program Origin 6 (Microcal). The apparent dissociation constant (Kd), corresponding to the concentration of N protein required to obtain half saturation in the binding profile, was calculated assuming that the complex formation obeys a simple bimolecular equilibrium.
Calculation of binding stoichiometry.
As described previously (21), the binding stoichiometry (expressed in terms of number of N molecules bound per RNA) was estimated from the intersection of two straight lines of a least-square fit plot of the percent increase of bound RNA against the ratio of input concentrations of N protein and RNA. We also used continuous-variation plots to verify the binding stoichiometry results. At a constant temperature (25°C), the fluorescence signal was recorded for the solutions where the concentrations of both RNA and N protein were varied, keeping the sum of their concentrations constant at 420 nM. ΔF330 (the difference between the fluorescence intensities of N protein in the absence and presence of RNA) was plotted as a function of the input mole fraction of N protein. The breakpoint in the resulting plot corresponds to the mole fraction of N in the N-RNA complex. The binding stoichiometry is obtained in terms of N:RNA [xligand:(1 − xligand)], where xligand represents the molar concentration of N divided by the total molar concentration of N and RNA.
RESULTS
Capped RNAs with viral UTRs exhibit preferential N-dependent translation.
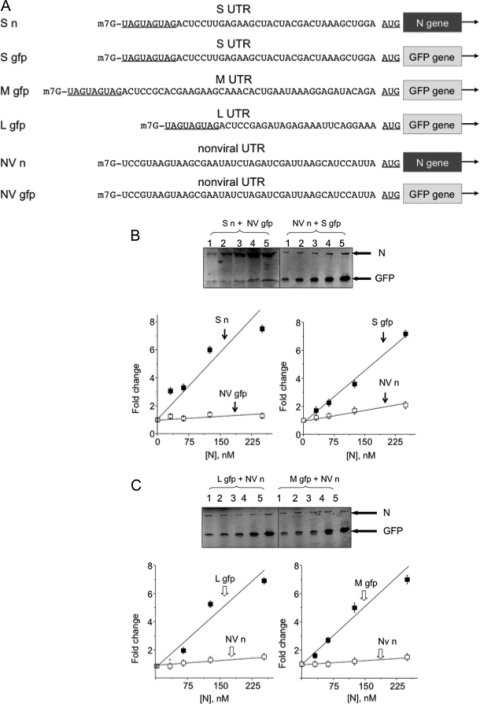
The 5′ UTRs of mRNAs from the SNV S, M, and L segments differ slightly in length but are similar in terminal sequence (Fig. 1). We first compared the N-mediated translation initiation efficiencies of capped mRNAs containing a viral 5′ UTR with that of an mRNA containing a nonviral 5′ UTR in competitive in vitro translation assays with capped synthetic RNAs. An RNA with the 5′ UTR of S segment mRNA, carrying the N gene as an reporter (S n) (Fig. 2A), was simultaneously translated in reticulocyte lysates with a second RNA containing a nonviral UTR with an identical length and similar nucleotide composition to those of the viral UTR. The mRNA with the nonviral UTR carried the GFP gene as an reporter (NV gfp) (Fig. 2A). Translation reaction mixtures contained increasing concentrations of bacterially expressed and purified N protein. The translation products were labeled with [35S]methionine during synthesis and fractionated on SDS-polyacrylamide gels, and N and GFP expression was quantified using phosphorimage analysis. With increasing concentrations of N, there was an increase in translation of the RNA containing the viral UTR relative to the RNA with the nonviral UTR (Fig. 2B). We also carried out reactions with complementary capped reporter RNAs containing reciprocal UTRs; the viral UTR preceded the GFP gene (N gfp) and the nonviral UTR preceded the N gene (NV n) (Fig. 2A). The results of this experiment again revealed an N-dependent increase in expression of the RNA containing the viral UTR (Fig. 2B). These data corroborate our previous observations indicating that N-mediated translation is more efficient for mRNAs harboring a viral 5′ UTR than for those harboring a nonviral UTR (18). It should be noted that in this translation assay, expression of N from the synthetic mRNA was used simply as a reporter, analogous to GFP, and did not contribute significantly to the amount of N in the reaction. Additionally, we carried out reactions with several nonviral mRNAs with alternative UTRs. Data from these experiments were similar to those displayed in Fig. 2 (data not shown). We carried out analogous experiments using RNAs harboring the UTRs from M and L segment mRNAs (M gfp and L gfp) in competition with NV n (Fig. 2C). Again we observed an N-dependent increase in translation owing to the presence of either the M or L mRNA 5′ UTR.
FIG. 2.
N enhances the translational expression of RNAs with the 5′ UTR from S, M, and L mRNAs. (A) Sequences of viral and nonviral reporters. (B) Translational expression of capped reporter RNAs S n and NV gfp (left) and from S gfp and NV n (right) with increasing concentrations of N. (C) Expression from reporters L gfp (left) and NV n (right) with increasing concentrations of N. Error bars indicate standard deviations.
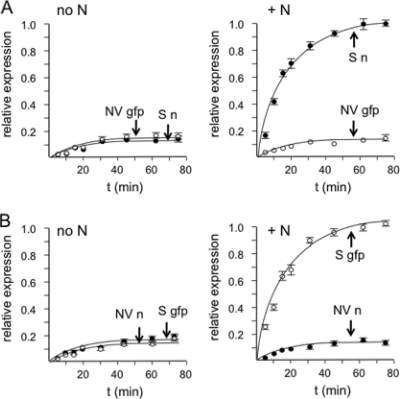
We performed a time course experiment to further examine the competitive translation of capped RNAs with a viral or nonviral UTR in the presence and absence of N. In reaction mixtures without N, translational expressions of reporter genes from RNAs containing or lacking a viral UTR were similar (Fig. 3). In contrast, in translation reaction mixtures supplemented with N, expression from RNAs harboring a viral UTR was significantly more robust that that from RNA with a nonviral UTR.
FIG. 3.
Time course of translation of RNA reporters with viral and nonviral UTRs in the absence or presence of N. (A) Translation of capped S n and NV gfp in the absence (left) or presence (right) of 250 nM N as a function of time. (B) Translational expression of S gfp and NV n in the absence (left) or presence (right) of N. See the Fig. 2A for descriptions of S n, NV gfp, S gfp, and NV n. Error bars indicate standard deviations.
In bone fide viral mRNA, the UTR is preceded by a capped oligomeric RNA of cellular origin, 8 to 17 nucleotides in length, that arises through cap snatching (5). Moreover, the 3′ nucleotide of this short nonviral RNA, which is joined to the viral UTR, is usually a G residue (5). To mimic viral mRNA with such a cellular cap, we synthesized an RNA with a nonviral cap nine nucleotides in length preceding the UTR transcribed from the viral S segment. The sequence of this cap was identical to that of an authentic cell-derived oligonucleotide observed to have been acquired during virus infection (5). This nonviral oligonucleotide terminated in a 3′ G (RNA 2) (Fig. 4). Additionally, we generated an RNA identical to RNA 2 but lacking a terminal G (RNA 3). Both of these mRNAs were used in competitive translation assays with NV n. Competitive translation reactions indicated that the presence of a nonviral cap had no apparent effect on preferential N-mediated translation of RNA harboring a viral UTR (RNA 2) (Fig. 5). Similarly, the absence of a 3′ G in the nonviral sequence did not detectibly affect N-mediated translation (RNA 3) (Fig. 5)
FIG. 4.
GFP reporter RNAs containing derivatives of the 5′ UTR from S segment mRNA. Nucleotides that would be derived from the S segment template are shown in black, and the triplet repeats and the start codon are underlined. As described in the text, viral mRNA contains a nonviral oligonucleotide that arises from cap snatching. RNAs 2 through 11 all contain such a nonviral sequence (shown in gray). Triangles highlight nucleotides or regions that are deleted relative to the full-length viral UTR.
FIG. 5.
Competition-translation analysis of reporter RNAs with deletions or rearrangements in the 5′ UTR. Each of the GFP reporter RNAs shown in Fig. 4 was used in competitive translation reactions with a reporter RNA containing a nonviral 5′ UTR (NV n) (Fig. 2) and increasing concentrations of SNV N. For clarity, the data for pairs of the viral UTR reporters are displayed on five parallel graphs. The expression data for each of the plotted competitor NV n RNA used for each pair were compiled together since their values were similar. Error bars indicate standard deviations.
The terminal triplet repeat is required for N-mediated translation of viral mRNA.
To identify the nucleotides required in cis for N-mediated translation, we generated a panel of capped S segment RNAs with deletions in the viral UTR (Fig. 4). The deletions ranged in length from 3 to 18 nucleotides and affected various portions of the UTR. All of these mutants were used in competitive translation assays with NV n mRNA. Most of these UTR derivatives were still able to support N-mediated translation. Notably, this included RNA 6, which lacks 27 nucleotides of the UTR but retains two of the terminal triplet repeats of the UTR. However, RNA 7 and RNA 8 were unable to support N-mediated translation. RNA 7 is identical to RNA 6 except that only one of the triplet repeats is retained, while RNA 8 retains most of the UTR but lacks two of the three triplet repeats. The straightforward interpretation of these data is that the terminal triplet repeats are necessary for N-mediated translation. Further, retention of two of the three repeats is necessary for function.
As described above, 5′ cellular caps arising from cap snatching usually end in a G residue. RNA 10 and RNA 9 both contain two copies of the triplet repeat. However, RNA 9 contains a terminal G residue in the nonviral oligomeric cap, whereas RNA 10 does not (Fig. 4). Interestingly, RNA 9 was translated efficiently, while translation of RNA 10 was inefficient (Fig. 5). RNA 3 also lacks the terminal G residue but retains all three of the triplet repeats and was translated efficiently. The sequence at the junction between the nonviral cap and the viral UTR is G+1UAGUAGUAG. Taken together, these data indicate that the actual sequence required for N-mediated translation is GUAGUAG. In viral mRNA this sequence is present both when all three triplet repeats are present or when there are two triplet repeats along with a G in the minus 1 position arising from cap snatching.
We next changed the position of the conserved terminal triplet repeat. In RNA 11, the nucleotides UAGUAGUAG were moved to a position 27 nucleotides from the 5′ terminus (Fig. 4). RNA 11 was not efficiently translated in competitive translation assays with NV n mRNA, indicating that the triplet repeats must be situated near the terminus for efficient N-mediated translation (Fig. 5).
Interaction of N with the triplet repeat.
We synthesized as capped, labeled oligonucleotides each of the UTRs used in the competitive translation experiments. These UTRs contained the same sequences displayed in Fig. 2 and 4 but were devoid of reporter genes or other distal nucleotides. We then carried out RNA filter binding assays to examine the interaction affinity of each of these oligoribonucleotides with SNV N. Binding profiles for the 5′ UTR corresponding to the S segment mRNA and for the nonviral UTR shown in Fig. 2 are displayed in Fig. 6A and D, respectively. Similar binding experiments for each of the UTRs were carried out, and their binding affinities with SNV N are shown in Table 1. Significantly, the UTRs that enabled high-efficiency N-mediated translation also exhibited higher-affinity binding to N. In particular, N interacted at high affinity with the UTR from S mRNA lacking or containing a pseudo-cellular cap as well as RNAs 3, 4, 5, 6, and 9. These UTRs also mediated superior N-mediated translation initiation. Conversely, N associated with low affinity to the RNAs corresponding to nonviral UTR as well as UTRs 7, 8, and 10. These UTRs were also unable to facilitate efficient N-mediated translation initiation. RNA 11, which harbors the triplet repeat transposed to a position 27 nucleotides from its normal site in the UTR, was still bound at high affinity by N but is unable to promote efficient translation initiation by N. This observation is consistent with the idea that utilization of the triplet repeat in translation by N is position dependent and requires close proximity to the 5′ cap.
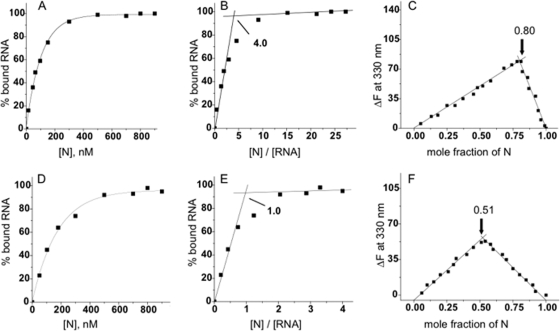
FIG. 6.
Binding affinity and binding stoichiometry between 5′ UTR oligoribonucleotides and N. (A and D) Representative RNA binding curves with capped RNAs corresponding to mRNA from the S segment 5′ UTR and to a nonviral RNA with increasing concentrations of N, respectively. The determined dissociation constants based on these and additional experiments are presented in Table 1. Stoichiometry was calculated using two complementary methods. (B and E) Representative least-square fit plots derived for the interaction between the UTR from the S segment and nonviral UTR, respectively. The binding stoichiometry derived from the intersection of the two pertinent lines is depicted. These calculations were verified using continuous-variation plots, which were generated based on the change in fluorescence emission with various concentrations RNA and N. (C and F) Plots for the UTR from S segment mRNA and for the nonviral UTR, respectively. Each of the UTRs in this study was similarly analyzed by both methods to determine stoichiometry. These data are summarized in Table 1.
TABLE 1.
Binding of SNV N with the 5′ UTRs shown in Fig. 6
| RNA | Kd, nM (mean ± SD) | Binding stoichiometrya (mean ± SD) |
|---|---|---|
| NV | 130 ± 3.0 | 1.0 ± 0.10 |
| S mRNA | 41 ± 2.0 | 4.0 ± 0.20 |
| RNA 2 | 43 ± 1.5 | 4.2 ± 0.15 |
| RNA 3 | 42 ± 3.0 | 3.9 ± 0.21 |
| RNA 4 | 45 ± 2.5 | 4.3 ± 0.23 |
| RNA 5 | 47 ± 1.7 | 3.8 ± 0.27 |
| RNA 6 | 48 ± 2.5 | 4.1 ± 0.21 |
| RNA 7 | 132 ± 3.2 | 1.0 ± 0.12 |
| RNA 8 | 135 ± 2.8 | 1.0 ± 0.15 |
| RNA 9 | 39 ± 4.8 | 4.1 ± 0.15 |
| RNA 10 | 139 ± 4.7 | 1.0 ± 0.15 |
| RNA 11 | 40 ± 2.1 | 4.2 ± 0.25 |
The binding stoichiometry is expressed in terms of number of molecules of N bound per RNA (UTR) molecule.
We next determined the binding stoichiometry between the various UTRs and N. We generated least-square and continuous-variation plots of the interactions of the S segment viral UTR and a nonviral UTR with N. The least-square plots are derived directly from the RNA binding data used to determine binding affinity (Fig. 6B and E). Binding of N to RNA results in a conformational change in N and a corresponding alteration in fluorescence emission at a wavelength of 330 nm (21). Thus, fluorescence emission was measured in reaction mixtures with various concentrations of N and RNA and plotted as a function of the mole fraction of N (Fig. 6C and F). Both methods to determine binding stoichiometry between UTR RNAs and N were in accord with each other and indicated that four molecules of N bind to the viral UTR while a single molecule of N binds to the nonviral UTR. The binding stoichiometry for each of the UTRs based on analogous plots is presented in Table 1. As with the direct-binding plots, the data indicate that the UTRs can be placed in one of two sets. One set is composed of RNAs that facilitate efficient N-mediated translation, high-affinity binding to N, and interaction with four molecules of N. The second set of UTRs do not efficiently facilitate translation, bind N weakly, and associate solely with N monomers.
DISCUSSION
In competitive translation reactions, we find that N promotes the expression of capped RNAs with a viral 5′ UTR at the expense of RNA with a nonviral UTR. This is consistent with the idea that the translation initiation activity of N serves to enhance or ensure viral gene expression. N binds strongly to the 5′ UTR of viral mRNA owing to the presence of the triplet repeat, and this N-RNA interaction provides a straightforward explanation to account for preferential translation of viral mRNA.
A priori, the reason for selection of virus-mediated translation initiation would be to ensure expression of viral mRNA. Hantaviruses do not overtly block host cell translation. However, other species of the bunyavirus family have established mechanisms for abrogation of host cell gene expression. It is extremely unlikely that N-mediated translation initiation arose capriciously in hantaviruses and is without biological benefit for the virus. In nature hantaviruses are harbored in rodent populations and incidentally infect humans through aerosolized excreta. The preferential translational expression of viral mRNA by N is consistent with a role of N to ensure viral protein expression. However, the context in which this activity of N comes into play has yet to be identified.
Transcription initiation via cap snatching results in an oligomeric cap terminating in a G residue followed by the viral UTR bearing either three or two copies of the templated triplet repeat, UAGUAGUAG or UAGUAG (5, 17). The G residue at the 3′ end of the cellular cap ostensibly arises from a viral endonuclease activity that cleaves at G residues near the 5′ ends of capped mRNAs. Based on a set of RNAs harboring alterations in the sequence in and around the triplet repeat UAGUAGUAG, the sequence sufficient for high-affinity interaction and N-mediated translation initiation is GUAGUAG. This sequence is embedded within the mRNAs with three copies of the triplet repeat and would also be present in mRNAs with only two copies of the repeat with the 5′ G arising from the oligomeric cellular cap.
N binds to the viral UTR with a stoichiometry of 4:1 and to a nonviral UTR with a stoichiometry of 1:1. The majority of SNV N molecules in N preparations are trimeric, but there are also detectable amounts of monomer and dimer species (22). Since monomeric N associates with a nonviral UTR, we favor the possibility that a monomer binds indiscriminately to the 5′ caps of mRNAs and that trimeric N binds with specificity to the triplet repeat. As discussed above, the latter interaction appears to be crucial for preferential N-mediated translation initiation.
As diagrammed in Fig. 1, mRNA derived from the S segment lacks a poly(A) tail, while mRNA encoded by the M segment is polyadenylated. Based on multiple criteria, it is unlikely that polyadenylation is important for efficient N-facilitated translation initiation, and translation of viral mRNA is unlikely to require a closed-loop complex arising from protein-mediated juxtaposition of the mRNA termini. Whereas eIF4G binds with poly(A)-binding protein (PABP) to generate cellular mRNA in a closed-loop configuration, there is not detectable analogous association of N with PABP (18). Moreover, translation of viral mRNA in a prototypic bunyavirus, bunyamwera virus, arises from coupled transcription [i.e., translation initiation ensues on nascent transcripts that lack poly(A) tails] (1, 2). This is made possible since bunyavirus transcription is cytoplasmic. It is reasonable to assume that gene expression of other genera of the family, including the hantaviruses, is similarly coupled. As might be expected, S segment mRNA containing the full-length 5′ UTR but devoid of a poly(A) tail was preferentially translated in competitive in vitro translation reactions similar to those presented in Fig. 2 (data not shown).
It is worth comparing the association of SNV N with the 5′ viral UTR during translation initiation with the interaction that takes place between N and the minus-strand viral genome. Each of the negative-sense viral RNA (vRNA) genome segments can form “panhandles” that arise through complementary base pairing between the 5′ and 3′ ends of the genome. The hydrogen-bonded panhandle region includes the same triplet repeat sequences found in the UTR of mRNA, and trimeric SNV N binds preferentially with the vRNA panhandle (21). Consequently, the triplet repeats serve as a high-affinity binding site during both vRNA recognition and translation initiation by N. However, binding of N to the vRNA panhandle is of higher affinity than is binding of N to the viral UTR. Moreover, initial binding to the vRNA panhandle involves interaction with approximately 20 nucleotides from both the 5′ and 3′ termini of the genome (21), whereas, as described here, association with the 5′ UTR of viral mRNA involves only seven nucleotides. Inhibition of the association of N with either the minus-strand vRNA during the encapsidation process or viral mRNA during translation initiation would be expected to attenuate replication, and therefore these are viable targets for antiviral molecules.
SNV N is intimately involved in vRNA genome recognition, transcription initiation, and translation initiation. vRNA panhandle binding by N is followed by N-mediated unwinding of the panhandle, with N remaining associated with the 5′ terminus of vRNA (19). N also facilitates cap snatching and transcription initiation in coordination with the viral polymerase. It is likely that panhandle unwinding occurs in concert with cap snatching and transcription initiation, since unwinding would be expected to make accessible the 3′ end of the vRNA template for transcription initiation. Significantly, in accord with this general model, it has recently been observed that SNV N contains separate binding sites for the vRNA panhandle and 5′ caps (23).
The majority of detectable N in cells is found in association with processing bodies (P bodies) (17). These cytoplasmic foci contain significant portions of the cellular machinery involved in mRNA turnover, and N sequesters cellular caps for use in transcription initiation. Thus, it is likely that the vRNA template is also found in association with P bodies and that mRNA synthesis takes place in this cytoplasmic locale. As discussed previously, transcription and translation during bunyavirus replication are coupled in a manner that is unusual for eukaryotic gene expression (1, 2). Consequently, a spatial model for hantaviral transcription and translation likely involves coordination of gene expression in P bodies. In this context, the triplet repeats would be recognized by N, and translation of the nascent viral mRNA could ensue in the cytoplasmic space immediately adjacent to P bodies.
Acknowledgments
This work was supported by research grant R01AI074011 from the NIH.
Footnotes
Published ahead of print on 23 June 2010.
REFERENCES
- 1.Barr, J. N. 2007. Bunyavirus mRNA synthesis is coupled to translation to prevent premature transcription termination. RNA 13:731-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellocq, C., and D. Kolakofsky. 1987. Translational requirement for La Crosse virus S-mRNA synthesis: a possible mechanism. J. Virol. 61:3960-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dever, T. E. 1999. Translation initiation: adept at adapting. Trends Biochem. Sci. 24:398-403. [DOI] [PubMed] [Google Scholar]
- 4.Dunn, E. F., D. C. Pritlove, H. Jin, and R. M. Elliott. 1995. Transcription of a recombinant bunyavirus RNA template by transiently expressed bunyavirus proteins. Virology 211:133-143. [DOI] [PubMed] [Google Scholar]
- 5.Garcin, D., M. Lezzi, M. Dobbs, R. M. Elliott, C. Schmaljohn, C. Y. Kang, and D. Kolakofsky. 1995. The 5′ ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J. Virol. 69:5754-5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 7.Hellen, C. U., and P. Sarnow. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593-1612. [DOI] [PubMed] [Google Scholar]
- 8.Hentze, M. W. 1997. eIF4G: a multipurpose ribosome adapter? Science 275:500-501. [DOI] [PubMed] [Google Scholar]
- 9.Hernandez, G., and P. Vazquez-Pianzola. 2005. Functional diversity of the eukaryotic translation initiation factors belonging to eIF4 families. Mech. Dev. 122:865-876. [DOI] [PubMed] [Google Scholar]
- 10.Hershey, J. W. B., and W. C. Merrick. 2000. Pathway and mechanism of initiation of protein synthesis, p. 33-88. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 11.Hinnebusch, A. G. 2006. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 31:553-562. [DOI] [PubMed] [Google Scholar]
- 12.Hutchinson, K. L., C. J. Peters, and S. T. Nichol. 1996. Sin Nombre virus mRNA synthesis. Virology 224:139-149. [DOI] [PubMed] [Google Scholar]
- 13.Kozak, M. 1992. Regulation of translation in eukaryotic systems. Annu. Rev. Cell Biol. 8:197-225. [DOI] [PubMed] [Google Scholar]
- 14.Kozak, M. 1991. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 266:19867-19870. [PubMed] [Google Scholar]
- 15.Mader, S., H. Lee, A. Pause, and N. Sonenberg. 1995. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 15:4990-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mir, M. A., B. Brown, B. Hjelle, W. A. Duran, and A. T. Panganiban. 2006. Hantavirus N protein exhibits genus-specific recognition of the viral RNA panhandle. J. Virol. 80:11283-11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mir, M. A., W. A. Duran, B. L. Hjelle, C. Ye, and A. T. Panganiban. 2008. Storage of cellular 5′ mRNA caps in P bodies for viral cap-snatching. Proc. Natl. Acad. Sci. U. S. A. 105:19294-19299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mir, M. A., and A. T. Panganiban. 2008. A protein that replaces the entire cellular eIF4F complex. EMBO J. 27:3129-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mir, M. A., and A. T. Panganiban. 2006. Characterization of the RNA chaperone activity of hantavirus nucleocapsid protein. J. Virol. 80:6276-6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mir, M. A., and A. T. Panganiban. 2006. The bunyavirus nucleocapsid protein is an RNA chaperone: possible roles in viral RNA panhandle formation and genome replication. RNA 12:272-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mir, M. A., and A. T. Panganiban. 2005. The hantavirus nucleocapsid protein recognizes specific features of the viral RNA panhandle and is altered in conformation upon RNA binding. J. Virol. 79:1824-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mir, M. A., and A. T. Panganiban. 2004. Trimeric hantavirus nucleocapsid protein binds specifically to the viral RNA panhandle. J. Virol. 78:8281-8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mir, M. A., S. Sheema, A. Haseeb, and A. Haque. 2010. Hantavirus nucleocapsid protein has distinct m7G cap- and RNA-binding sites. J. Biol. Chem. 285:11357-11368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obijeski, J. F., D. H. Bishop, F. A. Murphy, and E. L. Palmer. 1976. Structural proteins of La Crosse virus. J. Virol. 19:985-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pestova, T. V., I. N. Shatsky, and C. U. Hellen. 1996. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 16:6870-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pettersson, R. F., and C. H. von Bonsdorff. 1975. Ribonucleoproteins of Uukuniemi virus are circular. J. Virol. 15:386-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raju, R., and D. Kolakofsky. 1989. The ends of La Crosse virus genome and antigenome RNAs within nucleocapsids are base paired. J. Virol. 63:122-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richter, J. D., and N. Sonenberg. 2005. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433:477-480. [DOI] [PubMed] [Google Scholar]
- 29.Rogers, G. W., Jr., A. A. Komar, and W. C. Merrick. 2002. eIF4A: the godfather of the DEAD box helicases. Prog. Nucleic Acid Res. Mol. Biol. 72:307-331. [DOI] [PubMed] [Google Scholar]
- 30.Schmaljohn, C., and B. Hjelle. 1997. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 3:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmaljohn, C. M. 1996. Molecular biology of hantaviruses. Plenum Press, New York, NY.
- 32.Taylor, S. L., N. Frias-Staheli, A. Garcia-Sastre, and C. S. Schmaljohn. 2009. Hantaan virus nucleocapsid protein binds to importin alpha proteins and inhibits tumor necrosis factor alpha-induced activation of nuclear factor kappa B. J. Virol. 83:1271-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von der Haar, T., J. D. Gross, G. Wagner, and J. E. McCarthy. 2004. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat. Struct. Mol. Biol. 11:503-511. [DOI] [PubMed] [Google Scholar]