Abstract
The linear DNA genomes of recombinant adeno-associated virus (rAAV) gene delivery vectors are acted upon by multiple DNA repair and recombination pathways upon release into the host nucleus, resulting in circularization, concatemer formation, or chromosomal integration. We have compared the fates of single-strand rAAV (ssAAV) and self-complementary AAV (scAAV) genomes in cell lines deficient in each of three signaling factors, ATM, ATR, and DNA-PKCS, orchestrating major DNA double-strand break (DSB) repair pathways. In cells deficient in ATM, transduction as scored by green fluorescent protein (GFP) expression is increased relative to that in wild-type (wt) cells by 2.6-fold for ssAAV and 6.6-fold for scAAV vectors, arguing against a mechanism related to second-strand synthesis. The augmented transduction is not reflected in Southern blots of nuclear vector DNA, suggesting that interactions with ATM lead to silencing in normal cells. The additional functional genomes in ATM−/− cells remain linear, and the number of circularized genomes is not affected by the mutation, consistent with compartmentalization of genomes into different DNA repair pathways. A similar effect is observed in ATR-deficient cells but is specific for ssAAV vector. Conversely, a large decrease in transduction is observed in cells deficient in DNA-PKCS, which is involved in DSB repair by nonhomologous end joining rather than homologous recombination. The mutations also have differential effects on chromosomal integration of ssAAV versus scAAV vector genomes. Integration of ssAAV was specifically reduced in ATM−/− cells, while scAAV integration was more profoundly inhibited in DNA-PKCS−/− cells. Taken together, the results suggest that productive rAAV genome circularization is mediated primarily by nonhomologous end joining.
Recombinant adeno-associated virus (rAAV) vectors are an important tool for gene transfer, with a growing presence in gene therapy clinical applications. The single-stranded DNA genome of conventional rAAV vectors (ssAAV) contains only the desired transgene and the 145-bp palindromic inverted terminal repeat (ITR) sequences, which function as the viral origins of replication, packaging sequences, and priming sites for synthesis of the cDNA strand. A derivative rAAV vector, termed self-complementary AAV (scAAV), is packaged as a single-stranded inverted repeat, which can fold into double-stranded DNA (dsDNA) without the requirement for DNA synthesis, leading to more efficient transduction (20, 21).
Once released into the nucleus, the free DNA ends of rAAV genomes are targets for DNA repair pathways, leading to circularization or concatemerization and, infrequently, chromosomal DNA integration. At low multiplicities of infection (MOIs), monomeric circular episomes are the predominant stable form of the rAAV genome, and at higher MOIs, increased opportunities for end-to-end joining lead to the formation of concatemeric episomes, which also circularize. These recombination events appear to follow the conversion of the ssDNA genome to dsDNA (41). We have found similar processes acting on scAAV vectors, demonstrating that ssDNA is not a prerequisite for the formation of circles and concatemers (5).
Our primary interest in rAAV circularization is how it relates to the persistence of rAAV vector genomes in the nucleus and to chromosomal integration, with its potential for genotoxicity and oncogenesis (9, 23, 24, 29). It is currently not known whether the same DNA recombination processes that mediate circularization also lead to integration. However, the most likely starting substrate for either fate is a linear rAAV genome that has been converted to dsDNA, with an ITR in either a covalently closed hairpin or an open duplex configuration at each end. Either structure is subject to recombination, though the efficiencies and host factor requirements differ (5). The genome ends interact with each other to form circles and concatemers or with preexisting chromosomal DNA double-strand breaks (DSBs), leading to integration (22).
Circularization of ssAAV is highly dependent on the nonhomologous end-joining (NHEJ) pathway for DSB repair in some animal tissues but not others (12, 16a, 25, 34, 35). While NHEJ plays an important role in processing ssAAV genomes, it also appears to have negative effects on transduction efficiency. A deficiency in Ku86, an essential core component of NHEJ-mediated repair, leads to higher levels of transduction from ssAAV vectors in cultured cells (42). The NHEJ pathway also participates in AAV Rep protein-mediated site-specific integration into the chromosome 19 AAV-S1 site, though with differential effects on scAAV and ssAAV vector genomes (8).
The conversion of the ssAAV genome to dsDNA may be intricately linked to its subsequent recombination to circular or concatemeric forms. Transduction from ssAAV vectors is significantly increased by treatments that induce DNA damage repair pathways (1). Transduction is also increased by the adenovirus (Ad) E4ORF6 gene product, which disrupts an important DSB repair function through the segregation of cellular Mre11 (10, 13, 15, 31). Mre11 is a component of the MRN complex (Mre11, Rad50, and NbsI), which promotes DSB repair by both homologous recombination (HR) and the NHEJ pathway (28). The MRN complex colocalizes with ssAAV genomes through interaction either with the ssDNA or with the ITR hairpin (3, 31). Consistent with the positive effects of Ad E4ORF6 on ssAAV transduction, MRN interactions with the rAAV genome inhibit transduction, possibly at the step of conversion to dsDNA. This is also consistent with reports that mutations in ATM (ataxia telangiectasia mutated) increase transduction from ssAAV vectors (30, 42). ATM is the primary signaling kinase responding to DSB in association with the MRN complex, and it is involved in the repair of DSB caused by ionizing radiation. These studies cumulatively suggest that interactions with the ATM-mediated pathway of DSB repair block conversion of the ssAAV genome to dsDNA.
Because scAAV vectors do not require complementary strand synthesis, they can be used to specifically test the contributions of DNA repair factors to recombination independent of dsDNA conversion. We measured the ratio of circularized to linear vector using scAAV-GFP reporters in 18 different DNA repair-deficient cell lines and found significant differences in only 6 (4). Circularization was never completely eliminated in any cell type, suggesting a great deal of redundancy in the DSB repair pathways that can operate on rAAV genomes. However, the greatest inhibition came from mutations in ATM, which resulted in an apparent 70 to 80% decrease in the ratio of circularized to linear genomes in cultured cells. A similar decrease was observed when the vectors were injected into the muscles of ATM-deficient mice. Mutations in other DNA repair factors had significant effects in either muscle (DNA-PKCS) or cultured cells (NBS1), but not both, possibly reflecting the relative dominance of DSB repair by NHEJ in quiescent cells and by HR in dividing cells. Homologous recombination has also been implicated in rAAV end joining and concatemer formation in HeLa cells through the use of vectors with nonhomologous ITRs derived from AAV serotype 2 (AAV2) and AAV5 (40).
In contrast to the case for ATM, mutations in ATR (ATM and rad3 related), another factor orchestrating DSB repair by HR, did not affect scAAV circularization in dividing cells, suggesting that this pathway was not utilized. The ATR complex recognizes single-stranded DNA bound by replication protein A (RPA), the cellular ssDNA binding factor required for DNA replication. In association with ATR-interacting protein (ATRIP), this mediates cell cycle checkpoint activation in the presence of significant ssDNA gaps or stalled replication forks (32). The ATR protein has previously been reported to associate with infecting wild-type (wt) AAV, and this association induces an ATR-dependent DNA damage response in infected cells (17). A more recent study using recombinant ssAAV and scAAV vectors suggested that the damage response was specific not to the ssDNA or the hairpin ITR but to a cis-acting sequence within the wt AAV P5 promoter region (16). While rAAV vectors lacking the P5 promoter element do not provoke a pan-nuclear DNA damage response and G2/M cell cycle arrest, it remains likely that the ssDNA associates with RPA in the context of dsDNA conversion, making it a potential substrate for recombination through the ATR pathway. In contrast, the scAAV genome folds rapidly into double-stranded DNA, without the requirement for DNA synthesis, and therefore lacks significant regions of ssDNA. This raises the possibility that ssAAV and scAAV vector genomes could undergo differential processing to reach their stable isoforms.
In order to determine how the various DNA repair pathways interact with ssAAV and scAAV vector genomes to promote circularization or chromosomal integration, we used reporter vector-based assays with cells deficient in three phosphatidylinositol 3-kinase (PI3)-like kinase activities that orchestrate DNA repair (ATM, ATR, and DNA-PKCS).
MATERIALS AND METHODS
Cell lines, maintenance, and treatments.
Cell lines overexpressing wt ATR (GW33) or kinase-dead ATR (ATRKD) (GK41) under the control of a tetracycline (Tet)-inducible promoter were a kind gift from Paul Nghiem (26). wt ATR and ATRKD cells were grown in alpha minimal essential medium (MEM) (GIBCO) supplemented with 10% fetal bovine serum (FBS), 200 μg/ml G418, and 200 μg/ml hygromycin B. For vector transduction assays, overexpression of the transgene was induced with1 μg/ml doxycycline for 48 h prior to viral infection and maintained until green fluorescent protein (GFP) expression was evaluated at 24 h postinfection.
Simian virus 40 (SV40)-transformed fibroblast cell lines from a normal individual (wt cells, GM00637 J) and from an ataxia telangiectasia (AT) homozygous patient (ATM−/− cells, GM05849 E) were purchased from Coriell Institute (Camden, NJ). wt and ATM−/− cells were grown in Eagle MEM with Earle's balanced salt solution (BSS) (Lonza BioWhittaker) supplemented with 10% FBS and 1× GlutaMAX (GIBCO). To inhibit ATM or DNA-PKCS activity in the transformed fibroblasts, cultures were treated with 10 μM KU55933 (Tocris) or 10 μM NU7441 (Tocris), respectively, for 24 h before infection, and the treatment was maintained for the duration of the experiment.
Human malignant glioblastoma cells lines expressing normal levels of DNA-PK (DNA-PK+ cells, M059K) or lacking DNA-PK activity (DNA-PK− cells, M059J) were purchased from ATCC (Manassas, VA). DNA-PK+ and DNA-PK− cells were grown in Dulbecco modified Eagle medium (DMEM)-F-12 supplemented with 10% FBS and 1× nonessential amino acids. All cell lines were cultured as monolayer cultures at 37°C in a 5% CO2 humidified incubator.
Viral vector construction and vector purification.
The intact and circularization-dependent scAAV-GFP vectors have been described previously (5). The vectors with the intact GFP cassette, which can be expressed equally well from linear or circular genomes, are designated scAAV-GFP and ssAAV-GFP for self-complementary and single-strand vector, respectively. The circularization-dependent vectors, with the split GFP cassette, which must be circularized or concatemerized in order to express GFP, are designated scAAV-GFP-CD and ssAAV-GFP-CD. The ssAAV vector versions of these constructs were cloned into the pSSV9 background and contained functional ITRs at each end of the vector genome. In ssAAV-GFP-CD, a stuffer fragment of lambda DNA was inserted in the middle of the genome, between the two halves of the GFP cassette. In the intact ssAAV-GFP construct, two lambda (MluI 956 and 1268) stuffer fragments were inserted to flank the GFP cassette. Viruses were generated using the triple-transfection method with AAV serotype 2 capsid (14, 38) and purified by discontinuous iodixanol gradient separation and heparin chromatography (36). The viral vectors were quantified by dot blot hybridization, and infectious titers were determined by replication center assays in HeLa C12 cells (6).
Infection and quantification by flow cytometry.
For transduction and circularization assays with the inducible wt ATR and ATRKD cell lines (GW33 and GK41), cells were seeded on 12-well plates on day 1 at 12.5% confluence and treated with doxycycline (1 μg/ml). At day 3, the cells were infected with each rAAV vector (ssAAV-GFP, ssAAV-GFP-CD, scAAV-GFP, and scAAV-GFP-CD) at a multiplicity of infection (MOI) of 0.5 HeLa C12 infectious unit per cell. Medium with virus was replaced with fresh medium at 2 h postinfection. Doxycycline treatment was maintained until cells were harvested. wt, ATM−/−, DNA-PK+, and DNA-PK− cells were seeded on 12-well plates on day 1 at 25% confluence and infected on day 2 at an MOI of 1. All cell lines were harvested at 24 h postinfection, washed with ice-cold phosphate-buffered saline, and fixed with 4% paraformaldehyde in phosphate-buffered saline on ice. GFP expression was detected by flow cytometry (Coulter XL cytometer).
For integration assays, wt ATR and ATRKD cell lines were seeded and treated with doxycycline as described above. At day 3, cells were infected at an MOI of 260 for 2 h. Medium with virus was removed and replaced with fresh medium with doxycycline until day 2 postinfection, when doxycycline treatment was removed to maintain cell viability. wt, ATM−/−, DNA-PK+, and DNA-PK− cell lines were seeded on 24-well plates on day 1 at 25% confluence and infected at day 2 at an MOI of 1,400 for 2 h. At day 2 postinfection, cells were transferred to 12-well plates. All cells lines were passed at 1:5 or 1:10 every 4 to 7 days. At each passage, cell samples were collected and fixed as described above for flow cytometry analysis. Cells were passed continuously until the percentage of GFP-positive cells reached a plateau.
Southern blotting.
wt and ATM−/− cell lines were seeded on 10-cm plates on day 1 at 30% confluence and infected at day 2 with scAAV-GFP or scAAV-GFP-CD vector at an MOI of 250 (determined on HeLa C12 cells). Medium with virus was replaced with fresh medium at 5 h postinfection. At day 3, cells were harvested and cytoplasmic (C) and nuclear (N) fractions were isolated as described previously (37). Nuclear and cytoplasmic fractions were resuspended in 10 mM Tris-10 mM EDTA (pH 8.0) plus 1% SDS and incubated with 1 μl RNase (10 mg/ml) for 0.5 h at 37°C, followed by 2 h incubation with protease K (20 μg/ml) at 37°C. After the addition of NaCl (1.1 M), samples were refrigerated at 4°C overnight. Following centrifugation at 16,000 × g, supernatants were phenol-chloroform extracted and ethanol precipitated in the presence of 10 μg of yeast RNA carrier. All samples were digested with SphI and BsaBI (noncutters in AAV vector) to digest episomal SV40 DNA in the SV40-transformed fibroblasts. Nuclear fractions were divided for further digestion with restriction enzymes (XbaI, MluI, and Plasmid Safe PS-DNase). Samples were electrophoresed on 0.8% neutral agarose gels, blotted onto a nylon membrane, and probed with 32P-labeled probe specific for the cytomegalovirus (CMV) promoter and the GFP protein-coding region. Vector DNA excised from plasmids was subjected to the same digestions for molecular weight markers.
All statistical analyses were performed using Student's t test (GraphPad Prism 5 software). Samples for flow cytometry were run in triplicate. Error bars indicate standard deviations.
RESULTS
Comparison of ssAAV and scAAV transduction and genome circularization in ATM-deficient cells.
We had previously determined that ATM played a role in the circularization of scAAV vector genomes but that ATR did not. To determine whether ATM had differential effects on ssAAV and scAAV genomes, we compared transduction and circularization in a transformed ATM−/− fibroblast cell line (GM05849E) and transformed normal fibroblasts (GM00637J).
Vector transduction and circularization were determined by quantifying expression from vectors containing either an intact GFP or a split GFP gene cassette. The circularization-dependent split GFP (GFP-CD) has the left and right halves situated at the ends of the genome and fused to a split intron such that it can make the gene product only when two vector genome ends have been joined together by DNA recombination. The multiplicity of infection (MOI) was kept low (<1) in these experiments to ensure that expression from GFP-CD would be the result of circularization, rather than end-to-end joining of two vector genomes within the same nucleus, and that each GFP-positive cell was the result of a single recombination event involving a single, productively infecting rAAV genome.
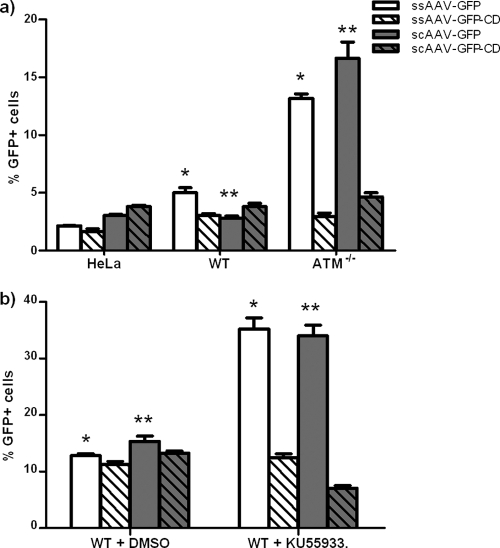
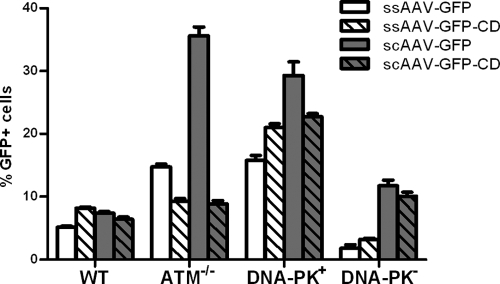
In ATM−/− cells, the overall levels of transduction from the ssAAV and scAAV were increased by 2.6- and 6.6-fold, respectively, compared to those in normal fibroblasts, as evidenced by the increase in GFP positive cells from the intact GFP vector (Fig. 1 a [open bars]; Table 1). This was consistent with a previously described effect on ssAAV vectors in ATM−/− cells, which had been attributed to increased second-strand synthesis (30). However, transduction from the scAAV vector was increased to an even greater extent, suggesting that the previously observed effect on ssAAV may not have been entirely due to increased second-strand synthesis, since this would not have affected transduction from the already-double-stranded scAAV vector.
FIG. 1.
Transduction and circularization with ssAAV and scAAV in ATM−/− cells. (a) Normal or ATM−/− transformed fibroblasts were infected with four different viral vectors containing either an intact GFP gene (ssAAV-GFP and scAAV-GFP) or a circularization-dependent GFP gene (ssAAV-GFP-CD and scAAV-GFP-CD) at an MOI of 1 for 2 h. GFP expression was quantified by flow cytometry at 24 h postinfection. The indicated (* and **) differences between wt and ATM−/− cells were significant (P < 0.0001). (b) Drug inhibition of ATM kinase activity. Transformed wt fibroblasts were treated with 10 μM KU 55933, a selective inhibitor of ATM kinase activity, for 24 h before infection with the four reporter vectors. Dimethyl sulfoxide (DMSO) was added in negative-control cells at the same final concentration. Transduction was assayed at 24 h postinfection. The indicated (* and **) differences were significant (P < 0.0001); P = 0.0768 for ssAAV-CD, and P = 0.0001 for scAAV-CD. Error bars indicate standard deviations.
TABLE 1.
Comparison of ssAAV and scAAV transduction, genome circularization, and integration
| Mutation | Fold differencea |
|||||
|---|---|---|---|---|---|---|
| ssAAV |
scAAV |
|||||
| Transductionb | Circularizationc | Integrationd | Transduction | Circularization | Integration | |
| ATM | +2.6 | 1.0 | −3.0 | +6.6 | +1.2 | −1.1 |
| ATR | +1.3 | 1.0 | +1.2 | 1.0 | 1.0 | +1.3 |
| DNA-PKCS | −8.0 | −8.4 | −8.5 | −2.5 | −2.2 | −10.5 |
Positive values indicate an increased percentage of GFP-positive cells relative to the wt. Negative values indicate a decrease relative to the wt. A value of 1.0 indicates no change.
Fold difference in intact GFP vector transduction between wt and mutant cell lines (data are from Fig. 1, 4, and 5).
Fold difference in circularized genomes between wt and mutant cell lines (data are from Fig. 1, 4, and 5).
Fold difference in integrated genomes between wt and mutant cell lines (data are from Fig. 8).
While transduction levels increased dramatically in ATM−/− cells, the numbers of circularized genomes did not change significantly for either the ssAAV or scAAV vector, suggesting that ATM is not directly involved in productive circularization (Fig. 1a, hatched bars). These results further suggest that previous interpretations of increased transduction, or decreased circularization relative to transduction, in the absence of ATM may instead be viewed as loss of functional rAAV genomes in normal cells through interaction with ATM-mediated recombination pathways. However, the pool of genomes that ultimately becomes circularized and expressed in normal cells does not appear to be affected by ATM.
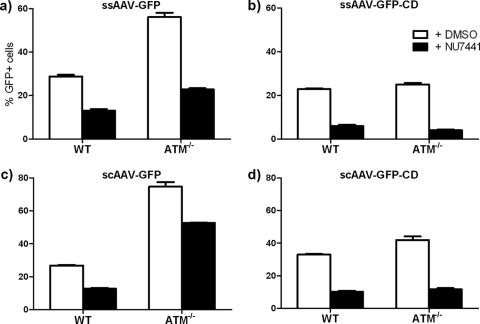
Because the transformed fibroblasts used in the experiment described above are not isogenic, we also tested the effect of a specific ATM inhibitor (KU55933) in a single wt fibroblast cell line. Similar to the effect of the ATM-deficient cells, inhibition of ATM activation led to increased transduction from intact GFP vectors (Fig. 1b, open bars). Again, the vector genomes mediating the increased transduction did not appear to be circularized (Fig. 1b, hatched bars).
Increased transduction in ATM−/− cells was not reflected at the level of vector DNA.
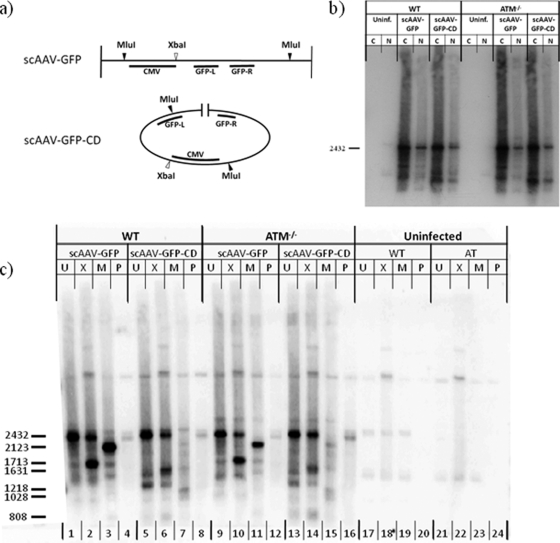
In order to directly observe the effects of ATM on the numbers of rAAV genomes in the nucleus, we infected wt and ATM−/− cells with scAAV-GFP and scAAV-GFP-CD at an MOI 250 and harvested at 24 h postinfection for Southern blot analysis. To decrease the signal from vector remaining in the cytoplasm, the cells were processed into nuclear and cytoplasmic fractions, and low-molecular-weight DNA was extracted from each (Fig. 2b). The amounts of vector DNA recovered from nuclear and cytoplasmic fractions were equivalent for the ATM−/− and wt cells, indicating that they are equally permissive for AAV infection. Nuclear DNA was divided into four aliquots. Uncut nuclear fractions were compared to digests with singly or multiply cut enzymes to delineate linear, circular, and total vector DNA (Fig. 2a). The fourth fraction was treated with plasmid-safe DNase, which degrades linear dsDNA or ssDNA but not supercoiled or nicked circular dsDNA, to confirm the identity of the circular form.
FIG. 2.
Southern blot of scAAV vector DNA from normal and ATM−/− cells. (a) Diagrams of intact GFP and GFP-CD vector genomes with relevant restriction sites indicated. (b) Southern blot of uncut low-molecular-weight DNA extracted from cytoplasmic (C) and nuclear (N) fractions from vector-infected or control uninfected wt and ATM−/− cells. Cells were infected at an MOI of 250 with intact GFP (scAAV-GFP) or circularization-dependent GFP (scAAV-GFP-CD) vectors and harvested at 24 h postinfection. The position of the linear vector genome is indicated (2,432 bp). (c) Southern blot of nuclear fractions from the same infections as described above. Nuclear fractions from wt or ATM−/− cells were divided into four aliquots and digested with XbaI (X), MluI (M), or Plasmid Safe nuclease (P) or were left undigested (U). The sizes in bp of the intact linear (2,432 bp) and digested vector DNA bands are indicated.
While our transduction-based assay showed a >5-fold increase exclusively from linear scAAV genomes in ATM−/− cells compared to the wt, we observed no corresponding increase at the molecular level, in either total vector DNA in the nucleus, linear DNA, or circular forms (Fig. 2c, compare lanes 1 to 4 with lanes 9 to 12 for scAAV-GFP vector and lanes 5 to 8 with lanes 13 to 16 for scAAV-GFP-CD vector). Uncut unit-length linear genomes ran at approximately 2,432 bp (lanes 1, 5, 9, and 13), with uncut circular genomes just below (lanes 4, 8, 12, and 16). Single-cut linear genomes dropped to 1,713 and 1,631 bp, respectively, for scAAV-GFP and scAAV-GFP-CD (lanes 2, 6, 10, and 14). Single-cut circular genomes were converted to unit-length linear DNA, running at 2,432 bp (head-to-head and tail-to-tail concatemers were not detected in this experiment). Linear and circular genomes were quantified from the single-cut vector DNA in lanes 2, 6, 10, and 14. There was no evidence of a 5-fold increase in the linear form between the wt and ATM−/− cells. The ratio of linear to circular genomes was approximately 2.6:1 in wt cells and 3.6:1 in the ATM−/− cells, which was consistent with the expression levels in ATM−/− cells but not in wt cells, where the majority of the expressed genomes were circular. Again, no differences between wt and ATM−/− cells were observed in these products. Digestion with the double-cut enzyme converts all vector DNA to single bands of 2,123 and 1,218 bp for scAAV-GFP and scAAV-GFP-CD, respectively (lanes 3, 7, 11, and 15). While it was possible that some of the linear genomes detected were derived from vector DNA that had remained encapsidated within the nucleus, the circular DNA could be formed only by interaction with cellular factors, and the ratio of circular to linear DNA was consistent with the excess transduction by linear vector genomes in ATM−/− cells as determined by GFP expression.
Similarly, Southern hybridization of DNA from ssAAV-infected cells showed no significant differences between ATM−/− and wt cells in the ratios of circular to linear genomes (data not shown). Because the amounts and ratios of circular and linear genomes did not change between wt and ATM−/− cells, even though there were clearly more linear genomes being expressed in ATM−/− cells, this suggested that the differential GFP expression may be a consequence of silencing of ATM-interacting vector genomes in normal cells.
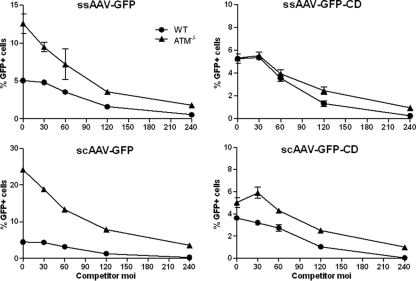
An alternate possibility was that the high vector dose (MOI, 250) required for Southern blot detection saturated the effects of the ATM mutation such that there was no increase in transduction under these condition. While we could not assess increased transduction simply by GFP expression at this dose because all of the cells are multiply transduced, we were able to test this notion in a competition experiment, where the doses of the GFP reporter vectors were kept constant at an MOI of 1 and increasing amounts of competing scAAV vector were added (Fig. 3). The competitor contained an irrelevant transgene and a liver-specific promoter, which was not active in these cells, to preclude transcriptional quenching effects. We observed competition for GFP transduction in wt cells beginning at a competitor MOI of 60. At MOIs of 120 and 240, GFP expression was reduced by half with each doubling of competitor, indicating that we had reached saturation for the number of genomes that can be productively delivered to the nucleus. The previously observed increases in transduction by linear vector genomes in ATM−/− cells relative to wt cells was clearly evident at all concentrations of competitor vector, up to the dose (MOI, 250) used in the Southern blot experiment. Thus, if there had been an increase in linear vector DNA in the nucleus matching the increased GFP expression from linear vector in ATM−/− cells, it should have been detected in the Southern blot.
FIG. 3.
Transduction response to ATM−/− mutation with increasing vector dose. wt and ATM−/− cells were infected at an MOI of 1 with each of the four reporter vectors plus the indicated increasing amounts of competitor vector containing an inactive (liver-specific) promoter and irrelevant (mouse PD-L1) transgene. Expression of GFP was assayed at 24 h postinfection. Error bars indicate standard deviations.
Interestingly, although the increased transduction in ATM−/− cells was evident at all vector concentrations, competition for transduction in ATM−/− cells began earlier and exhibited a higher slope than transduction in wt cells or transduction with genomes that would ultimately be circularized. This suggests that the pathway that leads to interaction with ATM is more readily saturated than the pathway that leads to productive circularization.
Transduction and circularization in ATR-deficient cells.
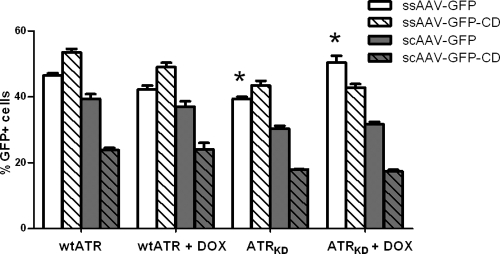
The PI3-like kinase ATR is activated by DNA damage involving significant regions of ssDNA, including UV lesions, significantly resected DSBs, and stalled replication forks. We therefore reasoned that the ssAAV was more likely to be a substrate for ATR-mediated recombination than scAAV, because the latter folds quickly into dsDNA after uncoating. The recombination efficiencies of the two kinds of vectors were compared in isogenic cell lines that overexpressed either a kinase-dead dominant negative ATR (ATRKD) or the wt ATR from a Tet-inducible promoter. Doxycycline induction in the ATRKD cell line, GK41, leads to effective inhibition of ATR activity, while induction of wt ATR in GW33 cells has no effect (26).
Doxycycline-induced overexpression of control wt ATR did not affect transduction or circularization of either type of vector (Fig. 4, wt ATR and wt ATR + DOX). As we had previously observed, overexpression of the dominant-negative ATRKD had no significant effect on the circularization of scAAV vectors, and the overall transduction efficiency was unchanged (Fig. 4, ATRKD and ATRKD + DOX, gray bars). In contrast, induction of ATRKD led to a significant (30%) increase in transduction from the ssAAV vector, similar to the effects of ATM on both vectors. Again, the number of circularized ssAAV genomes did not reflect the increase in active genomes available (Fig. 4, ATRKD and ATRKD + DOX, white bars). We concluded that ATR does not recognize the scAAV vector but does recognize and interact with the ssAAV vector. Similar to the effects of ATM, this interaction leads to a loss of functional genomes.
FIG. 4.
Transduction and circularization with ssAAV and scAAV in ATR-deficient cells. Cells containing doxycycline-inducible wt (wt ATR) or dominant-negative (ATRKD) ATR transgenes were infected with the four reporter vectors used for Fig. 1 at an MOI of 0.5 for 2 h. Doxycycline (DOX) treatment was initiated 48 h before infection and maintained through the experiment. GFP expression was quantified by flow cytometry at 24 h postinfection. The indicated (*) differences between wt and ATR-deficient cells were significant (P < 0.0010). Error bars indicate standard deviations.
Transduction and circularization in DNA-PKCS-deficient cells.
In order to determine whether the NHEJ pathway had differential effects on ssAAV and scAAV vectors, we also compared transduction and circularization in two cell lines with or without DNA-PKCS activity (M059K and M0597J, respectively) (Fig. 5). In contrast to the case for deficiencies in the HR pathways (ATM and ATR), transduction levels were greatly decreased for both ssAAV and scAAV intact GFP vectors in DNA-PKCS-deficient cells. Also in contrast to the case for cells deficient in HR pathways, most of the GFP-expressing genomes were circularized in the DNA-PKCS −/− cell line.
FIG. 5.
ssAAV and scAAV circularization assay with DNA-PKCS-deficient cells. Cells were infected with the four reporter vectors (see Fig. 1) at an MOI of 1 for 2 h. GFP expression was quantified by flow cytometry at 24 h postinfection. Means decreases for transduction in DNA-PK+ and DNA-PK− cells for each virus are all significantly different (P < 0.0001 for ssAAV-GFP, ssAAV-GFP-CD, and scAAV-GFP-CD; P = 0.0002 for scAAV-GFP). Error bars indicate standard deviations.
Southern blots from infected M059K and M059J cells were inconclusive as to whether the decreased transduction in the DNA-PKCS cell line was reflected in the amount of vector DNA in the nucleus, largely due to a larger amount of vector taken up in the cytoplasm of the mutant cells (data not shown). This led to various levels of potential contamination of the nuclear fraction with cytoplasmic vector. However, it should be noted that while the DNA-PKCS mutant cells took up more vector in the cytoplasm, they displayed less transduction than the wt cells.
Because the M059K and M059J cell lines were not isogenic, we tested the effects of NU7441, a specific inhibitor of DNA-PKCS activity, on transduction and circularization in normal and ATM−/− fibroblasts to ensure that the observed differences in transduction were not due to unrelated differences in the infectivities of the two cell lines (Fig. 6) (18). In each case, NU7441 treatment reduced transduction from both ssAAV and scAAV vectors, as had been observed in the mutant cell lines, although the change was not as great. We also tested the DNA-PKCS inhibitor in ATM−/− cells to determine how the two DSB repair pathways interacted (Fig. 6a to d). In ATM−/− cells, the DNA-PKCS inhibitor caused a decrease in transduction relative to that in untreated ATM−/− cells, similar to its effect in normal cells. Further, the inhibitor-treated ATM−/− cells showed the characteristic increase in transduction relative to inhibitor-treated wt cells. Again, the excess transduction was coming from vector genomes that remained linear. This suggested that that there was little overlap between the effects of ATM and DNA-PKCS on rAAV vector genomes; i.e., there appears to be a commitment to one or the other pathway early in infection, and the two pathways do not appear to complement or compete with each other, at least through the next 24 h.
FIG. 6.
Drug inhibition of DNA-PKCS activity. Transformed normal and ATM−/− cells were treated with 10 μM NU7441, a specific inhibitor of DNA-PKCS activity, for 24 h before infection with the four reporter vectors. In negative-control cells, DMSO was added at the same final concentration. Transduction was assayed at 24 h postinfection. Mean decreases for transduction in untreated and NU7441-treated cells for each vector are all significantly different (P = 0.0002 for scAAV-GFP in ATM−/− cells; P < 0.0001 for all the others). Error bars indicate standard deviations.
Chromosomal integration in DNA repair-deficient cells.
Having determined the effects of deficiencies in each of the three DSB signaling PI3-like kinases on circularization, we tested whether there were corollary effects on rAAV integration. For each cell line, integration was assayed by infection at a high MOI (approximately 260 to 1,400 HeLa cell infectious units per cell), followed by continuous passage until a stable percentage of GFP-expressing cells was reached, which represented the percentage of cells with integrated vector (Fig. 7).
FIG. 7.
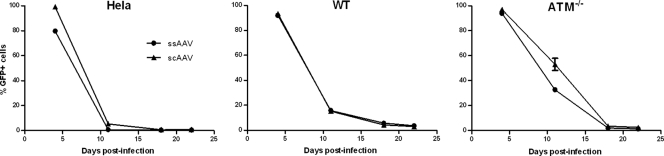
Integration assay for ssAAV-GFP and scAAV-GFP vectors in ATM−/− cells. Cell lines (HeLa, wt fibroblast, and ATM −/− fibroblast) were infected at an MOI of 1,400 for 2 h and passed every 4 to 7 days. Samples were taken at passage on the indicated days postinfection and fixed for quantification of GFP expression by flow cytometry. Infected cultures were passed until a stable percentage of cells expressed GFP, indicating that the vector was chromosomally integrated. Error bars indicate standard deviations.
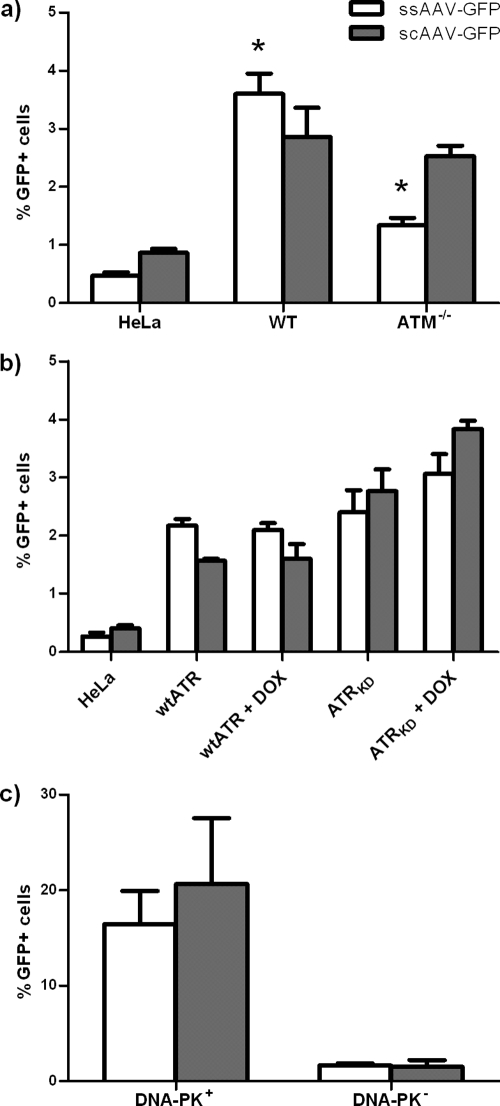
The increased GFP transduction with both scAAV and ssAAV linear vectors in ATM−/− cells was not reflected in a corresponding increase in integrated vector (Fig. 8a). Indeed, the integration of ssAAV was significantly decreased compared to that in wt cells, while scAAV vector integration was not significantly changed. This decrease was surprising because the numbers of circularized and linear genomes, as determined by Southern blotting, had remained unchanged for both vectors in the presence and absence of ATM, suggesting that circularization and integration were not mediated through the same processes.
FIG. 8.
Integration of ssAAV-GFP and scAAV-GFP vectors in ATR-, ATM-, and DNA-PKCS-deficient cells. (a) wt and ATM−/− cells were infected at an MOI of 1,400 for 2 h. Cells were passed and GFP expression was quantified by flow cytometry until the percentage of GFP-positive cells reached a plateau at 22 days postinfection. The indicated (*) differences between wt and ATM-deficient cells were significant (P < 0.0038). (b) Doxycycline-treated and untreated wt ATR and ATRKD cells were infected for 2 h at an MOI of 260 Treatment with DOX was maintained until 2 days postinfection. Cells were passed and GFP expression was quantified by flow cytometry until the percentage of GFP-positive cells reached a plateau at 25 days postinfection. Means for percent GFP-positive cells for ssAAV-GFP and scAAV-GFP vectors were not significantly different for wt ATR and wt ATR + DOX cells or for ATRKD and ATRKD + DOX cells. (c) DNA-PK+ and DNA-PK− cells were infected at an MOI of 1,400 for 2 h. Cells were passed and GFP expression was quantified by flow cytometry until percentage of GFP-positive cells reached a plateau at 48 days postinfection. Means for percent GFP-positive cells for ssAAV-GFP and scAAV-GFP vectors are significantly different (P = 0.0018 and P = 0.0088, respectively). Error bars indicate standard deviations.
To test the effects of ATR on integration, either the wt ATR or ATRKD was induced with doxycycline at 48 h prior to infection and maintained for an additional 48 h postinfection. Doxycycline was then discontinued, because ATR activity is essential for long-term cell viability. Induction of wt ATR had no effect on the rates of integration of either vector, while induction of ATRKD lead to a modest increase in integration for both the ssAAV and scAAV vectors (Fig. 8b). This suggested that ATR did not participate directly in the integration of either rAAV vector, despite its effects on the ssAAV genome at the transduction level.
Finally, the effect of DNA-PKCS deficiency on integration was tested in the M059K and M059J cells. Integration was reduced by 8.5-fold and 10.5-fold for the ssAAV and scAAV vectors, respectively, in the DNA-PKCS-deficient cells (Fig. 8c). While this generally followed the decreased transduction observed for both vectors in these cells, the effect on transduction with ssAAV vector had been greater than that with scAAV, converse to the effects on integration.
DISCUSSION
We have examined the roles of three principal factors orchestrating DSB repair in the circularization and integration of rAAV genomes. Our results suggest that there are qualitative and quantitative differences between ssAAV and scAAV vectors in terms of their interactions with different host DNA repair and recombination pathways. A striking and unexpected observation from these experiments is that deficiencies in DNA repair factors lead to significant changes in transduction efficiencies independent of dsDNA conversion and circularization. In the absence of ATM, transduction from the ssAAV and scAAV vectors increased by 2.6- and 6.6-fold, respectively (Table 1). In contrast, in the absence of DNA-PK, transduction is decreased 8.0- and 2.5-fold for the ssAAV and scAAV vectors, respectively.
Under the conditions used in these experiments, that is, in rapidly dividing cell cultures, rAAV genomes are acted upon by either HR or NHEJ pathways. Interactions with HR appear to lead to significant loss of functional genomes, as evidenced by the large increase in transduction with intact GFP vectors in ATM-deficient cells. This effect is greater for scAAV than ssAAV, arguing against a mechanism based strictly on conversion to dsDNA genomes, as had been previously suggested (30, 42). In the absence of ATM, the rescued genomes remain linear, seemingly trapped in this form and inaccessible to the NHEJ pathway, at least over the next 24 h. This suggests that the interaction choice between the major DNA repair pathways is fixed early after infection and is not easily altered. We also noted in our competition assay that the pathway that leads to interaction with ATM appears to be more easily saturated than the pathway that leads to productive circularization. Whether this suggests that the fate of the genome is determined through chance interactions with DSB repair factors that reside in separate compartments or that the different pathways are available at different stages of the cell cycle has not been determined.
A similar, though smaller, effect on transduction, specific for ssAAV genomes, is observed in ATR-deficient cells, suggesting a general negative outcome for interactions with HR pathways. Alternatively, it remains possible that ATR blocks conversion of ssAAV to dsDNA, since it does not affect scAAV genomes. However, it is clear that the fate of the majority of ssAAV vector genomes is determined by ATM, as evidenced by the greater increase in transduction in ATM-deficient cells than in ATR-deficient cells. This suggests that most interactions with HR repair pathways occur after dsDNA conversion, even in ATR-competent cells.
It is unlikely that the loss of GFP expression in HR-competent cells was due to excessive loss of sequence near the DNA ends, because the intact ssGFP vector used in these experiments contained 900 to 1,000 bp of stuffer sequences at each end, which would buffer the GFP cassette against imprecise end joining. Further, most circular rAAV junctions show end-to-end joining within the ITR sequences (11). Further, our Southern blot results are not consistent with extensive degradation of vector DNA, suggesting that the loss of GFP signal is due to silencing of gene expression in genomes interacting with the HR pathway. The possibility of a silencing mechanism is consistent with observations that ATM-mediated phosphorylation of histone H2AX is associated with reduced transcription in regions undergoing DSB repair (33). Further, parvovirus and rAAV vector genomes have been demonstrated to associate with chromatin, and rAAV vector infection induces a slight H2AX phosphorylation (2, 7, 19, 27). Future experiments employing immunoprecipitation of rAAV with specific chromatin components may address these issues.
We had previously reported that ATM and related HR factors were important for scAAV circularization, based on the ratio of circularized to noncircularized genomes in wt versus mutant cells and on similar observations with vectors injected into muscle of mutant mice in vivo (4). While we still observe the decreased ratio of circularized to noncircularized genomes, our previous interpretation did not take into account the present observation that deficiency in HR factors leads to a net gain in functional genomes which do not circularize. Because the presence or absence of either ATM or ATR did not substantially alter the number of circular genomes, we now conclude that NHEJ is likely to be the principal pathway for rAAV genome circularization.
Converse to the effects of HR-mediated DNA repair pathways, interactions with the NHEJ pathway lead to circularization without loss of functional genomes, as evidenced by the large decrease in transduction efficiency in the absence of DNA-PKCS activity. However, while the majority of functional genomes are lost in these cells, most of those remaining are circularized. It is not clear what mechanism is circularizing these genomes in the absence of DNA-PKCS activity, but it does not appear to be ATM, since similar numbers of genomes are circularized in ATM−/− cells in the presence of a DNA-PKCS inhibitor as in normal cells with the inhibitor. This may be a consequence of nonclassical NHEJ, which is DNA-PK independent (39). Taken together, these results suggest that interaction of rAAV genomes with HR DNA repair pathways is costly in terms of functional genomes, while circularization through NHEJ is more sparing. This has important implications for gene therapy, because quiescent cells rely predominantly on the NHEJ pathway for DNA repair and make up the majority of relevant target cell types for rAAV applications.
Interpretation of the effects of the various PI3-like kinases on integration is hampered somewhat by the effects of the mutations on the number of functional genomes available in the nucleus and their circularization status, leaving few fixed points of reference. However, our results suggest that there is not a simple relationship between rAAV vector genome circularization and integration, as might have been expected if they were competing parallel processes mediated by the same factors. In ATM-deficient cells, the excess of functional, noncircularized genomes, with free ends available for recombination with chromosomal DSBs, does not lead to increased integration. However, if transcriptional silencing is indeed the mechanism responsible for the loss of functional genomes in normal cells, it remains possible that silencing of episomal vector DNA is relieved over time, and these genomes can subsequently be scored in the integration assay.
There are clearly differences between ssAAV and scAAV vector integration in terms of their responses to ATM or DNA-PKCS deficiency. There is a significant decrease in the integration of ssAAV in ATM-deficient cells, while scAAV vector integration remains unchanged, despite the fact that ATM deficiency has a greater effect on the number of functional linear scAAV genomes in the nucleus. This suggests that ATM-mediated HR contributes productively to ssAAV integration but not scAAV integration. In DNA-PKCS-deficient cells, there are decreases in both the number of functional genomes and the number of integrations. For ssAAV, the decrease in integration is similar to the decrease in functional genomes (8.5-fold and 8.0-fold, respectively). In contrast, the decrease in integrated scAAV genomes is greater than the decrease in transduction (10.5-fold and 2.5-fold, respectively). While it is not clear why there is a disproportionately large decrease in scAAV integration, it is consistent with the observations of Daya et al., who noted an overall decrease in integration of scAAV in the absence of DNA-PKCS (8).
Based on these comparisons, we conclude that there are qualitative differences in the DNA recombination mechanisms that lead to rAAV genome circularization versus integration into the host chromosome and that ssAAV and scAAV vectors can follow different pathways.
Acknowledgments
We thank Kimberly Zaraspe for technical assistance with this research and Amy Lehman, OSU Biostatistics, and the RI-NCH Biostatistics Center for analysis.
This work was supported by NIH grant R01 AI070244 to D.M.M.
Footnotes
Published ahead of print on 10 June 2010.
REFERENCES
- 1.Alexander, I. E., D. W. Russell, and A. D. Miller. 1994. DNA-damaging agents greatly increase the transduction of nondividing cells by adeno-associated virus vectors. J. Virol. 68:8282-8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Asher, E., S. Bratosin, and Y. Aloni. 1982. Intracellular DNA of the parvovirus minute virus of mice is organized in a minichromosome structure. J. Virol. 41:1044-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cervelli, T., J. A. Palacios, L. Zentilin, M. Mano, R. A. Schwartz, M. D. Weitzman, and M. Giacca. 2008. Processing of recombinant AAV genomes occurs in specific nuclear structures that overlap with foci of DNA-damage-response proteins. J. Cell Sci. 121:349-357. [DOI] [PubMed] [Google Scholar]
- 4.Choi, V. W., D. M. McCarty, and R. J. Samulski. 2006. Host cell DNA repair pathways in adeno-associated viral genome processing. J. Virol. 80:10346-10356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, V. W., R. J. Samulski, and D. M. McCarty. 2005. Effects of adeno-associated virus DNA hairpin structure on recombination. J. Virol. 79:6801-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark, K. R., F. Voulgaropoulou, and P. R. Johnson. 1996. A stable cell line carrying adenovirus-inducible rep and cap genes allows for infectivity titration of adeno-associated virus vectors. Gene Ther. 3:1124-1132. [PubMed] [Google Scholar]
- 7.Collaco, R. F., J. M. Bevington, V. Bhrigu, V. Kalman-Maltese, and J. P. Trempe. 2009. Adeno-associated virus and adenovirus coinfection induces a cellular DNA damage and repair response via redundant phosphatidylinositol 3-like kinase pathways. Virology 392:24-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daya, S., N. Cortez, and K. I. Berns. 2009. Adeno-associated virus site-specific integration is mediated by proteins of the nonhomologous end-joining pathway. J. Virol. 83:11655-11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donsante, A., D. G. Miller, Y. Li, C. Vogler, E. M. Brunt, D. W. Russell, and M. S. Sands. 2007. AAV vector integration sites in mouse hepatocellular carcinoma. Science 317:477. [DOI] [PubMed] [Google Scholar]
- 10.Duan, D., P. Sharma, L. Dudus, Y. Zhang, S. Sanlioglu, Z. Yan, Y. Yue, Y. Ye, R. Lester, J. Yang, K. J. Fisher, and J. F. Engelhardt. 1999. Formation of adeno-associated virus circular genomes is differentially regulated by adenovirus E4 ORF6 and E2a gene expression. J. Virol. 73:161-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duan, D., Z. Yan, Y. Yue, and J. F. Engelhardt. 1999. Structural analysis of adeno-associated virus transduction circular intermediates. Virology 261:8-14. [DOI] [PubMed] [Google Scholar]
- 12.Duan, D., Y. Yue, and J. F. Engelhardt. 2003. Consequences of DNA-dependent protein kinase catalytic subunit deficiency on recombinant adeno-associated virus genome circularization and heterodimerization in muscle tissue. J. Virol. 77:4751-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrari, F. K., T. Samulski, T. Shenk, and R. J. Samulski. 1996. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J. Virol. 70:3227-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrari, F. K., X. Xiao, D. McCarty, and R. J. Samulski. 1997. New developments in the generation of Ad-free, high-titer rAAV gene therapy vectors. Nat. Med. 3:1295-1297. [DOI] [PubMed] [Google Scholar]
- 15.Fisher, K. J., G. P. Gao, M. D. Weitzman, R. DeMatteo, J. F. Burda, and J. M. Wilson. 1996. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J. Virol. 70:520-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fragkos, M., M. Breuleux, N. Clement, and P. Beard. 2008. Recombinant adeno-associated viral vectors are deficient in provoking a DNA damage response. J. Virol. 82:7379-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Inagaki, K., C. Ma, T. A. Storm, M. A. Kay, and H. Nakai. 2007. The role of DNA-PKcs and Artemis in opening viral DNA hairpin termini in various tissues in mice. J. Virol. 81:11304-11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jurvansuu, J., M. Fragkos, C. Ingemarsdotter, and P. Beard. 2007. Chk1 instability is coupled to mitotic cell death of p53-deficient cells in response to virus-induced DNA damage signaling. J. Mol. Biol. 372:397-406. [DOI] [PubMed] [Google Scholar]
- 18.Leahy, J. J., B. T. Golding, R. J. Griffin, I. R. Hardcastle, C. Richardson, L. Rigoreau, and G. C. Smith. 2004. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg. Med. Chem. Lett. 14:6083-6087. [DOI] [PubMed] [Google Scholar]
- 19.Marcus-Sekura, C. J., and B. J. Carter. 1983. Chromatin-like structure of adeno-associated virus DNA in infected cells. J. Virol. 48:79-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarty, D. M. 2008. Self-complementary AAV vectors; advances and applications. Mol. Ther. 16:1648-1656. [DOI] [PubMed] [Google Scholar]
- 21.McCarty, D. M., P. E. Monahan, and R. J. Samulski. 2001. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 8:1248-1254. [DOI] [PubMed] [Google Scholar]
- 22.Miller, D. G., L. M. Petek, and D. W. Russell. 2004. Adeno-associated virus vectors integrate at chromosome breakage sites. Nat. Genet. 36:767-773. [DOI] [PubMed] [Google Scholar]
- 23.Miller, D. G., E. A. Rutledge, and D. W. Russell. 2002. Chromosomal effects of adeno-associated virus vector integration. Nat. Genet. 30:147-148. [DOI] [PubMed] [Google Scholar]
- 24.Nakai, H., E. Montini, S. Fuess, T. A. Storm, M. Grompe, and M. A. Kay. 2003. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat. Genet. 34:297-302. [DOI] [PubMed] [Google Scholar]
- 25.Nakai, H., T. A. Storm, S. Fuess, and M. A. Kay. 2003. Pathways of removal of free DNA vector ends in normal and DNA-PKcs-deficient SCID mouse hepatocytes transduced with rAAV vectors. Hum. Gene Ther. 14:871-881. [DOI] [PubMed] [Google Scholar]
- 26.Nghiem, P., P. K. Park, Y. S. Kim Ys, B. N. Desai, and S. L. Schreiber. 2002. ATR is not required for p53 activation but synergizes with p53 in the replication checkpoint. J. Biol. Chem. 277:4428-4434. [DOI] [PubMed] [Google Scholar]
- 27.Penaud-Budloo, M., C. Le Guiner, A. Nowrouzi, A. Toromanoff, Y. Cherel, P. Chenuaud, M. Schmidt, C. von Kalle, F. Rolling, P. Moullier, and R. O. Snyder. 2008. Adeno-associated virus vector genomes persist as episomal chromatin in primate muscle. J. Virol. 82:7875-7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rass, E., A. Grabarz, I. Plo, J. Gautier, P. Bertrand, and B. S. Lopez. 2009. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 16:819-824. [DOI] [PubMed] [Google Scholar]
- 29.Russell, D. W. 2007. AAV vectors, insertional mutagenesis, and cancer. Mol. Ther. 15:1740-1743. [DOI] [PubMed] [Google Scholar]
- 30.Sanlioglu, S., P. Benson, and J. F. Engelhardt. 2000. Loss of ATM function enhances recombinant adeno-associated virus transduction and integration through pathways similar to UV irradiation. Virology 268:68-78. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz, R. A., J. A. Palacios, G. D. Cassell, S. Adam, M. Giacca, and M. D. Weitzman. 2007. The Mre11/Rad50/Nbs1 complex limits adeno-associated virus transduction and replication. J. Virol. 81:12936-12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shechter, D., V. Costanzo, and J. Gautier. 2004. Regulation of DNA replication by ATR: signaling in response to DNA intermediates. DNA Repair 3:901-908. [DOI] [PubMed] [Google Scholar]
- 33.Solovjeva, L. V., M. P. Svetlova, V. O. Chagin, and N. V. Tomilin. 2007. Inhibition of transcription at radiation-induced nuclear foci of phosphorylated histone H2AX in mammalian cells. Chromosome Res. 15:787-797. [DOI] [PubMed] [Google Scholar]
- 34.Song, S., P. J. Laipis, K. I. Berns, and T. R. Flotte. 2001. Effect of DNA-dependent protein kinase on the molecular fate of the rAAV2 genome in skeletal muscle. Proc. Natl. Acad. Sci. U. S. A. 98:4084-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song, S., Y. Lu, Y. K. Choi, Y. Han, Q. Tang, G. Zhao, K. I. Berns, and T. R. Flotte. 2004. DNA-dependent PK inhibits adeno-associated virus DNA integration. Proc. Natl. Acad. Sci. U. S. A. 101:2112-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Summerford, C., J. S. Bartlett, and R. J. Samulski. 1999. AlphaVbeta5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat. Med. 5:78-82. [DOI] [PubMed] [Google Scholar]
- 37.Wang, Y., W. Zhu, and D. E. Levy. 2006. Nuclear and cytoplasmic mRNA quantification by SYBR green based real-time RT-PCR. Methods 39:356-362. [DOI] [PubMed] [Google Scholar]
- 38.Xiao, X., J. Li, and R. J. Samulski. 1998. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 72:2224-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan, C. T., C. Boboila, E. K. Souza, S. Franco, T. R. Hickernell, M. Murphy, S. Gumaste, M. Geyer, A. A. Zarrin, J. P. Manis, K. Rajewsky, and F. W. Alt. 2007. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 449:478-482. [DOI] [PubMed] [Google Scholar]
- 40.Yan, Z., R. Zak, Y. Zhang, and J. F. Engelhardt. 2005. Inverted terminal repeat sequences are important for intermolecular recombination and circularization of adeno-associated virus genomes. J. Virol. 79:364-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yue, Y., and D. Duan. 2003. Double strand interaction is the predominant pathway for intermolecular recombination of adeno-associated viral genomes. Virology 313:1-7. [DOI] [PubMed] [Google Scholar]
- 42.Zentilin, L., A. Marcello, and M. Giacca. 2001. Involvement of cellular double-stranded DNA break binding proteins in processing of the recombinant adeno-associated virus genome. J. Virol. 75:12279-12287. [DOI] [PMC free article] [PubMed] [Google Scholar]