Abstract
Although the action of interferons (IFNs) has been extensively studied in vitro, limited information is available on the spatial and temporal activation pattern of IFN-induced genes in vivo. We created BAC transgenic mice expressing firefly luciferase under transcriptional control of the Mx2 gene promoter. Expression of the reporter with regard to onset and kinetics of induction parallels that of Mx2 and is thus a hallmark for the host response. Substantial constitutive expression of the reporter gene was observed in the liver and most other tissues of transgenic mice, whereas this expression was strongly reduced in animals lacking functional type I IFN receptors. As expected, the reporter gene was induced not only in response to type I (α and β) and type III (λ) IFNs but also in response to a variety of IFN inducers such as double-stranded RNA, lipopolysaccharide (LPS), and viruses. In vivo IFN subtypes show clear differences with respect to their kinetics of action and to their spatial activation pattern: while the type I IFN response was strong in liver, spleen, and kidney, type III IFN reactivity was most prominent in organs with mucosal surfaces. Infection of reporter mice with virus strains that differ in their pathogenicity shows that the IFN response is significantly altered in the strength of IFN action at sites which are not primarily infected as well as by the onset and duration of gene induction.
Type I interferons (IFNs), a single IFN-β and more than 13 α subtypes in humans and mice, were discovered as antiviral proteins that are typically induced by viruses (38). Other members of the type I IFN family were discovered later and comprise IFN-ω, -ɛ, and -κ (37). These IFNs all use the same heterodimeric receptor (IFNAR1/2) (47). Another class of antiviral IFNs, the type III IFNs that comprise three λ subtypes, signal through a different receptor complex composed of the interleukin (IL)-10Rβ and the IL-28Rα subunits (29, 40). Despite the use of different receptors, both IFN types share an interferon regulatory factor (IRF)-dependent induction pathway, induce a similar set of genes (ISGs), and exhibit similar biological activities (1). This is due to their ability to activate the transcription factors STAT1 and STAT2 (10, 43). Type I IFNs have been shown to activate natural killer cells, modulate the activity of T cells as part of their innate immune activity, and also serve as activators of the adaptive immune system (4, 5). The receptor for type I IFNs is expressed on essentially all cell types (with a few exceptions, like red blood cells). Thus, all cells can respond to type I IFNs and their actual response is thought to depend on the accessibility of the respective tissue (33, 38). In contrast, recent work revealed a distinct set of cells and organs that is dominantly affected by IFN-λ. Prominent IFN-λ responses were found in stomach, intestine, and lungs (10, 41). At the cellular level the response in kidney and brain was restricted to epithelial cells. From these data Sommereyns et al. (41) concluded that IFN-λ might contribute to the prevention of viral invasion through skin and mucosal surfaces.
Substantial amounts of IFN are produced upon infection. At least in vitro, all cell types, when exposed to viruses, can act as IFN producer cells (IPCs) (3, 33). However, this production differs significantly in vivo depending on inducer and responding cell type (3). While RNA viruses are very strong inducers, other viruses and nonviral pathogens induce IFNs to various extents (3, 6, 18). Main IFN-producing cells have been identified for several pathogens as being plasmacytoid dentritic cells (pDCs) and macrophages (30, 32, 39), but less attention has been drawn to the target cells of IFNs. This induction follows recognition of pathogen-associated molecular patterns (PAMPs) of respective microorganisms by RIG-I-like helicases (RLHs) or Toll-like receptors (TLRs) (27, 46). Downstream signaling makes use of NF-κB and IRF activation, ending with transcriptional activation of the IFN genes (23). Primary induction leads predominantly to the upregulation of IFN-β and IFN-α4 in most cell types. However, in cells that have been pretreated with IFNs and which have induced ISGs, other IFNs, mainly IFNs of the α subtype, are induced as well by infection, leading to a super-stimulation of ISGs, a phenomenon that is called “IFN type I receptor mediated feed-forward” (34). This action relies on the ISG IRF-7 (22, 34). pDCs constitutively express IRF-7 and secrete a large amount of IFNs upon primary infection. Recently, Kumagai et al. (30) analyzed the IFN-α response to viral infection using a reporter system in which a green fluorescent protein gene was knocked into the IFN-α6 gene. They found that both the infection site and the virus strain define the cell types that become dominant IFN producers. The fact that in all infections mobile cells from the hematopoietic compartment are involved predicts a distribution of the IFNs to different sites in the body, with blood and lymphoid tissue being specifically affected.
In addition to virus-induced IFN production, low constitutive expression of type I IFN has been reported (11, 21). A recent publication making use of a reporter mouse, in which the IFN-β structural gene is replaced by the firefly luciferase gene, confirms this constitutive expression and demonstrates its origin in the thymus and other sites (31).
IFNs are produced at different positions and by different cells in the body depending on the nature and site of infection. To identify not only IPC but subsequently cells and tissues reacting to IFN, highly sensitive methods are needed to dissect the spatial resolution of the gene expression triggered by IFNs in vivo. At least as important is the dynamic behavior of these IFN responses. The molecular and physiological consequences of IFNs depend on the nature (affinity), the concentration, and the time course of exposure to these cytokines. It may induce either antiviral activity (short exposure and weak binders) or antiproliferative activity (long exposure, strong binders) (26). Further, the target cells and organs will define the biological activity. Therefore, we have created reporter mice by which several of these questions can be solved, including local versus systemic distribution of IFNs, the definition of the main target cells and organs, the dispersion and availability dynamics of IFNs, and the course of the IFN response during infection of individual living animals.
To generate reporter mice for type I as well as type III IFNs we chose the Mx2 gene. Both Mx genes, Mx1 and Mx2, are specifically induced by type I and type III IFNs (2, 19, 20, 35). It is noteworthy that the Mx genes in inbred mouse strains are not functionally translated and, thus, the disruption or manipulation of these genes should not significantly alter cellular functions (17, 42). For high sensitivity we chose the optimized firefly luciferase 2 gene (Promega Corporation) as a marker. This allows sensitive in vivo monitoring as well as measurement of luciferase activity in organ homogenates and individual cell types. To guarantee specific reporter gene regulation, BAC constructs were generated in which the Mx2 gene was replaced by the luciferase gene. BAC transgenic mice were created, and whole-body live imaging was performed. Also, organs from in vivo stimulated animals were excised and luciferase activity was quantified in vitro. Further, information regarding the extent and targets of the constitutive IFN response as well as the impact of type III IFNs on the localization of antiviral response was derived from experiments with such mice. With these methods a number of new and surprising insights into the dynamics and distribution of IFNs in the whole animal were obtained.
MATERIALS AND METHODS
Generation of Mx2Luc BAC transgenic mice.
The BAC clone RP24-71I6 containing the murine Mx2 locus was obtained from the BACPAC Resource Center. Homologue recombination was performed using the bacteriophage λ recombination system (7). Thereby the open reading frame (ORF) of the murine Mx2 gene was replaced by a linear fragment containing the amplified reporter firefly luciferase (pGL4.10; Promega), followed by the simian virus 40 (SV40) polyadenylation signal and an FRT (FLP recognition target)-flanked cassette harboring a prokaryotic promoter, the PGK promoter, the kanamycin/neomycin phosphotransferase, and the bovine growth hormone polyadenylation signal. Primers used were Mx2Phom+Fluc2 (5′-TTATAATATTCATTTCCCACAGAGTACCCAACTGAGAGAAGAAATAAAAGATGGAAGATGCCAAAAACATTAAGA-3′) and Mx2Exon14hom+BamHI (5′-AAAGAAAAGTGGTTTATTAAGGAATGCAACAGGCAGCTCCCATTTGTACACTCAAGGGCATCGGTCGACGGATCC-3′), where italics represent homology to the Mx2 sequence in the BAC and roman indicates homology to the Luc2 gene and to part of the second FRT site at the 3′ end of the targeting construct, respectively. Modified BAC DNA was isolated using the NucleoBond BAC100 (Macherey-Nagel) and applied onto a Sepharose column for purification. Pronuclear microinjection into C57BL/6 oocytes was performed at the Max Planck Institute Dresden as described earlier (50). Founder mice carrying the BAC transgene were identified by PCR analysis of genomic DNA. Southern blot analysis of the Mx2 core promoter region was done as described previously (49). A 1,652-bp luciferase 2 fragment was used as a hybridization probe. BAC DNA used for pronuclear injection and genomic DNA of adult ear fibroblasts derived from Mx2 transgenic mice were digested with BamHI.
Mice and ethics statement.
All animals used were of a C57BL/6 genetic background. Mx2-Luc transgenic mice were crossed to IFNAR1−/− (36). Animals were handled in strict accordance with good animal practice as defined by the relevant local animal welfare bodies, and all animal work was approved by the appropriate committee (Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit [LAVES], Oldenburg, Germany). All mice were bred under standard conditions at the Helmholtz Centre for Infection Research (Braunschweig, Germany).
Viruses and viral infections.
We used variants of influenza virus strain hvPR8 with complete (hvPR8-delNS1) or partial (hvPR8-NS1(1-126) deletion of the IFN antagonistic factor NS1 (28). Further, mutant Thogoto virus THOV-delML lacking the IFN-antagonistic factor ML (16) was applied. All these viruses are classified as biosafety level 2 pathogens in Germany. Animals were anesthetized by intraperitoneal injection of a mixture of ketamine (100 μg per gram of body weight) and xylazine (5 μg per gram of body weight) before intranasal infection with indicated doses of the various influenza A viruses in 50 μl phosphate-buffered saline (PBS) containing 0.3% bovine serum albumin (BSA). For THOV-delML infections, 100-μl samples of diluted virus stocks were applied intraperitoneally without anesthesia. Animals were euthanized if severe symptoms developed or body weight loss approached 30% of the initial value.
Induction of IFN response.
Mice were treated intravenously with indicated concentrations of either IFN-α4, IFN-β, IFN-λ3, or IFN-γ (all purchased from PBL Interferonsource), with 200 μg poly(I:C) (Fluka), or with 50 μg lipopolysaccharide (LPS) (Sigma). For in vitro stimulation of cells IFN-β produced by a recombinant mammalian cell line was used.
Isolation of cells.
Adult fibroblasts were prepared from ear biopsy specimens and cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS), 100 μg/ml of penicillin/streptomycin, and nonessential amino acids. For isolation of hepatocytes, livers were perfused with liver perfusion medium (Invitrogen Life Technologies) and the perfusate was collected. For separation, cells were centrifuged at 500 rpm for 5 min. Hepatocytes in the pellet were washed twice with PBS + 1% FCS, passed through a cell strainer, and resuspended in RPMI 1640 medium supplemented with 5% FCS (Invitrogen Life Technologies). Bone marrow-derived cells were cultured in Iscove modified Dulbecco medium (IMDM) supplemented with 10% FCS and 20% supernatant from a granulocyte-macrophage colony-stimulating factor (GM-CSF)-expressing cell line (20 to 30 ng GM-CSF/ml). After 7 days of cultivation, macrophages were used for further analysis. For isolation of T cells, spleens from Mx2-Luc mice were mashed through a 70-μm-pore-size strainer. T cells were purified using a pan-T-cell isolation kit and AutoMacs magnetic separation system (Miltenyi Biotec) according to the manufacturer's instructions.
RNA isolation and qPCR.
For quantitative PCR (qPCR), total RNA was extracted from cells or tissues using the RNeasy minikit (Qiagen) and reverse transcribed into cDNA with the Ready-To-Go You-Prime first-strand beads kit (Amersham Biosciences) using oligo(dT)18 primers according to the manufacturer's instructions. Real-time PCR on the LightCycler (Roche) was performed in a total volume of 20 μl using the QuantiTect SYBR green PCR kit (Qiagen).
Detection of luciferase.
For quantification of the enzymatic activity of luciferase, cells were lysed in passive lysis buffer (Promega). For luciferase activity assays from tissues, the weights of respective tissue portions were determined and portions were homogenized in proportional volumes of PBS using the FastPrep system (MP Biomedicals). Cell and tissue extracts were assayed for luciferase activity using standard reaction buffer (20 mM glycylglycine, 12 mM MgSO4, 1 mM ATP) containing luciferin (SynChem OHG) and a single-tube luminometer (Berthold). For in vivo imaging, mice were injected intraperitoneally with 3 mg of d-luciferin (Synchem OHG) in PBS and anesthetized using 2 to 2.5% isoflurane (Abbott). After 5 min, the mice were placed in the imaging chamber of the Caliper in vivo imaging system (IVIS-200) and gray-scale images followed by bioluminescent images were acquired. Relative intensities of emitted light were presented as pseudocolor images ranging from red (most intense) to blue (least intense). Gray-scale photographs and the corresponding pseudocolor images were superimposed with the Living Image 3.1 software (Caliper). Signal emitted by regions of interest was measured, and data were expressed as photon flux, quantified as photons s−1 cm−2 sr−1 (Living Image 3.1 software; Caliper). Steradian (sr) refers to the photons emitted from a unit solid angle of a sphere.
RESULTS
Generation of Mx2 transgenic reporter mice and their validation.
To investigate which cell populations or organs react to type I and type III IFNs, we generated BAC transgenic mouse strains that contain the optimized firefly luciferase 2 reporter gene under transcriptional control of a characteristic type I IFN-regulated gene. Therefore, we chose the mouse Mx gene locus. In the BAC clone RP24-71I6, the Mx2 gene is flanked by 5′ and 3′ sequences of 103 and 75 kb, respectively. In our BAC construct the coding region of the Mx2 gene was replaced by the sequence of the reporter gene (Fig. 1 A). Three transgenic mouse lines (designated Mx2-Luc) were obtained by pronuclear injection. A molecular characterization of the core regulatory region of the integrated BACs, including the proximal promoter region and the reporter gene, revealed that an intact reporter locus is present in all three mouse lines (Fig. 1B). To show specificity of reporter gene induction, macrophages from Mx2-Luc reporter mice were isolated and stimulated with different types of IFN as well as with IL-6 and tumor necrosis factor alpha (TNF-α) (Fig. 1C). Quantitative determination of luciferase expression showed strongly increased reporter gene activity in cells stimulated with IFN-α and IFN-β for 12 h. In contrast, stimulation of macrophages with IFN-λ3, TNF-α, and IL-6 did not induce significant Mx2 promoter induction while treatment of cells with IFN-γ resulted in a marginal induction of the reporter gene. Next, we studied the dose response of IFN-β on primary macrophages (Fig. 1D). IFN-β markedly stimulated the reporter, depending on the concentration. Dose-dependent induction of luciferase activity was also observed in fibroblasts derived from ear biopsy specimens of adult mice (data not shown). Thus, we conclude that the Mx2-Luc BAC construct represents a specific and highly sensitive marker to quantify the response toward IFNs.
FIG. 1.
Characterization of transgenic Mx2-Luc reporter mice. (A) Flow scheme of Mx2-Luc recombineering. The BAC RP24-71I6 harboring the mouse Mx2 gene (upper line) was targeted by homologous recombination with a cassette carrying the gene encoding firefly luciferase 2 (open box) followed by the SV40 virus poly(A) signal, the FRT-flanked (gray ovals) fragment of prokaryotic and eukaryotic (PGK) promoter (black arrow) controlling expression of the kanamycin/neomycin phosphotransferase gene (light gray box), and the poly(A) signal of the bovine growth hormone gene (dark gray box). The complete Mx2 ORF located on the BAC was replaced by this cassette. (B) Integration of Mx2-Luc into genomes of three transgenic mouse lines. BamHI endonuclease-restricted mouse genomic DNA, isolated from adult ear fibroblasts, was blotted on nylon membrane and probed with a [32P]dCTP-labeled luc2-ORF probe. BAC DNA used for pronuclear injection is shown as a control. A single 4.1-kb fragment shows complete insertion of luc2 and at least 2.2 kb of the upstream promoter region of Mx2 into the mice's genomes. HindIII-digested λ DNA was used as a marker (M). (C) Bone marrow-derived macrophages from Mx2-Luc mice (line 2) were treated with IFN-α4 (500 U/ml), IFN-β (500 U/ml), IFN-γ (500 U/ml), IFN-λ3 (0.1 μg/ml), TNF-α (0.1 μg/ml), and IL-6 (0.1 μg/ml). Fold induction represents relative luminescence units of stimulated compared to untreated cells (cont.). All values were expressed as means ± standard deviation (SD) (n = 3). (D) Bone marrow-derived macrophages from Mx2-Luc mice (line 2) were incubated with the indicated concentrations of IFN-β, and luciferase activity was measured 12 h later. Relative luminescence units (RLU) are shown as mean values ± SD (n = 3). (E) Comparison of relative RNA levels of luc2 (open bars) and endogenous Mx2 (black bars). Bone marrow-derived macrophages (Mx2-Luc line 1) were treated with IFN-β (500 U/ml) for the indicated times. Total RNA was extracted and reverse transcribed into cDNA, and qPCR was performed by using specific primers for amplification of luciferase or Mx2 RNA. Displayed RNA levels were calculated relative to the RNA levels of β-actin.
We further examined whether the transgenic Mx2-Luc allele recapitulates endogenous Mx2 mRNA expression. We correlated the response of macrophages from transgenic mice to IFN stimulation by comparing mRNA levels of the endogenous Mx2 gene and the luciferase reporter at different time points (Fig. 1E). In these cells Mx2 and luciferase mRNA levels peaked around 8 h after IFN treatment. Afterwards, the levels of both mRNAs declined. Importantly, the luciferase mRNA expression pattern resembles regulation of the endogenous Mx2 gene. Taken as a whole, these results showed that expression of luciferase faithfully mimics Mx2 induction. Furthermore, due to the relatively short lifetime of the luciferase protein, the transcriptional activation period of the Mx2 gene can be deduced in the reporter mice.
Tissue-specific response to interferons in vivo.
To analyze the distribution of the IFN response in the whole organism, IFN-β as the major “initial” IFN was administered into the tail vein of reporter mice. At several time points after IFN injection, the mice were anesthetized and luciferase activity was imaged after administration of luciferin (Fig. 2A). Two observations deserve attention. First, the reporter gene activity had already reached a maximum at 3 h posttreatment and started to decline afterwards. Second, most of the signal was focused on the area of the liver. When various organs from untreated and IFN-β-treated mice were excised and imaged individually, the liver was confirmed to be the major responder to intravenously (i.v.) injected IFN-β (data not shown). Comparison of the three Mx2-Luc reporter lines in this experimental setting showed identical induction ratios for all lines, although the absolute levels of light emission differed significantly (see Fig. S1 in the supplemental material).
FIG. 2.
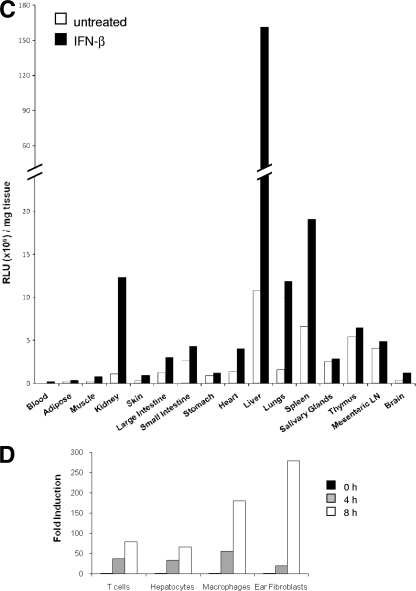
IFN responses in vivo and in vitro. (A and B) Whole-body imaging of Mx2-Luc reporter mice (line 2) after i.v. injection of 1,000 U IFN-β (A) and 1,000 U IFN-α4 (B). Mice were imaged before treatment (0 h) and followed over time as indicated, starting 3 h postinjection. Images from representative Mx2-Luc mice are shown. The rainbow scales indicate the number of photons measured per second per cm2 per steradian (sr). (C) Mx2-Luc mice (line 2) were i.v. injected with IFN-β (5,000 U). At 3 h postinjection, mice were sacrificed and selected organs were rapidly isolated and homogenized for luciferase activity measurement. The mean values (n = 2) of RLU per mg tissue (μl blood) for untreated (open bars) and treated (black bars) mice are shown. (D) Primary cultures of T cells, hepatocytes, bone marrow-derived macrophages, and ear-derived fibroblasts from Mx2-Luc mice were assayed for their ability to respond to IFN-β treatment (500 U/ml) in vitro. Fold induction represents relative luminescence units of stimulated compared to untreated cells for the indicated time points. All values were expressed as means (n = 3).
Despite the fact that the type I IFNs constitute a family of related cytokines that all recognize the same receptor, differences in action have been shown in vitro and in vivo (48). We compared the in vivo action of IFN-α4 with that of IFN-β (Fig. 2B and A, respectively). Although the main target organs seemed to be identical, the kinetics of reporter induction by IFN-α4 was slower, with a delay of about 3 h. Thus, we conclude that type I IFNs show different response kinetics in the same target organs. This conclusion is compatible with the fact that all IFNs bind to the same receptor but with different properties (24).
Several organs from untreated mice and from mice treated for 3 h with IFN-β were isolated and homogenized, and luciferase activity was quantified by standard luminometric measurement (Fig. 2C). By far, the highest level of luciferase activity (per mg tissue) was found in the liver. All other organs investigated showed significantly lower absolute levels of the luciferase reporter. Within those organs, high induction levels (>10-fold) were revealed in blood, liver, and kidney (compare with Fig. 3D). Some other organs, including lung and spleen, were found to be moderately induced. All other organs showed a comparably low induction ratio or absolute luciferase activity. Thus, it seems that the strongest response toward exogenously applied IFN can be detected in organs which are intensively supplied with blood.
FIG. 3.
Responses to constitutive type I IFN and exogenous IFN-λ. (A) Mx2-Luc mice were crossed to IFNAR−/− mice. Untreated Mx2-Luc mice heterozygous or homozygous for the IFNAR deletion were imaged. (B) Mx2-Luc IFNAR−/− mice were injected i.v. with 5 μg of IFN-λ3. Kinetics of the response from a representative mouse is shown. (C) Mx2-Luc IFNAR−/− mice injected i.v. with 5 μg of IFN-λ3 were sacrificed 4 h postinjection, and selected organs were rapidly harvested. Luciferase activity was determined in homogenized organs. The mean values (n = 3) of RLU per mg of tissue (μl blood) for untreated (open bars) and treated (black bars) mice are shown. (D) Fold induction of luciferase activity in homogenized organs for IFN-β (from Fig. 2C) and IFN-λ3 (from Fig. 3C). Note that the high induction ratio of blood cells results from low values of the uninduced animals and that the induced levels are rather low (see panel C). n.d., not done.
The different magnitudes and rates of induction during in vivo stimulation raised the question to which extent the investigated organs and cells would respond to IFN if they were equally exposed. It was of particular interest to estimate the strong response of the liver compared to other organs: whether this is a consequence of high sensitivity toward type I IFN or due to an organ-specific accumulation of the inducer. To answer this question, primary cells from different organs were prepared and cultivated in the presence of IFN-β for 8 h. The response was determined at different time points (Fig. 2D). All examined cell types are able to react to IFN-β by activating the reporter gene. The fold induction was found to vary between different cell types. Reproducibly, fibroblasts and macrophages showed significantly higher induction levels than splenic T cells and hepatocytes. These results suggest that the strong in vivo reaction in the liver is not due to highly sensitive cells but rather depends on increased exposure to IFN. Taken together, the observed luciferase activities in the reporter mouse reflect a real-life IFN response.
Monitoring the IFN type III response.
Reporter mice from all three lines showed constitutive reporter activity (see Fig. S1 and S2 in the supplemental material). This activity is unevenly distributed in the mouse and has, similar to the situation after IFN administration, a focus on the liver. Quantification of this prevalence of IFN activity in different organs is depicted in Fig. 2C. Systemic administration of an antibody directed against mouse IFN-β reduced the reporter activity to about 50% without changing the overall distribution (data not shown). Further, a comparison of reporter mice with animals crossed to IFNAR deficiency demonstrated that less than 20% of the constitutive response is independent of functional type I IFN signaling (Fig. 3A). This indicates that 80% of the reporter activity in unstimulated mice has to be attributed to constitutively produced type I IFN. Since the half-life time of the reporter signal is in the range of a few hours, we conclude that the constitutive activity is permanently available. According to the finding of Lienenklaus et al. (31) that the main source of constitutive IFN-β is the thymus, our data indicate that this IFN is also rapidly distributed and arrives in the same target organs as the injected IFN-β.
Though the activity was significantly reduced, the IFNAR−/− reporter mice still showed some constitutive luciferase activity (Fig. 3A, right panel). The distribution of this activity differs from that in IFNAR+/+ reporter mice, as shown by analysis of the IFN response in various organs (Fig. 3C). Constitutive expression of IFN-λ is presumably not responsible for the remaining activity, as similar background values were observed in reporter mice lacking functional type I and type III IFN receptors (data not shown). However, the differential organ responses argue against unspecific reporter gene activity. Since type III IFNs, like type I IFNs, can induce Mx proteins, it is possible to use the Mx2-Luc reporter mouse for monitoring IFN-λ responses. To demonstrate this, IFN-λ was injected into the tail vein of Mx2-Luc reporter mice and whole-animal imaging was carried out at different time points after injection (see Fig. S3A in the supplemental material). The response was fast (3 h), and the maximum was reached by 9 h after injection. In order to avoid overlaps with type I IFN signaling, the same experiment was carried out in IFNAR−/− Mx2 reporter mice (Fig. 3B). A detailed analysis of the organs showed that IFN-λ mainly targets large and small intestine, lung, and salivary gland. Stomach, kidney, spleen, and heart showed moderate responses. Thus, the dominant target organs of type I and type III IFNs are clearly distinct (Fig. 3D).
Production and fate of interferon after virus infection.
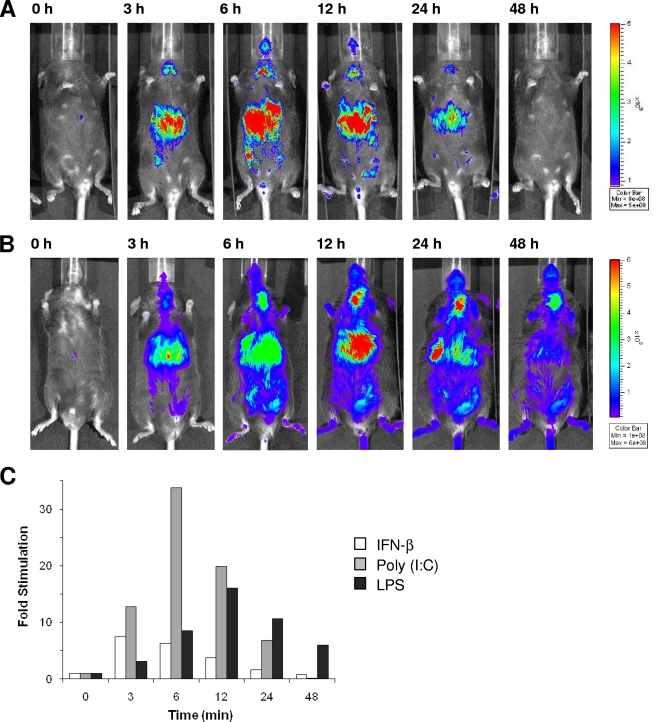
Several stimuli induce IFN production which, in turn, leads to specific IFN responses. In order to assess the time course of this process, we first used the double-stranded RNA (dsRNA) analogue poly(I:C) as a ubiquitous inducer of the IFN system. Poly(I:C) was injected into the tail vein of Mx2-Luc reporter mice (Fig. 4A). Furthermore, the response dynamics to LPS, a bacterially derived inducer of IFN-β, was examined (Fig. 4B). Treatment of mice with poly(I:C) or LPS induced an IFN response in the liver that successively increased from the very beginning to reach a maximum at 6 or 12 h, respectively. The IFN response to poly(I:C) and LPS was compared to the response with direct injection of IFN-β. The data showed that about 3 h for poly(I:C) and 6 h for LPS distinguish the maximal reporter activation from IFN-β injection experiments. This is seen in the quantification graphs for the luminescence of the whole animal (Fig. 4C). The rapid appearance of the IFN response in peripheral organs after poly(I:C) injection suggests that in situ synthesized IFN must rapidly enter the blood circulation and confer antiviral effects to the organs. However, the delay in maximal induction of the Mx2 reporter depends on the kinetics of type I IFNs production upon stimulation with the different inducers.
FIG. 4.
Whole-body imaging kinetics of the response to IFN and IFN inducers. (A and B) Mx2-Luc reporter mice (line 2) were injected i.v. with 200 μg of poly(I:C) (A) and 50 μg of LPS (B). Mice were imaged before treatment (0 h) and then followed over time as indicated, starting 3 h postinjection. Images from representative Mx2-Luc mice are shown. (C) Signal progression of luciferase activity from mice treated with IFN-β (from Fig. 2A) (open bars), poly(I:C) (from Fig. 4A) (gray bars), and LPS (from Fig. 4B) (black bars) are presented as fold induction of the reporter signal.
To investigate the kinetics of the IFN response after viral infection Mx2-Luc reporter mice were infected with viruses which are known to induce a strong IFN response. We used two different viruses that infect either locally or systemically. Thogoto virus (THOV) is known to replicate and spread to different sites in the mouse body and will eventually kill the mouse. A THOV mutant lacking the IFN-antagonistic protein ML (THOV-delML) exhibits similar virulence but induces higher levels of IFN (25). If injected intraperitoneally into a reporter mouse (IFN-β-luc) that specifically reports IFN-β gene activation by expressing luciferase (31), THOV-delML elicited a strong IFN-β response starting 24 h postinfection (Fig. 5A). The IFN-β response was most pronounced in the peritoneum at early times postinfection and then became most prominent in the liver, the site at which the virus replicates to very high titers. In a parallel experiment the Mx2-Luc reporter mouse was infected and imaged. Interestingly, initially, even after 24 h postinfection a strong induction of Mx2 reporter can be detected and most of this IFN response appeared in the liver. Compared to the induction of IFN-β no significant retardation in the IFN response was obvious (Fig. 5B). After 2 days postinfection, the IFN response further increased and was also detectable at other sites. To more quantitatively determine the contribution of different organs, extracts from various tissues at 0, 24, 48, and 72 h postinfection were analyzed (Fig. 5C). These data confirmed the early appearance of the IFN response and its predominance in the liver. However, other organs like spleen, kidney, and lung contribute to the IFN response picture obtained by whole-body imaging and Mx2 induction continuously increases in these organs as well as in the liver.
FIG. 5.
Comparison of IFN-β induction and IFN response after Thogoto virus infection. Luciferase activity in IFN-β+/Δβ-luc (A) and Mx2-Luc (B) reporter mice infected with THOV-delML (1 × 104 PFU per mouse intraperitoneally). At the indicated time points mice were life-imaged. Note that the animals are critically ill 72 h after infection and would have died within the next days. This might explain the high variability of the imaging data at 72 h. Representative mouse images are shown. (C) Signal progression in organs from Mx2-Luc mice before and after infection with THOV-delML. Bars from left to right: 0 (ctl), 24, 48, and 72 h. Organs were isolated and homogenized, and luciferase activity was determined. Relative luminescence units were calculated per mg of tissue. All values were expressed as mean ± standard deviation (n = 4).
We next analyzed the IFN response in mice infected with influenza virus strain hvPR8 that replicates exclusively in cells of the respiratory tract (15). We first used hvPR8-delNS1, a mutant virus that lacks the IFN antagonistic factor NS1. hvPR8-delNS1 is a powerful IFN inducer that can hardly replicate in the lung of IFN-competent mice (28). The virus was intranasally applied to Mx2-Luc reporter mice, and the response toward IFN was imaged at different time points. Imaging showed a strong signal in the upper chest but did not allow us to clearly distinguish lung and liver responses (see Fig. S4 in the supplemental material). The IFN response was quantitatively measured in extracts from various tissues at 0, 24, 48, and 72 h postinfection (Fig. 6A). In accordance with the fact that type I IFN is produced in the lung, a fast induction of the Mx2 reporter can be measured at the site of production. However, a low response toward IFN is also detectable in spleen and liver. Taking into account that the liver has a 6 to 10 times higher mass than the lung, the pictures obtained from in vivo imaging point out that substantial amounts of IFN are distributed over the whole body.
FIG. 6.
IFN response upon infection with influenza virus. (A and B) Mx2-Luc reporter mice were intranasally infected with 1 × 104 PFU per mouse of influenza virus strains hvPR8-delNS1 (A) and hvPR8-NS1(1-126) (B). At the indicated time points organs were isolated and homogenized and luciferase activity was determined. Relative luminescence units were calculated per mg of tissue. All values were expressed as mean ± standard deviation (n = 4). Bars from left to right: 0, 24, 48, and 72 h.
It is of particular interest to compare infection with the nonpathogenic hvPR8-delNS1 strain to a pathogenic situation. Therefore, we infected mice with the hvPR8-NS1(1-126) strain and quantified luciferase activity in tissue extracts. This strain leads to death of most infected animals within 4 to 5 days (28). In general, Mx2 reporter mice infected with hvPR8-NS1(1-126) revealed a much stronger luciferase activity than that of hvPR8-delNS1-infected animals. Interestingly, there was a 24-h delay in the maximal response toward IFN (Fig. 6B). While Mx2-Luc induction in the different organs was comparable for the first 24 h postinfection, at later time points luciferase expression increased dramatically not only in the lung but even more in liver and the other organs. Furthermore, after 48 h, luciferase activity stayed maximal until the animals had to be euthanized. The data also indicate that at later time points during infection, induction of the reporter gene in the liver is stronger than in the lung and even activity in the kidney is at comparable levels. Thus, it seems that the IFN response from both virus strains differs significantly in the strength of IFN action at sites which are not primarily infected and by the duration of ISG induction.
DISCUSSION
In this study, BAC transgenic mice carrying recombinant Mx2 locus containing firefly luciferase under the control of natural promoter elements were generated. Although the absolute expression strength of IFN-induced luciferase in three independent founder lines differs, the relative levels and induction kinetics are highly comparable. Our validation experiments in primary cells of the Mx2 reporter mice perfectly recapitulated the kinetics of endogenous Mx2 mRNA expression. The Mx2 gene as well as the Mx1 gene is regarded as a bona fide ISG that is representative for a typical IFN-induced antiviral response. We note that depending on the cell type not all ISGs are strictly coregulated and thus, the Mx2 induction cannot report truly the full spectrum of an antiviral activity in any cell type (8, 14). However, in all primary cell types tested so far the reporter system was activated by type I IFN and therefore allows a quantitative determination of at which organs and cells an IFN response is elicited. Furthermore, this work showed that the Mx2 reporter system is valid not only for the response to type I IFNs but also for the response to type III IFNs. In particular, the use of the IFNAR-deficient reporter mouse is a valuable tool to distinguish the type III from type I IFN responses.
Regarding the kinetics of luciferase expression, as with all reporter genes, the measured responses primarily reflect the onset of gene induction, assuming that transcription and translation of the replaced gene are similar to those of the reporter gene. Figure 1E suggests that the mRNA level of luciferase is always higher than that of the endogenous Mx2 but the decline of both mRNAs during the continuous presence of IFN-β goes in parallel. Since the lifetimes of luciferase and Mx2 protein are different, reporter gene activity was not taken as a measure of a particular endogenous protein. However, the fact that in vivo luciferase expression after IFN-β injection declines rapidly after an initial onset (Fig. 2A) indicates that the reporter follows the presence of IFN in the organism. This is confirmed by analysis of the IFN concentration in the serum after a single i.v. injection, showing that most of the introduced dosage disappears from the bloodstream within a few minutes (see Fig. S5 in the supplemental material). Thus, if the reporter response in the animal is sustained it indicates ongoing IFN production, e.g., as seen in the virus infection experiments.
From previous publications it is known that the expression density of the IFN-λ receptor corresponds to the main targets of an IFN-λ response. For the liver-specific expression, conflicting results were published. While Doyle et al. (9) find significant functional IFN-λ receptors expressed in human hepatocytic cells in vitro, Mordstein et al. (35) and Sommereyns et al. (41) find essentially no IFN-λ receptor expression and very little response in the mouse liver. According to the results from this work, mouse liver tissue indeed responds to IFN-λ; however, induction of Mx2 reporter in IFNAR+/+ mice is only 2-fold (see Fig. S3B in the supplemental material). Clinical data suggest an important role of the IFN-λ in the clearance of hepatitis C virus (HCV). A genetic polymorphism near the IFNλ3 gene associates with spontaneous clearance and treatment response (13, 45). With respect to the response of the liver to IFN-λ in mice containing functional type I IFN receptor, indirect or priming effects may be considered. Additional work is needed to clarify that indeed substantial type I IFN priming is required for an IFN-λ response of the liver.
So far, published results suggest that the respective receptor expression is the main determinant for the site of the biological IFN response. Consequently, concentration and affinity of the ligand to the receptor as well as receptor density would be additional parameters. In this line, IFN-α, which has recently been shown to exhibit lower affinity to the IFNAR than IFN-β, should have a retarded response (24). Indeed, this could be verified by following the response by whole-animal imaging (compare Fig. 2A and B).
In this work we confirmed that constitutively produced type I IFN targets organs like the liver. The question of which function this effect may have for new infections remains to be elucidated. The IFNAR−/− reporter mice show a reduced but still significant constitutive activity of reporter gene action. At this stage we cannot fully exclude unspecific background elicited by the BAC transgenes. However, since the “background values” response is not evenly distributed in the mouse body, it is tempting to speculate that IFN receptor-independent inflammatory mechanisms are responsible for this activity.
The injection of poly(I:C), a ubiquitous IFN inducer, leads to a maximal IFN response 6 h after administration. Based on the observation that IFN-β elicits an earlier maximum response and assuming that this subtype is the main type I IFN produced upon poly(I:C) administration, the maximal IFN production appears to occur at 3 h after injection. Interestingly, the response to LPS is retarded and the response upon infection with the studied viruses is found even much later. The detailed understanding of the kinetics will be of importance for the understanding of infections.
The finding that the liver is the main response organ for type I IFNs is surprising. We defined parenchymal cells as the main responding cells in this organ. Nonparenchymal cells showed a significantly lower response (data not shown). Independent of the inducing agent the type I IFN response was rapidly detectable in the liver. This is explained by two mechanisms. First, secreted IFN entering the blood circulation is rapidly absorbed by the liver, as shown by the i.v. injection of IFN-β and IFN-α. This rapid elimination from the bloodstream explains why little IFN, even after massive infections, is found in the serum. Second, IFN-producing cells (IPCs) that become activated at the site of infection might circulate and become trapped in the interstitial space of the liver to release their load close to the parenchymal cells. These consist of virus-specific target cells, such as the lung epithelial cells for influenza virus, but as well of certain immune cells that are typically found in infected tissue. Kumagai et al. (30) concluded that in epithelial lung infection three different cell species are sequentially activated. These are the main IPCs: alveolar macrophages (AMs), conventional DCs (cDCs), and pDCs. Also, responses to the constitutively expressed type I IFN were predominantly found in the liver. According to these findings, liver tissue should be permanently in a mild antiviral state. Since IFNs induce activities other than only antiviral effects (5, 44) the constitutively expressed IFN is expected to have a profound influence on the overall immune status of the liver as well as on immune cells that are affected by the liver.
The focus of the constitutive and induced type I IFN response to the liver motivated us to speculate that a specific protection of this organ from virus propagation might have advantages for the host. It is known that many circulating pathogens are preferentially trapped in the liver (12), and the IFN-induced status might help to control massive infection. Another, although less obvious, explanation is that the liver would have the function to eliminate overflowing IFN levels from the organism and thus eliminate the well-known site effects.
Primary and established cell lines, when exposed to IFNs, typically show an induction of their ISGs with a maximum expression between 6 and 24 h. It was interesting to see that the IFN preparations injected i.v. elicited a response that was maximal between 3 and 6 h for IFN-β and -α4, respectively. One possible explanation would be that the liver parenchymal cells as major IFN targets in vivo would respond more rapidly than other cell types. However, primary cultures of liver parenchymal cells did not confirm this assumption (see Fig. S6 in the supplemental material). Another explanation concerns the short time for which the organs are exposed to a single injection. This would simulate a pulsed response with an earlier maximum activity.
Our observations and the new mouse lines will have implications for basic research, for clinical translation, and for the use of IFNs in therapy. The reporter mice will help to elucidate the complex kinetics and cell-specific function of type I and III interferons in normal and diseased states. Among the potential applications are mouse models for certain autoimmune diseases in which excessive amounts of IFNs are produced. Another application concerns the screening for new compounds that serve as agonists or antagonists for the IFN system. Cells derived from the reporter mouse and the mouse itself could be used to test and validate such compounds for their activity. Currently, the major use of type I IFNs concerns the treatment of multiple sclerosis and chronic hepatitis induced by hepatitis viruses B and C. The systems might help to find better injection and dosage schemes and evaluate the modification of IFN preparations. Also, new IFN subtypes could be directly compared and their kinetics and accessibilities could be defined. The differences shown in IFN-α4 and -β might explain the differences in treatment with these IFN species. Since human IFNs are species specific, the expression of human receptor proteins for such experiments in the mouse might be essential.
Supplementary Material
Acknowledgments
We thank Maren Freund for technical assistance, Upneet Sandhu for assistance with isolation of hepatocytes, and members of the lab for helpful discussions. We thank Martin Messerle and Eva Borst (Hannover Medical School) for their help with the BAC recombination system.
This work was supported in part by grants from the German Ministry of Research and Education (BMBF) (FORSYS-Partner), the German Research Foundation (SFB 566 and 599), the Helmholtz Society (Systems Biology of Signaling in Cancer), and the European Community (LSHB-CT-2006-018933; MEST-CT-2004-504990).
Footnotes
Published ahead of print on 23 June 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ank, N., H. West, and S. R. Paludan. 2006. IFN-lambda: novel antiviral cytokines. J. Interferon Cytokine Res. 26:373-379. [DOI] [PubMed] [Google Scholar]
- 2.Asano, A., H. K. Jin, and T. Watanabe. 2003. Mouse Mx2 gene: organization, mRNA expression and the role of the interferon-response promoter in its regulation. Gene 306:105-113. [DOI] [PubMed] [Google Scholar]
- 3.Baig, E., and E. N. Fish. 2008. Distinct signature type I interferon responses are determined by the infecting virus and the target cell. Antivir. Ther. 13:409-422. [PubMed] [Google Scholar]
- 4.Biron, C. A. 2001. Interferons alpha and beta as immune regulators—a new look. Immunity 14:661-664. [DOI] [PubMed] [Google Scholar]
- 5.Bogdan, C., J. Mattner, and U. Schleicher. 2004. The role of type I interferons in non-viral infections. Immunol. Rev. 202:33-48. [DOI] [PubMed] [Google Scholar]
- 6.Colonna, M., A. Krug, and M. Cella. 2002. Interferon-producing cells: on the front line in immune responses against pathogens. Curr. Opin. Immunol. 14:373-379. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Veer, M. J., M. Holko, M. Frevel, E. Walker, S. Der, J. M. Paranjape, R. H. Silverman, and B. R. Williams. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69:912-920. [PubMed] [Google Scholar]
- 9.Doyle, S. E., H. Schreckhise, K. Khuu-Duong, K. Henderson, R. Rosler, H. Storey, L. Yao, H. Liu, F. Barahmand-pour, P. Sivakumar, C. Chan, C. Birks, D. Foster, C. H. Clegg, P. Wietzke-Braun, S. Mihm, and K. M. Klucher. 2006. Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology 44:896-906. [DOI] [PubMed] [Google Scholar]
- 10.Dumoutier, L., A. Tounsi, T. Michiels, C. Sommereyns, S. V. Kotenko, and J. C. Renauld. 2004. Role of the interleukin (IL)-28 receptor tyrosine residues for antiviral and antiproliferative activity of IL-29/interferon-lambda 1: similarities with type I interferon signaling. J. Biol. Chem. 279:32269-32274. [DOI] [PubMed] [Google Scholar]
- 11.Galabru, J., N. Robert, C. Buffet-Janvresse, Y. Rivière, and A. G. Hovanessian. 1985. Continuous production of interferon in normal mice: effect of anti-interferon globulin, sex, age, strain and environment on the levels of 2-5A synthetase and p67K kinase. J. Gen. Virol. 66:711-718. [DOI] [PubMed] [Google Scholar]
- 12.Gao, B., W. I. Jeong, and Z. Tian. 2008. Liver: an organ with predominant innate immunity. Hepatology 47:729-736. [DOI] [PubMed] [Google Scholar]
- 13.Ge, D., J. Fellay, A. J. Thompson, J. S. Simon, K. V. Shianna, T. J. Urban, E. L. Heinzen, P. Qiu, A. H. Bertelsen, A. J. Muir, M. Sulkowski, J. G. McHutchison, and D. B. Goldstein. 2009. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461:399-401. [DOI] [PubMed] [Google Scholar]
- 14.Geiss, G. K., V. S. Carter, Y. He, B. K. Kwieciszewski, T. Holzman, M. J. Korth, C. A. Lazaro, N. Fausto, R. E. Bumgarner, and M. G. Katze. 2003. Gene expression profiling of the cellular transcriptional network regulated by alpha/beta interferon and its partial attenuation by the hepatitis C virus nonstructural 5A protein. J. Virol. 77:6367-6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimm, D., P. Staeheli, M. Hufbauer, I. Koerner, L. Martinez-Sobrido, A. Solorzano, A. Garcia-Sastre, O. Haller, and G. Kochs. 2007. Replication fitness determines high virulence of influenza A virus in mice carrying functional Mx1 resistance gene. Proc. Natl. Acad. Sci. U. S. A. 104:6806-6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagmaier, K., S. Jennings, J. Buse, F. Weber, and G. Kochs. 2003. Novel gene product of Thogoto virus segment 6 codes for an interferon antagonist. J. Virol. 77:2747-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haller, O., M. Acklin, and P. Staeheli. 1987. Influenza virus resistance of wild mice: wild-type and mutant Mx alleles occur at comparable frequencies. J. Interferon Res. 7:647-656. [DOI] [PubMed] [Google Scholar]
- 18.Haller, O., G. Kochs, and F. Weber. 2007. Interferon, Mx, and viral countermeasures. Cytokine Growth Factor Rev. 18:425-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haller, O., P. Staeheli, and G. Kochs. 2007. Interferon-induced Mx proteins in antiviral host defense. Biochimie 89:812-818. [DOI] [PubMed] [Google Scholar]
- 20.Haller, O., S. Stertz, and G. Kochs. 2007. The Mx GTPase family of interferon-induced antiviral proteins. Microbes Infect. 9:1636-1643. [DOI] [PubMed] [Google Scholar]
- 21.Hida, S., K. Ogasawara, K. Sato, M. Abe, H. Takayanagi, T. Yokochi, T. Sato, S. Hirose, T. Shirai, S. Taki, and T. Taniguchi. 2000. CD8(+) T cell-mediated skin disease in mice lacking IRF-2, the transcriptional attenuator of interferon-alpha/beta signaling. Immunity 13:643-655. [DOI] [PubMed] [Google Scholar]
- 22.Honda, K., H. Yanai, H. Negishi, M. Asagiri, M. Sato, T. Mizutani, N. Shimada, Y. Ohba, A. Takaoka, N. Yoshida, and T. Taniguchi. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772-777. [DOI] [PubMed] [Google Scholar]
- 23.Honda, K., H. Yanai, A. Takaoka, and T. Taniguchi. 2005. Regulation of the type I IFN induction: a current view. Int. Immunol. 17:1367-1378. [DOI] [PubMed] [Google Scholar]
- 24.Jaks, E., M. Gavutis, G. Uze, J. Martal, and J. Piehler. 2007. Differential receptor subunit affinities of type I interferons govern differential signal activation. J. Mol. Biol. 366:525-539. [DOI] [PubMed] [Google Scholar]
- 25.Jennings, S., L. Martinez-Sobrido, A. Garcia-Sastre, F. Weber, and G. Kochs. 2005. Thogoto virus ML protein suppresses IRF3 function. Virology 331:63-72. [DOI] [PubMed] [Google Scholar]
- 26.Kalie, E., D. A. Jaitin, Y. Podoplelova, J. Piehler, and G. Schreiber. 2008. The stability of the ternary interferon-receptor complex rather than the affinity to the individual subunits dictates differential biological activities. J. Biol. Chem. 283:32925-32936. [DOI] [PubMed] [Google Scholar]
- 27.Kawai, T., and S. Akira. 2009. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 21:317-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kochs, G., L. Martinez-Sobrido, S. Lienenklaus, S. Weiss, A. Garcia-Sastre, and P. Staeheli. 2009. Strong interferon-inducing capacity of a highly virulent variant of influenza A virus strain PR8 with deletions in the NS1 gene. J. Gen. Virol. 90:2990-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kotenko, S. V., G. Gallagher, V. V. Baurin, A. Lewis-Antes, M. Shen, N. K. Shah, J. A. Langer, F. Sheikh, H. Dickensheets, and R. P. Donnelly. 2003. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 4:69-77. [DOI] [PubMed] [Google Scholar]
- 30.Kumagai, Y., O. Takeuchi, H. Kato, H. Kumar, K. Matsui, E. Morii, K. Aozasa, T. Kawai, and S. Akira. 2007. Alveolar macrophages are the primary interferon-alpha producer in pulmonary infection with RNA viruses. Immunity 27:240-252. [DOI] [PubMed] [Google Scholar]
- 31.Lienenklaus, S., M. Cornitescu, N. Zietara, M. Lyszkiewicz, N. Gekara, J. Jablonska, F. Edenhofer, K. Rajewsky, D. Bruder, M. Hafner, P. Staeheli, and S. Weiss. 2009. Novel reporter mouse reveals constitutive and inflammatory expression of IFN-beta in vivo. J. Immunol. 183:3229-3236. [DOI] [PubMed] [Google Scholar]
- 32.Liu, Y. J. 2005. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu. Rev. Immunol. 23:275-306. [DOI] [PubMed] [Google Scholar]
- 33.Malmgaard, L. 2004. Induction and regulation of IFNs during viral infections. J. Interferon Cytokine Res. 24:439-454. [DOI] [PubMed] [Google Scholar]
- 34.Marie, I., J. E. Durbin, and D. E. Levy. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mordstein, M., G. Kochs, L. Dumoutier, J. C. Renauld, S. R. Paludan, K. Klucher, and P. Staeheli. 2008. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 4:e1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller, U., U. Steinhoff, L. F. Reis, S. Hemmi, J. Pavlovic, R. M. Zinkernagel, and M. Aguet. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918-1921. [DOI] [PubMed] [Google Scholar]
- 37.Oritani, K., P. W. Kincade, C. Zhang, Y. Tomiyama, and Y. Matsuzawa. 2001. Type I interferons and limitin: a comparison of structures, receptors, and functions. Cytokine Growth Factor Rev. 12:337-348. [DOI] [PubMed] [Google Scholar]
- 38.Pestka, S., C. D. Krause, and M. R. Walter. 2004. Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 202:8-32. [DOI] [PubMed] [Google Scholar]
- 39.Scheu, S., P. Dresing, and R. M. Locksley. 2008. Visualization of IFNbeta production by plasmacytoid versus conventional dendritic cells under specific stimulation conditions in vivo. Proc. Natl. Acad. Sci. U. S. A. 105:20416-20421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheppard, P., W. Kindsvogel, W. Xu, K. Henderson, S. Schlutsmeyer, T. E. Whitmore, R. Kuestner, U. Garrigues, C. Birks, J. Roraback, C. Ostrander, D. Dong, J. Shin, S. Presnell, B. Fox, B. Haldeman, E. Cooper, D. Taft, T. Gilbert, F. J. Grant, M. Tackett, W. Krivan, G. McKnight, C. Clegg, D. Foster, and K. M. Klucher. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 4:63-68. [DOI] [PubMed] [Google Scholar]
- 41.Sommereyns, C., S. Paul, P. Staeheli, and T. Michiels. 2008. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 4:e1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Staeheli, P., R. Grob, E. Meier, J. G. Sutcliffe, and O. Haller. 1988. Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Mol. Cell. Biol. 8:4518-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 44.Takaoka, A., and T. Taniguchi. 2003. New aspects of IFN-alpha/beta signalling in immunity, oncogenesis and bone metabolism. Cancer Sci. 94:405-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas, D. L., C. L. Thio, M. P. Martin, Y. Qi, D. Ge, C. O'Huigin, J. Kidd, K. Kidd, S. I. Khakoo, G. Alexander, J. J. Goedert, G. D. Kirk, S. M. Donfield, H. R. Rosen, L. H. Tobler, M. P. Busch, J. G. McHutchison, D. B. Goldstein, and M. Carrington. 2009. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 461:798-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson, A. J., and S. A. Locarnini. 2007. Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol. Cell Biol. 85:435-445. [DOI] [PubMed] [Google Scholar]
- 47.Uze, G., G. Schreiber, J. Piehler, and S. Pellegrini. 2007. The receptor of the type I interferon family. Curr. Top. Microbiol. Immunol. 316:71-95. [DOI] [PubMed] [Google Scholar]
- 48.van Boxel-Dezaire, A. H., M. R. Rani, and G. R. Stark. 2006. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity 25:361-372. [DOI] [PubMed] [Google Scholar]
- 49.Verhoeyen, E., H. Hauser, and D. Wirth. 2001. Evaluation of retroviral vector design in defined chromosomal loci by Flp-mediated cassette replacement. Hum. Gene Ther. 12:933-944. [DOI] [PubMed] [Google Scholar]
- 50.Vintersten, K., G. Testa, R. Naumann, K. Anastassiadis, and A. F. Stewart. 2008. Bacterial artificial chromosome transgenesis through pronuclear injection of fertilized mouse oocytes. Methods Mol. Biol. 415:83-100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.