Abstract
Species C adenovirus establishes a latent infection in lymphocytes of the tonsils and adenoids. To understand how this lytic virus is maintained in these cells, four human lymphocytic cell lines that support the entire virus life cycle were examined. The T-cell line Jurkat ceased proliferation and died shortly after virus infection. BJAB, Ramos (B cells), and KE37 (T cells) continued to divide at nearly normal rates while replicating the virus genome. Viral genome numbers peaked and then declined in BJAB cells below one genome per cell at 130 to 150 days postinfection. Ramos and KE37 cells maintained the virus genome at over 100 copies per cell over a comparable period of time. BJAB cells maintained the viral DNA as a monomeric episome. All three persistently infected cells lost expression of the cell surface coxsackie and adenovirus receptor (CAR) within 24 h postinfection, and CAR expression remained low for at least 340 days postinfection. CAR loss proceeded via a two-stage process. First, an initial loss of cell surface staining for CAR required virus late gene expression and a CAR-binding fiber protein even while CAR protein and mRNA levels remained high. Second, CAR mRNA disappeared at around 30 days postinfection and remained low even after virus DNA was lost from the cells. At late times postinfection (day 180), BJAB cells could not be reinfected with adenovirus, even when CAR was reintroduced to the cells via retroviral transduction, suggesting that the expression of multiple genes had been stably altered in these cells following infection.
The adenovirus species C serotypes (Ad1, Ad2, Ad5, and Ad6) are commonly associated with upper and lower respiratory tract infections in children less than 5 years of age (1, 6, 9). These viruses are only weakly pathogenic in normal hosts, where the rate of asymptomatic primary infection ranges from 50 to 90% (2, 6, 9). Following primary infection, species C adenoviruses often enter a persistent stage characterized by intermittent release of live virus in stool long after nasopharyngeal shedding has ceased (9, 10).
Early reports found small quantities of replication-competent adenoviruses in mucosal-associated lymphoid tissues, notably tonsils and adenoids, in the absence of acute infection (7, 15, 36). Studies also reported quantities of nonreplicating adenoviral DNA in tonsils, leading to speculation that these viruses maintain a latent infection in lymphoid tissues (22). We found that the vast majority of adenovirus DNA in tonsil and adenoids is quiescent and preferentially localized in the T-lymphocyte population (12). More recently, we reported that while only a small minority of samples from tonsil donors contain replicating virus (<15%), most tissue samples (85%) could be stimulated in vitro to activate virus gene expression and replication (13). Together, these findings reveal infection by species C adenoviruses as a two-step process of acute replication in epithelial cells of the nasopharyngeal mucosa, followed by latent infection of lymphocytes in mucosa-associated lymphoid tissues. Epidemiology suggests that virus reactivation from latency and intermittent shedding in stool maintain these endemic viruses in the population (10).
In naturally infected tonsil lymphocyte populations, the frequency of adenovirus-bearing cells is very low, on the order of 1 cell in 104 to 105 (13). As an initial step in developing a picture of adenovirus infections in lymphocytes, a number of groups have reported success at infecting human lymphocyte cell lines with species C adenoviruses, and most infections appeared to be nonlytic (5, 17, 19, 20, 33), at least over the short term. In this report we explore adenovirus species C infection of several T- and B-lymphocyte cell lines to determine the nature of the virus-host cell interaction over extended periods of time and to unravel the cellular mechanisms controlling the virus replicative cycle as well as changes in the host cell initiated by the virus.
MATERIALS AND METHODS
Cell lines.
The human cell lines A549 (lung carcinoma), 293 (Ad5-transformed epithelia), Jurkat (acute T-cell leukemia), Ramos (Epstein-Barr virus [EBV]-negative Burkitt's lymphoma), and REH (precursor B-cell acute lymphoblastic leukemia [ALL]) cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA). BJAB (EBV-negative Burkitt's lymphoma) cells were obtained from Carl Ware (La Jolla Institute for Allergy and Immunology). KE37-CARhi was generated by transducing an isolate of KE37 cells, obtained from Periasamy Selveraj (Emory University), with a coxsackie and adenovirus receptor (CAR)-expressing retrovirus, as reported previously (19). With the exception of the use of KE37-CARhi cells as a control in the experiment shown in Fig. 8, all other experiments were performed with KE37 (immature T-cell ALL) cells that were purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ). These cells are designated KE37(DSMZ) to distinguish them from the KE37 cells used in our earlier studies that are resistant to adenovirus infection (19). The packaging cell line Phoenix for retrovirus production (25) was obtained from Joshy Jacob (Emory University). BJAB, Ramos, REH, Jurkat, KE37-CARhi, and KE37(DSMZ) cells were grown in RPMI medium supplemented with 10% fetal calf serum (FCS) and 10 mM glutamine. A549, 293, and Phoenix cells were grown in Dulbecco's modified Eagle medium (DMEM) with 4.5 μg of glucose/ml, 10% FCS, and 10 mM glutamine.
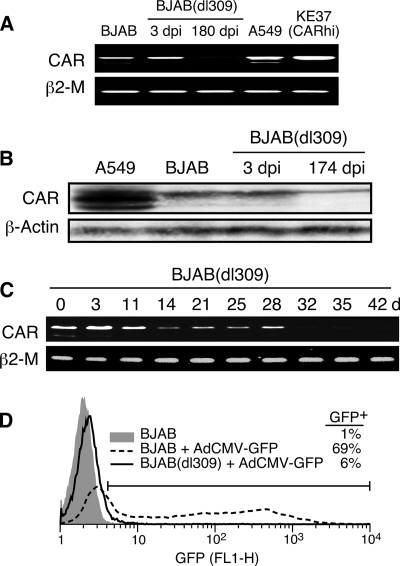
FIG. 8.
CAR gene expression is downregulated days after infection of BJAB cells. (A) Cellular RNA was extracted from Ad5dl309-infected BJAB cells at days 3 and 180 postinfection or from uninfected BJAB, A549, or KE37 cells transduced with the CAR gene (KE37-CARhi). Reverse transcription followed by PCR was used to detect mRNA for CAR and β2-microglobulin (β2-M). (B) Cellular lysates were prepared from equal numbers of uninfected A549 and BJAB cells or Ad5dl309-infected BJAB cells at 3 and 174 days postinfection and analyzed by immunoblotting for CAR or actin as a loading control. (C) BJAB cells were infected with Ad5dl309 at 100 PFU/cell. Cellular RNA was extracted at various days after infection and analyzed by reverse transcription followed by PCR to detect mRNA for CAR and β2-microglobulin. (D) BJAB cells were infected with Ad5dl309 at 100 PFU/cell. The persistently infected cells were maintained in culture medium for 75 days and then superinfected with AdCMV-GFP. GFP-positive cells were detected by flow cytometry. dpi, days postinfection.
Adenoviruses.
Wild-type (WT) species C Ad2 and Ad5 adenoviruses were provided by Dean Erdman (Centers for Disease Control and Prevention, Atlanta, GA). Mutant viruses Ad5dl309 and Ad5dl327 were obtained from Tom Shenk (Princeton University, Princeton, NJ) (16, 34). Ad5dl309 is an Ad5 mutant that lacks the genes for E3 RIDα and RIDβ proteins and for the 14,700-molecular-weight protein (14.7K protein) and was used as the wild-type virus in most experiments. Ad5dl327 is also an Ad5 mutant that lacks all of the E3 region except E3 12.5K and 3.6K proteins. Ad11 is a serotype B adenovirus and was generously provided by André Lieber, University of Washington. Ad5/35-GFP (where GFP is green fluorescent protein), also provided by André Lieber, is a first-generation Ad5 vector that has deletions of E1 and E3 and contains a chimeric Ad5/35 fiber gene that retains noncoding sequences from Ad5 and the amino-terminal tail (first 44 amino acids) of the Ad5 fiber. The remaining 323 amino acids of the fiber (shaft and knob) are derived from Ad35 (32). AdCMV-GFP, a replication-defective virus in which the immediate early gene E1A is deleted and replaced with the GFP gene driven by the cytomegalovirus (CMV) promoter, was obtained from James DeGregori (18). Ad007EGFP, a replication-competent Ad5 virus with an intact E1 region in which the enhanced GFP (EGFP) cDNA has replaced all of E3 except for the adenovirus death protein and the 12.5K protein, was the generous gift of Baoling Ying and William Wold (St. Louis University) (B. Ying and W. Wold, unpublished data). 12S-WT, an Ad5dl309 mutant that expresses only 12S but not the 13S isoform of E1A (21), was the generous gift of E. Moran (Cold Spring Harbor Laboratories, NY). The H5wt300-ΔpTP mutant adenovirus (the generous gift of Jerry Schaack, University of Denver) expresses all early adenovirus genes except E2B terminal protein and therefore does not replicate viral DNA or produce late viral proteins (24, 30). Virus titer was determined by plaque assay with 293 cells.
Infection of lymphocytes with adenovirus.
Infection of lymphocyte cell lines with adenovirus was performed as described previously (19) with minor modifications. Lymphocytes were collected and washed in serum-free (SF) RPMI medium. Cell density was adjusted to 107 cells/ml in SF-RPMI medium. Virus was added to the cell suspension at 100 PFU/cell and incubated at 37°C for 3 h. The infected cells were washed three times with RPMI complete medium and then resuspended in RPMI complete medium at 5 × 105 cells/ml.
PCR quantitation of viral DNA.
A total of 1 × 106 to 5 × 106 infected or control uninfected cells were washed in phosphate-buffered saline (PBS) and lysed in 100 μl of NP-40-Tween lysis buffer containing proteinase K, as described previously (13). Aliquots of 5 μl were tested by quantitative real-time PCR specific for a region of the hexon gene that is conserved among species C adenovirus serotypes by a previously described method (12) with primers P1 and P3 and probe P3 as described by Garnett et al. (13). Serial 10-fold dilutions (from 5 × 107 to 1 copy) of Ad2 DNA (Invitrogen, Carlsbad, CA) were included in each run to generate a standard curve. Analyses were done in triplicate and are reported as average values.
Limiting dilution analysis.
Limiting dilution analysis was used to estimate the frequency of viral DNA-containing cells. Adenovirus-infected lymphocytes were washed and diluted with uninfected cells to a final concentration of 1 × 105 cells/well in a ThermoGrid PCR plate (Denville Scientific, Metuchen NJ). DNA was extracted from individual wells as described above. A sensitive nested PCR assay for species C hexon was used to detect the presence of viral DNA in individual wells as described previously (13). Outer and inner primers were the pair P7 and P8 and the pair P1 and P2 as described previously (13).
Antibodies and flow cytometry.
The monoclonal mouse antibody (clone RmcB) to CAR was obtained from James DeGregori (18). The mouse monoclonal antibody to the adenovirus hexon protein (catalog number MAB8051) was purchased from Chemicon International/Millipore (Billerica, MA). The mouse monoclonal antibody to adenovirus fiber (ab3233) was purchased from Abcam, Inc. (Cambridge, MA). For flow cytometry, the secondary antibody was a goat F(ab′)2 fragment specific for mouse immunoglobulin conjugated to phycoerythrin (PE) (catalog number 1032-09) and was purchased from Southern Biotech (Birmingham, AL). Surface staining for CAR expression was performed by incubating live cells with an isotype control or the anti-CAR antibody at a 1:20 dilution on ice for 30 min. After being washed, cells were incubated with the PE-conjugated secondary antibody (2.5 μg/ml) for 30 min on ice, washed, and then analyzed on a FACSCalibur flow cytometer (fluorescence-activated cell sorter [FACS]) with Cell Quest software (Becton Dickinson Biosciences, San Jose, CA). Intracellular staining for hexon and fiber was performed essentially as described by Weaver and Kadan (37). Primary antibodies were diluted into 100 μl of FACS buffer (PBS with 2% FCS) before being added to the permeabilized cells. Staining was performed at 25°C for 30 min, followed by two washes with FACS buffer. PE-conjugated secondary antibody was added to the cells and incubated at room temperature for 30 min, followed by washing with FACS buffer and analysis by flow cytometry as described above. Hexon-positive cells were tabulated in the region defined by M2 as shown in the representative analysis of infected A549 cells (see Fig. S1 in the supplemental material).
Transduction of lymphocyte cell lines with CAR-expressing retrovirus.
Transduction of human lymphocyte cell lines with a CAR-expressing retrovirus was performed essentially as described before with a few modifications (19). In brief, Phoenix packaging cells were subcultured onto 100-mm culture dishes the day before transduction. Control vector LXSN or expression vector LXSN-CAR (from James DeGregori, University of Colorado Health Sciences) (18), both containing the neo resistance gene, were transfected into Phoenix cells using Trans IT-293 (Mirus Bio, Madison, WI) according to the manufacturer's instructions. Forty-eight hours after transduction, supernatant containing the CAR-expressing retrovirus was collected, filtered through a 0.45-μm-pore-size filter, and stored at −70°C. A total of 2 × 106 BJAB or REH cells in six-well culture plates were transduced with 0.5 ml of CAR-expressing retrovirus stock containing 8 μg/ml Polybrene (H9268; Sigma, St. Louis, MO) by centrifugation at 500 × g for 45 min at room temperature. Cells were returned to culture, and G418 (CellGro, Manassas, VA) was added to 1 mg/ml after 48 h. The transduced cells were kept in selection medium for at least 2 weeks. Cell surface CAR expression was confirmed with RmcB antibody staining. CAR-expressing REH cells were sorted to select for the population producing high levels of CAR by using a MoFlo cell sorter (Dako North America, Carpinteria, CA) at the Yerkes Regional Primate Center. Postsorting analysis showed that 97.6% of the cells were CAR positive. Transduced BJAB cells were >95% CAR positive and were used without further selection.
Southern blot analysis of viral DNA.
BJAB cells, infected as described above, were subcultured every 3 to 5 days in growth medium. At the indicated times postinfection, 5 × 106 infected cells were harvested, and total cellular DNA was collected using a GenElute Mammalian Genomic DNA miniprep kit (Sigma). Viral DNA (Ad5dl309 or Ad2) was purified from cesium-banded virus as a positive control. Total cellular DNA from infected BJAB cells (10 μg) or 500 ng of purified viral DNA was cleaved with restriction enzymes, as indicated on the figure, and the reaction mixture was separated by electrophoresis through a 0.7% agarose gel. The digested DNA fragments were transferred to a nylon support (Zeta-Probe; Bio-Rad, Hercules, CA) and UV cross-linked to immobilize the DNA. Fifty nanograms of viral DNA was radioactively labeled using a Prime-It II random primer labeling kit in the presence of [α-32P]dCTP and purified by a QiaQuick Nucleotide Removal Kit (Qiagen, Valencia, CA). Hybridization was performed by using Stratagene's Quickhyb hybridization solution. The viral DNA bound to the radioactive probe was visualized by exposure to X-ray film.
RT-PCR analysis of CAR expression.
BJAB cells were infected with Ad5dl309 [BJAB(dl309) cells] as described above. Total RNA was extracted from infected cells using TRIzol reagent (Invitrogen). RNA was converted to cDNA and amplified using a One-Step RT-PCR (reverse transcription-PCR) kit (Qiagen). One microgram of total RNA from uninfected or infected BJAB cells was reverse transcribed into cDNA, followed by PCR to amplify the CAR transcript. The primers used for the human CAR mRNA were the following: forward, 5′-AGGAGCGAGAGCCGCCTAC-3′, and reverse, 5′-ACGGAGAGCACAGATGAGACA-3′. The expected size of the CAR transcript was 1.1 kb (35). β2-Microglobulin mRNA was amplified to serve as an RNA input control yielding a 295-bp product. The primers for β2-microglobulin were the following: forward, 5′-TGCCTGCCGTGTGAAC-3′, and reverse, 5′-AGCCCTCCTAGAGCTAC-3′. RNA extracted from A549 cells or CAR gene-transduced KE37 cells (19) was used for a positive control.
Western blot analysis.
A total of 5 × 106 uninfected or infected BJAB cells were collected and washed in PBS, cell pellet was resuspended in 1 ml of Laemmli sample buffer (Bio-Rad) and sonicated briefly. The lysate was boiled for 10 min before separation by electrophoresis through an SDS-PAGE gel. The separated proteins were transferred to an Immobilon polyvinylidene difluoride (PVDF) membrane (IPFL00010; Millipore). Immunoblotting of CAR protein was carried out by using a CAR specific-antibody from Santa Cruz (sc-15405). Antibody to actin (sc-8432; Santa Cruz) was used as a loading control. Proteins were visualized with ECL Western Blotting Detection Reagents (RPN2109; GE Healthcare) used in accordance with the manufacturer's instructions.
RESULTS
Human B- and T-cell lines are permissive for adenovirus infection.
Our previous studies have demonstrated that T lymphocytes in human tonsil and adenoid tissues harbor adenovirus DNA (12). To further understand the infectious cycle of adenovirus in human lymphocytes, two T-cell lines, Jurkat and KE37(DSMZ), and two B-cell lines, BJAB and Ramos, were infected with the phenotypically wild-type Ad5dl309 at 100 PFU/cell. Growth medium was replenished twice during the 10 days of infection. Virus infection was monitored by flow cytometry for expression of the late protein hexon (Fig. 1 A; see Fig. S2 in the supplemental material). All four lymphocyte cell lines expressed hexon in >95% of cells, reaching this level of expression within 3 to 7 days after infection. Viral DNA levels were measured by quantitative, real-time PCR during the same 10-day period postinfection (Fig. 1B). At the peak of expression, viral DNA had increased 2 to 3 orders of magnitude. In three of the four cell lines, viral DNA content reached 100,000 or more genome copies per cell. The fourth cell line, KE37(DSMZ), replicated virus DNA to about 5,000 copies per cell from an initial 200 copies at the time of infection. However, in other experiments, this cell line accumulated adenovirus DNA up to levels in the range of 30,000 to 50,000 copies per cell. Thus, robust viral DNA replication was observed in all four lymphocyte cell lines.
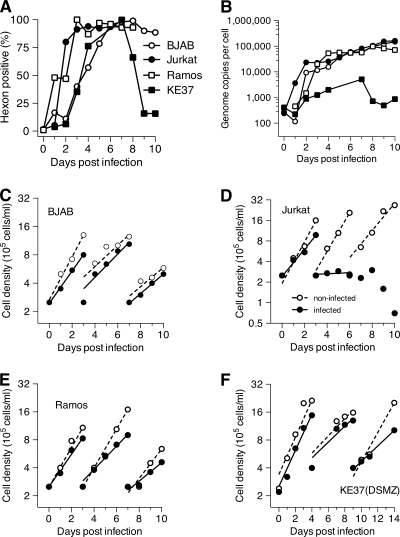
FIG. 1.
T- and B-lymphocyte cell lines support adenovirus productive infection. Two human T-cell lines, Jurkat and KE37(DSMZ), and two human B-cell lines, BJAB and Ramos, were infected with adenovirus Ad5dl309 at 100 PFU/cell. (A) Viral late gene expression was monitored for 10 days postinfection by intracellular staining for the hexon protein using antihexon antibody from Chemicon (MAB8051) and a secondary antibody, R-PE-goat F(ab′)2 anti-mouse IgG from Southern Biotechnologies. (B) Viral DNA replication was quantified by real-time PCR. (C to F) Growth and viability of infected cells (closed symbols) and uninfected cells (open symbols) were monitored over the 10-day period. The number of viable cells was determined daily. Cells were subcultured at 2.5 × 105 viable cells/ml on the indicated day. Lines represent a best-fit exponential regression.
The growth rates and viability of the four infected cell lines, along with uninfected control cultures, were monitored continuously. Cell density was adjusted to the initial 2.5 × 105 cells/ml every 3 to 4 days. Three out of four productively infected cell lines, BJAB (Fig. 1C), Ramos (Fig. 1E), and KE37(DSMZ) (Fig. 1F), remained viable and continued to divide at nearly the same rate as the mock-infected cells. Jurkat cells (Fig. 1D) showed a substantial decline in growth rate at 3 days postinfection, had ceased to divide shortly after the onset of late viral gene expression, and had begun to die before the end of the 10-day period.
Ad5dl309-infected lymphocyte cell lines can be maintained for long periods in vitro.
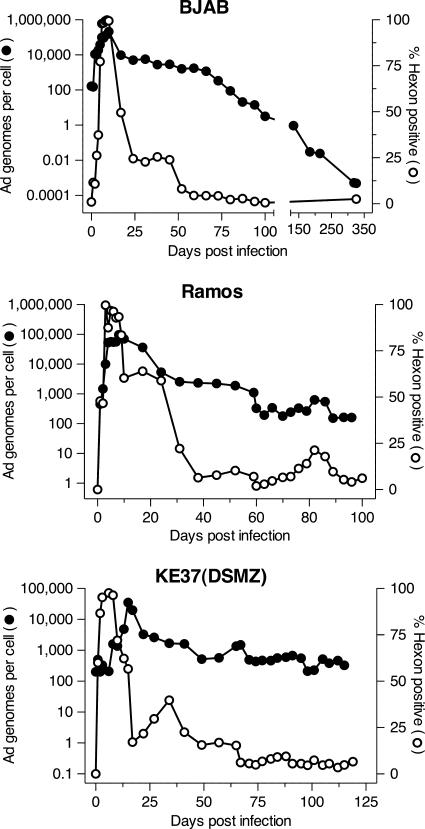
The three infected cell lines that continued to divide following adenovirus infection, BJAB, Ramos, and KE37(DSMZ), were maintained in culture and monitored periodically for viral DNA levels and hexon expression (Fig. 2). In BJAB cells, viral DNA levels dropped precipitously within 2 weeks of initial infection, followed by a gradual loss of genomes over the following 3 months. Similarly, hexon protein expression dropped abruptly by day 23 after peaking around day 10. One-quarter to one-fifth of cells continued to express hexon through day 45 to 50, after which hexon levels fell below the limits of detection and did not return throughout the observation period (up to day 325 postinfection) (Fig. 2 and data not shown). Similar levels of another viral late protein fiber showed a similar time course of expression in infected BJAB cells (data not shown). Thus, there was an extended period of time, from day 50 to 88 postinfection, when the cultures contained between 10 and 1,000 copies of the viral genome per cell and no detectable expression of at least two late proteins, hexon and fiber. In contrast, following the initial peak of viral DNA, infected Ramos and KE37(DSMZ) cells settled to a higher level of genome copies (between 100 and 1,000/cell) and maintained that level for the entire observation period without a perceptible trend downward. In addition, both infected Ramos and KE37(DSMZ) cells experienced brief periods of rising hexon expression at times after expression had fallen below the limit of detection of the FACS assay. This phenomenon was never observed with infected BJAB cells.
FIG. 2.
Long-term monitoring of Ad5dl309-infected lymphocyte cell lines. Ad5dl309-infected BJAB, Ramos, and KE37(DSMZ) cells were maintained in complete growth medium and subcultured every 3 to 4 days. Periodically, late viral protein hexon expression was measured by flow cytometry, and viral DNA levels were measured by real-time PCR as described in Materials and Methods.
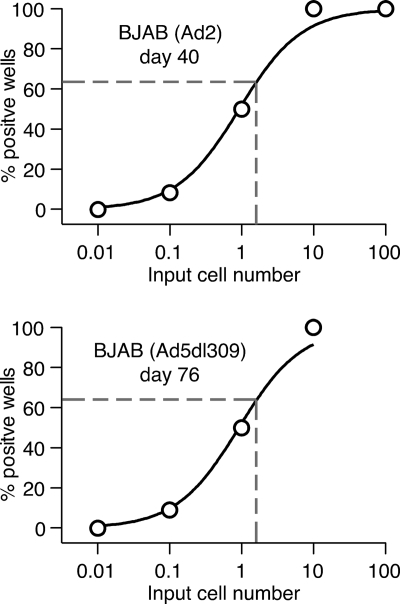
The finding that adenovirus-infected lymphocytes maintain viral DNA over prolonged periods of time in the absence of measurable late viral gene expression suggests that in these cells the viral genome enters a quiescent state, and virion protein synthesis is shut off, establishing a persistently infected state. Alternatively, it is possible that following initial infection, the majority of infected cells died, and the surviving cell population was dominated by uninfected cells, with a small population of infected cells containing large amounts of viral DNA. To differentiate between these two possibilities, a limiting dilution assay was used to determine the proportion of viral DNA-bearing cells. BJAB cells were previously infected with wild-type Ad2 (40 days) or Ad5dl309 (76 days) (Fig. 3). From Poisson considerations, the dilution at which viral DNA is detected in 63% of the wells provided an average of one viral DNA-positive cell per well. The data shown in Fig. 3 were fit by logistic regression to determine this value. For both cell lines analyzed in Fig. 3, approximately 1 in 1.8 cells (95% confidence interval [CI] ∼0.3 to 9.8) was determined to contain adenovirus DNA. These results led us to conclude that most infected BJAB cells harbor viral DNA for extended periods of time postinfection.
FIG. 3.
Most infected BJAB cells harbor viral DNA months following initial infection. BJAB cells were infected with Ad2 or Ad5dl309 for 40 days or 76 days, respectively. The infected cells were used for limiting dilution analysis to estimate the frequency of virus-bearing cells. A master lysate plate was prepared by diluting infected BJAB cells at 1,000, 100, 10, 1.0, and 0.1 cells per well with uninfected cells so that the total equaled 1 × 105 cells/well. Twelve wells for each dilution were tested. The total sample in each well was digested with proteinase K at 55°C for 9 h, followed by inactivation at 95°C for 15 min. Nested PCR for the hexon gene was performed, and the PCR products were visualized on a 1% agarose gel. The percentage of hexon-positive wells was plotted with a best-fit three-parameter logistic fit constrained between 0% and 100%. Dashed lines indicate the number of cells added per well that yield 63% positive wells, which corresponds to one adenovirus DNA-positive cell per well.
Most adenovirus genomes remain episomal in persistently infected BJAB cells.
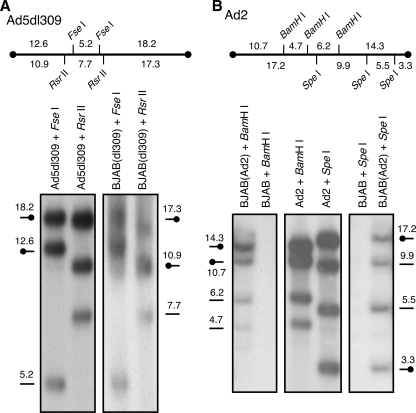
One possible way in which the adenovirus genome could persist over a long period in cultured cells is for all or part of the genome to become integrated into the host cell genome. To test whether the adenovirus genome remains free and monomeric or is integrated in persistently infected BJAB cells, Southern blot analysis was performed on BJAB cells infected with Ad2 for 234 days or Ad5dl309 for 180 days. DNA was extracted, digested with restriction endonucleases, and separated by electrophoresis as described in Materials and Methods. Purified DNA from Ad2 or Ad5dl309 was radiolabeled with [32P]dCTP and used as the probe for hybridization. As illustrated in Fig. 4, viral DNA fragments extracted from two independently derived, persistently infected cell lines are indistinguishable from fragments produced by purified viral DNA and predicted by the sequence of the genome. This finding indicates several things about the viral DNA in these cells. First, because all predicted fragments were detected, the viral genome appeared to be generally complete, with no large regions missing or duplicated. In addition, the majority of the DNA had not been rearranged, with restriction fragments remaining the same size as the free virus. Because the end fragments are discrete and of the predicted size, most of the DNA is not covalently connected to cellular DNA. If integration had occurred at a unique site and if that cell population had spread through the culture, then a new fragment would appear that contains both viral and cellular DNA. If multiple integration events had occurred, then the end fragments would occur as additional discrete fragments or be too heterogeneous to detect. Finally, the viral DNA remains monomeric and is not concatenated, which would have resulted in new fragments appearing due to head-to-head, tail-to-tail, or head-to-tail multimers. These results indicate that, even after more than 6 months postinfection, the viral DNA appears to remain episomal in persistently infected BJAB cells.
FIG. 4.
Southern blot analysis of adenovirus DNA in persistently infected BJAB cells. BJAB cells were infected with wild-type adenovirus Ad2 for 234 days or Ad5dl309 for 180 days before total infected cell DNA was extracted. (A) For Ad5dl309-infected BJAB cells, 500 ng of purified Ad5dl309 or 10 μg of total cellular DNA was digested with restriction enzymes FseI or RsrII, separated by electrophoresis, and hybridized with 32P-radiolabeled Ad5dl309 DNA. (B) For Ad2-infected BJAB cells, purified Ad2 DNA or 10 μg of total cellular DNA was cleaved with BamHI or SpeI before analysis. The predicted restriction fragments are indicated in kbp above each blot, and at the approximate position is indicated along the blots. Terminal fragments are indicated by symbols with the closed circle.
Persistently infected BJAB and Ramos cells produce small amounts of infectious virus.
Infected BJAB and Ramos cells were collected weekly, and plaque assays were performed on permissive A549 cells. Viral DNA replication was quantified in parallel by real-time PCR. Table 1 demonstrates that in BJAB cells, virus production decreased rapidly after 2 weeks of infection, and virus production continued for weeks at very low levels. The infected Ramos cells also produced infectious virus at a very low but detectable level even after a prolonged period of infection. In both cases, the low-level production of infectious virus could reflect a very low rate of virus assembly and release from many infected cells, or it could be due to a rare event that leads to production of large amounts of virus from a small number of cells. At present, these possibilities cannot be distinguished.
TABLE 1.
Virus production by adenovirus-infected B-cell linesa
| Cell line | No. of days postinfection | Virus production |
||
|---|---|---|---|---|
| No. of genomes/cell | No. of PFU/cell | No. of genomes/PFU | ||
| BJAB | 1 | 10 | 32 | 5 |
| 14 | 9,800 | 6 | 1,600 | |
| 21 | 4,400 | 6 | 700 | |
| 35 | 3,600 | 5 | 700 | |
| 42 | 2,600 | 2 | 1,300 | |
| 49 | 1,900 | 2 | 1,000 | |
| Ramos | 24 | 5,400 | 1 | 5,400 |
| 31 | 2,600 | 2 | 1,300 | |
| 45 | 2,200 | 2 | 1,100 | |
| 59 | 1,100 | 2 | 550 | |
| 73 | 4,100 | 1 | 4,100 | |
Cells were infected with Ad5dl309 at 100 PFU/cell. Infected cells were collected weekly. Plaque assays were performed on permissive A549 cells. Viral DNA was quantified by real-time PCR.
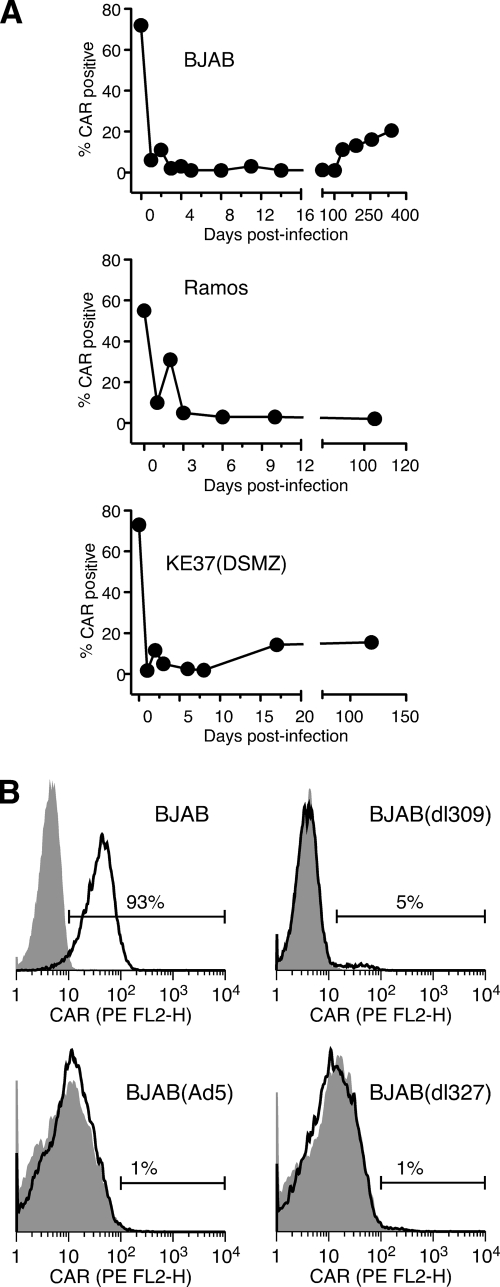
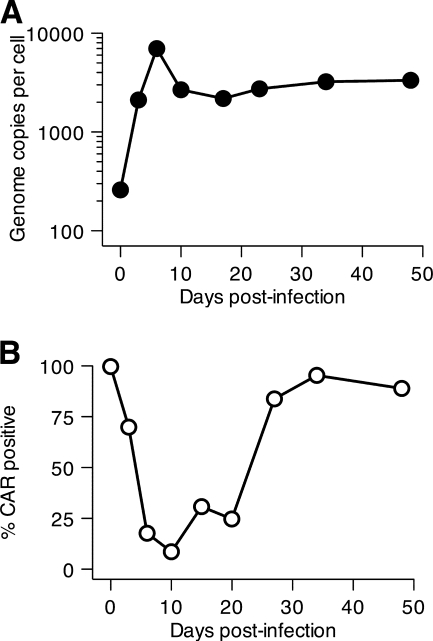
Cell surface CAR becomes rapidly undetectable from adenovirus-infected B and T lymphocytes.
As shown in Fig. 2, over time after infection with Ad5dl309, a decreasing fraction of BJAB cells retained viral DNA. This was surprising, given the presence of low levels of infectious virus, which should lead to reinfection of cells and prevent total loss of viral DNA from these cultures. Species C adenoviruses require the coxsackie B virus and adenovirus receptor (CAR) for infection of most cells (reviewed in reference 27). All four of the cell lines used in these studies express moderate to high levels of CAR, as measured by immunofluorescence on intact cells (Fig. 5 A) (19). Following infection with Ad5dl309, however, all four of the cell lines were observed to lose expression of CAR. As illustrated in Fig. 5A, loss of CAR was seen within 18 to 24 h postinfection, the first time point evaluated for all three cell lines that undergo persistent infection. For all cell lines analyzed, the fraction of CAR-positive cells remained low for at least 100 days after infection (Fig. 5A). The fraction of CAR-positive BJAB cells increased only after 100 days. Although the basis for this increase is unknown, it could reflect the expansion of a small fraction of CAR-positive cells with a growth advantage over CAR-negative cells.
FIG. 5.
Cell surface CAR is lost from cells infected with adenovirus. (A) BJAB, Ramos, and KE37(DSMZ) cells were infected with Ad5dl309 at 100 PFU/cell. The fraction of cells expressing CAR on their surface was monitored by flow cytometry at the indicated times postinfection. (B) BJAB cells were left uninfected or were infected with wild-type adenovirus Ad5 or Ad5dl327 for 20 days. Ad5dl309-infected BJAB cells were kept in culture medium for 75 days. Cell surface CAR was measured by flow cytometry using the RmcB mouse monoclonal antibody and a secondary antibody conjugated to phycoerythrin (PE), as described in Materials and Methods. The shaded area represents staining with secondary antibody alone; the solid line indicates staining with anti-CAR.
Ad5dl309 was used as the wild-type virus in these studies. This virus contains a deletion in the E3 region (16). To exclude the possibility that cell surface CAR loss was associated with loss of some E3 gene function, BJAB cells were infected with a true wild-type adenovirus, Ad5, or a mutant that lacks nearly the entire E3 region, Ad5dl327. Infected cells were stained for the cell surface CAR level on day 20 postinfection (Fig. 5B). Cells infected with either Ad5 or the E3 deletion mutant showed profound loss of surface CAR expression, indicating that the loss of CAR is independent of E3 gene activity.
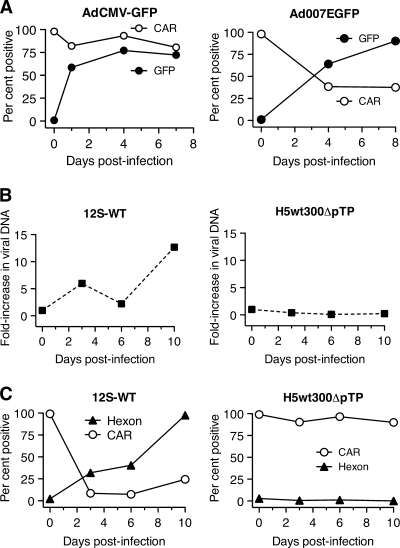
Attachment of species C adenoviruses to most cells occurs when the virion protein fiber binds to CAR on the cell surface (3). Thus, the rapid loss of CAR from the surface of infected cells could be due to binding and internalization of the CAR-fiber complex during infection. To explore this possibility, BJAB cells were infected with two GFP-expressing mutant viruses, AdCMV-GFP and Ad007EGFP. AdCMV-GFP is a replication-defective virus that lacks the E1 region (18). Ad007EGFP is a replication-competent virus in which the E1 region remains intact and the GFP gene replaces most of E3 (W. Wold, unpublished data). Figure 6 A shows that replication-competent Ad007EGFP resulted in decreased cell surface CAR in infected BJAB cells, but AdCMV-GFP did not. Both viruses were used at a multiplicity of 1,500 virus particles/cell, indicating that binding and internalization during initial infection are not sufficient to cause the loss of surface CAR.
FIG. 6.
Loss of cell surface CAR protein requires late events in the virus life cycle. (A) BJAB cells were infected with replication-defective AdCMV-GFP or replication-competent Ad007EGFP virus. The fraction of cells containing CAR on their surfaces was determined by flow cytometry. The fraction of cells that were productively infected was measured by GFP fluorescence using flow cytometry at the indicated times postinfection. (B) BJAB cells were infected with adenovirus 12S-WT or H5wt300-ΔpTP and harvested at 3, 6, or 10 days postinfection. Viral DNA replication was quantified by real-time PCR. (C) The infected BJAB cells analyzed in panel B were evaluated for hexon expression by intracellular staining and cell surface CAR expression with the RmcB antibody using flow cytometry.
Other potential mechanisms for the initial loss of CAR from infected BJAB cells are sequestration due to binding of CAR with newly synthesized fiber protein or downregulation of CAR synthesis by an E1 protein. The Ad007EGFP virus initiates viral DNA synthesis and late gene expression in these cells as expected, but the E1-deficient AdCMV-GFP virus does not (data not shown). To distinguish between these possibilities, BJAB cells were infected with an wild-type E1 virus that is deficient in virus DNA replication and late gene expression due to mutation of the terminal protein gene (H5wt300-ΔpTP) (24) (Fig. 6B and C). Cells infected with H5wt300-ΔpTP fail to express late genes (hexon in Fig. 6C) and fail to cause loss of surface CAR from the cells, supporting the hypothesis that sequestration or masking of CAR causes the apparent loss from the cell surface. Products of both the 12S and 13S E1A mRNAs promote viral gene expression and virus replication (39). However, the 13S product can modulate the transcription of cellular genes (26). A mutant virus that expresses only the 12S E1A product was evaluated to determine if changes in CAR levels were due to transcriptional effects that could be attributed to the 13S product. This mutant caused nearly complete loss of cell surface CAR. Together, these results suggest that late viral gene expression is essential for the apparent loss of cell surface CAR.
Ad11 infection did not result in loss of cell surface CAR.
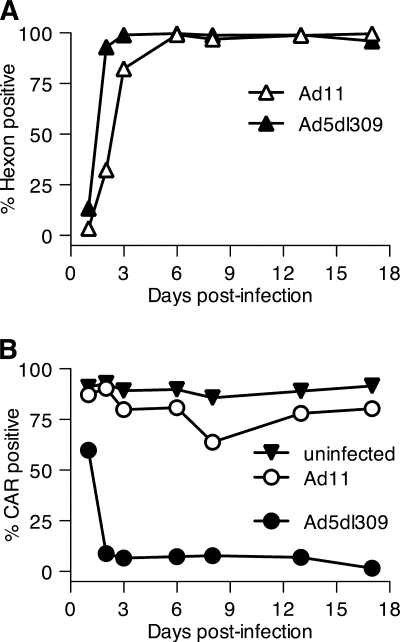
To test further the hypothesis that early loss of cell surface CAR was the result of binding by fiber protein, cells were infected with Ad11, a species B serotype whose fiber protein does not bind CAR (29). Instead, Ad11 uses CD46 as its receptor for cellular entry (11, 31). These experiments were performed in Ramos cells, which express abundant levels of both CAR and CD46 (data not shown). As shown in Fig. 7 A, Ad11 rapidly established productive infection of Ramos cells, as measured by late gene (hexon) expression. In contrast, Ad11 infection caused little or no loss of surface CAR during 17 days of infection (Fig. 7B). Thus, loss of surface CAR correlates with synthesis of a fiber protein that can bind CAR and either sequester it within the cell or mask recognition by the RmcB antibody.
FIG. 7.
Cell surface CAR level remains high when Ramos cells are infected with Ad11. Ramos cells were infected with Ad5dl309 or Ad11 at 100 PFU/cell. (A) Hexon expression was detected by intracellular staining with antihexon antibody and flow cytometry. (B) Cell surface CAR protein was detected with the RmcB antibody and flow cytometry.
CAR loss also occurs on 293 cells and requires a CAR-binding fiber protein.
To evaluate further the basis for rapid CAR loss from cells following infection, E1-expressing 293 cells were infected with Ad5/35-GFP, which is an Ad5 virus containing a deletion of E1 and a hybrid fiber protein that does not bind CAR (Table 2). Ad5/35-GFP-infected cells express GFP and enter the late phase of infection (as measured by hexon expression) but fail to cause loss of surface CAR expression. This is in contrast to a similar E1 deletion virus, AdCMV-GFP, which also expresses GFP and enters the late phase of infection in 293 cells but does induce loss of CAR. Note that AdCMV-GFP induces CAR loss on 293 cells but not on BJAB cells (Fig. 6A) because 293 cells produce the E1 gene products necessary to permit the virus to enter the late phase of infection and produce CAR-binding fiber protein. Therefore, the initial loss of CAR expression from the surface of infected cells requires synthesis of a CAR-binding fiber protein and is not limited to infected lymphocytes.
TABLE 2.
Virus infectivity, cell surface CAR, and hexon production in 293 cellsa
| Virus | No. of days postinfection | Protein level (% positive) |
||
|---|---|---|---|---|
| GFP | CAR | Hexon | ||
| AdCMV-GFP | 0 | ND | 98 | ND |
| 3 | 95 | 15 | 56 | |
| 4 | 86 | 5 | 51 | |
| Ad007EGFP | 0 | ND | 95 | ND |
| 2 | 35 | 21 | 15 | |
| 3 | 85 | 18 | 71 | |
| Ad5/35-GFP | 0 | ND | 100 | ND |
| 2 | 100 | 90 | 76 | |
| 3 | 100 | 91 | 83 | |
293 cells were infected with the indicated E1-deleted, GFP- or EGFP-expressing virus at 100 PFU/cell. Cells were collected at the indicated day after infection and analyzed directly by flow cytometry for GFP or EGFP expression or after cell surface staining for CAR or by intracellular staining for hexon as described in Materials and Methods. Values are expressed as the percentage of positively stained cells compared to noninfected cells or an isotype control. ND, not done.
CAR mRNA and protein levels are downregulated at late times postinfection.
Although binding of CAR to fiber protein in infected cell cultures can account for the initial loss of CAR from the surface of cells, limited recovery of CAR expression is observed on infected lymphocyte cells for up to 300 days after infection (Fig. 5A), long after viral late gene products have ceased to be detected (Fig. 2). This suggests that other mechanisms are involved, particularly at late times postinfection. To test for transcriptional regulation of CAR in persistently infected lymphocytes, BJAB cells were infected with Ad5dl309, total RNA was extracted on day 3 or day 180 postinfection, and RT-PCR was performed (Fig. 8 A). RNA extracted from A549 cells and CAR-transduced KE37 cells (KE37-CARhi) (19) were used as positive controls. Amplification of the β2-microglobulin transcript was used as an RNA input control (Fig. 8A). Compared to uninfected BJAB cells, the level of CAR transcripts was not affected by viral infection on day 3 postinfection; however, CAR gene transcripts were barely detectable from BJAB cells infected with Ad5dl309 for 180 days. Western blot analysis confirmed reduced cellular CAR protein levels at late times in persistently infected cells (Fig. 8B). To determine the kinetics of loss of CAR gene transcripts, CAR transcripts were monitored over time postinfection. Figure 8C shows that while CAR transcripts are nearly normal in the first few days after infection, by 14 days CAR transcripts are decreased and are nearly undetectable by 32 days. At late times postinfection, lymphocyte cells remain resistant to reinfection with adenovirus, as illustrated in Fig. 8D, where BJAB(dl309) cells at 75 days postinfection resist superinfection with AdCMV-GFP.
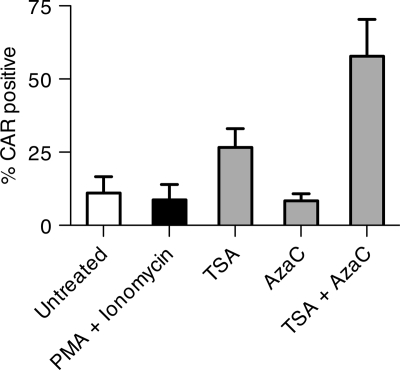
To test further the hypothesis that the CAR gene is transcriptionally downregulated, persistently infected BJAB cells were treated with several transcriptional regulators, including the histone deacetylase inhibitor trichostatin A (TSA), DNA methylase inhibitor 5′ azacytidine, and phorbol myristate acetate (PMA) and ionomycin for 40 h. Figure 9 shows that TSA alone causes some recovery of CAR expression, and a combination of TSA and 5′ azacytidine reverses most of the repression of CAR gene expression. This finding implies that epigenetic downregulation of the CAR gene promoter is involved in suppression of CAR expression in persistently infected lymphocytes.
FIG. 9.
Epigenetic control of CAR expression on persistently infected BJAB cells. Ad5dl309-infected BJAB cells at day 183 postinfection were treated separately with phorbol-12-myristate-13-acetate (PMA; 6 nM) plus ionomycin (0.75 μM), TSA (300 nM), 5′ azacytidine (AzaC; 10 μM), and TSA (300 nM) plus AzaC (10 μM). The fraction of CAR-positive cells after 40 h of treatment was determined with the RmcB antibody and flow cytometry and is expressed as the mean plus standard deviation of three experiments.
CAR expressed from an exogenous promoter is not repressed at late times after adenovirus infection.
The data presented above support a model in which CAR expression is suppressed on adenovirus-infected lymphocytes by two different mechanisms. At early times postinfection, CAR transcripts remain at or near normal levels, but the production and binding of viral fiber prevent CAR protein from being detected on the cell surface. At later times, when fiber production is lost in persistently infected cells, transcription of the CAR gene is suppressed, leading to loss of cell surface expression of CAR. To test this model further, a human lymphocyte cell line, REH, which expresses no CAR on the cell surface and is refractory to adenovirus infection (data not shown), was transduced with a retroviral vector containing the CAR gene driven by the mouse Moloney sarcoma virus long terminal repeat (LTR) promoter. REH-CAR cells were infected with Ad5dl309, and productive infection was confirmed by real-time PCR to quantify viral DNA replication (Fig. 10 A). Cell surface CAR expression was measured periodically for 50 days (Fig. 10B). As predicted, the cell surface CAR level was reduced immediately after infection, at a time when transcriptional control was not detected, presumably due to an interaction between newly synthesized fiber protein and CAR. However, unlike cells in which CAR is driven by the autologous promoter, CAR levels on REH-CAR cells recovered completely by 30 days after infection.
FIG. 10.
Cell surface CAR levels were not suppressed at late times postinfection with CAR expressed from a heterologous promoter. The precursor B-cell ALL line REH was transduced with a retrovirus expressing the human CAR gene from a heterologous promoter. The transduced REH-CAR cells were infected with Ad5dl309 at 100 PFU/cell, harvested at various times postinfection, and analyzed for viral genomes by real-time PCR (A) and cell surface CAR expression by flow cytometry (B).
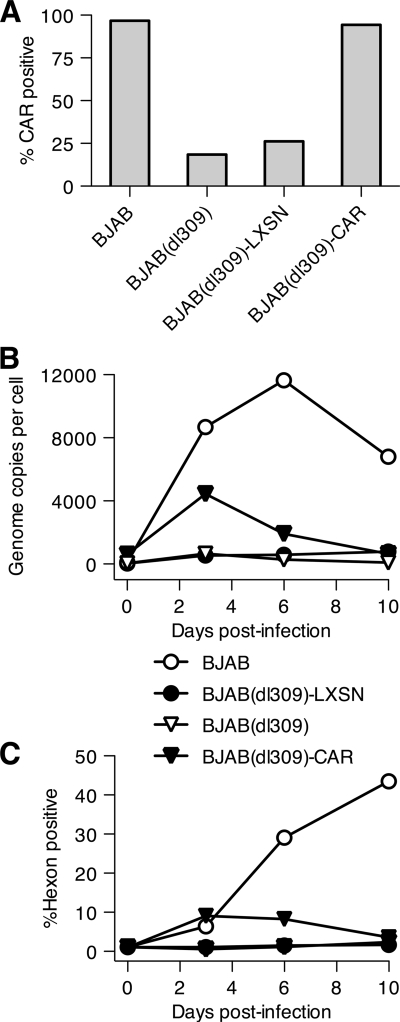
Reestablishing CAR expression in persistently infected BJAB cells did not restore productive infection.
The absence of CAR from persistently infected BJAB cells could be sufficient to prevent reinfection of the cells. To determine whether loss of CAR is the only cellular change leading to resistance to infection, persistently infected BJAB cells were transduced with either an empty retroviral vector (LXSN) or the same vector encoding CAR. The transduced cells were maintained under selection for the integrated retrovirus for 40 days. CAR expression was confirmed by staining with the RmcB anti-CAR antibody. Figure 11 A shows that 99% of the transduced BJAB(dl309) cells were CAR positive, compared to less than 20% for untransduced BJAB(dl309) cells or BJAB(dl309) cells transduced with the control vector. Cells were infected with adenovirus, and viral DNA replication and late gene (hexon) expression were monitored (Fig. 11B and C). BJAB cells supported robust viral replication and late gene expression as expected, and BJAB(dl309) cells did not. Expression of CAR from the retroviral transgene produced only a small increase in virus replication and gene expression, both of which were rapidly terminated. This suggests that other cellular changes in addition to CAR downregulation in BJAB cells have occurred to transform these persistently infected BJAB cells into cells that can no longer support adenovirus replication.
FIG. 11.
Restoration of cell surface CAR levels in persistently infected BJAB cells does not render the cells able to support a productive infection. BJAB cells were infected with Ad5dl309 and maintained as a persistently infected culture for 190 days before being transduced with an empty retrovirus vector (LXSN) or a CAR-expressing retrovirus vector. The transduced BJAB cells were maintained under selection for the integrated retrovirus for an additional 40 days before being reinfected with Ad5dl309 at 100 PFU/cell. (A) The fraction of CAR-positive cells at the time of reinfection was determined by flow cytometry. (B) Reinfected cells were harvested at 3, 6, and 10 days after reinfection, and the level of viral DNA was quantified by real-time PCR. (C) Viral late protein expression was measured at the times indicated by intracellular staining for hexon and flow cytometry.
DISCUSSION
Following infection with a species C adenovirus, many human lymphoid cell lines, of both B- and T-cell origin, continue to divide while establishing control of virus gene expression. In the current study, when infected BJAB cells were maintained in culture long enough, the viral genome was eventually lost (Fig. 2). Between 10 and 24 days of infection, the viral genome was lost at a rate that could not be distinguished from dilution due to cell division. However, because limiting dilution measurements (Fig. 3) showed that each cell contained viral DNA after 76 days, the persistence of the viral genome at this time implies that viral DNA continued to replicate in the persistently infected BJAB cells. These findings suggest that adenovirus could persist in mucosal lymphoid tissues by at least two means. First, the quiescent viral genome could persist in nondividing lymphocytes indefinitely. In dividing lymphocytes, the viral genome could persist by replicating sufficiently to counter loss due to dilution.
One compelling reason to gain an understanding of this nonlytic infection is the likelihood that adenovirus gene products cause damage to the host cell genome. Adenovirus inactivates the cellular activities that preserve the integrity of the cellular genome via inhibition of double-stranded DNA break repair (38). Double-stranded DNA breaks arise during development of the immune system, as an error during DNA replication, or following exposure to a DNA-damaging agent. Species C adenoviruses express redundant and overlapping means of disrupting double-stranded DNA break repair, and it is undoubtedly these mechanisms that lead to the demonstrated “hit-and-run” transforming ability whereby adenoviral genes lead to cellular transformation without maintenance of the viral genome in the resultant tumor cells (23). While these functions are irrelevant to the lytic infection of epithelial cells where all infected cells die, they are of serious concern when infected lymphocytes have carried the viral genome and survived.
There are at least two scenarios in which long-lived adenovirus-infected lymphocytes might be encountered in human hosts. We have previously reported that normal human tonsils contain species C adenovirus DNA in the T-lymphocyte population (12). The half-life of virus DNA in children is 2.6 years, which probably means that virus-containing cells live for years in the host (13). Episodic shedding of virus in stool years after primary infection is likely to be the result of periodic activation of latently infected lymphocytes, leading to renewed viral replication and release (9, 10). Culture of adenovirus-containing tonsil lymphocytes with T-cell stimuli leads to activation of virus gene expression and DNA replication (13). In addition, we have postulated that adenovirus infection of lymphocytes is also encountered during the initiating step in development of acute lymphoblastic leukemia (ALL) in children (14). In this model, adenovirus infection of lymphoid precursor cells, with its attendant inhibition of DNA repair, leads to the genetic abnormalities commonly encountered in childhood ALL. These cancers are mostly of B-lymphocyte origin, providing the rationale for investigating adenovirus infection in human lymphocyte cell lines of both B- and T-cell origin.
The present study investigates the long-term interaction between infecting species C adenovirus and four lymphoid cell lines, two B-cell lines (BJAB and Ramos) and two T-cell lines [KE37(DSMZ) and Jurkat]. Among these four cell lines, three different phenotypes are observed. Following infection of Jurkat, cells rapidly cease division and die while producing large amounts of infectious virus (Fig. 1) (19). This infectious cycle is similar to that of fully permissive epithelial cells except that it occurs at a significantly slower pace, with cell death complete by 8 to 10 days. In the second phenotype, characterized by BJAB, cells are infected to high levels while continuing to divide at the same rate as uninfected cells. These cells produce large numbers of viral genomes but very little infectious virus and appear able to completely shut down the viral genome, as monitored by hexon gene expression and genome copy numbers per cell. When maintained for several months in culture, these cells lose the viral genome altogether.
The third phenotype of adenovirus-infected lymphocytes described here is seen in Ramos and KE37(DSMZ) cells. These cells continue to divide at nearly normal levels, despite rapid replication of the viral genome at early times postinfection. Like BJAB, these cells at their peak contain tens of thousands of copies of the viral genome but produce only very small amounts of infectious virus. Unlike BJAB, these cells maintain a steady-state level of the viral genome and low, but detectable levels of hexon expression over long periods of time. One possible reason for this difference is that these cells may be able to replicate the viral genome at a rate sufficient to maintain from a few hundred up to a thousand genomes per cell over prolonged periods. It is also possible that, unlike BJAB, Ramos and KE37(DSMZ) cells can be reinfected with virus that is released into the culture medium. In experiments similar to that shown in Fig. 5A with BJAB cells, Ramos and KE37(DSMZ) cells, at days 72 and 90 postinfection with Ad5dl309, respectively, were exposed to AdCMV-GFP, and GFP expression was followed by flow cytometry. KE37(DSMZ) cells, like BJAB, showed no measurable reinfection. Ad5dl309-infected Ramos cells, however, expressed GFP at a level only slightly lower than that of uninfected Ramos cells (data not shown). At present we do not know which of these scenarios is active in maintaining the levels of virus genome seen in these cells, and it is possible that both mechanisms contribute to maintenance of the viral genome in these cells.
This third phenotype of adenovirus-infected lymphocyte cell lines has been described previously by others. Flomenberg et al. isolated a lymphoblastoid cell line from a patient with a B-cell lymphoproliferative disease and adenovirus pneumonia (8). The cell line continued to proliferate normally while producing large quantities of infectious adenovirus (∼103 per cell) over a period of 6 months. Chu et al. (4) infected the human promonocytic cell line U937 with Ad5 and followed the resultant persistently infected cell line for more than a year. Like Ramos cells, U937 cells maintained a steady-state level of the viral genome while producing only very small quantities of infectious virus. Hence this phenotype, characterized by maintenance of the viral genome over long periods of time accompanied by production of low levels of infectious virus, has been encountered in cell lines of myeloid, B-lymphocyte, and T-lymphocyte origin.
The four human lymphocyte cell lines in this study were chosen because all four expressed high levels of CAR and all four were permissive for the entire virus life cycle. We have examined 29 human myeloid and lymphoid cell lines chosen at random for infection with species C adenoviruses and find that more than half (16, or 56%) permit virus DNA replication and late gene expression (data not shown). Of the 16 permissive cell lines, 6 die within a few days following virus infection, and 10 survive (data not shown). Hence, the behavior described here of nonlytic infection followed by persistence of the viral genome is common among human lymphoid and myeloid cell lines.
Within 24 h of infection, CAR can no longer be detected on the surface of all three persistently infected cell lines described here (Fig. 5A). CAR disappearance requires infection with a replicating virus that contains a CAR-binding fiber protein (Fig. 6 and 7 and Table 2). Because CAR protein and mRNA continued to be detected on day 3 postinfection (Fig. 8), the apparent loss of CAR on the cell surface may be due to intracellular sequestration or epitope masking. Although it is not known whether fiber protein can bind CAR within the cell and prevent its externalization, Rebetz and associates have recently characterized the disappearance of cell surface CAR and the species B adenovirus receptor, CD46, shortly after infection (28). These investigators suggested that secreted fiber protein from a small fraction of infected cells masks the cognate receptor in neighboring cells. These findings suggest one possible mechanism by which cell-surface CAR becomes undetectable following infection with adenovirus. In contrast, the failure of cell surface CAR to reappear at late times postinfection, when little or no late gene expression (either hexon or fiber [data not shown]) is detected, was due to shutdown of CAR gene transcription that was detected at around 30 days postinfection in BJAB cells (Fig. 8), i.e., at around the same time that viral late gene expression was being lost in this cell population (Fig. 2). CAR synthesis and expression remained repressed even after the viral genome was lost (Fig. 8 and data not shown), suggesting a virus-induced epigenetic change to the cells that does not require the continued presence of the virus. This was confirmed by treating BJAB cells at 183 days postinfection with inhibitors of DNA methylation and histone deacetylation, which in combination restored CAR to nearly normal levels (Fig. 9).
In Ad5dl309-infected BJAB cells, the fraction of CAR-positive cells began to increase only after about 100 days postinfection. It is possible that in the absence of sustained viral gene expression, epigenetic changes imposed by the virus are gradually reversed. However, this effect also could result from the outgrowth of a fraction of cells that expressed CAR. Data obtained with Ad5dl309-infected BJAB cells (Fig. 5A and data not shown) were modeled by an equation in which an initial fraction of CAR-positive cells was assumed to possess a growth advantage over CAR-negative cells. This analysis revealed that the observed outcome could occur if 1.6% of the cells were CAR positive (95% CI, 0.4% to 3.9%), and these CAR-positive cells required 3% less time (95% CI, 1.6% to 3.6%) to double than the CAR-negative cells. The results described here do not exclude either possibility.
CAR is not the only gene whose expression is modified following adenovirus infection of BJAB cells. Even when CAR levels were restored by transduction with a CAR-containing retrovirus, the previously infected cells could not be reinfected (Fig. 11). This suggests other changes to cellular gene expression in addition to suppression of CAR expression. At present we do not know the extent to which cellular gene expression was modified in these cells. We have previously shown that there are multiple mechanisms by which lymphoid cells resist adenovirus infection, some of which act after the virus has entered the cell (19). What is unique about BJAB cell cultures that have “recovered” from adenovirus infection (e.g., that have lost the viral genome) is that the nonpermissive phenotype of these cells was induced by the virus itself.
At present, the extent of changes in gene expression mediated by adenovirus in lymphocytes is not known. However, in preliminary experiments, postinfection BJAB cells not only lost CAR expression but also gained expression of CD20, a cell surface differentiation marker of B lymphocytes that is normally not expressed on BJAB cells (data not shown). If, indeed, lymphocyte differentiation markers are among the proteins whose expression is altered following adenovirus infection, then it follows that techniques that rely on these markers to identify virus-containing cells are of questionable significance. We have reported that adenovirus DNA can be detected in the T-cell subpopulation of lymphocytes in tonsils based on expression of CD4 and CD8 or CD3 (12). Adenovirus DNA was not detected in the B-cell population identified by expression of CD19 (12). If in B cells the virus inhibits expression of CD19, then this population of virus-bearing cells would have been missed in the cell separation analysis. Given that adenovirus forms persistent/latent infection in B-cell, T-cell, and myeloid cell lines in vitro, it remains possible that many different cell populations may carry the viral genome in naturally infected tonsils.
The picture that emerges from this and our previous study (19) is of a complex of possible interactions between species C adenoviruses and host cells of lymphoid origin. Many lymphoid cells lack CAR on the cell surface. Even after the virus enters the cell, some lymphoid cells can prevent even early virus gene expression (19). What is perhaps most startling is that some lymphoid cell lines permit full virus gene expression even to the point of virion production, but the cells themselves continue to proliferate. Despite this normal appearance, the cells display altered gene expression long after the virus is lost. The precise changes that have occurred that quiet the viral genome and alter cellular gene expression will be the subject of future investigation.
Supplementary Material
Acknowledgments
We thank Dean Erdman, Tom Shenk, André Lieber, Baoling Ying, William Wold, Betty Moran, and Jerry Schaack for kindly providing viruses and James DeGregori for providing antibodies and viruses that were used in this study.
This research was supported by grants R01 AI052280 from the National Institute of Allergy and Infectious Diseases and RO1 CA127621 from the National Cancer Institute.
The content of this report is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases, the National Cancer Institute, or the National Institutes of Health.
Footnotes
Published ahead of print on 23 June 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Avila, M., G. Carballal, H. Rovalett, et al. 1989. Viral etiology in acute lower respiratory infections in children from a closed community. Am. Rev. Respir. Dis. 140:634-637. [DOI] [PubMed] [Google Scholar]
- 2.Bell, J. A., R. J. Huebner, R. S. Paffenbarger, Jr., W. P. Rowe, R. G. Suskind, and T. G. Ward. 1956. Studies of adenoviruses (APC) in volunteers. Am. J. Public Health Nations Health 46:1130-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 4.Chu, Y., K. Sperber, L. Mayer, and M. T. Hsu. 1992. Persistent infection of human adenovirus type 5 in human monocyte cell lines. Virology 188:793-800. [DOI] [PubMed] [Google Scholar]
- 5.Colin, M., L. Renaut, L. Mailly, and J. C. D'Halluin. 2004. Factors involved in the sensitivity of different hematopoietic cell lines to infection by subgroup C adenovirus: implication for gene therapy of human lymphocytic malignancies. Virology 320:23-39. [DOI] [PubMed] [Google Scholar]
- 6.Edwards, K. M., J. Thompson, J. Paolini, and P. F. Wright. 1985. Adenovirus infections in young children. Pediatrics 76:420-424. [PubMed] [Google Scholar]
- 7.Evans, A. S. 1958. Latent adenovirus infections of the human respiratory tract. Am. J. Hyg. 67:256-266. [DOI] [PubMed] [Google Scholar]
- 8.Flomenberg, P., V. Piaskowski, J. Harb, A. Segura, and J. T. Casper. 1996. Spontaneous, persistent infection of a B-cell lymphoma with adenovirus. J. Med. Virol. 48:267-272. [DOI] [PubMed] [Google Scholar]
- 9.Fox, J. P., C. D. Brandt, F. E. Wassermann, C. E. Hall, I. Spigland, A. Kogon, and L. R. Elveback. 1969. The virus watch program: a continuing surveillance of viral infections in metropolitan New York families. VI. Observations of adenovirus infections: virus excretion patterns, antibody response, efficiency of surveillance, patterns of infections, and relation to illness. Am. J. Epidemiol. 89:25-50. [DOI] [PubMed] [Google Scholar]
- 10.Fox, J. P., C. E. Hall, and M. K. Cooney. 1977. The Seattle Virus Watch. VII. Observations of adenovirus infections. Am. J. Epidemiol. 105:362-386. [DOI] [PubMed] [Google Scholar]
- 11.Gaggar, A., D. M. Shayakhmetov, and A. Lieber. 2003. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 9:1408-1412. [DOI] [PubMed] [Google Scholar]
- 12.Garnett, C. T., D. Erdman, W. Xu, and L. R. Gooding. 2002. Prevalence and quantitation of species C adenovirus DNA in human mucosal lymphocytes. J. Virol. 76:10608-10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garnett, C. T., G. Talekar, J. A. Mahr, W. Huang, Y. Zhang, D. A. Ornelles, and L. R. Gooding. 2009. Latent species C adenoviruses in human tonsil tissues. J. Virol. 83:2417-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gustafsson, B., W. Huang, G. Bogdanovic, F. Gauffin, A. Nordgren, G. Talekar, D. A. Ornelles, and L. R. Gooding. 2007. Adenovirus DNA is detected at increased frequency in Guthrie cards from children who develop acute lymphoblastic leukaemia. Br. J. Cancer 97:992-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Israel, M. 1962. The viral flora of enlarged tonsils and adenoids. J. Pathol. Bacteriol. 84:169-176. [Google Scholar]
- 16.Jones, N., and T. Shenk. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17:683-689. [DOI] [PubMed] [Google Scholar]
- 17.Lavery, D., S. M. Fu, T. Lufkin, and S. Chen-Kiang. 1987. Productive infection of cultured human lymphoid cells by adenovirus. J. Virol. 61:1466-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leon, R. P., T. Hedlund, S. J. Meech, S. Li, J. Schaack, S. P. Hunger, R. C. Duke, and J. DeGregori. 1998. Adenoviral-mediated gene transfer in lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 95:13159-13164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNees, A. L., J. A. Mahr, D. Ornelles, and L. R. Gooding. 2004. Postinternalization inhibition of adenovirus gene expression and infectious virus production in human T-cell lines. J. Virol. 78:6955-6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meeker, T. C., L. T. Lay, J. M. Wroblewski, F. Turturro, Z. Li, and P. Seth. 1997. Adenoviral vectors efficiently target cell lines derived from selected lymphocytic malignancies, including anaplastic large cell lymphoma and Hodgkin's disease. Clin. Cancer Res. 3:357-364. [PubMed] [Google Scholar]
- 21.Moran, B., and B. Zerler. 1988. Interactions between cell growth-regulating domains in the products of the adenovirus E1A oncogene. Mol. Cell. Biol. 8:1756-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neumann, R., E. Genersch, and H. J. Eggers. 1987. Detection of adenovirus nucleic acid sequences in human tonsils in the absence of infectious virus. Virus Res. 7:93-97. [DOI] [PubMed] [Google Scholar]
- 23.Nevels, M., B. Tauber, T. Spruss, H. Wolf, and T. Dobner. 2001. “Hit-and-run” transformation by adenovirus oncogenes. J. Virol. 75:3089-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nichols, G. J., J. Schaack, and D. A. Ornelles. 2009. Widespread phosphorylation of histone H2AX by species C adenovirus infection requires viral DNA replication. J. Virol. 83:5987-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. U. S. A. 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pelka, P., J. N. Ablack, J. Torchia, A. S. Turnell, R. J. Grand, and J. S. Mymryk. 2009. Transcriptional control by adenovirus E1A conserved region 3 via p300/CBP. Nucleic Acids Res. 37:1095-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Philipson, L., and R. F. Pettersson. 2004. The coxsackie-adenovirus receptor—a new receptor in the immunoglobulin family involved in cell adhesion. Curr. Top. Microbiol. Immunol. 273:87-111. [DOI] [PubMed] [Google Scholar]
- 28.Rebetz, J., M. Na, C. Su, B. Holmqvist, A. Edqvist, C. Nyberg, B. Widegren, L. G. Salford, H. O. Sjogren, N. Arnberg, Q. Qian, and X. Fan. 2009. Fiber mediated receptor masking in non-infected bystander cells restricts adenovirus cell killing effect but promotes adenovirus host co-existence. PLoS One 4:e8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roelvink, P. W., A. Lizonova, J. G. Lee, Y. Li, J. M. Bergelson, R. W. Finberg, D. E. Brough, I. Kovesdi, and T. J. Wickham. 1998. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J. Virol. 72:7909-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaack, J., X. Guo, and S. J. Langer. 1996. Characterization of a replication-incompetent adenovirus type 5 mutant deleted for the preterminal protein gene. Proc. Natl. Acad. Sci. U. S. A. 93:14686-14691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Segerman, A., J. P. Atkinson, M. Marttila, V. Dennerquist, G. Wadell, and N. Arnberg. 2003. Adenovirus type 11 uses CD46 as a cellular receptor. J. Virol. 77:9183-9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shayakhmetov, D. M., T. Papayannopoulou, G. Stamatoyannopoulos, and A. Lieber. 2000. Efficient gene transfer into human CD34+ cells by a retargeted adenovirus vector. J. Virol. 74:2567-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silver, L., and C. W. Anderson. 1988. Interaction of human adenovirus serotype 2 with human lymphoid cells. Virology 165:377-387. [DOI] [PubMed] [Google Scholar]
- 34.Thimmappaya, B., C. Weinberger, R. J. Schneider, and T. Shenk. 1982. Adenovirus VAI RNA is required for efficient translation of viral mRNAs at late times after infection. Cell 31:543-551. [DOI] [PubMed] [Google Scholar]
- 35.Thoelen, I., C. Magnusson, S. Tagerud, C. Polacek, M. Lindberg, and M. Van Ranst. 2001. Identification of alternative splice products encoded by the human coxsackie-adenovirus receptor gene. Biochem. Biophys. Res. Commun. 287:216-222. [DOI] [PubMed] [Google Scholar]
- 36.van der Veen, J., and M. Lambriex. 1973. Relationship of adenovirus to lymphocytes in naturally infected human tonsils and adenoids. Infect. Immun. 7:604-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weaver, L. S., and M. J. Kadan. 2000. Evaluation of adenoviral vectors by flow cytometry. Methods 21:297-312. [DOI] [PubMed] [Google Scholar]
- 38.Weitzman, M. D., and D. A. Ornelles. 2005. Inactivating intracellular antiviral responses during adenovirus infection. Oncogene 24:7686-7696. [DOI] [PubMed] [Google Scholar]
- 39.Winberg, G., and T. Shenk. 1984. Dissection of overlapping functions within the adenovirus type 5 E1A gene. EMBO J. 3:1907-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.