Abstract
Tick-borne encephalitis virus (TBEV) (family Flaviviridae, genus Flavivirus) accounts for approximately 10,000 annual cases of severe encephalitis in Europe and Asia. Here, we investigated the induction of the antiviral type I interferons (IFNs) (alpha/beta IFN [IFN-α/β]) by TBEV. Using strains Neudörfl, Hypr, and Absettarov, we demonstrate that levels of IFN-β transcripts and viral RNA are strictly correlated. Moreover, IFN induction by TBEV was dependent on the transcription factor IFN regulatory factor 3 (IRF-3). However, even strain Hypr, which displayed the strongest IFN-inducing activity and the highest RNA levels, substantially delayed the activation of IRF-3. As a consequence, TBEV can keep the level of IFN transcripts below the threshold value that would permit the release of IFN by the cell. Only after 24 h of infection have cells accumulated sufficient IFN transcripts to produce detectable amounts of secreted IFNs. The delay in IFN induction appears not to be caused by a specific viral protein, since the individual expressions of TBEV C, E, NS2A, NS2B, NS3, NS4A, NS4B, NS5, and NS2B-NS3, as well as TBEV infection itself, had no apparent influence on specific IFN-β induction. We noted, however, that viral double-stranded RNA (dsRNA), an important trigger of the IFN response, is immunodetectable only inside intracellular membrane compartments. Nonetheless, the dependency of IFN induction on IFN promoter stimulator 1 (IPS-1) as well as the phosphorylation of the alpha subunit of eukaryotic initiation factor 2 (eIF2α) suggest the cytoplasmic exposure of some viral dsRNA late in infection. Using ultrathin-section electron microscopy, we demonstrate that, similar to other flaviviruses, TBEV rearranges intracellular membranes. Virus particles and membrane-connected vesicles (which most likely represent sites of virus RNA synthesis) were observed inside the endoplasmic reticulum. Thus, apparently, TBEV rearranges internal cell membranes to provide a compartment for its dsRNA, which is largely inaccessible for detection by cytoplasmic pathogen receptors. This delays the onset of IFN induction sufficiently to give progeny particle production a head start of approximately 24 h.
Tick-borne encephalitis virus (TBEV) is one of the most important arboviruses in Europe and Asia. In humans, it can cause severe encephalitis with a mortality rate of up to 30% and neurological sequelae in 30 to 60% of survivors (19, 23, 35, 45). The natural route of infection is by the bite of an infected tick or by the consumption of infected milk products. In regions of Europe where TBEV is endemic, between 0.5 and 5% of ticks carry the virus, but these numbers can be as high as 40% in certain regions of Russia (8, 19). TBEV belongs to the family Flaviviridae of the genus Flavivirus. Taxonomically, the species was recently divided into four virus types, namely, Louping ill virus, Western TBEV, Eastern TBEV, and Turkish sheep encephalitis virus (17). For unknown reasons, disease symptoms are strain dependent and vary from asymptomatic infection to severe encephalitis and meningitis (8). Although there is an effective vaccine against TBEV, the number of cases is on the upsurge and currently in the range of approximately 10,000 per year (8, 67).
Despite the tremendous medical importance of TBEV, many aspects of infection and pathogenesis remain unsolved so far. For example, the interactions of TBEV with the innate immune system, in particular the antiviral type I interferon (IFN) (alpha/beta IFN [IFN-α/β]) system, are not well characterized, although TBEV served as a model system in early pioneer studies of IFN (70). IFNs are synthesized and secreted by infected cells and cause neighboring cells to express antiviral factors. Tissue cells recognize virus infection mainly by intracellular pathways (50). Viral signature molecules such as double-stranded RNA (dsRNA) or RNAs bearing a 5′-triphosphate group are detected by cytoplasmic pathogen recognition receptors (PRRs) (55, 57, 80). Activated PRRs trigger a signaling chain that eventually results in the phosphorylation of the transcription factor IFN regulatory factor 3 (IRF-3) (24), a member of the IRF family, and phosphorylated IRF-3 moves into the nucleus, where it initiates IFN-β mRNA synthesis. Secreted IFNs bind to and activate a receptor that is present on virtually all host cells. IFN signaling via the so-called JAK/STAT pathway initiates the transcription of more than 300 IFN-stimulated genes (ISGs), which inhibit virus multiplication at the level of transcription, translation, genome replication, assembly, and exit and stimulate subsequent adaptive immune responses (61).
For several flaviviruses, including hepatitis C virus (HCV), West Nile virus (WNV), and dengue virus (DENV), it is known that the clearance of infection by the host is dependent on the IFN system (12, 16, 34, 38, 62). Consequently, these viruses express gene products that interfere with induction, signaling, or the action of IFNs, similar to what is known for other viruses (2, 11, 30, 51, 52, 58). For TBEV, however, neither the details of IFN induction nor potential anti-IFN strategies are known, with the notable exception of the NS5 protein, which was identified previously as an inhibitor of IFN-dependent signaling (3, 76). Here, we compared three different Western strains of TBEV for their abilities to induce IFNs. We observed that the activation of IRF-3 and the induction of IFN-β were not completely abrogated but were substantially delayed after infection and that the IFN induction by different strains was strictly coupled to the extent of viral RNA replication. Moreover, we were unable to identify a specific viral gene product that could explain the impediment of IRF-3 activation. Rather, viral dsRNA was found to be largely unavailable for cytoplasmic detection, most likely because TBEV rearranges intracellular membrane compartments and replicates inside them. Thus, TBEV appears to rely on a passive escape strategy in order to avoid premature IFN induction.
MATERIALS AND METHODS
Reagents, cells, and viruses.
Streptolysin-O (SL-O), poly(I:C), and brefeldin A (BFA) were purchased from Sigma. Microcrystalline cellulose Avicel RC/CL was purchased from FMC BioPolymer. Simian Vero B4 and Vero E6 cells, human lung carcinoma cells (A549), human embryonic kidney cells (293T), baby hamster kidney cells (BHK-21), and mouse embryonic fibroblasts (MEFs) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal calf serum (FCS). Human 293T cells transgenic for the Mx1 promoter-driven firefly luciferase gene (kindly provided by George Kochs) (28) were propagated in DMEM supplemented with 5% FCS and 0.5 μM G418. TBEV strains (Neudörfl, Hypr, and Absettarov) and Rift Valley fever virus strain clone 13 were propagated in Vero B4 or Vero E6 cells, respectively, under biosafety laboratory 3 (BSL-3) conditions.
Plasmid constructs.
The firefly luciferase (FF-Luc) reporter plasmid for monitoring IFN-β promoter activation (p-125Luc) was kindly provided by Takashi Fujita, Institute for Virus Research, Kyoto University, Kyoto, Japan (81). The control plasmid pRL-SV40 (Promega) contains the Renilla luciferase (Ren-Luc) gene under the control of the constitutive simian virus 40 (SV40) promoter. cDNA expression plasmids encoding each component of the viral polyprotein were constructed by using standard PCR cloning methods. KOD Hot Start polymerase (Novagen), restriction enzymes, and T4 DNA ligase (Fermentas) were used according to the manufacturers' recommendations. cDNA from TBEV Hypr-infected cells (71) was used as a template, and the PCR products encoding the different precursor TBEV proteins were cloned into the eukaryotic cloning vector pI.18 (kindly provided by Jim Robertson, National Institute for Biological Standards and Control, Hertfordshire, United Kingdom). All plasmids were sequenced to ensure correctness, and oligonucleotide primer sequences are available upon request.
Viral infection and dsRNA transfection.
Monolayers of cells grown in 12-well plates were incubated with viruses for 1 h at 37°C by using Opti-MEM (Invitrogen). The virus inoculum was removed, 1 ml of DMEM-2% FCS was added, and the incubation was continued at 37°C. For the transfection of cells with dsRNA, 5 μg of poly(I:C) was prepared with 5 μl of Metafectene liposomes (Biontex) in 200 μl of Opti-MEM according to the manufacturers' instructions. After 15 min of incubation, the dsRNA-liposome mixture was dropped onto cells without changing the medium.
RNA extraction and real-time RT-PCR.
Total cellular RNA was extracted at the indicated times postinfection (p.i.) by using the Nucleospin RNA II kit (Macherey-Nagel) according to the manufacturer's recommendations. Aliquots of 600 ng to 1 μg of RNA were used to synthesize cDNA with the Quantitect reverse transcription (RT) kit (Qiagen). mRNA levels of human γ-actin, IFN-β, ISG15, ISG56, interferon-inducible protein 10 (IP-10), and RANTES were detected with validated QuantiTect primers (Qiagen) and the QuantiTect SYBR green RT-PCR kit (Qiagen) by using a LightCycler 1.5 instrument (Roche). Viral RNAs were detected by using previously described TaqMan probes for TBEV (65) and Rift Valley fever virus (5) and the QuantiFast probe PCR kit (Qiagen). Signals of inducible cellular mRNAs or viral RNAs were normalized to the γ-actin mRNA signal.
Virus titrations.
Viral titers were determined by using a focus-forming assay (47). BHK-21 cells were seeded into 96-well dishes and infected with a 10-fold serial dilution of TBEV or clone 13 in a total volume of 50 μl of Opti-MEM. After 1 h the inoculum was removed, and a 100-μl overlay containing 1.25% Avicel RC/CL, 1× DMEM, and 0.5% bovine serum albumin (BSA) was added. The plates were incubated at 37°C for 48 h (TBEV) or 24 h (clone 13) before the fixation of cells with 3% paraformaldehyde dissolved in phosphate-buffered saline (PBS). Cells were permeabilized with PBS containing 0.5% Triton X-100 and 20 mM glycine. Viral foci were detected by using primary mouse antibodies directed against the TBEV E protein (monoclonal antiserum 1493 [53]) or the clone 13 N protein (polyclonal antiserum [20]) diluted 1:1,000 and 1:800, respectively, in PBS supplemented with 10% FCS and 0.05% Tween 80. Secondary horseradish peroxidase (HRP)-conjugated anti-mouse antibody (Dako) was diluted 1:2,000 in PBS supplemented with 10% FCS-0.05% Tween 80. Antigen-positive cell foci were detected with TrueBlue peroxidase substrate (KPL, Gaithersburg, MD) as described previously (47).
IFN assays.
Total amounts of IFNs in cell supernatants were measured by using 293T cells stably expressing the firefly luciferase gene under the control of the mouse Mx1 promoter (Mx1-luc reporter cells) (28). Cell supernatants were harvested and virus particles were removed by using Amicon spin columns with a cutoff of 100 kDa (Millipore) according to the manufacturer's instructions. The absence of infectious virus particles was verified by plaque assay. Mx1-luc reporter cells were seeded into 48-well dishes and were treated 24 h later with filtered supernatants diluted 1:10 in DMEM-5% FCS. At 16 h postincubation, cells were lysed with passive lysis buffer (Promega), and luciferase activity was measured with a Sirius luminometer (Berthold detection system). The assay sensitivity was between 0.5 and 5 U/ml IFN-β and was determined by a standard curve.
For determinations of the ratio between intracellular and extracellular IFN, an enzyme-linked immunosorbent assay (ELISA) specific for human IFN-β (PBL InterferonSource) was employed. The level of the intracellular IFN-β protein was measured in lysates of cells treated with buffer containing 0.5 M NaCl, 5 mM EDTA, 10 mM Tris-HCl (pH 7.6), and 1% Triton X-100 (59), and the amount of extracellular IFN-β was determined directly in cell supernatants.
Immunofluorescence analysis.
Cells were grown on coverslips to 30 to 50% confluence and infected and incubated for the indicated times. Cells were fixed with 3% paraformaldehyde, permeabilized with 0.5% Triton X-100 dissolved in PBS, and washed three times with PBS containing 1% FCS. Primary antibodies were diluted in PBS containing 1% FCS. The TBEV E protein was detected by using mouse monoclonal antibody 1493 diluted 1:1,000 (53), the N protein of clone 13 was detected with a mouse polyclonal antibody diluted 1:500 (20), and dsRNA was detected with mouse monoclonal antibody J2 (English & Scientific Consulting, Szirak, Hungary) diluted 1:200. Rabbit polyclonal antisera were used to detect IRF-3 (BD Biosciences Pharmingen) and tubulin (Abcam), diluted 1:200 and 1:500, respectively. For the sensitive detection of dsRNA or IRF-3, the fluorophore signal was visualized by using the tyramide signal amplification (TSA)-cyanine 3 (Cy3) system (Perkin-Elmer). After incubation at room temperature for 1 h, the coverslips were washed three times in PBS and then treated with goat anti-mouse Cy2 secondary antibody conjugated at a dilution of 1:200. Cells were again washed three times in PBS and mounted by using Fluorsave solution (Calbiochem). Stained cell samples were examined by using a Zeiss confocal microscope.
Plasma membrane permeabilization.
SL-O was used to selectively permeabilize the plasma membrane (7, 9). First, cells were washed with permeabilization buffer (125 mM potassium acetate, 25 mM HEPES [pH 7.2], 10 mM glucose, 2.5 mM magnesium acetate, 10 mM NaCl, 1.8 mM CaCl2, and 5 mM EGTA) and treated with 1 U/ml SL-O in permeabilization buffer for 15 min on ice. The SL-O was washed away with permeabilization buffer, and cells were incubated at 37°C for 5 min with prewarmed permeabilization buffer, followed by an additional washing step with PBS and fixation with 3% paraformaldehyde dissolved in PBS. Immunodetection of specific antigens was performed as described above.
IRF-3 dimerization assay.
Cells were lysed in buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% NP-40, protease inhibitors, and phosphatase inhibitors; incubated on ice for 10 min; and then centrifuged at 4°C for 5 min at 10,000 × g. An aliquot of 7 μg of protein was then separated by electrophoresis for 2 h at 60 mA in a 7.5% nondenaturing polyacrylamide gel with 1% deoxycholate in the cathode buffer (26). The proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane (Amersham), followed by incubation in saturation buffer (PBS containing 5% nonfat dry milk and 0.05% Tween). Membranes were incubated overnight with polyclonal anti-IRF-3 antibody FL-425 (Santa Cruz Biotechnology) diluted 1:1,000 in saturation buffer and then washed three times with 0.05% PBS-Tween followed by incubation with an HRP-conjugated secondary antibody. After three additional washing steps, detection was performed by using the SuperSignal West Femto chemiluminescence kit (Pierce).
Western blot analyses.
Cells were lysed in radioimmunoprecipitation assay buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 0.05% SDS) containing protease inhibitors (Complete protease inhibitor; Roche) and phosphatase inhibitors (Phosphatase Inhibitor Cocktail II; Calbiochem). A total of 10 μg of protein was separated by SDS-PAGE and transferred onto an Immobilon-P PVDF membrane (Millipore), followed by incubation in saturation buffer (PBS containing 5% nonfat dry milk and 0.05% Tween). The membrane was first incubated for 1 h with primary antibodies and washed three times with 0.05% PBS-Tween, followed by incubation with horseradish peroxidase-conjugated secondary antibodies (Pierce). After an additional three washing steps, detection was performed by using the SuperSignal West Femto kit (Pierce). Primary antibodies used were directed against phosphorylated eIF2α (alpha subunit of eukaryotic initiation factor 2) (rabbit polyclonal anti-Ser51, diluted 1:200; Biosource), eIF2α (mouse monoclonal, diluted 1:1,000; Cell Signaling), TBEV E (mouse monoclonal, diluted 1:5,000), and actin (rabbit polyclonal anti-actin, diluted 1:5,000; Sigma).
Transient transfections and reporter gene assays.
Subconfluent monolayers of 293T cells grown in 12-well dishes were transfected with 0.25 μg p125-luc reporter plasmid (FF-Luc gene under the control of the IFN-β promoter), 0.025 μg control plasmid pRL-SV40 (Ren-Luc gene under the control of the SV40 promoter), and 0.5 μg of expression plasmid with Nanofectin (PAA) according to the manufacturer's recommendations. At 24 h posttransfection, cells were infected with clone 13 at a multiplicity of infection (MOI) of 5. At 16 h p.i., cells were harvested and lysed in 100 μl of passive lysis buffer (Promega). An aliquot of 10 μl of lysate was used to measure FF-Luc and Ren-Luc activity as described by the manufacturer of the Dual luciferase assay kit (Promega), and the activity was measured with a Sirius luminometer (Berthold detection system).
EM.
293T cells were seeded onto 10-cm plates and infected with TBEV strain Hypr (MOI of 5). The cells were either left untreated or treated with BFA (2.8 ng/ml) at 12 h p.i. At 24 h p.i., cells were treated with modified Ito's fixative (2.5% formaldehyde, 0.1% glutaraldehyde, 0.03% trinitrophenol, and 0.03% CaCl2 in 0.05 M cacodylate buffer [pH 7.2]) (54) for 1 h at room temperature, washed with 0.1 M cacodylate buffer, scraped off, pelleted (10 min at 10,000 × g), and kept at 4°C until further processing. Parallel cell pellets were processed either for regular transmission electron microscopy (EM) or for immuno-EM. For transmission EM, pellets were postfixed with 1% OsO4 in 0.1 M cacodylate buffer for 1 h at room temperature, stained en bloc with 2% aqueous uranyl acetate (20 min at 60°C), dehydrated in ethanol followed by propylene oxide, and embedded in Poly/Bed 812 (Polysciences). Ultrathin sections were cut on a Reichert-Leica Ultracut S ultramicrotome, placed onto uncoated copper grids, and stained with lead citrate. For immuno-EM, cell pellets were stained en bloc with 2% aqueous uranyl acetate (20 min at 60°C), dehydrated through 75% ethanol, and embedded into LR White resin (Electron Microscopy Sciences [EMS], Hatfield, PA). Ultrathin sections were cut on the same ultramicrotome and placed onto nickel Formvar-carbon-coated grids. The grids were incubated first on drops of blocking buffer (0.1% BSA and 0.01 M glycine in 0.05 M Tris-buffered saline [TBS]) and then with anti-dsRNA antibody J2 (0.5 mg/ml) dissolved 1:10 in diluting buffer (1% BSA in 0.05 M TBS) first for 1 h at room temperature and then overnight at 4°C. After washing in blocking buffer, grids were incubated with the secondary antibody (goat anti-mouse IgG/IgM conjugated with 10-nm colloidal gold) (Aurion, catalog number 25169; EMS) diluted 1:20 in diluting buffer for 1 h at room temperature. After washing first in blocking buffer, then in TBS, and then in distilled water, grids were fixed in 2% aqueous glutaraldehyde, washed, stained with 2% aqueous uranyl acetate and lead citrate, and examined with a Philips 201 or CM-100 electron microscope at 60 kV.
RESULTS
Differential induction of IFN-β and ISG transcription by TBEV strains.
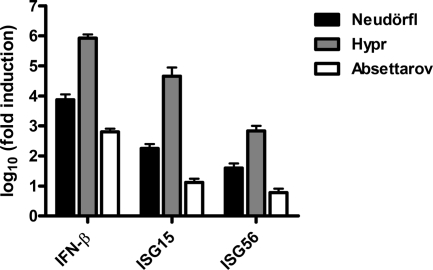
We studied the antiviral host response to TBEV using A549 cells, a human lung carcinoma line that is fully IFN competent (33). Three strains of Western TBEV, namely, Neudörfl, Hypr, and Absettarov, were employed in parallel. Low-pathogenicity strain Neudörfl was originally isolated in 1971 from Ixodes ricinus ticks in Burgenland, Austria (71). High-pathogenicity strain Hypr was originally isolated in 1953 from the blood of a deceased child in Moravia, Czech Republic (71). TBEV strain Absettarov, with an unclear pathogenicity level compared to those of the other two strains, was isolated in 1951 from the blood of an ill patient in St. Petersburg, Russia (14). A549 cells were infected with the viruses at an MOI of 1, and total RNA was extracted at 24 h p.i. Cellular transcript levels were determined by using real-time RT-PCR, normalized to an internal cellular control mRNA (γ-actin), and set in relation to mock-infected cells. Figure 1 shows that the transcriptions of IFN-β as well as the virus-inducible genes ISG15 and ISG56 (72) were upregulated by several orders of magnitude after TBEV infection. For all host genes tested, strain Hypr was the strongest inducer, whereas strain Absettarov was the weakest inducer and strain Neudörfl had an intermediate phenotype.
FIG. 1.
Induction of IFN and ISGs. A549 cells were infected with TBEV strains Neudörfl, Hypr, and Absettarov at an MOI 1, and total cell RNA was extracted at 24 h p.i. Levels of IFN-β, ISG15, and ISG56 mRNAs were measured by real-time RT-PCR analysis, normalized to the cellular γ-actin mRNA, and set in relation to mRNA levels of mock-infected cells. Mean values and standard deviations from three independent experiments are shown.
TBEV strains differ in RNA levels and speeds of replication.
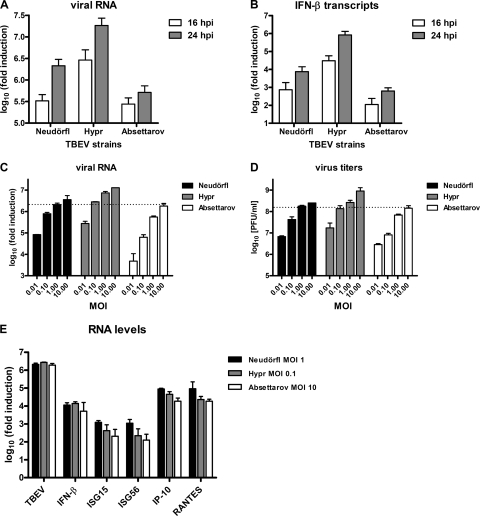
The observed differences in the host response to the three virus strains could be based either on qualitative traits, i.e., stronger or weaker viral anti-IFN factors, or on quantitative traits, i.e., different speeds of RNA replication. To distinguish between these possibilities, we first compared viral RNA levels at two different time points postinfection. A549 cells were infected with the three TBEV strains at an MOI of 1, and viral RNA was analyzed at 16 and 24 h p.i. by real-time RT-PCR. Clear differences could be detected (Fig. 2 A). Hypr accumulated faster and at much higher RNA levels than did Absettarov, and Neudörfl was intermediate. Moreover, real-time RT-PCR revealed that IFN-β mRNA levels paralleled the viral RNA levels; again, Hypr was the best inducer, followed by Neudörfl, and Absettarov was a comparatively low inducer (Fig. 2B).
FIG. 2.
TBEV strains differ in replication speeds, endpoint titers, and IFN induction capabilities. (A and B) A549 cells were infected with TBEV strains Neudörfl, Hypr, and Absettarov at an MOI of 1, and cellular RNA was extracted either at 16 h p.i. or at 24 h p.i. Viral RNA (A) and IFN-β mRNA levels (B) were quantified by real-time RT-PCR analysis as indicated in the legend of Fig. 1. (C and D) Multiplication of virus strains is dependent on the input MOI. A549 cells were infected with the different TBEV strains by using MOIs varying in 10-fold steps from 0.01 to 10. Total cell RNA and cell culture supernatants were harvested at 24 h p.i., and viral RNA levels (C) and titers (D) were determined by real-time RT-PCR and plaque assays, respectively. (E) Levels of viral RNA and innate immunity genes after infection with TBEV strains using normalized input MOIs. Total RNA preparations from the cells infected for 24 h with the different TBEV strains at MOIs of 1 (Neudörfl), 0.1 (Hypr), and 10 (Absettarov) (see C and D) were used to measure levels of viral RNA and mRNAs for IFN-β, ISG15, ISG56, IP-10, and RANTES by real-time RT-PCR. In all cases, mean values and standard deviations from three independent experiments are shown.
The data collected so far suggest that the three TBEV strains replicate with different speeds and efficiencies and that there might be a correlation between levels of viral RNA and the height of IFN induction. To follow this up, we sought conditions under which RNA levels and particle production of the strains were comparable. Indeed, when the infection outcomes of different amounts of input virus were compared, we observed that input MOIs of 1 for Neudörfl, 0.1 for Hypr, and 10 for Absettarov resulted in similar RNA levels and virus titers after 24 h of infection (Fig. 2C and D). We used the RNAs of A549 cells infected with these normalized MOIs to investigate IFN induction. All antiviral host genes investigated, namely, IFN-β, ISG15, and ISG56, as well as the chemokines IP-10 and RANTES, were induced by the normalized virus infections with comparable efficiencies (Fig. 2E). Thus, taken together, these results imply that it is the speed of virus replication and the amount of accumulated RNA that determine the strength of the innate immune response to TBEV.
TBEV delays the onset of IFN induction.
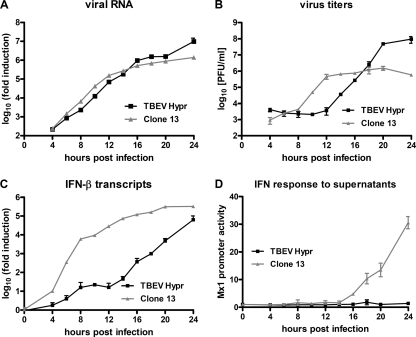
We were intrigued by the fact that TBEV strain Hypr, which is a highly infectious human pathogen, activates the antiviral host response with such amplitude. To obtain a more detailed picture, we performed a time course analysis of TBEV infection and the IFN response over a period of 24 h. As positive control, we used the Rift Valley fever virus mutant clone 13, a well-established inducer of IFN (4). A549 cells were infected with an MOI of 1, and total cell RNA and supernatants were harvested every 2 h from 4 to 20 h p.i. and then again at 24 h p.i. The results of the real-time RT-PCR analysis showed that intracellular RNA levels of both viruses rose with surprisingly similar kinetics for the first 20 h (Fig. 3 A). At 24 h p.i. TBEV RNA levels further increased, whereas this was less pronounced for clone 13. In contrast, virus titers in the cell supernatants sharply differed in their kinetics. Clone 13 rapidly produced progeny particles, whereas TBEV particles were released with delayed kinetics (Fig. 3B). A similar eclipse phase with very little particle production was previously described for the flavivirus Kunjin virus (KUNV) (77). Clone 13 virus release, however, reached a plateau already at 12 h p.i., with no further augmentation in particle production from then on. TBEV titers steadily increased and surpassed clone 13 titers at 18 h p.i. For TBEV, a plateau was reached at 20 h p.i. with a final titer of 9.5 × 107 PFU/ml, whereas clone 13 reached a plateau at 12 h p.i. with approximately 1 × 106 PFU/ml, i.e., almost 100-fold less. The induction of IFN also differed between the viruses. Real-time RT-PCR analyses showed that the activation of IFN-β transcription by TBEV was delayed by 8 to 12 h compared to that of clone 13 (Fig. 3C). To detect IFNs in the supernatants, we incubated an IFN-responsive 293T reporter cell line (28) with supernatants of infected cells. We found the sensitivity of this assay system to be in the range of 0.5 to 5 U/ml of IFN-β (data not shown). The reporter cells revealed that clone 13-infected cells released increasing levels of IFNs from 16 h p.i. on, while for TBEV-infected cells, no IFNs were detected at 24 h p.i. Thus, although intracellular virus RNA levels increased for both viruses with almost identical kinetics, particle production was more efficient for TBEV than for clone 13, most likely because TBEV substantially delays the transcriptional induction of the IFN-β gene.
FIG. 3.
Time course of viral multiplication, IFN transcription, and IFN production. A549 cells were infected with TBEV strain Hypr and the IFN-inducing control virus clone 13 at an MOI of 1. Total cell RNA and cell culture supernatants were collected at different time points p.i. (A) Quantification of intracellular levels of virus RNA determined by real-time RT-PCR analysis. (B) Viral titers in cell culture supernatants determined by plaque assays. (C) Levels of IFN-β mRNA determined by real-time RT-PCR analysis. (D) Presence of type I IFN in the supernatants of infected cells measured using a 293T cell line carrying the firefly luciferase gene under the control of the IFN-responsive Mx1 promoter (Mx1-luc reporter cells). Mean values and standard deviations from three independent experiments are shown.
Slow accumulation of IFN-β mRNAs results in late secretion of IFNs.
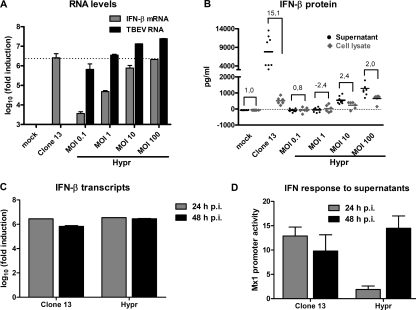
Despite a significant upregulation of IFN-β transcripts by TBEV late in infection, no IFN activity could be detected in the supernatants of those cells (24 h p.i.) (Fig. 3C and D). This raised the possibility that TBEV may interfere with the translation of IFN mRNAs or with the secretion of IFNs. To address these issues, we set up an assay to correlate levels of IFN-β transcripts, intracellular IFN-β protein, and secreted IFN-β protein. First, to permit such a comparison, we searched for conditions under which TBEV would induce amounts of IFN-β transcripts similar to those of clone 13. Figure 4 A shows that increasing the input MOI of TBEV to 100 PFU/cell forced the upregulation of the IFN-β gene at 24 h p.i. to levels that were comparable to the IFN induction by clone 13 at an MOI of 5. In parallel dishes, we quantified levels of intracellular IFN-β protein (from cell extracts) and secreted IFN-β protein (from cell culture supernatants) by ELISA. Intracellular IFN-β protein levels (Fig. 4B) were comparable for TBEV (MOI of 100) and clone 13 (MOI of 5) at 24 h p.i. This led us to conclude that TBEV does not inhibit mRNA translation. The level of extracellular IFN-β, however, was much lower for TBEV-infected cells than for clone 13-infected cells (Fig. 4B). Calculation of the ratios between secreted and intracellular IFN-β levels gave a factor of about 15 for clone 13-infected cells but gave a factor of only 2 for TBEV-infected cells (Fig. 4B).
FIG. 4.
Correlation between transcription, translation, and secretion of IFN. (A and B) Search for comparable IFN induction levels by clone 13 and TBEV. A549 cells were infected for 24 h either with clone 13 at an MOI of 5 or with TBEV strain Hypr at increasing MOIs. (A) Total RNA was extracted from cells, and levels of IFN-β mRNA and clone 13 and TBEV RNAs were determined by real-time RT-PCR analysis. (B) In parallel dishes, supernatants were harvested, and cellular proteins were extracted to measure extracellular (black dots) and intracellular (gray diamonds) IFN-β by ELISA. (C and D) Release of IFN by infected cells. A549 cells were infected with clone 13 (MOI of 5) or TBEV strain Hypr (MOI of 100). (C) Cellular RNA was extracted at 24 h p.i. or 48 h p.i., and levels of IFN-β mRNA were determined by real-time RT-PCR analysis. (D) Either supernatants were harvested at 24 h p.i. or medium was exchanged at 24 h p.i. and harvested at 48 h p.i. IFN levels were measured by using 293T Mx1-luc cells as described in the legend of Fig. 3. Mean values and standard deviations from three independent experiments are shown.
The reduced secretion of IFN-β in TBEV-infected cells could be caused by a direct inhibition of IFN-β secretion, but it could also be a consequence of the delayed upregulation of the IFN-β gene that we had observed. To distinguish between these possibilities, we performed a burst experiment to measure how much de novo IFN is released late in infection, when IFN-β mRNA levels are comparable between TBEV and the control virus clone 13. Two sets of cells were infected with either clone 13 or TBEV. For the first set of cells, total RNA and supernatants were harvested after 24 h of infection. For the second set of cells, the medium was changed at 24 h p.i., and RNA and supernatants were harvested at 48 h p.i. Any IFN measured in the supernatants of the second set of cells was hence released between 24 h and 48 h p.i. Figure 4C shows that comparable IFN-β transcript levels were induced by the two viruses at 24 h p.i. IFN-β transcript levels remained virtually unchanged between 24 h and 48 h p.i, suggesting that the observed 106-fold upregulation represents an upper limit. IFNs in the supernatants were analyzed by using the sensitive Mx1-luc reporter cell system (Fig. 4D). At 24 h p.i., high levels of IFN were detected in supernatants of clone 13-infected cells, but much lower levels were detected in supernatants of TBEV-infected cells, as described above (Fig. 3D). Strikingly, however, when newly secreted IFNs were measured at 48 h p.i., similar IFN activities were present in the supernatants of clone 13 and TBEV cells. This finding indicates that at between 24 h and 48 h p.i., when IFN mRNA levels remain at comparable levels, cells also responded with comparable levels of IFN secretion. Thus, the difference between TBEV and the IFN inducer virus clone 13 is due solely to the delay in IFN transcript accumulation by TBEV and is not caused by a block in IFN secretion. To our knowledge, this is the first study in which levels of IFN-β mRNA, intracellular IFN-β protein, and secreted IFN-β protein were compared over a time course and in a quantitative manner. The results strongly suggest that IFN-β mRNA needs to accumulate 105- to 106-fold over an extended period of time until substantial levels of the IFN-β protein become secreted. With respect to TBEV, our data imply that this virus delays the transcriptional induction of IFN in order to remain below the threshold value for IFN-β protein secretion. Thus, a block in IFN secretion, as was shown previously for poliovirus (13) and mouse hepatitis virus (59), does not apply and is not required for TBEV.
Incomplete activation of IRF-3.
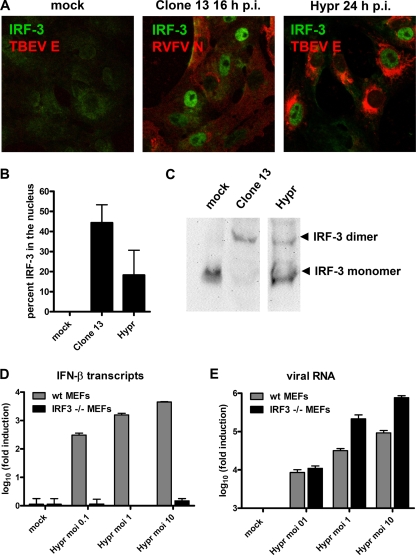
We wanted to characterize the mechanism behind the delay in IFN induction by TBEV. The activation of the IFN-β promoter involves the constitutively expressed transcription factor IRF-3. In uninfected cells, IRF-3 resides in the cytoplasm, but after phosphorylation by virus-activated kinases, it homodimerizes and translocates into the nucleus to transactivate the IFN-β promoter (24). We analyzed the activation state of IRF-3 in response to TBEV. To monitor the subcellular localization of IRF-3, cells were either mock infected or infected with clone 13 or TBEV and stained with specific antibodies. Fixation was done at 16 h p.i. for clone 13 and at 24 h p.i. for TBEV, since at these time points, IFN-β transcript levels were comparable (Fig. 3C). Despite this 8-h-longer infection period, infection with TBEV caused a much smaller percentage of IRF-3 to enter the nucleus than did infection with clone 13, whereas in mock-infected cells, IRF-3 remained in the cytoplasm, as expected (representative pictures are shown in Fig. 5 A, and quantification is shown in Fig. 5B). To examine IRF-3 homodimerization, we extracted cellular proteins at 16 h p.i. and 24 h p.i. for clone 13- and TBEV-infected cells, respectively, and performed nondenaturing polyacrylamide gel electrophoresis coupled to Western blot analysis (26). Again, the positive control clone 13 triggered a complete dimerization of IRF-3, whereas TBEV induced intermediate dimerization, and no IRF-3 dimers were detected after mock infection (Fig. 5C). Importantly, when TBEV infection was stopped at 16 h p.i., neither any nuclear localization nor any dimerization of IRF-3 was observed (data not shown).
FIG. 5.
Involvement of IRF-3. (A) Immunofluorescence analysis. Vero B4 cells were infected at an MOI of 1 with clone 13 or TBEV strain Hypr. At 16 h (clone 13) or 24 h (TBEV) p.i., cells were fixed and immunostained using antisera specific for IRF-3 and viral N antigens, respectively. (B) Percentage of infected cells with nuclear IRF-3 calculated from three independent experiments (100 to 200 cells counted in each experiment), performed as described above (A). (C) Homodimerization assay. Extracts from A549 cells infected with either clone 13 for 16 h or TBEV for 24 h were subjected to nondenaturing gel electrophoresis followed by immunoblotting to detect IRF-3. (D and E) Dependence of IFN induction on IRF-3. wt MEFs (gray bars) and IRF-3 knockout MEFs (black bars) were infected with TBEV at different MOIs, and cellular RNA was extracted at 24 h p.i. Levels of IFN-β transcripts (D) and viral RNA (E) were determined by real-time RT-PCR. Mean values and standard deviations from three independent experiments are shown.
To clarify whether the delay of IRF-3 activation is of any relevance for IFN induction by TBEV, we employed mouse embryonic fibroblasts (MEFs) with a knockout in the IRF-3 gene (63). As illustrated in Fig. 5D, IFN-β transcription was found to be overwhelmingly dependent on IRF-3, even when input virus levels were raised to an MOI of 10. Importantly, the knockout of IRF-3 had no negative influence on the growth of TBEV but rather increased viral RNA levels (Fig. 5E).
Collectively, these results indicate that TBEV infection causes an only suboptimal activation of IRF-3 in order to substantially delay the onset of IFN-β transcription.
IRF-3 inhibition appears not to be caused by a specific virus protein.
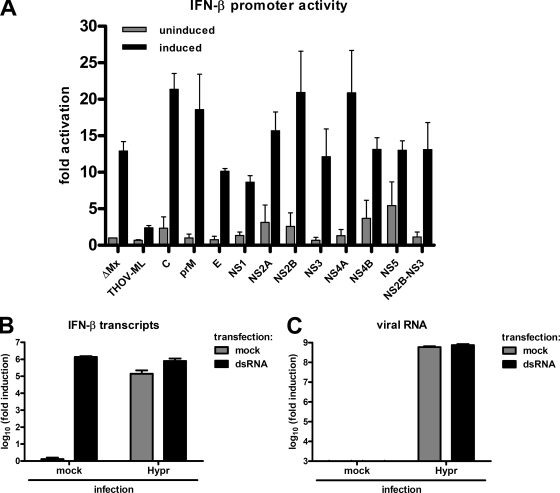
Many viruses express IFN-antagonistic proteins able to specifically inhibit the IRF-3 activation pathway (57, 73). To test whether TBEV would also encode such a factor, we set up a reporter assay for measuring the influence of individual virus proteins on the activation of the IFN-β promoter. To achieve this goal, we first constructed cDNA expression plasmids for each component of the viral polyprotein (see Materials and Methods). The individual TBEV cDNA plasmids were then transfected into A549 cells along with two reporter constructs containing either the firefly luciferase (FF-Luc) gene under the control of the IFN-β promoter (81) or the Renilla luciferase (Ren-Luc) gene under the control of the constitutively active SV40 promoter. Individual TBEV constructs expressed the viral proteins C, prM, E, NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5. Moreover, we also expressed the NS2B-NS3 fusion protein, the viral protease responsible for processing parts of the polyprotein. In parallel with these constructs, cDNA plasmids for the N terminus of the human MxA protein (ΔMx) and for the IRF-3 inhibitor ML of Thogoto virus (27) were used as negative and positive controls, respectively. After 24 h of incubation to allow for the expression of the transfected cDNAs, IFN induction was stimulated by superinfection with clone 13, and cells were incubated for another 16 h until lysis and the subsequent measurement of luciferase activities. Values for FF-Luc activities (reflecting IFN-β promoter activation) were normalized to the corresponding Ren-Luc activities (reflecting constitutive polymerase II [Pol II]-driven gene expression) in order to determine specific IFN induction or suppression, respectively. Figure 6 A shows that in cells expressing the negative control ΔMx, infection with clone 13 was able to stimulate the IFN-β reporter, while the positive control ML suppressed IFN induction substantially. The individual expression of the TBEV proteins C, E, NS2A, NS2B, NS3, NS4A, NS4B, NS5, and NS2B-NS3, in contrast, had no apparent influence on the specific IFN-β promoter activation. The addition of an N-terminal Flag tag to detect recombinant gene expression by Western blotting led to similar observations (data not shown). It should be noted that prM, NS1, and NS4B had a certain suppressive effect on general reporter expression and that NS2A had a strong general suppressive effect (data not shown). However, none of the individually expressed TBEV proteins mediated a specific effect on the IFN-β promoter.
FIG. 6.
Screen of TBEV gene products for inhibition of the IFN-β promoter. (A) Reporter assay for testing overexpressed TBEV gene products. A549 cells were transfected with an FF-Luc construct under the control of the IFN-β promoter and a Ren-Luc construct carrying the constitutively active SV40 promoter. In addition, cDNA plasmids expressing individual TBEV genes or a positive (THOV-ML) or a negative (ΔMx) control were transfected. At 24 h posttransfection, cells were infected with clone 13 (MOI of 1) to stimulate the IFN-β promoter. After a further incubation period of 16 h, cells were lysed, and reporter activities were determined. Specific IFN-β promoter activity was determined by normalizing FF-Luc to Ren-Luc activities and setting the mock-infected uninduced ΔMx control as 1. Unstimulated and stimulated values are depicted as gray bars and black bars, respectively. Mean values and standard deviations from three independent experiments are shown. (B and C) Real-time RT-PCR assay of TBEV-infected cells induced with dsRNA. A549 cells were infected with TBEV strain Hypr for 16 h at an MOI of 5 and then transfected with poly(I:C) (dsRNA) for another 6 h before total RNA extraction. Mock infection and mock transfections were performed in parallel as controls. IFN-β mRNA levels (B) and TBEV RNA levels (C) were quantified by real-time RT-PCR analysis. Mean values and standard deviations from three independent experiments are shown.
It remained possible that several viral proteins inhibit IFN induction in a cooperative manner. To investigate this possibility, we determined whether ectopic IFN induction can occur in cells infected with TBEV. A549 cells were infected with TBEV for 16 h to obtain saturating viral gene expression. The cells were then transfected with the dsRNA analog poly(I:C) for another 6 h and monitored for IFN induction by real-time RT-PCR. Figure 6B shows that in mock-infected cells, dsRNA transfection strongly upregulated IFN induction, as expected. Infection with TBEV alone also upregulated IFN-β transcription, as described above, but to a lesser extent than dsRNA. When cells were first infected with TBEV and subsequently transfected with dsRNA, a clear increase in IFN induction was observed, indicating the absence of active IFN suppression. Since the 6-h dsRNA treatment apparently did not influence viral multiplication (Fig. 6C), we conclude from these results that the delay in IRF-3-dependent IFN induction imposed by TBEV is unlikely to be caused by the expression of a viral IFN antagonist.
dsRNA of TBEV is protected by intracellular membranes.
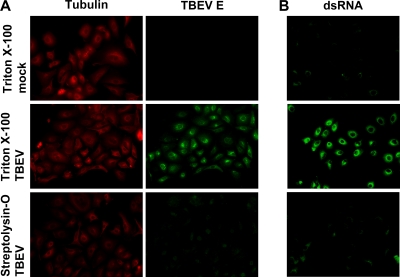
Positive-strand RNA viruses are known to produce substantial amounts of dsRNA, a strong inducer of type I IFNs (32, 49, 56, 68, 74, 78). Also, for some of these viruses, it was shown previously that their dsRNA is associated with or even located inside intracellular membrane compartments and, hence, possibly unavailable to the dsRNA-recognizing pathogen receptors in the cytoplasm (32, 39, 75). As these issues have not been addressed for TBEV, we designed a differential permeabilization protocol coupled to immunofluorescence analysis to (i) assess the presence of dsRNA in infected cells and (ii) simultaneously determine whether the dsRNA would be accessible to host cell factors in the cytoplasm. Cells grown on coverslips were infected with TBEV for 24 h, permeabilized with streptolysin-O (SL-O), and then fixed with paraformaldehyde. SL-O is a bacterial toxin that produces plasma membrane pores of 20 to 30 nm in diameter without affecting intracellular membranes (7, 9). As a control, fixed cells that had been grown and infected in parallel dishes were treated with Triton X-100, an agent that permeabilizes all cellular membranes indiscriminately. All fixed and permeabilized cells were then analyzed by immunostaining for the cytoplasmic protein tubulin, the endoplasmic reticulum (ER)-resident E glycoprotein of TBEV, and dsRNA. As shown in Fig. 7 A, tubulin was detected after both permeabilization regimens, demonstrating that SL-O treatment allows the entry of antibodies into the cytoplasm. The TBEV E protein, in contrast, was readily detected in infected cells but not in uninfected cells treated with Triton X-100, but only background staining was observed for infected cells treated with SL-O. This confirmed that antigens inside cytoplasmic membrane compartments are not accessible after SL-O treatment. Strikingly, for viral dsRNA, a strong signal was obtained only when Triton X-100 was used, whereas SL-O permeabilization prohibited dsRNA immunostaining. Thus, dsRNA is indeed a frequent by-product of TBEV RNA replication, as expected, but apparently, it is not accessible to cytoplasmic factors due to a shielding by intracellular membrane compartments.
FIG. 7.
Subcellular localization of viral dsRNA. Cells were infected with TBEV (MOI of 1) or left uninfected (mock) and fixed 24 h later with 3% paraformaldehyde. Fixed cells were treated either with Triton X-100, which permeabilizes all cellular membranes, or with SL-O, which selectively permeabilizes the plasma membrane. (A) Immunodetection of tubulin (located in the cytoplasm) and the TBEV E protein (located inside the ER). (B) Immunodetection of dsRNA in cells treated the same way as described above (A).
TBEV rearranges intracellular membranes.
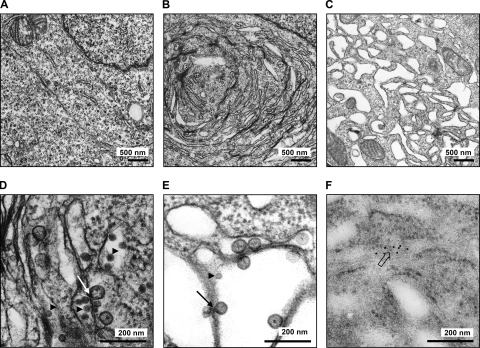
To obtain a better picture of the intracellular membrane compartments used by TBEV for replication, we performed ultrathin-section electron microscopy (EM) of cells that were infected for 24 h with TBEV and then fixed with Ito's modified fixative. As shown in Fig. 8 A and B, TBEV markedly changed the morphology of the ER to tightly packed stacks of laminar membranes in paracrystalline arrays, similar to what was described previously for the flaviviruses KUNV and DENV (40, 75).
FIG. 8.
Ultrathin-section EM of infected cells. 293T cells were infected with TBEV strain Hypr (MOI of 5) for 24 h before fixation and processing for regular transmission EM (A to E) or immuno-EM (F). (A) Transmission EM of mock-infected cells. (B) Transmission EM of TBEV-infected cells. (C) Transmission EM of TBEV-infected cells treated with BFA at 12 h p.i. (D and E) Close-up EM pictures of untreated and BFA-treated cells that were infected with TBEV. (F) Close-up immuno-EM picture of cells infected with TBEV. Immunogold staining was performed by using anti-dsRNA mouse monoclonal J2 as the primary antibody and goat anti-mouse IgG/IgM conjugated with 10-nm colloidal gold as the secondary antibody. Arrowheads indicate virus particles, lined arrows indicate virus-induced membrane vesicles, and the boxed arrow indicates immunogold-labeled dsRNA.
The results obtained so far did not allow us to conclude why TBEV dsRNA is unavailable for cytoplasmic detection. The dsRNA could be hiding between the tightly packaged membrane stacks, or it could be inside them. For KUNV, it was shown previously that the addition of brefeldin A (BFA), a drug which disrupts the Golgi apparatus (31), can inhibit the virally induced membrane rearrangements (40) and renders the virus vulnerable to the antiviral protein MxA (25). In analogy to these experiments, we attempted to disrupt the TBEV-induced membrane stacks with BFA to see whether the viral dsRNA would be exposed to the cytoplasm. TBEV-infected cells were treated with BFA at 12 h p.i. and fixed and processed for ultrathin-section EM at 24 h p.i. A clear change of the paracrystalline arrays was seen after BFA treatment, since membrane structures obtained a more vesicular appearance (Fig. 8C). Nonetheless, TBEV dsRNA was still not detectable by immunofluorescence analysis after SL-O treatment, and no increase in IFN induction was obtained after BFA treatment (data not shown). Interestingly, besides virus particles (Fig. 8D and E), we observed small vesicles inside the ER (Fig. 8D and E) in both untreated (Fig. 8D) and BFA-treated (Fig. 8E) infected cells. Similar ER-borne vesicles were previously identified in cells infected with KUNV (41, 43, 78) and with DENV (75). These vesicles were reported previously to contain viral dsRNA and replication complexes and to have small openings toward the cytoplasm for the exchange of metabolites and mRNAs (75). In TBEV vesicles, inklings of similar openings could also be seen for a limited number of vesicles (Fig. 8E). We also performed immunogold staining of TBEV-infected cells to detect dsRNA. Figure 8F shows that the gold staining localized to vesicular structures within the rough ER, indicating that TBEV dsRNA is located within ER-derived vesicles, similar to what was described previously for KUNV and DENV (41, 43, 75, 78).
Taken together, these results indicated that dsRNA of TBEV is stored inside BFA-resistant membrane vesicles to prevent detection by PRRs. This most likely causes the observed delay in IRF-3 activation and IFN-β induction.
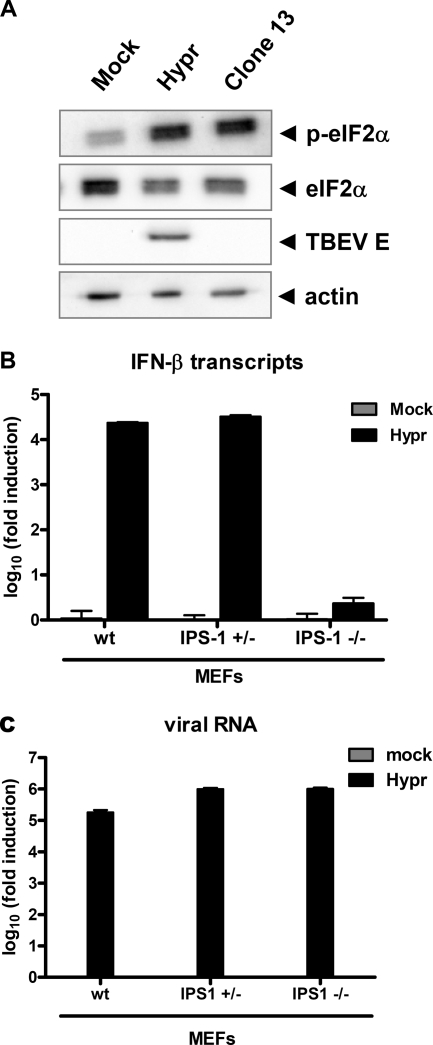
Indications of dsRNA leakage late in infection.
Despite the apparent inaccessibility of viral dsRNA inside membranous compartments, IFN induction by TBEV finally occurs. To address a possible exposure of viral dsRNA, we first investigated whether the dsRNA-activated antiviral kinase PKR would be relocalized to sites of TBEV dsRNA late in infection but found that this is not the case (data not shown). As a more sensitive method, we determined the phosphorylation of eIF2α, the main substrate of PKR. A459 cells were infected for 24 h with TBEV, and the phosphorylation state of eIF2α was monitored by Western blot analysis. Figure 9 A shows that TBEV infection upregulated eIF2α phosphorylation in a manner similar to that of clone 13, a known PKR activator (21). To more directly determine the presence of IFN-inducing viral RNAs in the cytoplasm, we measured IFN induction in MEF cells lacking interferon promoter stimulator 1 (IPS-1), the essential adaptor for the cytoplasmic virus RNA sensors RIG-I and MDA5 (29). The real-time RT-PCR analysis depicted in Fig. 9B demonstrates that 24 h of TBEV infection strongly upregulated IFN-β induction in wild-type (wt) MEFs as well as in MEFs expressing one genomic copy of IPS-1. MEFs with a complete knockout of IPS-1, in contrast, were barely able to induce any IFN transcription despite comparable levels of virus replication (Fig. 9C). These results demonstrate that the IFN induction by TBEV that occurs late in infection is mediated by the cytoplasmic virus RNA sensors RIG-I and/or MDA5. Thus, taken together, both the activation of PKR and the strict dependency of IFN induction on IPS-1 indicate that spurious amounts of TBEV dsRNA become accessible to cytoplasmic PRRs late in infection. This most likely explains the eventual upregulation of IFN induction by TBEV.
FIG. 9.
Cytoplasmic detection of viral RNA late in infection. (A) eIF2α activation. A549 cells were infected with TBEV strain Hypr for 24 h and then assayed by Western blot analysis using antisera recognizing phosphorylated eIF2α (p-eIF2α), eIF2α, TBEV E, or actin as a control. (B and C) Real-time RT-PCR assay. MEF cells expressing or lacking one or both genomic copies of the IPS-1 gene were infected with Hypr at an MOI of 1, and total cell RNA was extracted at 24 h p.i. Mock infections were performed in parallel as controls. IFN-β mRNA levels (B) and TBEV RNA levels (C) were quantified by real-time RT-PCR analysis. Mean values and standard deviations from three independent experiments are shown.
DISCUSSION
TBEV causes epidemics of acute encephalitis in forested regions of Europe and Asia. The disease takes a characteristic biphasic course: first an unspecific flu-like phase of approximately 5 days followed by a 7-day period of apparent recovery and a second, specific phase involving often severe neurological symptoms (35). Encephalitis is supposed to be caused by CD8+ T-cell-mediated immunopathology along with overshooting inflammatory responses and direct damage by the virus (22, 60). However, only about 20 to 30% of infections enter the second phase and result in full-blown disease (35, 45).
For the TBEV-related Langat virus as well as for other encephalitic flaviviruses, it is known that the pretreatment of cells with type I IFNs can hamper virus multiplication (3, 34). It was thus expected that TBEV disturbs the activation of the IFN system, i.e., IFN induction, in order to efficiently spread in the host. Our results suggest that TBEV does not entirely block the transcriptional induction of the IFN-β promoter but slows it down considerably. The delayed onset of IFN transcription results in a late synthesis of biologically active IFNs. No IFN was measured in supernatants of TBEV-infected cells even at up to 24 h p.i. Measurable IFN production by infected cells occurred between 24 h p.i. and 48 h p.i., when large amounts of infectious TBEV particles had already been produced. Thus, TBEV appears to employ a “runaway” strategy to escape the antiviral effects of the IFN system. This is in line with previously reported findings for WNV, where it was shown that the onset of the IFN response occurred at a late stage of infection (15). Moreover, for influenza A virus, it was previously demonstrated in vivo that accelerated virus multiplication is a viral strategy to outcompete the IFN response (18).
Our attempts to identify a specific TBEV protein responsible for the delay in IFN induction were unsuccessful. In a similar manner, for WNV, it was shown previously that infected cells can normally activate IRF-3 in response to Sendai virus superinfection, arguing against the presence of an active mechanism to inhibit cytoplasmic IFN induction by this flavivirus as well (15). It must be added, however, that WNV is able to inhibit IFN induction by ectopic dsRNA, most likely because the viral NS1 gene inhibits Toll-like receptor 3, an IFN-activating PRR that recognizes dsRNA in the extracellular phase (64, 79). Also, the NS2A protein of KUNV, which is a variant of WNV, was shown previously to directly suppress IFN induction (36, 37). For TBEV, however, neither NS1 nor NS2A or NS2B had any obvious inhibitory effect on IFN induction. Although we cannot exclude the existence of an IFN antagonist that has escaped our attention, our finding that no viral dsRNA can be immunodetected outside internal membranes favors the model that TBEV relies on hiding its dsRNA in membrane compartments. For intracellular PRRs, the access to this important pathogen marker is hence restricted. It is important that most, if not all, viruses with a positive-strand RNA genome like TBEV rearrange cytoplasmic membranes for the formation of virus factories (1, 10, 39, 48). Moreover, these viruses are known to produce significant amounts of dsRNA during their life cycle (32, 49, 56, 68, 74, 78). For some of the viruses, it was shown previously that viral dsRNA is indeed sequestered inside virus-induced membranous structures (32, 39, 75). Nonetheless, many of them express specific factors inhibiting IFN induction (57, 69, 73). Possible explanations for this discrepancy are that these viruses are vulnerable to PRR detection in the early phase of infection, when membrane reorganization is not yet completed, or that vesicles may rupture. TBEV also appears to expose certain amounts of biologically active dsRNA late in infection. In this context, it should be noted that one molecule of dsRNA is enough to trigger an IFN response under certain conditions (46) and that, at least for severe acute respiratory syndrome (SARS) coronavirus, dsRNA outside viral membrane compartments has been detected (32). SARS coronavirus expresses a set of IFN induction antagonists (69), which together completely block the activation of IRF-3 in infected cells (66). TBEV, which partially activates IRF-3, may outrun IFN induction by fast replication. It is feasible that the speed of replication also plays a role in IFN escape for other viruses besides TBEV, WNV, and influenza A virus (11, 18, 30). Quantitative time course measurements of IFN induction (transcription and IFN release) and virus multiplication, as we have performed for TBEV, could help gain a better understanding of the kinetic aspect of IFN escape by these viruses.
Using different strains of TBEV, we observed that the weak IFN induction by TBEV was directly proportional to the level of accumulated virus RNA. This finding is in agreement with data from a previous study of WNV that showed that IFN induction is dependent on genome replication (6). Moreover, IFN induction by TBEV is strongly dependent on IRF-3 and on IPS-1. Moreover, TBEV activates the phosphorylation of eIF2α, another signaling pathway triggered by viral dsRNA. It is thus conceivable that, at least late in infection, some viral molecules activate cytoplasmic PRRs. Whether the dsRNA that we have detected in large amounts in infected cells is the only viral molecule triggering IFN induction or whether 5′-triphosphorylated RNA, e.g., the viral antigenome, also plays a role remains to be solved. Also, the contribution of individual cytoplasmic PRRs and Toll-like receptors to IFN induction by TBEV will be the subject of future studies.
TBEV efficiently rearranges internal cytoplasmic membranes to host the viral replication factories. ER membrane rearrangements and the formation of replication factories were first described for KUNV (78). Small vesicle packets (VPs) inside the ER-derived compartments were reported previously to contain the viral proteins NS1, NS2A, NS3, and NS4A and dsRNA (41-43, 78). The ultrastructure of VPs was also studied for DENV, and the VP was identified as the site for RNA replication. These vesicles contained a small pore toward the cytoplasm where metabolites and RNA could be exchanged with the cytoplasm (75). For TBEV (this study) and KUNV (44), it was shown that the disruption of the membrane compartments with BFA at 12 h p.i. did not interfere with virus multiplication. However, the treatment rendered KUNV sensitive to the antiviral action of the IFN-induced protein MxA (25), suggesting a certain exposure. For TBEV, we observed that BFA did not alter the inaccessibility of dsRNA to cellular recognition and that the level of viral IFN induction is not elevated (data not shown). This finding indicates that the membrane compartments induced by TBEV are robustly protecting the viral replication complexes.
We found that at the 24-h time point of infection, levels of IFN transcripts were comparable between TBEV and the inducer virus clone 13, but levels of released IFN were different. Only a burst experiment measuring the IFN released de novo between 24 h and 48 h p.i. revealed that cells are able to secrete similar amounts of IFN in both cases. Time course analyses showed that TBEV-infected cells lagged behind the inducer control because final IFN-β mRNA levels were reached with a delay of at least 8 h. If we had measured only mRNAs and viral titers in supernatants at 24 h p.i., the wrong impression of a TBEV-imposed block of IFN secretion would have been produced. This demonstrates that in cases of such observations, rigorous quantitative and kinetic analyses are mandatory before conclusions can be drawn. In general, our quantitative time course analyses of virus multiplication, IFN-β mRNA levels, and IFN protein synthesis indicate that IFN-β mRNAs have to be synthesized over an extended period of time until detectable levels of IFN are released by infected cells. We are not aware that these dependencies have been previously investigated to this detail.
In summary, we propose a kinetic model of TBEV IFN escape. The virus rearranges internal cell membranes to provide a compartment for its dsRNA, which is inaccessible for PRRs. This delays the onset of IFN induction sufficiently to give progeny particle production a head start. Cells start to secrete IFN only later than 24 h after infection. At this time point, however, secondary infections of surrounding cells have already occurred. The well-documented ability of TBEV to block JAK/STAT signaling (3, 76) makes sure that late-point IFN is unable to develop its antiviral activity. This combination of an IFN induction delay and an IFN signaling block may allow the virus to enter the central nervous system before an efficient antiviral response is launched.
Acknowledgments
We thank Valentina Wagner for excellent technical assistance and Julie W. Wen for expert assistance in electron microscopy. Moreover, we are indebted to Gerhard Dobler, Georg Kochs, Takashi Fujita, Jim Robertson, Andrea Kröger, Hansjörg Hauser, Christoph Borner, and Shizuo Akira for providing essential reagents and to Otto Haller for constant support and advice.
Work in our laboratories is supported by grants 01 KI 0711 (F.W.) and 01 KI 0714 (M.N.) from the Bundesministerium für Bildung und Forschung (BMBF).
Footnotes
Published ahead of print on 16 June 2010.
REFERENCES
- 1.Ahlquist, P. 2006. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 4:371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashour, J., M. Laurent-Rolle, P. Y. Shi, and A. Garcia-Sastre. 2009. NS5 of dengue virus mediates STAT2 binding and degradation. J. Virol. 83:5408-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Best, S. M., K. L. Morris, J. G. Shannon, S. J. Robertson, D. N. Mitzel, G. S. Park, E. Boer, J. B. Wolfinbarger, and M. E. Bloom. 2005. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J. Virol. 79:12828-12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billecocq, A., M. Spiegel, P. Vialat, A. Kohl, F. Weber, M. Bouloy, and O. Haller. 2004. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J. Virol. 78:9798-9806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bird, B. H., D. A. Bawiec, T. G. Ksiazek, T. R. Shoemaker, and S. T. Nichol. 2007. Highly sensitive and broadly reactive quantitative reverse transcription-PCR assay for high-throughput detection of Rift Valley fever virus. J. Clin. Microbiol. 45:3506-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourne, N., F. Scholle, M. C. Silva, S. L. Rossi, N. Dewsbury, B. Judy, J. B. De Aguiar, M. A. Leon, D. M. Estes, R. Fayzulin, and P. W. Mason. 2007. Early production of type I interferon during West Nile virus infection: role for lymphoid tissues in IRF3-independent interferon production. J. Virol. 81:9100-9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, A. M., P. D. Kessler, and D. M. Fambrough. 1992. The alternative carboxyl termini of avian cardiac and brain sarcoplasmic reticulum/endoplasmic reticulum Ca(2+)-ATPases are on opposite sides of the membrane. J. Biol. Chem. 267:9321-9325. [PubMed] [Google Scholar]
- 8.Charrel, R. N., H. Attoui, A. M. Butenko, J. C. Clegg, V. Deubel, T. V. Frolova, E. A. Gould, T. S. Gritsun, F. X. Heinz, M. Labuda, V. A. Lashkevich, V. Loktev, A. Lundkvist, D. V. Lvov, C. W. Mandl, M. Niedrig, A. Papa, V. S. Petrov, A. Plyusnin, S. Randolph, J. Suss, V. I. Zlobin, and X. de Lamballerie. 2004. Tick-borne virus diseases of human interest in Europe. Clin. Microbiol. Infect. 10:1040-1055. [DOI] [PubMed] [Google Scholar]
- 9.Courageot, J., E. Fenouillet, P. Bastiani, and R. Miquelis. 1999. Intracellular degradation of the HIV-1 envelope glycoprotein. Evidence for, and some characteristics of, an endoplasmic reticulum degradation pathway. Eur. J. Biochem. 260:482-489. [DOI] [PubMed] [Google Scholar]
- 10.Denison, M. R. 2008. Seeking membranes: positive-strand RNA virus replication complexes. PLoS Biol. 6:e270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diamond, M. S. 2009. Mechanisms of evasion of the type I interferon antiviral response by flaviviruses. J. Interferon Cytokine Res. 29:521-530. [DOI] [PubMed] [Google Scholar]
- 12.Diamond, M. S., T. G. Roberts, D. Edgil, B. Lu, J. Ernst, and E. Harris. 2000. Modulation of dengue virus infection in human cells by alpha, beta, and gamma interferons. J. Virol. 74:4957-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dodd, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2001. Poliovirus 3A protein limits interleukin-6 (IL-6), IL-8, and beta interferon secretion during viral infection. J. Virol. 75:8158-8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ecker, M., S. L. Allison, T. Meixner, and F. X. Heinz. 1999. Sequence analysis and genetic classification of tick-borne encephalitis viruses from Europe and Asia. J. Gen. Virol. 80(Pt. 1):179-185. [DOI] [PubMed] [Google Scholar]
- 15.Fredericksen, B. L., and M. Gale, Jr. 2006. West Nile virus evades activation of interferon regulatory factor 3 through RIG-I-dependent and -independent pathways without antagonizing host defense signaling. J. Virol. 80:2913-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frese, M., T. Pietschmann, D. Moradpour, O. Haller, and R. Bartenschlager. 2001. Interferon-alpha inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J. Gen. Virol. 82:723-733. [DOI] [PubMed] [Google Scholar]
- 17.Grard, G., G. Moureau, R. N. Charrel, J. J. Lemasson, J. P. Gonzalez, P. Gallian, T. S. Gritsun, E. C. Holmes, E. A. Gould, and X. de Lamballerie. 2007. Genetic characterization of tick-borne flaviviruses: new insights into evolution, pathogenetic determinants and taxonomy. Virology 361:80-92. [DOI] [PubMed] [Google Scholar]
- 18.Grimm, D., P. Staeheli, M. Hufbauer, I. Koerner, L. Martinez-Sobrido, A. Solorzano, A. Garcia-Sastre, O. Haller, and G. Kochs. 2007. Replication fitness determines high virulence of influenza A virus in mice carrying functional Mx1 resistance gene. Proc. Natl. Acad. Sci. U. S. A. 104:6806-6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gritsun, T. S., V. A. Lashkevich, and E. A. Gould. 2003. Tick-borne encephalitis. Antiviral Res. 57:129-146. [DOI] [PubMed] [Google Scholar]
- 20.Habjan, M., N. Penski, V. Wagner, M. Spiegel, A. K. Overby, G. Kochs, J. T. Huiskonen, and F. Weber. 2009. Efficient production of Rift Valley fever virus-like particles: the antiviral protein MxA can inhibit primary transcription of bunyaviruses. Virology 385:400-408. [DOI] [PubMed] [Google Scholar]
- 21.Habjan, M., A. Pichlmair, R. M. Elliott, A. K. Overby, T. Glatter, M. Gstaiger, G. Superti-Furga, H. Unger, and F. Weber. 2009. NSs protein of rift valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J. Virol. 83:4365-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayasaka, D., N. Nagata, Y. Fujii, H. Hasegawa, T. Sata, R. Suzuki, E. A. Gould, I. Takashima, and S. Koike. 2009. Mortality following peripheral infection with tick-borne encephalitis virus results from a combination of central nervous system pathology, systemic inflammatory and stress responses. Virology 390:139-150. [DOI] [PubMed] [Google Scholar]
- 23.Heinz, F. X., and C. Kunz. 2004. Tick-borne encephalitis and the impact of vaccination. Arch. Virol. Suppl. 2004:201-205. [DOI] [PubMed] [Google Scholar]
- 24.Hiscott, J. 2007. Triggering the innate antiviral response through IRF-3 activation. J. Biol. Chem. 282:15325-15329. [DOI] [PubMed] [Google Scholar]
- 25.Hoenen, A., W. Liu, G. Kochs, A. A. Khromykh, and J. M. Mackenzie. 2007. West Nile virus-induced cytoplasmic membrane structures provide partial protection against the interferon-induced antiviral MxA protein. J. Gen. Virol. 88:3013-3017. [DOI] [PubMed] [Google Scholar]
- 26.Iwamura, T., M. Yoneyama, K. Yamaguchi, W. Suhara, W. Mori, K. Shiota, Y. Okabe, H. Namiki, and T. Fujita. 2001. Induction of IRF-3/-7 kinase and NF-kappaB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells 6:375-388. [DOI] [PubMed] [Google Scholar]
- 27.Jennings, S., L. Martínez-Sobrido, A. García-Sastre, F. Weber, and G. Kochs. 2005. Thogoto virus ML protein suppresses IRF3 function. Virology 331:63-72. [DOI] [PubMed] [Google Scholar]
- 28.Jorns, C., D. Holzinger, R. Thimme, H. C. Spangenberg, M. Weidmann, J. Rasenack, H. E. Blum, O. Haller, and G. Kochs. 2006. Rapid and simple detection of IFN-neutralizing antibodies in chronic hepatitis C non-responsive to IFN-alpha. J. Med. Virol. 78:74-82. [DOI] [PubMed] [Google Scholar]
- 29.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981-988. [DOI] [PubMed] [Google Scholar]
- 30.Keller, B. C., C. L. Johnson, A. K. Erickson, and M. Gale, Jr. 2007. Innate immune evasion by hepatitis C virus and West Nile virus. Cytokine Growth Factor Rev. 18:535-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klausner, R. D., J. G. Donaldson, and J. Lippincott-Schwartz. 1992. Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol. 116:1071-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knoops, K., M. Kikkert, S. H. Worm, J. C. Zevenhoven-Dobbe, Y. van der Meer, A. J. Koster, A. M. Mommaas, and E. J. Snijder. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kochs, G., A. Garcia-Sastre, and L. Martinez-Sobrido. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J. Virol. 81:7011-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leyssen, P., C. Drosten, M. Paning, N. Charlier, J. Paeshuyse, E. De Clercq, and J. Neyts. 2003. Interferons, interferon inducers, and interferon-ribavirin in treatment of flavivirus-induced encephalitis in mice. Antimicrob. Agents Chemother. 47:777-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindquist, L., and O. Vapalahti. 2008. Tick-borne encephalitis. Lancet 371:1861-1871. [DOI] [PubMed] [Google Scholar]
- 36.Liu, W. J., H. B. Chen, X. J. Wang, H. Huang, and A. A. Khromykh. 2004. Analysis of adaptive mutations in Kunjin virus replicon RNA reveals a novel role for the flavivirus nonstructural protein NS2A in inhibition of beta interferon promoter-driven transcription. J. Virol. 78:12225-12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, W. J., X. J. Wang, D. C. Clark, M. Lobigs, R. A. Hall, and A. A. Khromykh. 2006. A single amino acid substitution in the West Nile virus nonstructural protein NS2A disables its ability to inhibit alpha/beta interferon induction and attenuates virus virulence in mice. J. Virol. 80:2396-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lobigs, M., A. Mullbacher, Y. Wang, M. Pavy, and E. Lee. 2003. Role of type I and type II interferon responses in recovery from infection with an encephalitic flavivirus. J. Gen. Virol. 84:567-572. [DOI] [PubMed] [Google Scholar]
- 39.Mackenzie, J. 2005. Wrapping things up about virus RNA replication. Traffic 6:967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mackenzie, J. M., M. K. Jones, and E. G. Westaway. 1999. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J. Virol. 73:9555-9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mackenzie, J. M., M. K. Jones, and P. R. Young. 1996. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology 220:232-240. [DOI] [PubMed] [Google Scholar]
- 42.Mackenzie, J. M., M. K. Jones, and P. R. Young. 1996. Improved membrane preservation of flavivirus-infected cells with cryosectioning. J. Virol. Methods 56:67-75. [DOI] [PubMed] [Google Scholar]
- 43.Mackenzie, J. M., A. A. Khromykh, M. K. Jones, and E. G. Westaway. 1998. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 245:203-215. [DOI] [PubMed] [Google Scholar]
- 44.Mackenzie, J. M., and E. G. Westaway. 2001. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 75:10787-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mansfield, K. L., N. Johnson, L. P. Phipps, J. R. Stephenson, A. R. Fooks, and T. Solomon. 2009. Tick-borne encephalitis virus—a review of an emerging zoonosis. J. Gen. Virol. 90:1781-1794. [DOI] [PubMed] [Google Scholar]
- 46.Marcus, P. I., and M. J. Sekellick. 1977. Defective interfering particles with covalently linked [+/−]RNA induce interferon. Nature 266:815-819. [DOI] [PubMed] [Google Scholar]
- 47.Matrosovich, M., T. Matrosovich, W. Garten, and H. D. Klenk. 2006. New low-viscosity overlay medium for viral plaque assays. Virol. J. 3:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller, S., and J. Krijnse-Locker. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller, S., S. Sparacio, and R. Bartenschlager. 2006. Subcellular localization and membrane topology of the dengue virus type 2 non-structural protein 4B. J. Biol. Chem. 281:8854-8863. [DOI] [PubMed] [Google Scholar]
- 50.Mogensen, T. H. 2009. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 22:240-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Munoz-Jordan, J. L., M. Laurent-Rolle, J. Ashour, L. Martinez-Sobrido, M. Ashok, W. I. Lipkin, and A. Garcia-Sastre. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 79:8004-8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. U. S. A. 100:14333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niedrig, M., U. Klockmann, W. Lang, J. Roeder, S. Burk, S. Modrow, and G. Pauli. 1994. Monoclonal antibodies directed against tick-borne encephalitis virus with neutralizing activity in vivo. Acta Virol. 38:141-149. [PubMed] [Google Scholar]
- 54.Överby, A. K., V. Popov, E. P. Neve, and R. F. Pettersson. 2006. Generation and analysis of infectious virus-like particles of Uukuniemi virus (Bunyaviridae): a useful system for studying bunyaviral packaging and budding. J. Virol. 80:10428-10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pichlmair, A., and C. Reis e Sousa. 2007. Innate recognition of viruses. Immunity 27:370-383. [DOI] [PubMed] [Google Scholar]
- 56.Pichlmair, A., O. Schulz, C. P. Tan, J. Rehwinkel, H. Kato, O. Takeuchi, S. Akira, M. Way, G. Schiavo, and C. Reis e Sousa. 2009. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 83:10761-10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Randall, R. E., and S. Goodbourn. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1-47. [DOI] [PubMed] [Google Scholar]
- 58.Robertson, S. J., D. N. Mitzel, R. T. Taylor, S. M. Best, and M. E. Bloom. 2009. Tick-borne flaviviruses: dissecting host immune responses and virus countermeasures. Immunol. Res. 43:172-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roth-Cross, J. K., L. Martinez-Sobrido, E. P. Scott, A. Garcia-Sastre, and S. R. Weiss. 2007. Inhibition of the alpha/beta interferon response by mouse hepatitis virus at multiple levels. J. Virol. 81:7189-7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruzek, D., J. Salat, M. Palus, T. S. Gritsun, E. A. Gould, I. Dykova, A. Skallova, J. Jelinek, J. Kopecky, and L. Grubhoffer. 2009. CD8+ T-cells mediate immunopathology in tick-borne encephalitis. Virology 384:1-6. [DOI] [PubMed] [Google Scholar]
- 61.Sadler, A. J., and B. R. Williams. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Samuel, M. A., and M. S. Diamond. 2005. Alpha/beta interferon protects against lethal West Nile virus infection by restricting cellular tropism and enhancing neuronal survival. J. Virol. 79:13350-13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 64.Scholle, F., and P. W. Mason. 2005. West Nile virus replication interferes with both poly(I:C)-induced interferon gene transcription and response to interferon treatment. Virology 342:77-87. [DOI] [PubMed] [Google Scholar]
- 65.Schwaiger, M., and P. Cassinotti. 2003. Development of a quantitative real-time RT-PCR assay with internal control for the laboratory detection of tick borne encephalitis virus (TBEV) RNA. J. Clin. Virol. 27:136-145. [DOI] [PubMed] [Google Scholar]
- 66.Spiegel, M., A. Pichlmair, L. Martinez-Sobrido, J. Cros, A. Garcia-Sastre, O. Haller, and F. Weber. 2005. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 79:2079-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suss, J. 2008. Tick-borne encephalitis in Europe and beyond—the epidemiological situation as of 2007. Euro Surveill. 13:pii=18916. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=18916. [PubMed] [Google Scholar]
- 68.Targett-Adams, P., S. Boulant, and J. McLauchlan. 2008. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J. Virol. 82:2182-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thiel, V., and F. Weber. 2008. Interferon and cytokine responses to SARS-coronavirus infection. Cytokine Growth Factor Rev. 19:121-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vilcek, J. 1960. An interferon-like substance released from tickborne encephalitis virus-infected chick embryo fibroblast cells. Nature 187:73-74. [DOI] [PubMed] [Google Scholar]
- 71.Wallner, G., C. W. Mandl, M. Ecker, H. Holzmann, K. Stiasny, C. Kunz, and F. X. Heinz. 1996. Characterization and complete genome sequences of high- and low-virulence variants of tick-borne encephalitis virus. J. Gen. Virol. 77(Pt. 5):1035-1042. [DOI] [PubMed] [Google Scholar]
- 72.Wathelet, M. G., P. M. Berr, and G. A. Huez. 1992. Regulation of gene expression by cytokines and virus in human cells lacking the type-I interferon locus. Eur. J. Biochem. 206:901-910. [DOI] [PubMed] [Google Scholar]
- 73.Weber, F., and O. Haller. 2007. Viral suppression of the interferon system. Biochimie 89:836-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weber, F., V. Wagner, S. B. Rasmussen, R. Hartmann, and S. R. Paludan. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80:5059-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Welsch, S., S. Miller, I. Romero-Brey, A. Merz, C. K. Bleck, P. Walther, S. D. Fuller, C. Antony, J. Krijnse-Locker, and R. Bartenschlager. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Werme, K., M. Wigerius, and M. Johansson. 2008. Tick-borne encephalitis virus NS5 associates with membrane protein scribble and impairs interferon-stimulated JAK-STAT signalling. Cell. Microbiol. 10:696-712. [DOI] [PubMed] [Google Scholar]
- 77.Westaway, E. G. 1973. Proteins specified by group B togaviruses in mammalian cells during productive infections. Virology 51:454-465. [DOI] [PubMed] [Google Scholar]
- 78.Westaway, E. G., J. M. Mackenzie, M. T. Kenney, M. K. Jones, and A. A. Khromykh. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71:6650-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wilson, J. R., P. F. de Sessions, M. A. Leon, and F. Scholle. 2008. West Nile virus nonstructural protein 1 inhibits TLR3 signal transduction. J. Virol. 82:8262-8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yoneyama, M., and T. Fujita. 2009. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 227:54-65. [DOI] [PubMed] [Google Scholar]
- 81.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]