Abstract
The RNA polymerase of influenza A virus is a host range determinant and virulence factor. In particular, the PB2 subunit of the RNA polymerase has been implicated as a crucial factor that affects cell tropism as well as virulence in animal models. These findings suggest that host factors associating with the PB2 protein may play an important role during viral replication. In order to identify host factors that associate with the PB2 protein, we purified recombinant PB2 from transiently transfected mammalian cells and identified copurifying host proteins by mass spectrometry. We found that the PB2 protein associates with the cytosolic chaperonin containing TCP-1 (CCT), stress-induced phosphoprotein 1 (STIP1), FK506 binding protein 5 (FKBP5), α- and β-tubulin, Hsp60, and mitochondrial protein p32. Some of these binding partners associate with each other, suggesting that PB2 might interact with these proteins in multimeric complexes. More detailed analysis of the interaction of the PB2 protein with CCT revealed that PB2 associates with CCT as a monomer and that the CCT binding site is located in a central region of the PB2 protein. PB2 proteins from various influenza virus subtypes and origins can associate with CCT. Silencing of CCT resulted in reduced viral replication and reduced PB2 protein and viral RNA accumulation in a ribonucleoprotein reconstitution assay, suggesting an important function for CCT during the influenza virus life cycle. We propose that CCT might be acting as a chaperone for PB2 to aid its folding and possibly its incorporation into the trimeric RNA polymerase complex.
Influenza A viruses, members of the family of Orthomyxoviridae, contain a segmented RNA genome of negative polarity. The genomic RNA segments together with the three subunits of the viral RNA-dependent RNA polymerase (PB1, PB2, and PA protein) and the nucleoprotein (NP) form viral ribonucleoprotein complexes (vRNPs). The PB1 subunit is the polymerase itself, while the PB2 and PA subunits are involved in the generation of 5′ capped RNA primers through binding to and endonucleolytic cleavage of host pre-mRNAs (8, 10, 11, 41, 61). After the virus enters the cell via endocytosis, vRNPs are released into the cytoplasm and transported into the nucleus. In the nucleus, vRNPs catalyze the synthesis of viral mRNAs and complementary RNAs (cRNA) which, in turn, are used as templates for the synthesis of vRNAs. The newly formed vRNPs in association with other viral proteins (M1 and nonstructural protein 2/nuclear export factor [NS2/NEP]) are transported into the cytoplasm and subsequently to the cell membrane, where the assembly process takes place, followed by the release of progeny virions by budding (44).
The PB1, PB2, and PA proteins are synthesized in the cytoplasm whereupon PB1 and PA form a dimeric complex that is transported into the nucleus. In the nucleus the dimer assembles with the PB2 subunit, which is transported separately (7, 14). RanBP5 was identified as a factor that is involved in the import of the PB1-PA dimer into the nucleus (6), while PB2 uses the classical importin-α/β pathway for nuclear import (57). Recently, further support for this transport and assembly model was provided by using fluorescence cross-correlation spectroscopy (25). An alternative pathway proposed for the import of the RNA polymerase subunits into the nucleus involves the heat shock protein 90 (Hsp90) that was shown to interact with the PB1 and PB2 proteins (39). Heat shock protein 70 (Hsp70) was also found to interact with the influenza virus polymerase subunits and vRNPs, and it was implicated in blocking the nuclear export of vRNPs (22).
The RNA polymerase has been implicated as a host range determinant and pathogenicity factor of influenza viruses. In particular, amino acid residue 627 in the PB2 subunit was shown to determine the ability of certain influenza viruses to replicate in avian and mammalian cells (34, 54). A lysine at position 627, characteristic of most human influenza virus strains, appears to enhance replication in mammalian cells, while a glutamic acid, found in most avian isolates, attenuates virus replication in mammalian cells. The presence of a lysine was also shown to enhance virulence in mammalian models and has been associated with the lethality of H5N1 viruses in humans (20). It has been proposed that a negative factor, present in mammalian cells, specifically reduces the activity of a polymerase containing a glutamic acid (38). However, the identity of this factor remains to be determined. Interestingly, the 2009 H1N1 pandemic influenza virus encodes a glutamic acid at this position, and a second-site suppressor mutation has been identified in PB2 that promotes activity in mammalian cells (37). Introduction of a lysine at residue 627 in the 2009 H1N1 pandemic virus did not result in enhanced virulence (21, 62). Several other amino acid residues in the PB2 protein were also implicated in host range determination and virulence, suggesting that multiple amino acid substitutions are involved (15, 48). Collectively, these results suggest that the PB2 protein interacts with host factors and that these interactions have implications for host range and virulence.
Therefore, we set up a biochemical copurification assay followed by mass spectrometry to identify host factors that associate with the PB2 protein in mammalian cells. We confirmed the interaction with several previously identified host factors, e.g., Hsp70 and Hsp90, and identified novel host proteins that interact with the PB2 protein. Among these, we have identified the oligomeric chaperonin containing TCP-1 (CCT) (also known as TRiC [TCP-1 ring complex]) and investigated the significance of this interaction in more detail. We found that CCT interacts with the PB2 protein but not with the PB1 or PA protein. However, PB2 in association with PB1 or PB1 and PA did not interact with CCT. We also found that PB2 proteins of different influenza virus strains of different origins, hosts, and subtypes interact with CCT. Growth of influenza virus, as well as the accumulation of the PB2 protein and viral RNAs in a ribonucleoprotein reconstitution assay, was reduced in CCT-silenced cells compared to that in control cells. These results suggest a role for CCT in the influenza A virus life cycle, possibly acting as a chaperone for the PB2 protein.
MATERIALS AND METHODS
Plasmids.
The plasmids coding for untagged or tandem affinity purification (TAP)-tagged PB1, PB2, and PA of influenza A/WSN/33 virus (pcDNA-PB1, pcDNA-PB2, pcDNA-PA, pcDNA-PB1-TAP, pcDNA-PB2-TAP, and pcDNA-PA-TAP) and pPOLI-NA-RT have been described previously (7, 12-14, 45). pcDNA-PB2-GFP (where GFP is green fluorescent protein) plasmids encoding wild-type or deletion mutant PB2-GFP fusion proteins and pcDNA-PB2-TAP plasmids encoding PB2 proteins of A/NT/60/68 (H3N2), A/Hong Kong/156/97 (H5N1), and A/Vietnam/1194/04 (H5N1) viruses were also described previously (3, 30). pcDNA-PB2-TAP plasmids encoding PB2 proteins of A/Ann Arbor/6/60ca (H2N2), A/Hong Kong/483/97 (H5N1), A/Hong Kong/486/97 (H5N1), and A/Netherlands/219/03 (H7N7) viruses were constructed by replacing the PB2 open reading frame (ORF) of the pcDNA-PB2-TAP (WSN) plasmid with the corresponding PB2 ORFs that were PCR amplified from cDNAs or viral RNAs provided by K. Subbarao (NIAID, NIH). pcDNA-PB2-TAP encoding the PB2 of A/turkey/England/5092/91 (H5N1) that lacks the calmodulin-binding domain of the TAP tag was constructed by R. Harvey (University of Oxford) by PCR amplification of the PB2 ORF from a template provided by W. Barclay (Imperial College London). pcDNA-NS2-TAP was constructed by M. Smith (University of Oxford) by replacing the GFP ORF of the pcDNA-GFP-TAP plasmid (12) with the NS2 ORF PCR amplified from pcDNA-NS2 (47).
Purification of TAP-tagged proteins.
Human embryonic kidney cells (293T) were transfected with a combination of pcDNA plasmids coding for untagged or TAP-tagged PB1, PB2, and/or PA as required. Polymerase and interacting cellular proteins were purified using the TAP method as described previously (7, 14, 46). Briefly, 48 h after transfection 293T cells were lysed, cell lysates were incubated with immunoglobulin G (IgG)-Sepharose, and bound proteins were released by tobacco etch virus (TEV) protease (Invitrogen). Purified protein samples were analyzed by sodium dodecyl sulfate-8% polyacrylamide gel electrophoresis (SDS-PAGE), followed by Western blotting or staining with a SilverXpress kit (Invitrogen).
Identification of proteins by mass spectrometry.
Protein samples were analyzed by 8% PAGE, and proteins were stained with Bio-Safe Coomassie (Bio-Rad). Bands of interest were excised, cut into small pieces, and washed twice in 100 μl of 50 mM ammonium bicarbonate in 50% acetonitrile. Gel pieces were then soaked in 100% acetonitrile for 20 min, followed by freeze-drying. The dried gel pieces were rehydrated in 30 μl of sequencing-grade trypsin (Promega) solution (0.01 μg/μl) in 20 mM ammonium bicarbonate and incubated at 16°C for 18 h. The supernatants were collected, and the gel pieces were extracted in 100 μl of 50% acetonitrile in 0.1% formic acid. The supernatants were combined, and the extracted peptides were freeze-dried. Peptides were dissolved in 7 μl of 0.1% formic acid and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) at the Oxford Central Proteomics Facility, Sir William Dunn School of Pathology, University of Oxford. The data were searched against the International Protein Index (IPI) database, fixed modification was set as carbamidomethyl cysteine, and variable modification was set as oxidized methionine.
Immunoprecipitations.
293T cells were transfected with pcDNA-PB2-GFP plasmids coding for wild-type or deletion mutant PB2-GFP fusion proteins (3). Forty-eight hours after transfection the cells were lysed, and immunoprecipitations were performed with a monoclonal CCTβ-specific antibody (Serotec). Immunoprecipitated PB2-GFP proteins were detected with a monoclonal GFP-specific antibody (Santa Cruz).
Design of siRNAs and knockdown of CCT.
The small interfering RNA (siRNA) targeting CCTβ (UGAUCAUGGUUCCGAUGAAUU) was designed by using the Dharmacon siDESIGN Center and purchased from Sigma-Proligo. To knock down CCT, human lung carcinoma cells (A549) in 35-mm dishes were transfected in suspension with 5 μg of siRNA using Lipofectamine 2000 transfection reagent (Invitrogen). The medium was replaced with fresh medium 6 h later. The transfection was repeated 24 h later in cell monolayers. An siRNA targeting GFP was used as a negative control (6). Twenty-four hours later the cells were infected with influenza A/WSN/33 virus at a multiplicity of infection (MOI) of 0.001. At 24, 36, and 48 h postinfection supernatants were collected, and plaque titers were determined by plaque assay in MDBK cells. Alternatively, for ribonucleoprotein reconstitution assays, 293T cells were transfected in suspension with 5 μg of siRNA targeting CCTβ or GFP as a negative control.
Ribonucleoprotein reconstitution assay, primer extension, and Western blot analyses.
Ribonucleoprotein reconstitution assays were performed in 293T cells treated with siRNAs for 32 h as described above. Cell monolayers were transfected with 1 μg each of pcDNA-PB1, pcDNA-PB2, pcDNA-PA, pcDNA-NP, and pPOLI-NA-RT using Lipofectamine 2000. At 24 h posttransfection, total RNA was extracted using TRIzol (Invitrogen), and the accumulation levels of neuraminidase gene-specific mRNA, cRNA, and vRNA were determined by primer extension analysis as described previously (13, 59). Protein was also harvested and analyzed for PB2 and CCTβ levels by Western blotting. Signals were generated by using ECL reagent (Amersham) and detected by autoradiography. Images were quantitated by using Aida.
Cell viability assay.
After two consecutive transfections with siRNAs, A549 cells were trypsinized, resuspended in phosphate-buffered saline (PBS), and stained with 1% trypan blue. Numbers of viable and dead cells were determined using a light microscope.
RESULTS
Identification of host proteins that interact with the PB2 protein of influenza A virus.
In a search for host proteins that interact with the PB2 subunit of the influenza virus RNA polymerase complex, we transfected 293T cells with a plasmid expressing PB2 of influenza A/WSN/33 virus with a C-terminal TAP (tandem affinity purification) tag. The PB2 protein was purified from cell lysates by using IgG-Sepharose chromatography and analyzed by SDS-PAGE, followed by staining with silver (Fig. 1). We used the viral nonstructural protein 2/nuclear export factor (NS2/NEP) as a negative control. In the sample containing purified PB2, several distinct bands were detected between the 50- and 70-kDa marker and near the 37-kDa size marker which were not present in the control sample. For proteomic analysis, the protein sample was concentrated by precipitation and analyzed by SDS-PAGE, followed by staining with Bio-Safe Coomassie. Gel slices corresponding to PB2 and the previously identified Hsp90 and Hsp70 (7) as well as gel slices originating from the 50- to 70-kDa and the 37-kDa regions were subjected to in-gel trypsin digestion followed by LC-MS/MS. The identities of PB2, Hsp90, and Hsp70 were confirmed (Table 1). In addition, all subunits of the eight-subunit chaperonin containing TCP-1 (CCT) were identified as well as stress-induced phosphoprotein 1 (STIP1; also known as Hsp70/90 organizing protein or transformation-sensitive protein IEF SSP 3521), Hsp60, FK506-binding protein (FKBP5), α- and β-tubulin, and mitochondrial protein p32 (also known as pre-mRNA splicing factor SF2p32). The theoretical masses of the identified proteins were compatible with their locations on the gel (Table 1).
FIG. 1.
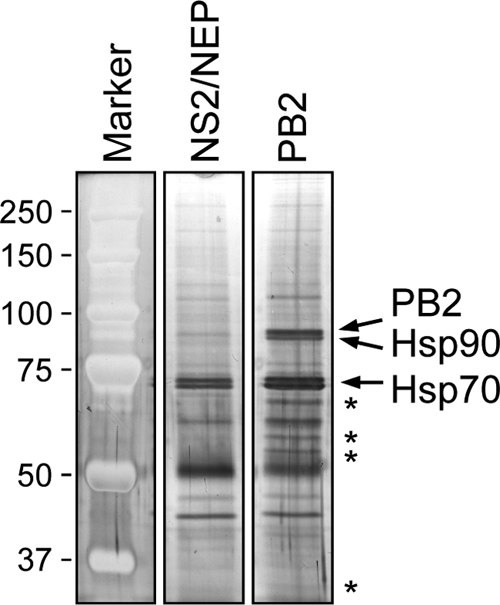
Analysis of purified influenza virus PB2 protein. Purified PB2-TAP and NS2/NEP-TAP were analyzed by SDS-8% PAGE and stained with silver. The positions of PB2, Hsp90, and Hsp70 are indicated on the right. Note that NS2/NEP-TAP after TEV cleavage has a molecular mass of approximately 17 kDa, and therefore it is not detectable in the gel system used. The positions of unique bands in the PB2 sample are indicated by asterisks on the right. Molecular mass markers (Bio-Rad) in kDa are indicated on the left.
TABLE 1.
Identification of PB2 and binding partners by mass spectrometry
| Accession code | Protein name | Mass (Da) | Mascot score | No. of unique peptides | Sequence coverage (%) |
|---|---|---|---|---|---|
| P3IV33 | PB2 | 86,027 | 926 | 40 | 39 |
| HHHU86 | Hsp90-α | 85,006 | 282 | 13 | 16 |
| HHHU84 | Hsp90-β | 83,584 | 392 | 16 | 20 |
| Q5SP17_HUMAN | Hsp70 1A | 70,280 | 372 | 16 | 31 |
| A38093 | STIP1 | 63,227 | 179 | 9 | 17 |
| A32800 | Hsp60 | 61,187 | 385 | 14 | 32 |
| S10486 | CCTα | 60,819 | 97 | 2 | 4 |
| AAH06543 | CCTɛ | 60,089 | 95 | 6 | 11 |
| TCPQ_HUMAN | CCTθ | 60,022 | 207 | 6 | 14 |
| Q6IBT3_HUMAN | CCTη | 59,804 | 150 | 6 | 16 |
| Q5SZY0_HUMAN | CCTγ | 58,505 | 52 | 3 | 9 |
| AAC96010 | CCTδ | 58,401 | 183 | 6 | 12 |
| TCPZ_HUMAN | CCTζ | 58,313 | 121 | 5 | 13 |
| TCPB_HUMAN | CCTβ | 57,663 | 270 | 10 | 23 |
| JC5422 | FKBP5 | 51,693 | 132 | 7 | 20 |
| I77403 | Tubulin-α-1 | 50,804 | 96 | 5 | 11 |
| A26561 | Tubulin-β | 50,240 | 200 | 11 | 28 |
| AAA73055 | Mitochondrial protein p32 | 31,287 | 98 | 3 | 9 |
CCT interacts with the monomeric PB2 protein.
CCT is a molecular chaperone belonging to the family of chaperonins, a conserved class of large double-ring complexes of ∼800 kDa enclosing a central cavity, which is found in the cytoplasm of all eukaryotic cells (19, 53). It is composed of two identical rings, each comprised of eight different subunits (CCTα, CCTβ, CCTγ, CCTδ, CCTɛ, CCTζ, CCTη, and CCTθ), and it was found to be a central mediator of cytosolic protein folding and assembly. The identification of all eight CCT subunits in the mass spectrometry analysis of interacting host proteins of PB2 strongly suggests that PB2 uses the CCT complex as a chaperone, and therefore we decided to investigate the characteristics and functional significance of this interaction in more detail.
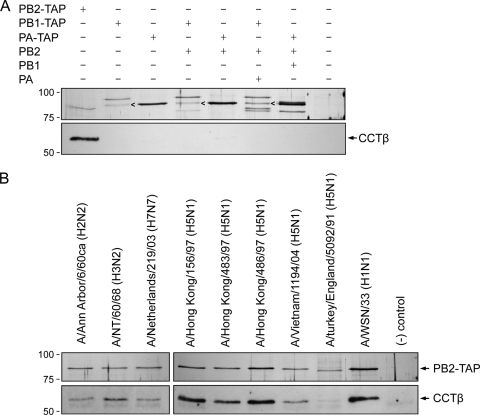
In order to address the specificity of the interaction of CCT with the influenza virus RNA polymerase, we purified monomers or dimers of polymerase subunits as well as the trimeric polymerase complex from cells transfected with combinations of expression plasmids encoding TAP-tagged or untagged polymerase subunits. Silver staining of SDS-PAGE gels confirmed the presence of the polymerase subunits (Fig. 2 A, top panel). To analyze for the presence of copurifying CCT, we performed Western blotting using an antibody specific for CCTβ (Fig. 2A, bottom panel). We found that CCTβ copurified only with monomeric PB2 and not with monomeric PB1 or PA. No CCTβ was observed copurifying with the polymerase trimer or dimers containing PB2. It should be noted that when expressing the polymerase trimer or dimers, we used TAP-tagged PB1 or PA to eliminate the possibility of purifying monomeric PB2 subunits interacting with CCT. The interaction of PB2 with CCT was also confirmed by using an antibody against CCTζ (results not shown).
FIG. 2.
Western blot analyses of the interaction of the influenza virus polymerase proteins with CCT. (A) Purification of TAP-tagged RNA polymerase subunits, dimers, and trimeric complexes from 293T cells transfected with the indicated plasmids. Purified proteins were analyzed by SDS-8% PAGE, followed by staining with silver (upper panel) or by Western blotting using a monoclonal CCTβ-specific antibody (Serotec) (lower panel). The arrowheads indicate the position of copurifying Hsp90. (B) Interaction of CCTβ with PB2 proteins of influenza viruses of various subtypes and origins. TAP-tagged PB2 proteins of the indicated influenza virus strains were purified from transfected 293T cells, and purified proteins were analyzed by SDS-8% PAGE, followed by staining with silver (upper panel) or by Western blotting using a CCTβ-specific antibody (lower panel). Positions of molecular mass markers (Bio-Rad) in kDa are indicated on the left.
CCT interacts with PB2 proteins originating from different influenza A virus subtypes.
To assess whether CCT binds to the PB2 protein of different influenza A viruses, we transfected 293T cells with plasmids expressing PB2-TAP from influenza A viruses of different subtypes (H1N1, H2N2, H3N2, H5N1, and H7N7) and origins (human and avian isolates) (Fig. 2B). The PB2 proteins from these viral strains are 94 to 99% identical but show differences at a number of key amino acid residues, e.g., 627, implicated in host range and virulence. Expression and purification of the PB2 proteins were confirmed by SDS-PAGE, followed by staining with silver (Fig. 2B, top panel). Copurification of CCT was analyzed by Western blotting using the CCTβ subunit-specific antibody (Fig. 2B, bottom panel). We found that CCT interacts with PB2 regardless the subtype, host, and origin of the virus. However, we found lower levels of CCT copurifying with the PB2 protein of A/turkey/England/5092/91 (H5N1), suggesting that there might be differences in the affinities of different PB2 proteins for CCT.
CCT interacts with a central region of PB2.
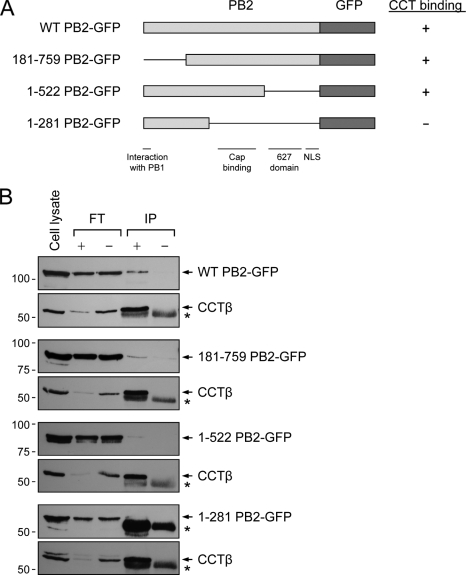
No structural information is available for a full-length PB2 protein although high-resolution three-dimensional (3D) structures have been reported for several PB2 fragments (17, 33, 55, 57, 58). The N terminus contains a binding site for PB1 while the C-terminal region contains a bipartite nuclear localization signal (NLS) that has been shown to bind importin-α. A cap-binding domain has been identified in the central region of PB2, and structural information has also been obtained on a so-called 627 domain that includes amino acid 627 implicated in host range determination and virulence. In order to address the question of which region of PB2 interacts with CCT, we used a set of previously described deletion mutants of PB2 fused to green fluorescent protein (GFP) (3) (Fig. 3 A). Full-length PB2-GFP or deletion mutants were expressed in transiently transfected 293T cells, and cell lysates were used to immunoprecipitate CCT with the antibody specific for CCTβ. Western blot analysis of CCTβ levels in cell lysates, flowthrough fractions, and immunoprecipitates confirmed that CCTβ was efficiently immunoprecipitated, as demonstrated by its depletion in the flowthrough fractions and enrichment in the immunoprecipitates in the presence of the antibody in comparison to results for the no-antibody control (Fig. 3B). Full-length PB2-GFP was coimmunoprecipitated with CCT, providing further evidence for their interaction (Fig. 3B). PB2 deletion mutants lacking the N-terminal PB1 interaction domain (residues 181 to 759 [181-759 PB2-GFP]) or the 627 domain and the C-terminal NLS (1-522 PB2-GFP) were also coimmunoprecipitated with CCT. In contrast, the PB2 deletion mutant encompassing the N-terminal PB1 interaction domain but lacking the central cap binding and 627 domains as well as the C-terminal NLS could not be coimmunoprecipitated. Taken together, these results suggest that a central region of PB2, possibly overlapping with the cap-binding domain, is involved in the interaction with CCT. However, it should be noted that only very small amounts of wild-type PB2-GFP or of the deletion mutants 181-759 PB2-GFP and 1-522 PB2-GFP coimmunoprecipitated with CCT, suggesting a transient interaction between the two proteins.
FIG. 3.
Mapping of the CCT interaction domain in PB2. (A) Diagrams of wild-type (WT) and deletion mutant PB2-GFP fusion constructs. The known domains of PB2 are indicated (49). (B) Coimmunoprecipitation of wild-type and deletion mutant PB2-GFP fusion proteins with CCT. Wild-type and the indicated deletion mutant PB2-GFP fusion proteins were expressed in transfected 293T cells, and immunoprecipitations were performed with a monoclonal CCTβ-specific antibody (Serotec) (+) or in the absence of an antibody (−). PB2-GFP and CCTβ proteins were detected with a monoclonal GFP-specific antibody (Santa Cruz) and the CCTβ-specific antibody, respectively, in cell lysates, flowthrough (FT), and immunoprecipitates (IP), as indicated. The star indicates the position of residual protein A released from the Sepharose and/or the position of the heavy chain of IgG that cross-reacts with the antibodies used in Western blotting. Positions of molecular mass markers (Bio-Rad) in kDa are indicated on the left.
Knockdown of CCT reduces influenza virus replication.
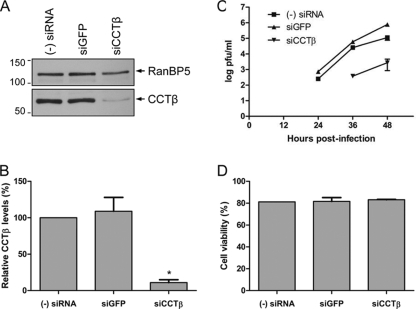
To assess the biological significance of CCT in the influenza A virus life cycle, we performed siRNA-mediated knockdown of CCT. An siRNA specific for the CCTβ subunit was transfected into A549 cells, and an siRNA targeting GFP was used as a negative control. Cell lysates were analyzed for the presence of CCTβ by Western blotting (Fig. 4 A). siRNA treatment resulted in about a 10-fold reduction in CCTβ levels (Fig. 4B).
FIG. 4.
Knockdown of CCT in A549 cells and its effect on virus growth. (A) Western blot analysis of CCTβ in A549 cells treated with no (−) siRNA or treated with GFP- or CCTβ-specific siRNA (siGFP or siCCTβ, respectively). RanBP5 detected with a polyclonal RanBP5-specific antibody (Santa Cruz) was used as a loading control. Positions of molecular mass markers (Bio-Rad) in kDa are indicated on the left. (B) Quantitation of Western blot analyses of the knockdown of CCTβ from panel A. CCTβ intensities in siRNA-treated cells were expressed as a percentage of intensities observed in cells not treated with siRNA, which was set to 100%. CCTβ levels were normalized to the levels of RanBP5. Bars represent standard deviations based on three independent experiments. *, P < 0.05, based on a one-sample Student's t test. (C) Growth curves of influenza A/WSN/33 virus in CCT-silenced A549 cells. Control cells not treated with siRNA (−) or cells treated with GFP- or CCTβ-specific siRNA were infected at an MOI of 0.001. At the indicated points postinfection (24, 36, and 48 h), samples were collected, and virus titers were determined by plaque assay in MDBK cells. The results shown represent an average of two independent experiments, with the range indicated. (D) Viability assay of CCT-silenced A549 cells. Cells treated as in panel A were stained with 1% trypan blue, and viable and dead cells were counted. Cell viability was expressed as the percentage of viable cells in the culture. Results shown are an average of two independent experiments, with the range indicated.
To investigate the effect of CCT knockdown on the virus life cycle, we infected A549 cells silenced for CCT as well as control knockdown cells with influenza A/WSN/33 virus at an MOI of 0.001. The amount of infectious virus released into the culture medium was determined by a plaque assay at 24, 36, and 48 h postinfection (Fig. 4C). At 24 h postinfection, no virus was detectable in the medium of cells treated with CCTβ-specific siRNA. In contrast, virus was detected in the medium of both the untreated and control siRNA-treated cells. At the 36- and 48-h time points there was at least a 2-log reduction in viral titers in CCT-silenced cells, suggesting an important role for CCT in the influenza virus life cycle.
As it has been previously reported that CCT knockdown affects cell viability (1), to ensure that the same numbers of viable cells were infected, we performed a cell viability assay. A549 cells were treated with CCTβ- or GFP-specific siRNA or were left untreated as with the infections above. The number of viable cells was determined by staining with 1% trypan blue (Fig. 4D). The results show that the numbers of viable cells used for infection were comparable.
Knockdown of CCT reduces PB2 and viral RNA accumulation levels in a ribonucleoprotein reconstitution assay.
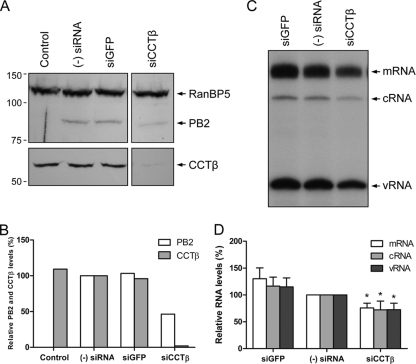
In order to investigate the mechanisms that resulted in the attenuation of viral growth in CCT knockdown cells, we used a ribonucleoprotein reconstitution assay to assess the effect of CCTβ knockdown on PB2 expression levels and the accumulation of viral RNAs. 293T cells were transfected with siRNA to silence CCTβ. siRNA against GFP was used as a negative control. Thirty-two hours later, siRNA-treated cells were transfected with plasmids to express the three viral RNA polymerase subunits, the nucleoprotein, and the neuraminidase vRNA as a reporter gene. Twenty-four hours later, total cell lysates were analyzed by Western blotting (Fig. 5 A). Analysis of CCTβ levels confirmed that treatment with CCTβ-specific siRNA resulted in the knockdown of CCTβ (Fig. 5B). Interestingly, there was also about a 2-fold reduction in the accumulation levels of the PB2 protein in the CCTβ-silenced cells. No reduction in PB2 levels was observed in cells treated with GFP-specific siRNA. The reduction of the PB2 protein levels in the CCTβ-silenced cells correlated with a reduction in viral RNA levels (Fig. 5C). Primer extension analysis revealed a statistically significant reduction in the accumulation of mRNA, cRNA, and vRNA of the neuraminidase reporter gene (Fig. 5D). No reduction was observed in viral RNA levels in cells treated with siRNA specific for GFP. Taken together, these results strongly suggest that the viral attenuation observed in CCTβ-silenced cells (Fig. 4C) could be due to reduced accumulation of PB2 and, consequently, a reduction in the accumulation of viral RNA levels.
FIG. 5.
The effect of CCT knockdown on PB2 protein and viral RNA levels. (A) Western blot analysis of PB2 and CCTβ in untreated 293T cells [(Control and (−) siRNA] or 293T cells treated with GFP- or CCTβ-specific siRNA (siGFP or siCCTβ, respectively). Cells were also transfected with pcDNA-PB1, pcDNA-PB2, pcDNA-PA, pcDNA-NP, and pPOLI-NA-RT plasmids to express recombinant ribonucleoproteins. No plasmids were transfected in the control cells. PB2 was detected with a polyclonal PB2-specific antibody (3). RanBP5 detected with a polyclonal RanBP5-specific antibody (Santa Cruz) was used as a loading control. Positions of molecular mass markers (Bio-Rad) in kDa are indicated on the left. (B) Quantitation of Western blot analysis of PB2 and CCTβ protein levels from panel A. PB2 and CCTβ intensities in siRNA-treated cells were expressed as a percentage of intensities observed in cells not treated with siRNA (−), which was set to 100%. PB2 and CCTβ levels were normalized to the levels of RanBP5. (C) Primer extension analysis of mRNA, cRNA, and vRNA of the neuraminidase gene in CCTβ-silenced 293T cells. Cells were treated with siRNAs, followed by the transfection of plasmids to reconstitute ribonucleoprotein complexes as in panel A. (D) Quantitation of primer extension analysis of viral mRNA, cRNA, and vRNA levels from panel C. RNA levels in siRNA-treated cells were expressed as a percentage of RNA levels observed in untreated cells [(−) siRNA], which was set to 100%. RNA levels were normalized to the total amount of RNA used in the primer extension assay. Bars represent standard deviations based on four independent experiments. *, P < 0.05, based on one-sample Student's t test.
DISCUSSION
In this study we investigated the interactions of the PB2 subunit of the influenza virus RNA polymerase with host proteins using copurification assays, followed by the identification of copurifying host proteins by mass spectrometry. We have identified the CCT complex as a binding partner of the PB2 subunit (Table 1). We confirmed the association of CCT with the PB2 protein by Western blot analysis of host proteins copurifying with PB2 as well as by immunoprecipitation assays of CCT following Western blot analysis of immunoprecipitates for the presence of PB2. Our results show that CCT specifically interacts with the PB2 subunit of the RNA polymerase as we failed to detect an interaction with the PB1 or PA subunits. We could not detect an interaction between CCT and PB2 in the context of the polymerase trimer and a PB1-PB2 dimer, suggesting that it is the PB2 monomer that associates with CCT. PB2 proteins of various influenza virus subtypes and origins were shown to interact with CCT, suggesting that the CCT interaction is a general property of PB2 proteins. A central region of PB2 that contains the cap-binding domain is required for interaction with CCT. However, further studies will be required to determine more precisely the interaction domain and the specific amino acid residues that participate in the interaction.
Knockdown of CCT resulted in reduced growth of influenza A/WSN/33 virus showing that CCT plays an important role in the viral life cycle. Our finding that PB2 proteins of various influenza virus subtypes and origins interact with CCT strongly suggests that CCT might have an important role in the life cycle of other influenza viruses. We also found a reduction in PB2 protein and viral mRNA, cRNA, and vRNA levels in a recombinant ribonucleoprotein reconstitution assay, suggesting that CCT might act as a chaperone for PB2 in the cytoplasm. However, our results do not exclude the possibility that CCT plays an additional, indirect role in the viral life cycle. CCT is a central mediator of cytosolic protein folding and assembly, and it assists the folding of many essential proteins, including actin, tubulin, and many cell cycle regulators and signaling molecules (5, 60). More recent studies suggest that the activity of CCT may well extend beyond the folding of newly synthesized polypeptides to include a role in the organization/polymerization of the actin- and tubulin-based cytoskeletal systems (1). Interaction of the viral ribonucleoproteins with actin filaments was shown to play a role in the regulation of RNP trafficking, and the actin cytoskeleton has been also implicated in virus budding (9, 51). Thus, it remains possible that CCT silencing affects viral growth also by affecting the cellular cytoskeleton, inhibiting vRNP transport and/or viral budding. Further studies will be required to determine whether CCT plays any additional roles in the influenza virus life cycle.
The CCT complex has also been implicated in playing a role in the life cycle of several other viruses. For example, it is involved in folding of newly synthesized Gag polyproteins of Mason-Pfitzer monkey virus (M-PMV) (23). CCT also plays a role in viral assembly of hepatitis B virus (35) and initial folding of EBNA-3 nuclear protein of Epstein-Barr virus (31). Interestingly, the CCT complex has not been identified by the recent genome-wide screening studies (2, 18, 29, 32, 50, 56) as a host factor involved in the influenza virus life cycle.
In addition to discovering the association of the CCT complex with PB2, we also confirmed the association of the PB2 protein with the previously identified binding partners Hsp90 and Hsp70 (7, 22, 36, 39). Treatment of cells with Hsp90 inhibitors, e.g., geldanamycin, resulted in the reduced half-life of the PB2 protein, suggesting that Hsp90 acts as a chaperone for PB2 (4). A role for Hsp90 in the assembly and nuclear import of the RNA polymerase has also been suggested (40). The interaction of the PB2 protein with several members of the chaperone family of proteins suggests that these chaperones might perform nonredundant functions. In fact, Hsp70 is known to act upstream of CCT by holding nascent polypeptides in a state competent for folding upon release into the cytosol before passing them on to CCT (19). CCT then provides a physically defined compartment inside which a complete protein or protein domain can fold while being sequestered from the cytosol. The heat shock proteins Hsp70 and Hsp90 are also functionally linked, and STIP1 is known to act as an adaptor protein that coordinates the functions of Hsp70 and Hsp90 in protein folding (24, 52). STIP1 is thought to assist in the transfer of proteins from Hsp70 to Hsp90 by binding both Hsp90 and substrate-bound Hsp70 (42). FKBP5, a member of the large family of immunophilins, is known to play a role in immunoregulation and basic cellular processes involving protein folding and trafficking by associating with Hsp90 and microtubules. In this study, we have identified STIP1, FKBP5, and α- and β-tubulin as binding partners of the PB2 protein (Table 1), suggesting that PB2 might be involved in multimeric complexes with several chaperones and associated proteins. These proteins were also identified in some of the recent genome-wide screening studies as important for the influenza virus life cycle (32, 50, 56). The association of the influenza virus RNA polymerase and vRNPs with tubulins has been described previously (28, 36), and it has been suggested that this interaction maybe involved in the long-distance transport of virus RNPs from the nucleus to the vicinity of the cell membrane, prior to virion formation and budding (28). Our findings that the PB2 protein interacts with multiple members of the chaperone family of proteins, cochaperones, adaptor proteins, and microtubules, whether directly or indirectly, suggests that a concerted action of several chaperones might be required to ensure proper folding of the PB2 protein, its assembly into a trimeric polymerase complex, and nuclear import.
Although most of the PB2 protein accumulates in the nucleus, some PB2 can be detected in the mitochondria, where it might play a role in regulating apoptotic and host innate immune responses during viral infection (3, 16). We found that PB2 associates with Hsp60 (Table 1), a chaperonin complex, that is primarily responsible for the folding and transportation of proteins from the cytoplasm into the mitochondrial matrix (27). This finding suggests that Hsp60 might be involved in the mitochondrial transport of the PB2 protein. In fact, Hsp60 has been identified as crucial for influenza virus replication in a genome-wide siRNA screen (29). We have also identified mitochondrial protein p32 as a binding partner of the PB2 protein (Table 1). The p32 protein localizes predominantly to the mitochondrial matrix, and it has been suggested to play a role in the regulation of apoptosis (26). Our findings suggest that PB2, via its association with Hsp60 and the mitochondrial p32 protein, might play a role in regulating apoptosis. However, p32 is a multifunctional protein, present also at the cell surface, cytoplasm, and the nucleus. It interacts with a large number of cellular and viral proteins, and in some cases the interaction between p32 and a target protein has been shown to regulate important cellular activities controlling gene expression (43). Thus, the p32 protein via its association with the PB2 polymerase subunit might also be involved in the regulation of viral transcription.
In summary, we have identified several binding partners of the PB2 subunit of the influenza virus RNA polymerase. Some of these binding partners associate with each other, suggesting that the PB2 protein might interact with these proteins in multimeric complexes that aid the folding, assembly, and transport of the RNA polymerase. In support of this, silencing of CCT resulted in the attenuation of viral growth, suggesting a functional role for CCT during the influenza virus life cycle.
Acknowledgments
We thank R. Harvey and M. Smith for constructing plasmids, A. Hodgson, G. Gabriel, and K. Bier for performing preliminary experiments, and K. Subbarao, W. Barclay, S. Carr, T. Kashiwagi, and G. G. Brownlee for providing reagents. We also thank G. G. Brownlee, K. Subbarao, and F. Vreede for helpful discussions and the critical reading of the manuscript.
This work was supported by grants from the European Commission (FLUINNATE) and the MRC (grant G0700848). K.M.G. was funded by the International Biomedical Research Alliance and the OxCam Graduate Partnership Programme (NIH).
Footnotes
Published ahead of print on 23 June 2010.
REFERENCES
- 1.Brackley, K. I., and J. Grantham. 2010. Subunits of the chaperonin CCT interact with F-actin and influence cell shape and cytoskeletal assembly. Exp. Cell Res. 316:543-553. [DOI] [PubMed] [Google Scholar]
- 2.Brass, A. L., I. C. Huang, Y. Benita, S. P. John, M. N. Krishnan, E. M. Feeley, B. J. Ryan, J. L. Weyer, L. van der Weyden, E. Fikrig, D. J. Adams, R. J. Xavier, M. Farzan, and S. J. Elledge. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carr, S. M., E. Carnero, A. Garcia-Sastre, G. G. Brownlee, and E. Fodor. 2006. Characterization of a mitochondrial-targeting signal in the PB2 protein of influenza viruses. Virology 344:492-508. [DOI] [PubMed] [Google Scholar]
- 4.Chase, G., T. Deng, E. Fodor, B. W. Leung, D. Mayer, M. Schwemmle, and G. Brownlee. 2008. Hsp90 inhibitors reduce influenza virus replication in cell culture. Virology 377:431-439. [DOI] [PubMed] [Google Scholar]
- 5.Dekker, C., P. C. Stirling, E. A. McCormack, H. Filmore, A. Paul, R. L. Brost, M. Costanzo, C. Boone, M. R. Leroux, and K. R. Willison. 2008. The interaction network of the chaperonin CCT. EMBO J. 27:1827-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng, T., O. G. Engelhardt, B. Thomas, A. V. Akoulitchev, G. G. Brownlee, and E. Fodor. 2006. The role of Ran binding protein 5 in the nuclear import and assembly of the influenza virus RNA polymerase complex. J. Virol. 80:11911-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng, T., J. Sharps, E. Fodor, and G. G. Brownlee. 2005. In vitro assembly of PB2 with a PB1-PA dimer supports a new model of assembly of influenza A virus polymerase subunits into a functional trimeric complex. J. Virol. 79:8669-8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dias, A., D. Bouvier, T. Crepin, A. A. McCarthy, D. J. Hart, F. Baudin, S. Cusack, and R. W. Ruigrok. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458:914-918. [DOI] [PubMed] [Google Scholar]
- 9.Digard, P., D. Elton, K. Bishop, E. Medcalf, A. Weeds, and B. Pope. 1999. Modulation of nuclear localization of the influenza virus nucleoprotein through interaction with actin filaments. J. Virol. 73:2222-2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elton, D., P. Digard, L. Tiley, and J. Ortin. 2006. Structure and function of the influenza virus RNP, p. 1-36. In Y. Kawaoka (ed.), Influenza virology: current topics. Caister Academic Press, Norwich, United Kingdom.
- 11.Engelhardt, O. G., and E. Fodor. 2006. Functional association between viral and cellular transcription during influenza virus infection. Rev. Med. Virol. 16:329-345. [DOI] [PubMed] [Google Scholar]
- 12.Engelhardt, O. G., M. Smith, and E. Fodor. 2005. Association of the influenza A virus RNA-dependent RNA polymerase with cellular RNA polymerase II. J. Virol. 79:5812-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fodor, E., M. Crow, L. J. Mingay, T. Deng, J. Sharps, P. Fechter, and G. G. Brownlee. 2002. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76:8989-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fodor, E., and M. Smith. 2004. The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex. J. Virol. 78:9144-9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabriel, G., A. Herwig, and H. D. Klenk. 2008. Interaction of polymerase subunit PB2 and NP with importin α1 is a determinant of host range of influenza A virus. PLoS Pathog. 4:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graef, K. M., F. T. Vreede, Y.-F. Lau, A. W. McCall, S. M. Carr, K. Subbarao, and E. Fodor. 10. June 2010. The PB2 subunit of the influenza virus RNA polymerase affects virulence by interacting with MAVS and inhibiting IFN-β expression. J. Virol. doi: 10.1128/jvi.00879-10. [DOI] [PMC free article] [PubMed]
- 17.Guilligay, D., F. Tarendeau, P. Resa-Infante, R. Coloma, T. Crepin, P. Sehr, J. Lewis, R. W. Ruigrok, J. Ortin, D. J. Hart, and S. Cusack. 2008. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat. Struct. Mol. Biol. 15:500-506. [DOI] [PubMed] [Google Scholar]
- 18.Hao, L., A. Sakurai, T. Watanabe, E. Sorensen, C. A. Nidom, M. A. Newton, P. Ahlquist, and Y. Kawaoka. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454:890-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartl, F. U., and M. Hayer-Hartl. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852-1858. [DOI] [PubMed] [Google Scholar]
- 20.Hatta, M., P. Gao, P. Halfmann, and Y. Kawaoka. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840-1842. [DOI] [PubMed] [Google Scholar]
- 21.Herfst, S., S. Chutinimitkul, J. Ye, E. de Wit, V. J. Munster, E. J. Schrauwen, T. M. Bestebroer, M. Jonges, A. Meijer, M. Koopmans, G. F. Rimmelzwaan, A. D. Osterhaus, D. R. Perez, and R. A. Fouchier. 2010. Introduction of virulence markers in PB2 of pandemic swine-origin influenza virus does not result in enhanced virulence or transmission. J. Virol. 84:3752-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirayama, E., H. Atagi, A. Hiraki, and J. Kim. 2004. Heat shock protein 70 is related to thermal inhibition of nuclear export of the influenza virus ribonucleoprotein complex. J. Virol. 78:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong, S., G. Choi, S. Park, A. S. Chung, E. Hunter, and S. S. Rhee. 2001. Type D retrovirus Gag polyprotein interacts with the cytosolic chaperonin TRiC. J. Virol. 75:2526-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honore, B., H. Leffers, P. Madsen, H. H. Rasmussen, J. Vandekerckhove, and J. E. Celis. 1992. Molecular cloning and expression of a transformation-sensitive human protein containing the TPR motif and sharing identity to the stress-inducible yeast protein STI1. J. Biol. Chem. 267:8485-8491. [PubMed] [Google Scholar]
- 25.Huet, S., S. V. Avilov, L. Ferbitz, N. Daigle, S. Cusack, and J. Ellenberg. 2010. Nuclear import and assembly of influenza A virus RNA polymerase studied in live cells by fluorescence cross-correlation spectroscopy. J. Virol. 84:1254-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Itahana, K., and Y. Zhang. 2008. Mitochondrial p32 is a critical mediator of ARF-induced apoptosis. Cancer Cell 13:542-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itoh, H., A. Komatsuda, H. Ohtani, H. Wakui, H. Imai, K. Sawada, M. Otaka, M. Ogura, A. Suzuki, and F. Hamada. 2002. Mammalian HSP60 is quickly sorted into the mitochondria under conditions of dehydration. Eur. J. Biochem. 269:5931-5938. [DOI] [PubMed] [Google Scholar]
- 28.Jorba, N., S. Juarez, E. Torreira, P. Gastaminza, N. Zamarreno, J. P. Albar, and J. Ortin. 2008. Analysis of the interaction of influenza virus polymerase complex with human cell factors. Proteomics 8:2077-2088. [DOI] [PubMed] [Google Scholar]
- 29.Karlas, A., N. Machuy, Y. Shin, K. P. Pleissner, A. Artarini, D. Heuer, D. Becker, H. Khalil, L. A. Ogilvie, S. Hess, A. P. Maurer, E. Muller, T. Wolff, T. Rudel, and T. F. Meyer. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818-822. [DOI] [PubMed] [Google Scholar]
- 30.Kashiwagi, T., B. W. Leung, T. Deng, H. Chen, and G. G. Brownlee. 2009. The N-terminal region of the PA subunit of the RNA polymerase of influenza A/HongKong/156/97 (H5N1) influences promoter binding. PLoS One 4:e5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kashuba, E., K. Pokrovskaja, G. Klein, and L. Szekely. 1999. Epstein-Barr virus-encoded nuclear protein EBNA-3 interacts with the epsilon-subunit of the T-complex protein 1 chaperonin complex. J. Hum. Virol. 2:33-37. [PubMed] [Google Scholar]
- 32.Konig, R., S. Stertz, Y. Zhou, A. Inoue, H. H. Hoffmann, S. Bhattacharyya, J. G. Alamares, D. M. Tscherne, M. B. Ortigoza, Y. Liang, Q. Gao, S. E. Andrews, S. Bandyopadhyay, P. De Jesus, B. P. Tu, L. Pache, C. Shih, A. Orth, G. Bonamy, L. Miraglia, T. Ideker, A. Garcia-Sastre, J. A. Young, P. Palese, M. L. Shaw, and S. K. Chanda. 2010. Human host factors required for influenza virus replication. Nature 463:813-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuzuhara, T., D. Kise, H. Yoshida, T. Horita, Y. Murazaki, A. Nishimura, N. Echigo, H. Utsunomiya, and H. Tsuge. 2009. Structural basis of the influenza A virus RNA polymerase PB2 RNA-binding domain containing the pathogenicity-determinant lysine 627 residue. J. Biol. Chem. 284:6855-6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Labadie, K., E. Dos Santos Afonso, M. A. Rameix-Welti, S. van der Werf, and N. Naffakh. 2007. Host-range determinants on the PB2 protein of influenza A viruses control the interaction between the viral polymerase and nucleoprotein in human cells. Virology 362:271-282. [DOI] [PubMed] [Google Scholar]
- 35.Lingappa, J. R., R. L. Martin, M. L. Wong, D. Ganem, W. J. Welch, and V. R. Lingappa. 1994. A eukaryotic cytosolic chaperonin is associated with a high molecular weight intermediate in the assembly of hepatitis B virus capsid, a multimeric particle. J. Cell Biol. 125:99-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayer, D., K. Molawi, L. Martinez-Sobrido, A. Ghanem, S. Thomas, S. Baginsky, J. Grossmann, A. Garcia-Sastre, and M. Schwemmle. 2007. Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J. Proteome Res. 6:672-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehle, A., and J. A. Doudna. 2009. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc. Natl. Acad. Sci. U. S. A. 106:21312-21316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mehle, A., and J. A. Doudna. 2008. An inhibitory activity in human cells restricts the function of an avian-like influenza virus polymerase. Cell Host Microbe 4:111-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Momose, F., T. Naito, K. Yano, S. Sugimoto, Y. Morikawa, and K. Nagata. 2002. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J. Biol. Chem. 277:45306-45314. [DOI] [PubMed] [Google Scholar]
- 40.Naito, T., F. Momose, A. Kawaguchi, and K. Nagata. 2007. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J. Virol. 81:1339-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neumann, G., G. G. Brownlee, E. Fodor, and Y. Kawaoka. 2004. Orthomyxovirus replication, transcription, and polyadenylation. Curr. Top. Microbiol. Immunol. 283:121-143. [DOI] [PubMed] [Google Scholar]
- 42.Odunuga, O. O., V. M. Longshaw, and G. L. Blatch. 2004. Hop: more than an Hsp70/Hsp90 adaptor protein. Bioessays 26:1058-1068. [DOI] [PubMed] [Google Scholar]
- 43.Ohrmalm, C., and G. Akusjarvi. 2006. Cellular splicing and transcription regulatory protein p32 represses adenovirus major late transcription and causes hyperphosphorylation of RNA polymerase II. J. Virol. 80:5010-5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palese, P., and M. L. Shaw. 2007. Orthomyxoviridae: the viruses and their replication, p. 1647-1689. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA.
- 45.Pleschka, S., R. Jaskunas, O. G. Engelhardt, T. Zurcher, P. Palese, and A. Garcia-Sastre. 1996. A plasmid-based reverse genetics system for influenza A virus. J. Virol. 70:4188-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 47.Robb, N. C., M. Smith, F. T. Vreede, and E. Fodor. 2009. NS2/NEP protein regulates transcription and replication of the influenza virus RNA genome. J. Gen. Virol. 90:1398-1407. [DOI] [PubMed] [Google Scholar]
- 48.Rolling, T., I. Koerner, P. Zimmermann, K. Holz, O. Haller, P. Staeheli, and G. Kochs. 2009. Adaptive mutations resulting in enhanced polymerase activity contribute to high virulence of influenza A virus in mice. J. Virol. 83:6673-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruigrok, R. W., T. Crepin, D. J. Hart, and S. Cusack. 2010. Towards an atomic resolution understanding of the influenza virus replication machinery. Curr. Opin. Struct. Biol. 20:104-113. [DOI] [PubMed] [Google Scholar]
- 50.Shapira, S. D., I. Gat-Viks, B. O. Shum, A. Dricot, M. M. de Grace, L. Wu, P. B. Gupta, T. Hao, S. J. Silver, D. E. Root, D. E. Hill, A. Regev, and N. Hacohen. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139:1255-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simpson-Holley, M., D. Ellis, D. Fisher, D. Elton, J. McCauley, and P. Digard. 2002. A functional link between the actin cytoskeleton and lipid rafts during budding of filamentous influenza virions. Virology 301:212-225. [DOI] [PubMed] [Google Scholar]
- 52.Song, Y., and D. C. Masison. 2005. Independent regulation of Hsp70 and Hsp90 chaperones by Hsp70/Hsp90-organizing protein Sti1 (Hop1). J. Biol. Chem. 280:34178-34185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spiess, C., A. S. Meyer, S. Reissmann, and J. Frydman. 2004. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 14:598-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Subbarao, E. K., W. London, and B. R. Murphy. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 67:1761-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sugiyama, K., E. Obayashi, A. Kawaguchi, Y. Suzuki, J. R. Tame, K. Nagata, and S. Y. Park. 2009. Structural insight into the essential PB1-PB2 subunit contact of the influenza virus RNA polymerase. EMBO J. 28:1803-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sui, B., D. Bamba, K. Weng, H. Ung, S. Chang, J. Van Dyke, M. Goldblatt, R. Duan, M. S. Kinch, and W. B. Li. 2009. The use of random homozygous gene perturbation to identify novel host-oriented targets for influenza. Virology 387:473-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tarendeau, F., J. Boudet, D. Guilligay, P. J. Mas, C. M. Bougault, S. Boulo, F. Baudin, R. W. Ruigrok, N. Daigle, J. Ellenberg, S. Cusack, J. P. Simorre, and D. J. Hart. 2007. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat. Struct. Mol. Biol. 14:229-233. [DOI] [PubMed] [Google Scholar]
- 58.Tarendeau, F., T. Crepin, D. Guilligay, R. W. Ruigrok, S. Cusack, and D. J. Hart. 2008. Host determinant residue lysine 627 lies on the surface of a discrete, folded domain of influenza virus polymerase PB2 subunit. PLoS Pathog. 4:e1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vreede, F. T., T. E. Jung, and G. G. Brownlee. 2004. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J. Virol. 78:9568-9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yam, A. Y., Y. Xia, H. T. Lin, A. Burlingame, M. Gerstein, and J. Frydman. 2008. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat. Struct. Mol. Biol. 15:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuan, P., M. Bartlam, Z. Lou, S. Chen, J. Zhou, X. He, Z. Lv, R. Ge, X. Li, T. Deng, E. Fodor, Z. Rao, and Y. Liu. 2009. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 458:909-913. [DOI] [PubMed] [Google Scholar]
- 62.Zhu, H., J. Wang, P. Wang, W. Song, Z. Zheng, R. Chen, K. Guo, T. Zhang, J. S. Peiris, H. Chen, and Y. Guan. 2010. Substitution of lysine at 627 position in PB2 protein does not change virulence of the 2009 pandemic H1N1 virus in mice. Virology 401:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]