Abstract
Mannose-binding lectin (MBL) is a serum protein that plays an important role in host defenses as an opsonin and through activation of the complement system. The objective of this study was to assess the interactions between MBL and severe acute respiratory syndrome-coronavirus (SARS-CoV) spike (S) glycoprotein (SARS-S). MBL was found to selectively bind to retroviral particles pseudotyped with SARS-S. Unlike several other viral envelopes to which MBL can bind, both recombinant and plasma-derived human MBL directly inhibited SARS-S-mediated viral infection. Moreover, the interaction between MBL and SARS-S blocked viral binding to the C-type lectin, DC-SIGN. Mutagenesis indicated that a single N-linked glycosylation site, N330, was critical for the specific interactions between MBL and SARS-S. Despite the proximity of N330 to the receptor-binding motif of SARS-S, MBL did not affect interactions with the ACE2 receptor or cathepsin L-mediated activation of SARS-S-driven membrane fusion. Thus, binding of MBL to SARS-S may interfere with other early pre- or postreceptor-binding events necessary for efficient viral entry.
A novel coronavirus (CoV), severe acute respiratory syndrome-CoV (SARS-CoV), is the causal agent of severe acute respiratory syndrome, which afflicted thousands of people worldwide in 2002 and 2003 (10, 39). SARS-CoV is an enveloped, single- and positive-strand RNA virus that encodes four major structural proteins: S, spike glycoprotein (GP); E, envelope protein; M, membrane glycoprotein; and N, nucleocapsid protein (46, 55). Similar to other coronaviruses, the S glycoprotein of the virus mediates the initial attachment of the virus to host cell receptors, angiotensin-converting enzyme 2 (ACE2) (44) and/or DC-SIGNR (dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin-related molecule; also CD209L or L-SIGN[liver/lymph node-SIGN]) (32) and subsequent fusion of the viral and cellular membranes to allow viral entry into susceptible target cells. The S glycoprotein of SARS-CoV (SARS-S) is a 1,255-amino-acid (aa) type I membrane glycoprotein (46) with 23 potential N-linked glycosylation sites (55). The S glycoproteins of some coronaviruses are translated as a large polypeptide that is subsequently proteolytically cleaved into two functional subunits, S1 (harboring the receptor-binding domain [RBD]) and S2 (containing the membrane fusion domains) (1, 31, 51), during biogenesis, but others are not. The S glycoprotein on mature SARS-CoV virions does not appear to be cleaved (50, 61), but sequence alignments with other coronavirus S glycoproteins allow definition of S1 and S2 regions (46, 55). More recently, studies showed the proteolysis of the S glycoprotein of SARS-CoV on mature virions by cathepsin L (CTSL) (28, 59), as well as trypsin (43, 61) and factor Xa (11), suggesting that a critical cleavage event may occur during cell entry rather than during virion biogenesis.
Mannose-binding lectin (MBL; also known as mannose-binding or mannan-binding protein [MBP]) is a Ca2+-dependent (C-type) serum lectin that plays an important role in innate immunity by binding to carbohydrates on the surface of a wide range of pathogens (including bacteria, viruses, fungi, and protozoa) (8, 14, 18), where it activates the complement system or acts directly as an opsonin (30, 40, 52). In order to activate the complement system, MBL must be in complex with a group of MBL-associated serine proteases (MASPs), MASP-1, -2, and -3. Currently, only the role of MASP-2 in complement activation has been clearly defined (65). The MBL-MASP-2 complex cleaves C4 and C2 to form C3 convertase (C4bC2a), which, in turn, activates the downstream complement cascade. MBL is a pattern recognition molecule (9), and surface recognition is mediated through its C-terminal carbohydrate recognition domains (CRDs), which are linked to collagenous stems by a short coiled-coil of alpha-helices. MBL is a mixture of oligomers assembled from subunits that are formed from three identical polypeptide chains (9) and usually has two to six clusters of CRDs. Within each of the clusters, the carbohydrate-binding sites have a fixed orientation, which allows selective recognition of patterns of carbohydrate residues on the surfaces of a wide range of microorganisms (8, 14, 18). The concentration of MBL in the serum varies greatly and is affected by mutations of the promoter and coding regions of the human MBL gene (45). MBL deficiency is associated with susceptibility to various infections, as well as autoimmune, metabolic, and cardiovascular diseases, although MBL-deficient individuals are generally healthy (13, 37, 67).
There are conflicting results with regard to the role of MBL in SARS-CoV infection (29, 42, 72, 73). While the association of MBL gene polymorphisms with susceptibility to SARS-CoV infection was reported in some studies (29, 73), Yuan et al. demonstrated that there were no significant differences in MBL genotypes and allele frequencies among SARS patients and controls (72). Ip et al. observed binding to, and inhibition of, SARS-CoV by MBL (29). However, in other studies, no binding of MBL to purified SARS-CoV S glycoprotein was detected (42).
In this study, retroviral particles pseudotyped with SARS-S and in vitro assays were used to characterize the role of MBL in SARS-CoV infection. The data indicated that MBL selectively bound to SARS-S and mediated inhibition of viral infection in susceptible cell lines. Moreover, we identified a single N-linked glycosylation site, N330, on SARS-S that is critical for the specific interactions with MBL.
MATERIALS AND METHODS
Mannose-binding lectin.
Affinity-purified, ∼90% pure plasma-derived human MBL (pdMBL) was obtained from Statens Serum Institute (Copenhagen, Denmark). The preparation allowed the partial copurification of MASPs along with MBL (41).
Recombinant human MBL (rMBL) produced in a mouse myeloma cell line, NS0, was purchased from R&D Systems, Minneapolis, MN.
Serum samples and measurement of MBL levels in serum.
The serum levels of MBL from 50 anonymous healthy humans (age, 20 to 68; race, Black [B], Hispanic [H], and Caucasian[C]) (Bioreclamation Inc., Westbury, NY) were measured by use of a human MBL enzyme-linked immunosorbent assay (ELISA) kit (Cell Sciences, Inc., Canton, MA) following the manufacturer's instructions.
Sera from three healthy male donors (ages of 29, 41, and 21 and races B, H, and C, respectively) with normal levels of MBL (2.96, 4.03, and 2.33 μg ml−1, respectively, as determined by ELISA) were pooled as a complement source (normal human pooled sera [NHPS]). Sera from three healthy MBL-deficient (below the cutoff 0.1 μg ml−1) male donors (ages of 24, 40, and 23 and races B, race H, C, respectively) were pooled as a source of MBL-deficient complement (MBL-deficient pooled sera [MDPS]). To inactive complement, pooled sera were heat inactivated at 56°C for 30 min.
Plasmids and site-directed mutagenesis.
Plasmids encoding S glycoprotein from SARS-CoV, human ACE2, Ebola virus GP, and vesicular stomatitis virus G protein (VSV-G), as well as avian sarcoma and leukosis virus (ASLV-A) envelope, have been described previously (20, 59, 61, 63). Hepatitis C virus (HCV) E1/E2 envelope was human codon optimized and synthesized based on the H77 published sequence and subcloned into pCAGGS.
To mutate the N-linked glycosylation sites of SARS-CoV S glycoprotein from Asn to Gln, a QuikChange site-directed mutagenesis system (Stratagene, La Jolla, CA) with Pfu Turbo DNA polymerase was used. Reactions were performed using pCDNA3.1 SARS-CoV S. The primer sequences were designed as previously described (23). All of the mutations were verified by sequencing and subcloned into pCAGGS.
Plasmid pNL4-3 Luc-R− E− (pNL-luc) encodes a replication-incompetent variant of the HIV-1 molecular clone NL4-3, in which the nef gene has been replaced by a firefly luciferase (luc) reporter, and the env and vpr genes were inactivated, as previously described (6). Similarly, pNL-gfp was constructed with replacement of the nef gene with the gfp reporter.
The DC-SIGN (CD209) and DC-SIGNR [collectively referred to as DC-SIGN(R)] lectin expression plasmids have been described previously (60).
Cell lines and reagents.
The human primary embryonic kidney cell line (293T) and the Huh-7.5 cell lines were obtained from the ATCC and Apath LLC, respectively, and were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and antibiotics. Vero E6 cells (Collection of Cell Lines in Veterinary Medicine, Friedrich Loeffler Institute, Insel Riems, Germany; kindly provided by G. Herrler, Hannover, Germany) were maintained and grown in DMEM containing 10% fetal calf serum (FCS; PAA, Pasching, Austria), 1 mM glutamine (PAA), 1 mM sodium pyruvate (PAA), 1% nonessential amino acids (PAA), 100 U/ml penicillin (PAA), and 100 μg/ml streptomycin (PAA). A HeLa/Tva cell line cultured in DMEM supplemented with 5% heat-inactivated FBS was produced by using pcDNA6-Tva and selection with blasticidin. The B-THP cell line expressing DC-SIGN and control DC-SIGN-negative parental cells was obtained through the AIDS Research and Reference Reagent Program (NIH, Rockville, MD) and grown in RPMI 1640 medium supplemented with 10% heat-inactivated FBS and antibiotics. T-REx cell lines, expressing DC-SIGN or DC-SIGNR, were grown in medium containing DMEM, 10% heat-inactivated FBS, zeocin (50 μg ml−1), and blasticidin (2.5 μg ml−1) as described previously (53). The T-REx parental cells were maintained in medium containing DMEM, 10% heat-inactivated FBS, and blasticidin (2.5 μg ml−1). Lectin expression was induced by addition of doxycycline (0.1 μg ml−1) to the medium. DC-SIGN(R) expression levels were verified by flow cytometry and Western blotting. The cells were seeded into 96-well plates 24 h before assays. All cells were grown at 37°C in 5% CO2.
DMEM, FBS, glutamine, and antibodies were obtained from Gibco Laboratories (Grand Island, NY). Antibodies for DC-SIGN(R) were obtained through the AIDS Research and Reference Reagent Program (NIH, Rockville, MD). The human anti-SARS-S antibody, 80R, was kindly provided by Wayne Marasco (Harvard Medical School). Rabbit anti-SARS-CoV S1 was raised by immunizing rabbits with a purified immunoadhesin fusion protein between S1 and the rabbit IgG heavy chain constant region.
Pseudotype production and Western blotting.
Pseudotyped viruses were generated by cotransfecting 293T cells with 30 μg of Env-encoding plasmid or ACE2 and 10 μg of plasmid pNL-luc or pNL-gfp per 10-cm dish by using calcium phosphate in the presence or absence of 1.0 mM mannosidase I inhibitor, deoxymannojirimycin (DMJ) (Calbiochem, San Diego, CA). Dual-envelope-expressing virions were made by transfecting cells with 10 μg of pNL-gfp, 15 μg of pCB6 ASLV-A envelope, and 20 μg of pCAGGS SARS-CoV S [termed HIV-gtp (SARS-S/ASLV-A)]. The next day, expression was induced with sodium butyrate (10 mM). Forty hours after transfection, the supernatant was filtered through a 0.45-μm-pore-size screen and then purified by ultracentrifugation (28,000 rpm in an SW28 rotor; Beckman) over a 20% sucrose cushion and stored at −80°C as aliquots (56). The amount of virus was assessed with a p24 antigen capture ELISA (Aalto Bio Reagents Ltd., Dublin, Ireland).
To measure the expression of wild-type or mutant S glycoproteins of SARS-CoV, the pseudoviruses with wild-type or mutant S glycoproteins were lysed, and protein bands were separated onto 3 to 8% Criterion XT Tris-acetate gels (Bio-Rad, Hercules, CA) with Tris-Tricine-SDS running buffer. Protein expression was verified with Western blot analysis with rabbit anti-SARS-S antiserum (1:200), followed by Alexa Fluor 488 goat anti-rabbit antibody (Invitrogen, Carlsbad, CA), and protein bands were visualized with a Molecular Dynamics Storm 860 imaging system in blue fluorescence mode.
Pseudotyped virus binding assay.
For the MBL binding assay, flat-bottomed, 96-well high-binding polystyrene plates (Costar, Corning, NY) were coated with 100 μl of bovine serum albumin (BSA), rMBL (10 μg ml−1 in most experiments), or pdMBL (0.1, 0.3, 1, 3, and 10 μg ml−1) diluted in veronal-buffer (Lonza, Rockland, ME) containing 10 mM CaCl2 (VBS-Ca). After overnight incubation at 4°C, wells were blocked with 2% BSA for 60 min at room temperature, washed with VBS-Ca, and incubated for 4 h with 100 μl of different pseudotyped viruses (100 ng of p24 ml−1 in most experiments) diluted in VBS-Ca. Wells were then washed with VBS-Ca, bound viruses were lysed with Tris-buffered saline (TBS) with 0.5% Empigen BB detergent (Calbiochem, San Diego, CA), and p24 was detected by p24 ELISA. The percentage of p24 bound for each virus was calculated as follows: percentage of p24 antigen bound = [(p24 bound in rMBL/pdMBL-coated wells − background binding to BSA-coated wells)/(input p24)] × 100.
Similarly, for the virus binding assays with anti-S antiserum, 96-well high-binding polystyrene plates were coated with 100 μl of rabbit anti-S antiserum (1:200) or rabbit preimmune serum (1:200) diluted in VBS-Ca buffer, and the percentage of p24 bound for each virus was calculated as follows: percentage of p24 antigen bound = [(p24 bound in rabbit anti-S antiserum-coated wells − background binding to rabbit preimmune serum-coated wells)/(input p24)] × 100.
Inhibition of the infectivity of pseudotyped viruses by MBL.
Pseudotyped viruses were incubated with pdMBL or rMBL (0.1, 0.3, 1, 3, or 10 μg ml−1) diluted in phosphate-buffered saline containing CaCl2 (PBS/+) at 37°C for 1 h. In some experiments, pseudoviruses were incubated with pdMBL (3 μg ml−1) diluted in PBS/− buffer (PBS without CaCl2) containing 20 mM EDTA, PBS/+ buffer with monoclonal anti-human MBL antibody (1.5 or 6 μg ml−1; R&D Systems), or PBS/+ buffer with mouse IgG2A (1.5 or 6 μg ml−1; R&D Systems) at 37°C for 1 h. Treated virus (100 μl) was transferred to 48-well flat-bottomed culture plates containing 7 × 104 cells per well, as follows: 293T cells expressing ACE2 (293T-ACE2) and Huh7.5 cells for pseudotyped viruses bearing SARS-CoV S, 293T or 293T-ACE2 cells for pseudotyped viruses with VSV-G or Ebola virus envelope glycoprotein, and Huh7.5 cells for pseudotyped viruses with HCV E1/E2. Plates were centrifuged at 1,000 × g for 1.5 h at 25°C to facilitate infection (61) and then incubated for 4 h at 37°C. The virus-MBL mixture was replaced with fresh medium, and cells were harvested and lysed in 0.5% Triton X-100 after 40 h of culture. The luciferase activities in cells were detected with reagents from Promega (Madison, WI), and the percentage of infection calculated.
Inhibition of the infectivity of live SARS-CoV by MBL with a plaque reduction assay.
The SARS-CoV strain Frankfurt-1 (10, 64, 66) was used in infection experiments. Virus titer was determined by plaque titration using an Avicel overlay as described previously (48). For plaque reduction assays, Vero E6 cells were seeded into 24-well tissue culture plates. Virus dilutions containing 50 PFU were prepared in DMEM and mixed with MBL, anti-MBL antibody, or PBS in triplicates. The total volume of each mixture was 200 μl, and mixtures were incubated at 37°C for 1 h. Cells were washed once with PBS, and virus dilutions were added. After 1 h of adsorption at 37°C, the supernatant was removed, and cells were overlaid with Avicel-containing medium. Following 48 h of incubation at 37°C, cells were fixed in 4% paraformaldehyde (Roth, Germany), and plaques were visualized by staining with crystal violet and counted thereafter.
Binding and transmission of pseudotyped virus by DC-SIGN.
Virus pseudotyped with SARS-CoV S glycoprotein was preincubated with pdMBL (0 to 10 μg ml−1) for 1 h at 37°C before incubation with 1 × 105 DC-SIGN+ or DC-SIGN− parental B-THP cells for 3 h at 37°C. Cells were washed and lysed with detergent (0.5% Empigen BB), and cell-bound virus was measured by p24 antigen ELISA. Binding of pseudotyped viruses to DC-SIGN was calculated as follows: p24-antigen bound to DC-SIGN+ B-THP1 cells minus p24 antigen bound to DC-SIGN− B-THP cells. The percentage of p24 bound for each virus was calculated as follows: percentage of p24 antigen bound = [(p24 bound in DC-SIGN+ B-THP1 cells − p24 bound in DC-SIGN− B-THP cells)/(input p24)] × 100.
The transmission analyses were carried out as described previously (4). Briefly, viruses pseudotyped with SARS-CoV S glycoprotein were preincubated with different concentrations of pdMBL (0 to 10 μg ml−1) or with mannan (100 μg ml−1; Sigma-Aldrich, Germany) and monoclonal anti-human MBL antibody (10 μg ml−1) or PBS for 1 h at 37°C. Subsequently, 5 × 104 DC-SIGN+ or DC-SIGN− B-THP cells were incubated with the pretreated viruses for 3 h at 37°C, and unbound virus was removed by washing twice with fresh medium. Cells were then incubated with 293T-ACE2 target cells, and luciferase activities in cellular lysates were determined 3 days after the start of the cocultivation by employing a commercially available system (Promega, Germany).
Complement neutralization of pseudotyped viruses.
Pseudotyped viruses were incubated with dilutions of NHPS, MDPS, or heat-inactivated sera at 37°C for 1 h. Treated viruses (100 μl) were transferred to 48-well flat-bottomed culture plates containing 7 × 104 293T-ACE2 cells per well, spin infected, and incubated for 4 h at 37°C. The virus-complement mixture was replaced with fresh medium, and cells were harvested after 40 h of culture. Luciferase activity in cells was measured, and the percentage of neutralization was calculated.
Binding of pseudotyped viruses or S1-Ig to ACE2.
Viruses pseudotyped with SARS-CoV wild-type or mutant S envelope glycoproteins were preincubated with pdMBL (0, 1.0, and 10 μg ml−1) or 80R antibody (5 μg ml−1) in VBS-Ca buffer for 1 h at 37°C before incubation with recombinant human ACE2 (2 μg ml−1; R&D Systems) or BSA-coated 96-well high-binding polystyrene plates for 4 h at room temperature. Wells were washed with VBS-Ca, bound viruses were lysed with TBS with 0.5% Empigen BB detergent, and p24 was detected by p24 ELISA. The percentage of p24 bound for each virus was calculated as follows: percentage of p24 antigen bound = [(p24 bound in ACE2-coated wells − background binding to BSA-coated wells)/(input p24)] × 100. Similar assays were assessed with purified soluble protein containing the predicted SARS-CoV S1 region fused to the Fc region of rabbit immunoglobulin (S1-Ig; 0.5 μg ml−1), and an Ebola virus envelope glycoprotein-Ig fusion (Ebola GP1-Ig) was used as a negative control. Binding of S1-Ig was detected using a goat-anti-rabbit Ig antibody conjugated to alkaline phosphatase (AP), followed by a chemiluminescent substrate for AP activity. Immunoglobulin fusion proteins were made by transient expression in 293T cells, followed by protein A purification from the resulting supernatant.
Intervirion fusion.
The intervirion fusion assays were carried out as previously described (59). Pseudovirus incorporating ACE2 [HIV-luc(ACE2)] (500 ng of p24) was mixed with 1,000 ng of p24 of HIV-gfp (SARS-S/ASLV-A) particles in HBSS buffer at 4°C for 30 min to allow binding. For first-step MBL inhibition assays, MBL was added to the virus mixture (final concentration, 50 μg ml−1 or 100 μg ml−1) at the same time. Samples were raised to 37°C for 15 min to allow for receptor-induced conformational rearrangements. Tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin (final concentration, 15 μg ml−1), recombinant human CTSL (final concentration, 50 μg ml−1; R&D Systems), MBL (final concentration, 50 μg ml−1), or a CTSL-MBL mixture (final concentration, 50 μg ml−1 each; used for second-step MBL inhibition) was then added. Recombinant CTSL was preactivated by incubation for 15 min at 250 μg ml−1 in 50 mM morpholineethanesulfonic acid (MES), 5 mM dithiothreitol (DTT), pH 6.0, on ice. Virions incubated with CTSL were adjusted to pH 6.0 with HCl. After a 30-min incubation at 25°C, proteolysis was halted by the addition of 300 μl of DMEM containing leupeptin (100 μg ml−1) and soybean trypsin inhibitor (STI; 50 μg ml−1). For the third-step MBL inhibition assays, MBL (final concentration, 50 μg ml−1) was added to the mixture at this step. Virions were then incubated at 37°C for 30 min to allow membrane fusion between the two types of virions. A total of 100 μl of the virion mixture was added in quadruplicate to HeLa/Tva cells pretreated for 1 h with leupeptin (100 μg ml−1). The cells were spin infected and incubated at 37°C for 5 h. The medium was replaced with fresh DMEM, and the cells were assayed for luciferase activity 40 h later.
RESULTS
Binding of pseudotyped viruses to MBL.
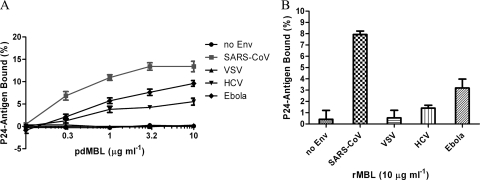
To determine whether SARS-S directly interacts with MBL, a microtiter capture assay was developed. HIV particles pseudotyped with no envelope protein (no Env), or envelope glycoprotein(s) from SARS-CoV, VSV, HCV, or Ebola virus were incubated in MBL-coated 96-well plates. The binding of the pseudotyped virus to MBL was assessed by lysing the virus with detergent and measuring released p24 core protein by ELISA (Fig. 1A). HIV particles lacking a viral envelope glycoprotein (no Env) or pseudotyped with VSV-G gave negligible binding to pdMBL (<0.5% of input), while HIV particles containing SARS-CoV, Ebola virus, and HCV envelope glycoproteins showed various levels of enhanced binding to pdMBL (5.5 to 13.4% of input with 10 μg ml−1 of pdMBL). While these results may imply differences in affinities for MBL, the variation in levels of binding may be also due to variations in the efficiency of envelope incorporation into pseudovirions. Similar results were observed for rMBL (Fig. 1B) although lower maximal levels of binding occurred for all envelopes.
FIG. 1.
Binding of pseudotyped viruses to mannose-binding lectin. Pseudovirions were bound to plates coated with various concentrations of pdMBL (A) or 10 μg ml−1 of rMBL (B). The binding of the pseudotyped virus to MBL was assessed by lysing the virus with detergent and measuring released p24 core protein by ELISA. The data are presented as percentage of recovered p24 antigen bound ± standard deviations. A single experiment carried out in triplicate is presented. Similar results were obtained in three independent experiments.
Inhibition of the infectivity of SARS-S-bearing pseudoviruses and live SARS-CoV by MBL.
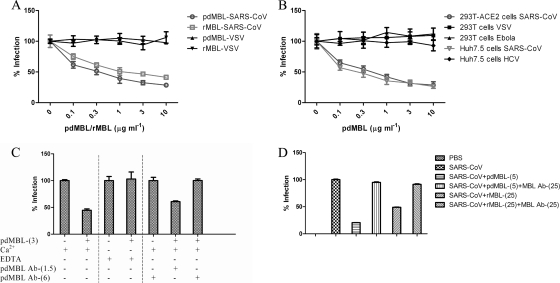
To investigate whether the binding of pseudotyped viruses to MBL could inhibit viral infectivity, we next examined the pseudovirion infection of different target cells (293T-ACE2 and Huh7.5 cells for SARS-S pseudoviruses, 293T or 293T-ACE2 cells for VSV and Ebola virus pseudoviruses, and Huh7.5 cells for HCV pseudoviruses). Preincubation of SARS-S pseudovirions with either pdMBL or rMBL resulted in a significant dose-dependent reduction of infectivity for both 293T-ACE2 and Huh7.5 cells (Fig. 2A and B), with 35 to 45% inhibition at 0.1 μg ml−1 and 60 to 70% inhibition at 3 or 10 μg ml−1 pdMBL (Fig. 2A, B, and C). rMBL was consistently slightly less potent than pdMBL in multiple experiments, even when concentrations were carefully equalized using a quantitative MBL ELISA based on mannan binding (Cell Sciences, Inc., Canton, MA). Interestingly, even physiologically unobtainable high concentrations of MBL could not reduce infection to background levels (data not shown), suggesting that a subset of virions does not interact correctly with MBL to allow inhibition. In contrast, no inhibition of viral infection was detected with VSV, Ebola virus, and HCV pseudoviruses (Fig. 2A and B). The results were highly reproducible, and the inhibition of the SARS-S pseudovirus infection by pdMBL could be abrogated by incubation of the virus with pdMBL (3 μg ml−1) in the presence of 20 mM EDTA in order to disrupt the calcium-dependent lectin (Fig. 2C). Similarly, incubation of the viral inoculum with pdMBL (3 μg ml−1) together with a monoclonal anti-human MBL antibody at 6 μg ml−1 completely abrogated inhibition (Fig. 2C). In controls, incubation of the viral inoculum with pdMBL with nonspecific mouse IgG2A (1.5 or 6 μg ml−1) did not affect the inhibition of the viral infection by pdMBL (with ∼60% inhibition observed) (data not shown). These results indicated that the specific interactions between MBL and SARS-S resulted in inhibition of virus infection in cell lines and that the interaction was calcium dependent and MBL antibody inhibitable. Similar inhibition assays were carried out with live SARS-CoV strain Frankfurt-1 using a plaque reduction assay. As shown in Fig. 2D, both pdMBL and rMBL inhibited virus infection and plaque formation in Vero E6 cells in a dose-dependent manner. While SARS-S pseudovirions demonstrated only a small increase in potency for pdMBL compared to rMBL (Fig. 2A), a greater effect was seen with replication-competent virus, with ∼80% inhibition with 5 μg ml−1 pdMBL versus ∼50% inhibition with 25 μg ml−1 rMBL. It is possible that the multiple rounds of infection required for plaque formation amplified the marginal difference in potency observed for pdMBL in single-round pseudotype assays or that this difference was due to technical differences between the two assays. The differences between abilities of pdMBL and rMBL to inhibit SARS-CoV suggest either a role for the MBL-associated proteins found in pdMBL or a small role for other non-MBL-associated serum proteins contaminating the nonrecombinant version.
FIG. 2.
Inhibition of the infectivity of SARS-S pseudoviruses and live SARS-CoV by mannose-binding lectin. Pseudoviruses preincubated with pdMBL or rMBL were added to 293T-ACE2 cells (A), and pseudoviruses preincubated with pdMBL were added to 293T-ACE2 and Huh7.5 cells for SARS-S pseudoviruses, 293T cells for VSV and Ebola virus pseudoviruses, and Huh7.5 cells for HCV pseudoviruses (B). The percentage of infection of no-MBL controls ± standard deviations is presented for a single experiment carried out in triplicate. Similar results were obtained in three independent experiments. (C) SARS-S pseudoviruses were incubated with pdMBL (3 μg ml−1) in the presence of either Ca2+, EDTA, or anti-human MBL antibody (1.5 or 6 μg ml−1), and infectivity was detected in 293T-ACE2 cells. (D) pdMBL and rMBL inhibition assays were carried out with live SARS-CoV strain Frankfurt-1 using a plaque reduction assay in Vero E6 cells. Following 48 h of incubation at 37°C, cells were fixed, and plaques were visualized by staining with crystal violet. Numbers in parentheses indicate the concentrations of pdMBL and rMBL (in μg/ml). The percentage of infection of no-MBL controls ± standard deviations is presented for a single experiment carried out in triplicate. Similar results were obtained in a further experiment. Ab, antibody.
MBL blocks binding of SARS-CoV to DC-SIGN.
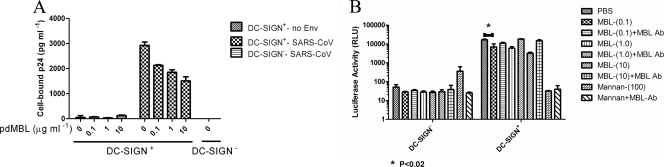
Previous studies have shown that SARS-S binds to the cell surface lectins DC-SIGN and DC-SIGNR (47, 69) and results in enhanced infectivity. The majority of binding to DC-SIGN(R) is mediated through recognition of the same high-mannose moieties that bind MBL. Thus, we next evaluated the capability of pdMBL to block binding of SARS-S pseudotyped particles to cells expressing DC-SIGN. As expected, SARS-S pseudotypes bound at dramatically higher levels to DC-SIGN-expressing B-THP cells than to parental B-THP cells, while pseudotyped particles with no envelope protein gave only background levels of binding (Fig. 3A). Preincubation of SARS-S pseudotypes with concentrations of pdMBL as low as 0.1 μg ml−1 reduced virus binding by ∼30% (Fig. 3A). However, addition of larger amounts of pdMBL increased inhibition of binding only marginally, with up to ∼50% inhibition at 10 μg ml−1 (Fig. 3A). Similarly, transmission of SARS-S bearing pseudotypes from DC-SIGN-expressing B-THP cells to target 293T-ACE2 cells could be partially, but nevertheless statistically significantly, inhibited by pdMBL in a dose-dependent manner, with 10 μg ml−1 of pdMBL leading to a 5-fold reduction in levels of transmission. This inhibition did not match the levels of inhibition seen by competing with the carbohydrate, mannan. Nevertheless, an antibody directed to human MBL fully reversed the inhibition (Fig. 3B). These data demonstrated that MBL binds to virus pseudotyped with SARS-S and at least partially blocks interactions between S glycoprotein and DC-SIGN. Thus, MBL and DC-SIGN may recognize a distinct, but highly overlapping, set of high-mannose moieties on SARS-S.
FIG. 3.
MBL blocks binding of SARS-CoV with DC-SIGN. (A) Virus pseudotyped with SARS-S or particles with no envelope protein (no Env) were preincubated with pdMBL (0 to 10 μg ml−1) before incubation with DC-SIGN+ or parental B-THP cells. Cells were washed and lysed, and cell-bound virus was measured by p24 ELISA. Binding of pseudotyped viruses to DC-SIGN ± standard deviations was calculated as follows: p24 bound to DC-SIGN+ B-THP cells minus p24 bound to DC-SIGN− parental B-THP cells. A single experiment carried out in triplicate is shown. Similar results were obtained in three independent experiments. (B) Transmission of the bound SARS-CoV pseudotypes to target 293T-ACE2 cells. Numbers in parentheses indicate the concentrations of MBL and mannan (in μg/ml). Differences with or without 0.1 μg ml−1 purified pdMBL were analyzed by pairwise t tests (*, P < 0.02). Ab, antibody.
Complement-mediated virus neutralization of pseudotyped viruses.
The above experiments indicated that MBL binds to SARS-S on the surface of pseudoviruses and could directly neutralize virus infection in different cell lines. However, for several viruses, MBL can also indirectly neutralize infectivity by complement fixation (33, 34). Thus, we next determined whether the lectin complement pathway was activated. Two pools of sera were used as sources of complement in neutralization assays: NHPS, a pool of sera from three individuals with normal levels of serum MBL, and MDPS, a pool of sera from three individuals deficient in serum MBL.
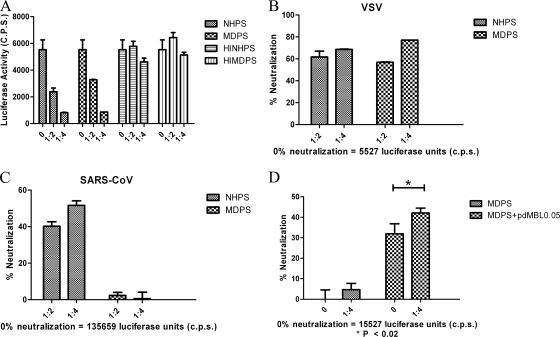
Neutralization of virus pseudotyped with VSV glycoprotein by serum was assessed as a positive control due to the presence of naturally occurring antibodies in human serum capable of directing complement lysis of VSV-G-positive particles (2, 7). Both NHPS and MDPS at low dilutions (1:2 and 1:4) significantly neutralized VSV pseudovirus particles compared with heat-inactivated NHPS and MDPS (Fig. 4A). When expressed as a percentage, virus neutralization was 61 and 68% at 1:2 and 1:4 dilutions of NHPS, respectively (Fig. 4B). MDPS neutralization of the virus was at levels similar to those of NHPS (Fig. 4B).
FIG. 4.
Complement-mediated neutralization of pseudotyped viruses. Pseudotyped viruses were incubated with 1:2 or 1:4 dilutions of pooled sera from either NHPS, MDPS, or heat-inactivated pooled sera of these two sources (HINHPS and HIMDPS, respectively). Treated viruses (100 μl) were then incubated with 293T-ACE2 cells. After 40 h, luciferase activity in cells was measured, and the percentage of neutralization ± standard deviations is presented for a single experiment carried out in triplicate. Raw luciferase values for no-serum controls are also presented. Similar results were obtained in three independent experiments. (A) Comparison of VSV-G luciferase activity in cells. A comparison of the percent neutralization of VSV-G (B) and SARS-S (C) pseudotyped viruses by NHPS and MDPS is also shown. (D) SARS-S pseudotyped viruses were incubated alone (0) or with a 1:4 dilution of MDPS and with or without 0.05 μg ml−1 purified pdMBL. Differences were analyzed by pairwise t tests (P < 0.02).
SARS-S pseudotypes were also neutralized by NHPS, with 40 and 52% neutralization with 1:2 and 1:4 dilutions of NHPS, respectively (Fig. 4C). However, MDPS was significantly less effective in neutralizing the virus (2.3% neutralization at a 1:2 dilution and 0.6% at a 1:4 dilution, respectively) (Fig. 4C). To confirm that MBL plays a role in directing complement lysis of the SARS-S pseudotyped virus, we assessed the effect on neutralization of reconstituting the lectin complement pathway in MDPS by adding exogenous MBL. Even the addition of very low concentrations of pdMBL (0.05 μg ml−1) to MDPS significantly neutralized the pseudovirus compared with MDPS alone (Fig. 4D). This level of neutralization was greater than that seen with the same concentrations of MBL in complement-deficient assays (P < 0.02) but not dramatically so. Thus, MBL-directed complement lysis of SARS-CoV occurs, but it is not clear how important this feature is, and further investigation would require the separation of the binding and neutralization effects.
Specific glycosylation sites are critical for MBL and SARS-CoV interaction.
Previous studies have shown that glycans on the S glycoprotein of SARS-CoV are important for DC-SIGN(R)-mediated infection, and site-directed mutagenesis analysis has identified several individual glycosylation sites directly involved in DC-SIGN(R)-mediated virus binding and entry (23, 58). Since MBL binds to virus pseudotyped with SARS-S and blocks S glycoprotein interaction with DC-SIGN, glycans on SARS-S are most likely to be the binding targets for MBL. We further investigated the role of glycosylation sites on S glycoprotein of SARS-CoV for MBL interaction by mutagenesis analysis.
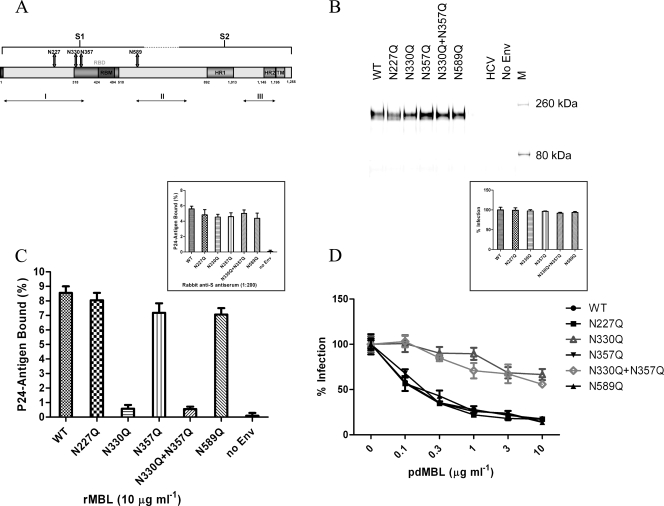
There are 23 potential asparagine (N)-linked glycosylation sites on SARS-S, and glycosylation at 13 of these sites (aa 118, 119, 227, 269, 318, 330, 357, 783, 1056, 1080, 1140, 1155, and 1,176) has been confirmed by either mass spectrometric (38, 71) or biochemical (5) analyses. On the linear map of S glycoprotein (Fig. 5A), these sites have been grouped into three distinct clusters: cluster I at the N terminus, cluster II near the border between the S1 and S2 regions, and cluster III at the C terminus (23). Four individual N-linked glycosylation sites (aa 227, 330, and 357 in cluster I and aa 589 in cluster II) have been described to be involved in DC-SIGN(R)-mediated virus entry (23, 58) and, thus, were selected for our study of MBL and SARS-CoV interactions. Five mutant SARS-S pseudoviruses were generated with mutations of the asparagine (N) residues to glutamines (Q) at the four sites: N227Q, N330Q, N357Q, and N589Q. As MBL directly inhibits SARS-CoV entry and as residues 330 and 357 lie within the RBD of S glycoprotein, it was hypothesized that MBL binding to aa 330 and 357 might be directly responsible for inhibition, and thus the double mutant N330Q N357Q was also made. The normal expression of mutant SARS-S on the pseudoviruses was confirmed by Western blot analyses with rabbit anti-S antiserum (Fig. 5B), and the binding of these mutant SARS-S pseudoviruses to the rabbit anti-S antiserum was comparable to that of the wild-type pseudovirus (Fig. 5C, inset). As shown in Fig. 5C, three of the mutant pseudoviruses (N227Q, N357Q, and N589Q) exhibited nearly wild-type levels of rMBL binding (∼7 to 8% of input). In contrast, N330Q and the N330Q N357Q double mutation exhibited significant reduction in their abilities to bind to rMBL, with binding of only ∼0.5% of input. To further characterize the effects of the five mutant pseudoviruses on virus infectivity and MBL-mediated viral infection neutralization, infectivity on ACE2-expressing 293T cells was compared with that of the wild-type SARS-S pseudovirus. All of the mutant pseudoviruses exhibited nearly wild-type levels of infectivity in 293T-ACE2 cells (Fig. 5D, inset). In the MBL-mediated viral infection neutralization assay, three of the mutant pseudoviruses (N227Q, N357Q, and N589Q) exhibited nearly wild-type pdMBL inhibition patterns, with 35 to 45% inhibition at 0.1 μg ml−1 and 70 to 80% inhibition at 3 or 10 μg ml−1 pdMBL (Fig. 5D). However, with the N330Q and the N330Q N357Q double mutation, no clear pdMBL inhibition was detected at 0.1 μg ml−1, and only ∼30 to 40% inhibition was detected at 3 or 10 μg ml−1 pdMBL (Fig. 5D). Thus, MBL interactions with SARS-S appear to be very specifically mediated by an N-glycan within the RBD at position 330. However, some residual binding and inhibitory capacity likely reside outside this region as some levels of inhibition were observed, even with the N330Q N357Q double mutant.
FIG. 5.
Specific glycosylation sites are critical for MBL and SARS-CoV interaction. (A) A schematic diagram of SARS-CoV S glycoprotein. Functional S1 and S2 domains, receptor-binding domain (RBD) and receptor-binding motif (RBM), heptad repeat regions HR1 and HR2, transmembrane domain TM, three clusters of potential N-linked glycosylation sites (I, II, and III), and four glycosylation sites used to make site mutations are indicated. (B) Western blot analysis of wild-type (WT) or mutant S glycoprotein expression performed under reducing and denaturing conditions and detected using rabbit anti-SARS-S antiserum. (C) Effects of glycosylation site mutations of SARS-S on binding of pseudotyped viruses to rMBL-coated plates (10 μg ml−1) (main panel) or rabbit anti-SARS S antiserum-coated plates (inset). The data are presented as the percentage of recovered p24 antigen bound ± standard deviations. (D) Effects of glycosylation site mutations on ACE2-mediated SARS-S pseudovirus infectivity (inset) or MBL-mediated inhibition (main panel). The percentage of infection ± standard deviations for a single experiment carried out in triplicate is presented. Similar results were obtained in three independent experiments.
Enhanced MBL binding and neutralization of SARS-S pseudovirions containing high-mannose-type N-glycans.
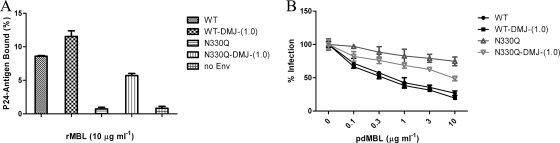
To better understand the specific interaction between SARS-S and MBL at the N-linked glycosylation site at position 330 and to test the hypothesis that N-linked glycosylation sites of S glycoprotein are important for virus interaction with MBL, we manipulated the carbohydrate composition of SARS-S by using a mannosidase I inhibitor, deoxymannojirimycin (DMJ), generating proteins containing only high-mannose content (36).
Production of wild-type and N330Q mutant SARS-S pseudoviruses in the presence of DMJ significantly enhanced binding of the pseudoviruses to rMBL (Fig. 6A) and resulted in increased MBL neutralization of the N330 mutant in ACE2-expressing 293T cells (Fig. 6B). Similar results were obtained with the N330Q N357Q double mutant pseudoviruses (data not shown). However, the levels of increased MBL binding and neutralization of the N330Q and N330Q N357Q double mutant pseudoviruses through DMJ treatment were still lower than those of the wild-type SARS-CoV (Fig. 6). These results indicated that while the DMJ-induced changes in the glycosylation pattern of SARS-S strengthened virus interactions with MBL, the specific interaction between S glycoprotein and MBL at the aa 330 glycosylation site is important for inhibition.
FIG. 6.
The effect of mannosidase I inhibitor, DMJ, on the interaction of MBL with SARS-CoV. (A) Binding of wild-type (WT) and N330Q mutant SARS-S pseudoviruses, produced in the presence or absence of 1 mM DMJ, to rMBL-coated plates (10 μg ml−1). (B) MBL-mediated neutralization of wild-type and N330Q mutant pseudoviruses, produced in the presence or absence of 1 mM DMJ. The percentage of infection ± standard deviations is presented for a single experiment carried out in triplicate. Similar results were obtained in three independent experiments.
Effects of MBL on virus interaction with ACE2 receptor.
The presence of N330 within the RBD of SARS-S led us to determine whether the virus-MBL interaction affected SARS-S binding to ACE2. However, preincubation of pdMBL with either virus pseudotyped with SARS-S or with purified soluble protein containing the predicted SARS-CoV S1 region fused to rabbit immunoglobulin Fc (S1-Ig) had no effect on SARS-S mediated binding to ACE2 (Fig. 7A and B). In contrast, a significant reduction of ACE2 binding was observed in both assays using the neutralizing antibody 80R (Fig. 7A and B). These results indicated that virus binding by MBL did not affect its interaction with ACE2.
FIG. 7.
Effect of MBL on virus interaction with ACE2 receptor and cathepsin L-mediated activation of SARS-S intervirion fusion. (A) Viruses pseudotyped with SARS-CoV wild-type or mutant S glycoproteins were preincubated with pdMBL (0, 1.0, and 10 μg ml−1) or 80R antibody (5 μg ml−1) in VBS-Ca buffer before incubation with recombinant human ACE2 (2 μg ml−1)- or BSA-coated plates. Wells were washed, and bound viruses were lysed and assayed for p24. A single experiment was carried out in triplicate. Similar results were obtained in three independent experiments. (B) Purified S1-Ig binding to ACE2-coated plates. S1-Ig was preincubated with pdMBL, and binding to ACE2-coated plates was detected using a goat anti-rabbit Ig antibody conjugated to alkaline phosphatase (AP) followed by a chemiluminescent substrate for AP activity. Ebola virus envelope glycoprotein with Ig conjugate (Ebola GP-Ig) was used as a negative control. (C) A diagram of intervirion assays with MBL, with the three steps indicated as S1, S2, and S3. (D) Intervirion assays with pdMBL. HIV-luc(ACE2) and HIV-gfp(SARS S/ASLV-A) particles were mixed and kept at 4°C for 30 min to allow binding. Samples were raised to 37°C for 15 min to allow for conformational rearrangements. Trypsin or CTSL was then added. The mixed viruses were used to infect HeLa/Tva cells pretreated with leupeptin. pdMBL (50 or 100 μg ml−1) was added to the mixture at three different steps: S1, concurrent with initial virion mixing; S2, concurrent with protease addition; and S3, after proteolysis. The percentage of infection achieved for mixed particles on untreated cells ± standard deviations is presented for a single experiment carried out in triplicate. Similar results were obtained in three independent experiments.
Effects of MBL on cathepsin L-mediated viral proteolysis.
SARS-CoV infectious entry proceeds via a four-step process: receptor binding and induced conformational changes in SARS-S, followed by virus uptake into endosomes and proteolysis of SARS-S by the endosomal protease cathepsin L (CTSL) (59), and finally membrane fusion. Since SARS-CoV interactions with MBL did not affect virus binding to ACE2, we hypothesized that the inhibition of viral infection could be due to the MBL effects on one of these postreceptor-binding events. To find out at which step MBL may play a role, intervirion fusion assays were performed as depicted in Fig. 7C. Briefly, HIV-luc(ACE2) and HIV-gfp(SARS S/ASLV-A) particles were mixed to allow viral binding to receptors and conformational rearrangement. Proteases such as trypsin or CTSL were then added. The mixed virus was used to infect HeLa/Tva cells pretreated with leupeptin in order to prevent cellular proteases mediating any effects. For our purpose, pdMBL or rMBL was added to the mixture at three different steps: step one, addition during the initial viral interactions; step two, addition with CTSL; and step three, addition after proteolysis (Fig. 7D, S1, S2, and S3, respectively). As expected, treatment of mixed HIV-luc(ACE2) and HIV-gfp(SARS-S/ASLV-A) particles with CTSL at pH 6.0 or with trypsin mediated intervirion fusion (Fig. 7D). In contrast, pdMBL alone did not increase intervirion fusion (Fig. 7D), suggesting that the MBL-associated serine proteases (MASPs) cannot mimic trypsin activation of S glycoprotein. Reduced levels of CTSL-mediated activation of membrane fusion was observed in the presence of high concentrations of pdMBL at the first step (Fig. 7D, S1, whereas no clear reduction of CTSL-mediated intervirion fusion with addition of MBL at the second or third steps was observed (Fig. 7D, S2 and S3). These results indicate that binding of MBL to SARS-S may interfere with an early step in postreceptor interactions during viral entry.
DISCUSSION
To date, the precise role of MBL in SARS-CoV infections is unclear and controversial (29, 42, 72, 73). In the present study, we demonstrated that MBL selectively binds to SARS-CoV pseudotyped virus and can potently inhibit SARS-CoV infection of susceptible cell lines at concentrations below those observed in the serum of healthy individuals.
Mannose-binding lectin is a serum protein produced in the liver that plays an important role for host defenses through activation of the complement system or acts directly as an opsonin (30, 40, 52). Although no detectable MBL mRNA was found in lung tissue by real-time quantitative PCR (57), a number of studies have showed the presence of MBL in bronchoalveolar lavage (BAL) fluid of patients with pneumonia but not in healthy controls, whose lungs were not inflamed (12, 17, 21). A number of studies have shown a role for MBL in the infection and pathogenesis of several human viral pathogens, including HIV (24, 25, 27) and Ebola virus (34). A large number of studies have also linked hepatitis B and C persistence and disease progression to MBL polymorphisms (reviewed in reference 3). As far as we are aware, this present study is the first to demonstrate a direct interaction between HCV E1/E2 glycoprotein and MBL and may thus begin to present a rationale for the role of MBL in HCV disease.
MBL binds to HIV via high-mannose moieties on N-linked glycans on the HIV envelope protein, gp120 (15, 24, 25), and hence activates the MBL-mediated complement pathway (27). While significant direct neutralization of a single cell line-adapted strain, HIVIIIB, has been reported (15), neutralization of HIV primary isolates in the absence of complement is poor, even at concentrations as high at 20 μg ml−1 (24, 70). Similarly, MBL binding to Ebola virus glycoprotein was observed but not direct inhibition of infection (34). In contrast, our data clearly showed direct MBL-mediated neutralization of SARS-CoV. Moreover, the concentrations required for inhibition of SARS-S-mediated entry were within or below the range commonly seen in serum from healthy individuals (1 to 3 μg ml−1). Similarly, MBL has also been shown to directly inhibit hemagglutination activity and infectivity of several strains of influenza A virus (26, 35).
The interaction between MBL and SARS-S also blocked viral binding to the membrane bound calcium-dependent (C-type) lectin DC-SIGN, suggesting that MBL most likely recognized an overlapping set of high-mannose-content glycans on S glycoprotein and thereby competed with DC-SIGN for binding. DC-SIGN and the closely related molecule DC-SIGNR have been shown to bind to, or facilitate infection by, a diverse array of viruses (reviewed in reference 19) and certain bacteria, yeasts, and parasites (68) by binding high-mannose carbohydrates and related surface glycans (16, 49). In particular, it has been suggested that DC-SIGNR acts directly as a receptor, rather than as an attachment-enhancing factor, for SARS-CoV entry into type II alveolar cells and endothelial cells (32). Thus, the ability of MBL to interfere with SARS-CoV interactions with C-type lectin receptors on these cells may play a major role in viral spread and pathogenicity.
Previous studies showed that mutating N330 and/or N357 to glutamine reduced SARS-CoV capture by DC-SIGN-expressing B-THP cells and subsequent trans-infection of HepG2 target cells that express ACE2 (58) but had no effect on DC-SIGNR-mediated SARS-CoV entry independent of ACE2 (23). In contrast, the N227Q and N589Q mutants exhibited partial loss in DC-SIGNR-mediated infection. In agreement with these findings, we observed that N330Q and/or N357Q mutations significantly reduced viral binding to both DC-SIGN-expressing B-THP cells and DC-SIGN(R)-expressing T-REx cells (data not shown), which suggested that glycosylation sites N330 and N357 are important for DC-SIGN(R) binding. Mutation of N330 significantly reduced both MBL binding and MBL-mediated viral neutralization, indicating that the glycan at N330 is critical for the specific interaction between MBL and SARS-S although some level of residual neutralization remained, even with the N330Q N357Q double mutant, suggesting that other glycans outside the RBD may also be involved in MBL inhibition, despite a lack of detectable binding. While MBL and DC-SIGN predominantly recognize the same ligand (high-mannose type), it is not surprising that differences in recognition patterns on complex proteins are observed due to the spacing of individual carbohydrate recognition domains and the relative inflexibility of the lectins (8, 14, 18). Alternately, differences in affinity for other sugars may explain the overlapping but distinct patterns of recognition; for example, DC-SIGN, but not DC-SIGNR, can also bind fucose-containing carbohydrates (22).
We also noted that generating high-mannose-content N-glycans on S glycoprotein through production of viruses in the presence of deoxymannojirimycin enhanced MBL binding and SARS-CoV infectivity neutralization by MBL. Previous studies have shown that production of HIV in the presence of DMJ significantly enhanced binding of HIV to MBL and increased MBL neutralization of an M-tropic HIV primary isolate (24). DMJ treatment of the N330Q mutants in our study resulted in a markedly increased MBL binding capacity and MBL-mediated infectivity neutralization. However, the level of MBL binding and neutralization was still lower than that with wild-type SARS-S pseudotyped virus. Recent studies demonstrate that the SARS-CoV S glycoprotein is highly glycosylated, with the major glycans of the SARS-CoV S glycoprotein produced in Vero E6 cells being high-mannose, hybrid, and bi-, tri-, and tetra-antennary complex moieties (54). These results imply that there is some, but not total, site specificity for the glycosylation at position N330 in the ability of MBL to inhibit SARS-CoV entry and that strength of binding may be a more important factor. It is relatively unusual to have this level of specificity for recognition of highly glycosylated viral envelopes such as SARS-S by a high-mannose-binding lectin.
Binding to MBL did not affect SARS-S interactions with the ACE2 receptor. Furthermore, MBL-mediated inhibition occurred at a step prior to CTSL-mediated activation of SARS-S fusion. Thus, we suggest that the binding of the MBL may interfere with the induction of conformational changes within the S glycoprotein and thus prevent an early, postreceptor-binding event. Interestingly, incubation of SARS-S with pdMBL resulted in partial proteolysis of SARS-S (data not shown), likely mediated by MBL-associated serine proteases. However, this cleavage was not sufficient to activate S glycoprotein in the manner mediated by CTSL and trypsin. Treatment of cell-free virus with trypsin abrogates viral infectivity (62), and it is possible that MBL-mediated proteolysis leads to some inactivation of SARS-CoV, explaining the subtle differences in neutralization between pdMBL and rMBL.
In summary, we have demonstrated that MBL selectively binds to SARS-S pseudotyped virus and can inhibit SARS-CoV infection in susceptible cell lines. Our results identified a single N-linked glycosylation site, N330, on S glycoprotein as the target for the specific interactions between MBL and SARS-CoV and provide evidence that the viral interaction with MBL did not affect its interaction with the ACE2 receptor.
Acknowledgments
This work was supported by grant R01AI074986 from the National Institute of Allergy And Infectious Diseases (to G.S.) and by grant 01KI 0703 from BMBF (to S.P.).
Footnotes
Published ahead of print on 23 June 2010.
REFERENCES
- 1.Abraham, S., T. E. Kienzle, W. Lapps, and D. A. Brian. 1990. Deduced sequence of the bovine coronavirus spike protein and identification of the internal proteolytic cleavage site. Virology 176:296-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beebe, D. P., and N. R. Cooper. 1981. Neutralization of vesicular stomatitis virus (VSV) by human complement requires a natural IgM antibody present in human serum. J. Immunol. 126:1562-1568. [PubMed] [Google Scholar]
- 3.Brown, K. S., S. D. Ryder, W. L. Irving, R. B. Sim, and T. P. Hickling. 2007. Mannan binding lectin and viral hepatitis. Immunol. Lett. 108:34-44. [DOI] [PubMed] [Google Scholar]
- 4.Chaipan, C., E. J. Soilleux, P. Simpson, H. Hofmann, T. Gramberg, A. Marzi, M. Geier, E. A. Stewart, J. Eisemann, A. Steinkasserer, K. Suzuki-Inoue, G. L. Fuller, A. C. Pearce, S. P. Watson, J. A. Hoxie, F. Baribaud, and S. Pohlmann. 2006. DC-SIGN and CLEC-2 mediate human immunodeficiency virus type 1 capture by platelets. J. Virol. 80:8951-8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakraborti, S., P. Prabakaran, X. Xiao, and D. S. Dimitrov. 2005. The SARS coronavirus S glycoprotein receptor binding domain: fine mapping and functional characterization. Virol. J. 2:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 7.DePolo, N. J., J. D. Reed, P. L. Sheridan, K. Townsend, S. L. Sauter, D. J. Jolly, and T. W. Dubensky, Jr. 2000. VSV-G pseudotyped lentiviral vector particles produced in human cells are inactivated by human serum. Mol. Ther. 2:218-222. [DOI] [PubMed] [Google Scholar]
- 8.Dommett, R. M., N. Klein, and M. W. Turner. 2006. Mannose-binding lectin in innate immunity: past, present and future. Tissue Antigens 68:193-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drickamer, K., and M. E. Taylor. 1993. Biology of animal lectins. Annu. Rev. Cell Biol. 9:237-264. [DOI] [PubMed] [Google Scholar]
- 10.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 11.Du, L., R. Y. Kao, Y. Zhou, Y. He, G. Zhao, C. Wong, S. Jiang, K. Y. Yuen, D. Y. Jin, and B. J. Zheng. 2007. Cleavage of spike protein of SARS coronavirus by protease factor Xa is associated with viral infectivity. Biochem. Biophys. Res. Commun. 359:174-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisen, D. P. Mannose-binding lectin deficiency and respiratory tract infection. 2010. J. Innate Immun. 2:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisen, D. P., and R. M. Minchinton. 2003. Impact of mannose-binding lectin on susceptibility to infectious diseases. Clin. Infect. Dis. 37:1496-1505. [DOI] [PubMed] [Google Scholar]
- 14.Epstein, J., Q. Eichbaum, S. Sheriff, and R. A. Ezekowitz. 1996. The collectins in innate immunity. Curr. Opin. Immunol. 8:29-35. [DOI] [PubMed] [Google Scholar]
- 15.Ezekowitz, R. A., M. Kuhlman, J. E. Groopman, and R. A. Byrn. 1989. A human serum mannose-binding protein inhibits in vitro infection by the human immunodeficiency virus. J. Exp. Med. 169:185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feinberg, H., D. A. Mitchell, K. Drickamer, and W. I. Weis. 2001. Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science 294:2163-2166. [DOI] [PubMed] [Google Scholar]
- 17.Fidler, K. J., T. N. Hilliard, A. Bush, M. Johnson, D. M. Geddes, M. W. Turner, E. W. Alton, N. J. Klein, and J. C. Davies. 2009. Mannose-binding lectin is present in the infected airway: a possible pulmonary defence mechanism. Thorax 64:150-155. [DOI] [PubMed] [Google Scholar]
- 18.Fraser, I. P., H. Koziel, and R. A. Ezekowitz. 1998. The serum mannose-binding protein and the macrophage mannose receptor are pattern recognition molecules that link innate and adaptive immunity. Semin. Immunol. 10:363-372. [DOI] [PubMed] [Google Scholar]
- 19.Geijtenbeek, T. B., J. den Dunnen, and S. I. Gringhuis. 2009. Pathogen recognition by DC-SIGN shapes adaptive immunity. Future Microbiol. 4:879-890. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert, J. M., P. Bates, H. E. Varmus, and J. M. White. 1994. The receptor for the subgroup A avian leukosis-sarcoma viruses binds to subgroup A but not to subgroup C envelope glycoprotein. J. Virol. 68:5623-5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomi, K., Y. Tokue, T. Kobayashi, H. Takahashi, A. Watanabe, T. Fujita, and T. Nukiwa. 2004. Mannose-binding lectin gene polymorphism is a modulating factor in repeated respiratory infections. Chest 126:95-99. [DOI] [PubMed] [Google Scholar]
- 22.Guo, Y., H. Feinberg, E. Conroy, D. A. Mitchell, R. Alvarez, O. Blixt, M. E. Taylor, W. I. Weis, and K. Drickamer. 2004. Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat. Struct. Mol. Biol. 11:591-598. [DOI] [PubMed] [Google Scholar]
- 23.Han, D. P., M. Lohani, and M. W. Cho. 2007. Specific asparagine-linked glycosylation sites are critical for DC-SIGN- and L-SIGN-mediated severe acute respiratory syndrome coronavirus entry. J. Virol. 81:12029-12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hart, M. L., M. Saifuddin, and G. T. Spear. 2003. Glycosylation inhibitors and neuraminidase enhance human immunodeficiency virus type 1 binding and neutralization by mannose-binding lectin. J. Gen. Virol. 84:353-360. [DOI] [PubMed] [Google Scholar]
- 25.Hart, M. L., M. Saifuddin, K. Uemura, E. G. Bremer, B. Hooker, T. Kawasaki, and G. T. Spear. 2002. High mannose glycans and sialic acid on gp120 regulate binding of mannose-binding lectin (MBL) to HIV type 1. AIDS Res. Hum. Retroviruses 18:1311-1317. [DOI] [PubMed] [Google Scholar]
- 26.Hartshorn, K. L., K. Sastry, M. R. White, E. M. Anders, M. Super, R. A. Ezekowitz, and A. I. Tauber. 1993. Human mannose-binding protein functions as an opsonin for influenza A viruses. J. Clin. Invest. 91:1414-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haurum, J. S., S. Thiel, I. M. Jones, P. B. Fischer, S. B. Laursen, and J. C. Jensenius. 1993. Complement activation upon binding of mannan-binding protein to HIV envelope glycoproteins. AIDS 7:1307-1313. [DOI] [PubMed] [Google Scholar]
- 28.Huang, I. C., B. J. Bosch, F. Li, W. Li, K. H. Lee, S. Ghiran, N. Vasilieva, T. S. Dermody, S. C. Harrison, P. R. Dormitzer, M. Farzan, P. J. Rottier, and H. Choe. 2006. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 281:3198-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ip, W. K., K. H. Chan, H. K. Law, G. H. Tso, E. K. Kong, W. H. Wong, Y. F. To, R. W. Yung, E. Y. Chow, K. L. Au, E. Y. Chan, W. Lim, J. C. Jensenius, M. W. Turner, J. S. Peiris, and Y. L. Lau. 2005. Mannose-binding lectin in severe acute respiratory syndrome coronavirus infection. J. Infect. Dis. 191:1697-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jack, D. L., N. J. Klein, and M. W. Turner. 2001. Mannose-binding lectin: targeting the microbial world for complement attack and opsonophagocytosis. Immunol. Rev. 180:86-99. [DOI] [PubMed] [Google Scholar]
- 31.Jackwood, M. W., D. A. Hilt, S. A. Callison, C. W. Lee, H. Plaza, and E. Wade. 2001. Spike glycoprotein cleavage recognition site analysis of infectious bronchitis virus. Avian Dis. 45:366-372. [PubMed] [Google Scholar]
- 32.Jeffers, S. A., S. M. Tusell, L. Gillim-Ross, E. M. Hemmila, J. E. Achenbach, G. J. Babcock, W. D. Thomas, Jr., L. B. Thackray, M. D. Young, R. J. Mason, D. M. Ambrosino, D. E. Wentworth, J. C. Demartini, and K. V. Holmes. 2004. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U. S. A. 101:15748-15753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji, X., H. Gewurz, and G. T. Spear. 2005. Mannose binding lectin (MBL) and HIV. Mol. Immunol. 42:145-152. [DOI] [PubMed] [Google Scholar]
- 34.Ji, X., G. G. Olinger, S. Aris, Y. Chen, H. Gewurz, and G. T. Spear. 2005. Mannose-binding lectin binds to Ebola and Marburg envelope glycoproteins, resulting in blocking of virus interaction with DC-SIGN and complement-mediated virus neutralization. J. Gen. Virol. 86:2535-2542. [DOI] [PubMed] [Google Scholar]
- 35.Kase, T., Y. Suzuki, T. Kawai, T. Sakamoto, K. Ohtani, S. Eda, A. Maeda, Y. Okuno, T. Kurimura, and N. Wakamiya. 1999. Human mannan-binding lectin inhibits the infection of influenza A virus without complement. Immunology 97:385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaushal, G. P., and A. D. Elbein. 1994. Glycosidase inhibitors in study of glycoconjugates. Methods Enzymol. 230:316-329. [DOI] [PubMed] [Google Scholar]
- 37.Kilpatrick, D. C. 2002. Mannan-binding lectin and its role in innate immunity. Transfus. Med. 12:335-352. [DOI] [PubMed] [Google Scholar]
- 38.Krokhin, O., Y. Li, A. Andonov, H. Feldmann, R. Flick, S. Jones, U. Stroeher, N. Bastien, K. V. Dasuri, K. Cheng, J. N. Simonsen, H. Perreault, J. Wilkins, W. Ens, F. Plummer, and K. G. Standing. 2003. Mass spectrometric characterization of proteins from the SARS virus: a preliminary report. Mol. Cell Proteomics 2:346-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 40.Kuhlman, M., K. Joiner, and R. A. Ezekowitz. 1989. The human mannose-binding protein functions as an opsonin. J. Exp. Med. 169:1733-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laursen, I. 2003. Mannan-binding lectin (MBL) production from human plasma. Biochem. Soc. Trans. 31:758-762. [DOI] [PubMed] [Google Scholar]
- 42.Leth-Larsen, R., F. Zhong, V. T. Chow, U. Holmskov, and J. Lu. 2007. The SARS coronavirus spike glycoprotein is selectively recognized by lung surfactant protein D and activates macrophages. Immunobiology 212:201-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, F., M. Berardi, W. Li, M. Farzan, P. R. Dormitzer, and S. C. Harrison. 2006. Conformational states of the severe acute respiratory syndrome coronavirus spike protein ectodomain. J. Virol. 80:6794-6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li, W., M. J. Moore, N. Vasilieva, J. Sui, S. K. Wong, M. A. Berne, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, T. C. Greenough, H. Choe, and M. Farzan. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madsen, H. O., P. Garred, S. Thiel, J. A. Kurtzhals, L. U. Lamm, L. P. Ryder, and A. Svejgaard. 1995. Interplay between promoter and structural gene variants control basal serum level of mannan-binding protein. J. Immunol. 155:3013-3020. [PubMed] [Google Scholar]
- 46.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 47.Marzi, A., T. Gramberg, G. Simmons, P. Moller, A. J. Rennekamp, M. Krumbiegel, M. Geier, J. Eisemann, N. Turza, B. Saunier, A. Steinkasserer, S. Becker, P. Bates, H. Hofmann, and S. Pohlmann. 2004. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J. Virol. 78:12090-12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matrosovich, M., T. Matrosovich, W. Garten, and H. D. Klenk. 2006. New low-viscosity overlay medium for viral plaque assays. Virol. J. 3:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitchell, D. A., A. J. Fadden, and K. Drickamer. 2001. A novel mechanism of carbohydrate recognition by the C-type lectins DC-SIGN and DC-SIGNR. Subunit organization and binding to multivalent ligands. J. Biol. Chem. 276:28939-28945. [DOI] [PubMed] [Google Scholar]
- 50.Moore, M. J., T. Dorfman, W. Li, S. K. Wong, Y. Li, J. H. Kuhn, J. Coderre, N. Vasilieva, Z. Han, T. C. Greenough, M. Farzan, and H. Choe. 2004. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 78:10628-10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mounir, S., and P. J. Talbot. 1993. Molecular characterization of the S protein gene of human coronavirus OC43. J. Gen. Virol. 74:1981-1987. [DOI] [PubMed] [Google Scholar]
- 52.Petersen, S. V., S. Thiel, and J. C. Jensenius. 2001. The mannan-binding lectin pathway of complement activation: biology and disease association. Mol. Immunol. 38:133-149. [DOI] [PubMed] [Google Scholar]
- 53.Pohlmann, S., F. Baribaud, B. Lee, G. J. Leslie, M. D. Sanchez, K. Hiebenthal-Millow, J. Munch, F. Kirchhoff, and R. W. Doms. 2001. DC-SIGN interactions with human immunodeficiency virus type 1 and 2 and simian immunodeficiency virus. J. Virol. 75:4664-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritchie, G., D. J. Harvey, F. Feldmann, U. Stroeher, H. Feldmann, L. Royle, R. A. Dwek, and P. M. Rudd. 2010. Identification of N-linked carbohydrates from severe acute respiratory syndrome (SARS) spike glycoprotein. Virology 399:257-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 56.Salvador, B., Y. Zhou, A. Michault, M. O. Muench, and G. Simmons. 2009. Characterization of Chikungunya pseudotyped viruses: Identification of refractory cell lines and demonstration of cellular tropism differences mediated by mutations in E1 glycoprotein. Virology 393:33-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seyfarth, J., P. Garred, and H. O. Madsen. 2006. Extra-hepatic transcription of the human mannose-binding lectin gene (mbl2) and the MBL-associated serine protease 1-3 genes. Mol. Immunol. 43:962-971. [DOI] [PubMed] [Google Scholar]
- 58.Shih, Y. P., C. Y. Chen, S. J. Liu, K. H. Chen, Y. M. Lee, Y. C. Chao, and Y. M. Chen. 2006. Identifying epitopes responsible for neutralizing antibody and DC-SIGN binding on the spike glycoprotein of the severe acute respiratory syndrome coronavirus. J. Virol. 80:10315-10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simmons, G., D. N. Gosalia, A. J. Rennekamp, J. D. Reeves, S. L. Diamond, and P. Bates. 2005. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U. S. A. 102:11876-11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simmons, G., J. D. Reeves, C. C. Grogan, L. H. Vandenberghe, F. Baribaud, J. C. Whitbeck, E. Burke, M. J. Buchmeier, E. J. Soilleux, J. L. Riley, R. W. Doms, P. Bates, and S. Pohlmann. 2003. DC-SIGN and DC-SIGNR bind Ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 305:115-123. [DOI] [PubMed] [Google Scholar]
- 61.Simmons, G., J. D. Reeves, A. J. Rennekamp, S. M. Amberg, A. J. Piefer, and P. Bates. 2004. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. U. S. A. 101:4240-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simmons, G., A. J. Rennekamp, and P. Bates. 2006. Proteolysis of SARS-associated coronavirus spike glycoprotein. Adv. Exp. Med. Biol. 581:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Simmons, G., R. J. Wool-Lewis, F. Baribaud, R. C. Netter, and P. Bates. 2002. Ebola virus glycoproteins induce global surface protein down-modulation and loss of cell adherence. J. Virol. 76:2518-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thiel, S., T. Vorup-Jensen, C. M. Stover, W. Schwaeble, S. B. Laursen, K. Poulsen, A. C. Willis, P. Eggleton, S. Hansen, U. Holmskov, K. B. Reid, and J. C. Jensenius. 1997. A second serine protease associated with mannan-binding lectin that activates complement. Nature 386:506-510. [DOI] [PubMed] [Google Scholar]
- 66.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 67.Turner, M. W., and R. M. Hamvas. 2000. Mannose-binding lectin: structure, function, genetics and disease associations. Rev. Immunogenet. 2:305-322. [PubMed] [Google Scholar]
- 68.van Kooyk, Y., and T. B. Geijtenbeek. 2003. DC-SIGN: escape mechanism for pathogens. Nat. Rev. Immunol. 3:697-709. [DOI] [PubMed] [Google Scholar]
- 69.Yang, Z. Y., Y. Huang, L. Ganesh, K. Leung, W. P. Kong, O. Schwartz, K. Subbarao, and G. J. Nabel. 2004. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 78:5642-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ying, H., X. Ji, M. L. Hart, K. Gupta, M. Saifuddin, M. R. Zariffard, and G. T. Spear. 2004. Interaction of mannose-binding lectin with HIV type 1 is sufficient for virus opsonization but not neutralization. AIDS Res. Hum. Retroviruses 20:327-335. [DOI] [PubMed] [Google Scholar]
- 71.Ying, W., Y. Hao, Y. Zhang, W. Peng, E. Qin, Y. Cai, K. Wei, J. Wang, G. Chang, W. Sun, S. Dai, X. Li, Y. Zhu, J. Li, S. Wu, L. Guo, J. Dai, P. Wan, T. Chen, C. Du, D. Li, J. Wan, X. Kuai, W. Li, R. Shi, H. Wei, C. Cao, M. Yu, H. Liu, F. Dong, D. Wang, X. Zhang, X. Qian, Q. Zhu, and F. He. 2004. Proteomic analysis on structural proteins of severe acute respiratory syndrome coronavirus. Proteomics 4:492-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuan, F. F., J. Tanner, P. K. Chan, S. Biffin, W. B. Dyer, A. F. Geczy, J. W. Tang, D. S. Hui, J. J. Sung, and J. S. Sullivan. 2005. Influence of FcγRIIA and MBL polymorphisms on severe acute respiratory syndrome. Tissue Antigens 66:291-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang, H., G. Zhou, L. Zhi, H. Yang, Y. Zhai, X. Dong, X. Zhang, X. Gao, Y. Zhu, and F. He. 2005. Association between mannose-binding lectin gene polymorphisms and susceptibility to severe acute respiratory syndrome coronavirus infection. J. Infect. Dis. 192:1355-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]