Abstract
The effects of the challenge dose and major histocompatibility complex (MHC) class IB alleles were analyzed in 112 Mauritian cynomolgus monkeys vaccinated (n = 67) or not vaccinated (n = 45) with Tat and challenged with simian/human immunodeficiency virus (SHIV) 89.6Pcy243. In the controls, the challenge dose (10 to 20 50% monkey infectious doses [MID50]) or MHC did not affect susceptibility to infection, peak viral load, or acute CD4 T-cell loss, whereas in the chronic phase of infection, the H1 haplotype correlated with a high viral load (P = 0.0280) and CD4 loss (P = 0.0343). Vaccination reduced the rate of infection acquisition at 10 MID50 (P < 0.0001), and contained acute CD4 loss at 15 MID50 (P = 0.0099). Haplotypes H2 and H6 were correlated with increased susceptibility (P = 0.0199) and resistance (P = 0.0087) to infection, respectively. Vaccination also contained CD4 depletion (P = 0.0391) during chronic infection, independently of the challenge dose or haplotype.
Advances in typing of the major histocompatibility complex (MHC) of Mauritian cynomolgus macaques (14, 20, 26) have provided the opportunity to address the influence of host factors on vaccine studies (13). Retrospective analysis of 22 macaques vaccinated with Tat or a Tat-expressing adenoviral vector revealed that monkeys with the H6 or H3 MHC class IB haplotype were overrepresented among aviremic or controller animals, whereas macaques with the H2 or H5 haplotype clustered in the noncontrollers (12). More recently, the H6 haplotype was reported to correlate with control of chronic infection with simian immunodeficiency virus (SIV) mac251, regardless of vaccination (18).
Here, we performed a retrospective analysis of 112 Mauritian cynomolgus macaques, which included the 22 animals studied previously (12), to evaluate the impact of the challenge dose and class IB haplotype on the acquisition and severity of simian/human immunodeficiency virus (SHIV) 89.6Pcy243 infection in 45 control monkeys and 67 monkeys vaccinated with Tat from different protocols (Table 1).
TABLE 1.
Summary of treatment, challenge dose, and outcome of infection in cynomolgus monkeys
| Protocol code | No. of monkeys | Immunogen (dose)a | Adjuvantb | Schedule of immunization (wk) | Routec | Challenged (MID50) | Virological outcomee |
Reference(s) or source | ||
|---|---|---|---|---|---|---|---|---|---|---|
| A | C | V | ||||||||
| ISS-ST | 6 | Tat (10) | Alum or RIBI | 0, 2, 6, 12, 15, 21, 28, 32, 36 | s.c., i.m. | 10 | 4 | 1 | 1 | 4, 17 |
| ISS-ST | 1 | Tat (6) | None | 0, 5, 12, 17, 22, 27, 32, 38, 42, 48 | i.d. | 10 | 1 | 0 | 0 | 4, 17 |
| ISS-PCV | 3 | pCV-tat (1 mg) | Bupivacaine + methylparaben | 0, 2, 6, 11, 15, 21, 28, 32, 36 | i.m. | 10 | 3 | 0 | 0 | 6 |
| ISS-ID | 3 | Tat (6) | none | 0, 4, 8, 12, 16, 20, 24, 28, 39, 43, 60 | i.d. | 10 | 1 | 1 | 1 | B. Ensoli, unpublished data |
| ISS-TR | 6 | Tat (10) | Alum-Iscom | 0, 2, 6, 11, 16, 21, 28, 32, 36 | s.c., i.d., i.m. | 10 | 4 | 2 | 0 | Ensoli, unpublished |
| ISS-TGf | 3 | Tat (10) | Alum | 0, 4, 12, 22 | s.c. | 15 | 0 | 3 | Ensoli, unpublished | |
| ISS-TG | 3 | Tatcys22 (10) | Alum | 15 | 0 | 3 | Ensoli, unpublished | |||
| ISS-TG | 4 | Tatcys22 (10) + Gag (60) | Alum | 15 | 0 | 4 | Ensoli, unpublished | |||
| ISS-TG | 4 | Tat (10) + Gag (60) | Alum | 15 | 0 | 4 | Ensoli, unpublished | |||
| ISS-MP | 3 | Tat (10) | H1D-Alum | 0, 4, 12, 18, 21, 38 | s.c., i.m. | 15 | 0 | 2 | 1 | Ensoli, unpublished |
| ISS-MP | 3 | Tat (10) | Alum | s.c. | 15 | 0 | 0 | 3 | Ensoli, unpublished | |
| ISS-GS | 6 | Tat (10) | H1D-Alum | 0, 4, 12, 18, 21, 36 | s.c., i.m. | 15 | 1 | 3 | 2 | Ensoli, unpublished |
| NCI-Ad-tat/Tat | 7 | Ad-tat (5 × 108 PFU), Tat (10) | Alum | 0, 12, 24, 36 | i.n., i.t., s.c. | 15 | 2 | 3 | 2 | Ensoli, unpublished |
| NCI-Tat | 9 | Tat (6 and 10) | Alum/Iscom | 0, 2, 6, 11, 15, 21, 28, 32, 36 | s.c., i.d., i.m. | 15 | 2 | 4 | 3 | 12 |
| ISS-NPT | 3 | pCV-tat (1 mg) | Bupivacaine + methylparaben-Iscom | 0, 2, 8, 13, 17, 22, 28, 46, 71 | i.m. | 20 | 0 | 0 | 3 | Ensoli, unpublished |
| ISS-NPT | 3 | pCV-tatcys22 (1 mg) | Bupivacaine + methylparaben-Iscom | 0, 2, 8, 13, 17, 22, 28, 46, 71 | i.m. | 20 | 1 | 1 | 1 | |
| Total vaccinated | 67 | 19 | 17 | 31 | ||||||
| Naive | 11 | None | None | NAg | NA | 10 or 15 | 1 | 3 | 7 | |
| Control | 34 | None, Ad, or pCV-0 | Alum, RIBI, H1D, Iscom or bupivacaine + methylparaben-Iscom | s.c., i.d., i.n., i.t., i.m. | 10, 15, or 20 | 5 | 13 | 16 | ||
| Total controls | 45 | 6 | 16 | 23 | ||||||
| Total | 112 | 25 | 33 | 54 | ||||||
All animals were inoculated with the indicated dose of Tat plasmid DNA (pCV-tat [8], adenovirus-tat [Ad-tat] [27]) or protein, Gag protein, or empty vectors (pCV-0, adenovirus [Ad]) by the indicated route. Doses are in micrograms unless indicated otherwise.
Alum, aluminum phosphate (4); RIBI oil-in-water emulsions containing squalene, bacterial monophosphoryl lipid A, and refined mycobacterial products (4); Iscom, immune-stimulating complex (4); H1D are biocompatible anionic polymeric microparticles used for vaccine delivery (10, 12, 25a).
s.c., subcutaneous; i.m., intramuscular; i.d., intradermal; i.n., intranasal; i.t., intratracheal.
All animals were inoculated intravenously with the indicated dose of the same SHIV89.6.Pcy243 stock.
According to the virological outcome upon challenge, monkeys were grouped as aviremic (A), controllers (C), or viremic (V).
Because of the short follow-up, controller status could not be determined and all infected monkeys of the ISS-TG protocol were therefore considered viremic.
NA, not applicable.
Study description.
All animals were challenged intravenously with 10 (n = 29), 15 (n = 68), or 20 (n = 15) monkey infectious doses (MID50) of SHIV86.6Pcy243 derived from a cynomolgus macaque inoculated with the original SHIV89.6P stock from rhesus monkeys (3, 5) (Table 1). The SIV RNA load in plasma and CD4 T-cell counts were determined as described previously (23).
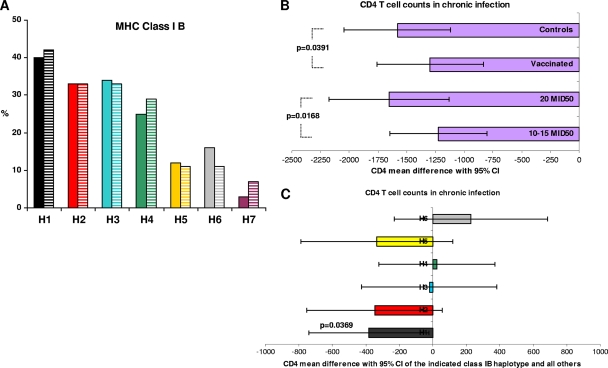
MHC allele predictions were generated on the basis of the microsatellite profiles (26), with the alleles for class I and class II regions being inferred on the basis of established haplotype-allele associations (20, 26). As shown in Fig. 1 A, the distribution of the seven major haplotypes (haplotypes H1 to H7) in vaccinated and control animals was balanced. Due to the presence of the H7 haplotype in only 3 out of 112 monkeys, it was excluded from the analyses.
FIG. 1.
(A) Frequency of MHC class IB haplotypes in 112 monkeys vaccinated with Tat (full-colored bars) or controls (zebra-striped bars). (B and C) Analysis of CD4 T cells in chronic infection in viremic macaques (n = 71) stratified by treatment (vaccinated, n = 37; controls, n = 34) and challenge dose (10 to 15 MID50, n = 58; 20 MID50, n = 13) (B) or class IB haplotype (H1, n = 26; H2, n = 24; H3, n = 25; H4, n = 25; H5, n = 8; H6, n = 9) (C). On the abscissa is reported the difference in CD4 T-cell counts pre- and postchallenge; least-squares means with 95% confidence intervals are shown (analysis of variance model).
As the factors investigated in this study may affect susceptibility to acute (1 to 4 weeks) or chronic (16 to 52 weeks) infection differently, these were analyzed separately. Since the plasma viral load is a good indicator of infection dynamics, monkeys were stratified into the aviremic group (<50 RNA eq/ml) or viremic group (>50 RNA eq/ml). During the chronic phase, the macaques were stratified as controllers (viremia of <5 × 103 RNA eq/ml and at least two determinations in which viral RNA was undetectable [<50 RNA eq/ml]) or viremic (persistent viremia, >5 × 103 RNA eq/ml). In chronic infection, the viral load set point was calculated as the mean of all values between the 16th and 52nd weeks, expressed as log10 number of copies/ml; a log value of log10 (50)/2 was assigned to viral loads below the cutoff (50 RNA eq/ml).
All 112 monkeys were evaluated for their susceptibility to infection, but only viremic animals were considered for the analyses of acute (n = 87) and chronic (n = 71) infection. In the chronic phase, monkeys with follow-up at ≥28 weeks were further categorized as viremic (n = 38) or controllers (n = 33).
Susceptibility to infection.
Upon challenge, 39 of 45 controls (87%) and 48 of 67 vaccinated animals (72%) became viremic. As shown in Table 2, the challenge dose did not affect susceptibility to infection in controls, while in vaccinated macaques, the percentage of aviremic monkeys increased to 68% after challenge with 10 MID50, remaining comparable to that for the controls after challenge with 15 (12%) or 20 (17%) MID50 (P < 0.0001) (Table 2).
TABLE 2.
Univariate analysis of the effects of challenge dose and class IB haplotype on susceptibility/resistance to infection (aviremic or viremic) in control and vaccinated monkeys
| Dose or haplotype | Controls |
Vaccinated |
||||||
|---|---|---|---|---|---|---|---|---|
| No. (%) of monkeys |
P valuea | No. (%) of monkeys |
P valuea | |||||
| Total (n = 45) | Aviremic | Viremic | Total (n = 67) | Aviremic | Viremic | |||
| MID50 | ||||||||
| 10 | 10 | 3 (30) | 7 (70) | 19 | 13 (68) | 6 (32) | ||
| 15 | 26 | 2 (8) | 24 (92) | 42 | 5 (12) | 37 (88) | ||
| 20 | 9 | 1 (11) | 8 (89) | 0.2239 | 6 | 1 (17) | 5 (83) | <0.0001 |
| H1 | 19 | 5 (26) | 14 (74) | 27 | 9 (33) | 18 (67) | ||
| Non-H1 | 26 | 1 (4) | 25 (96) | 0.0687 | 40 | 10 (25) | 30 (75) | 0.5821 |
| H2 | 15 | 0 (0) | 15 (100) | 22 | 2 (9) | 20 (91) | ||
| Non-H2 | 30 | 6 (20) | 24 (80) | 0.1575 | 45 | 17 (38) | 28 (62) | 0.0199 |
| H3 | 15 | 2 (13) | 13 (87) | 23 | 6 (26) | 17 (74) | ||
| Non-H3 | 30 | 4 (13) | 26 (87) | 1.0000 | 44 | 13 (30) | 31 (70) | 1.0000 |
| H4 | 13 | 2 (15) | 11 (85) | 17 | 3 (18) | 14 (82) | ||
| Non-H4 | 32 | 4 (12) | 28 (88) | 1.0000 | 50 | 16 (32) | 34 (68) | 0.3561 |
| H5 | 5 | 0 (0) | 2 (100) | 8 | 2 (25) | 6 (75) | ||
| Non-H5 | 40 | 6 (15) | 34 (85) | 1.0000 | 59 | 17 (29) | 42 (71) | 1.0000 |
| H6 | 5 | 0 (0) | 5 (100) | 11 | 7 (64) | 4 (36) | ||
| Non-H6 | 40 | 6 (15) | 34 (85) | 1.0000 | 56 | 12 (21) | 44 (79) | 0.0087 |
Fisher's exact test.
In the controls, the IB haplotype did not influence the infection acquisition rate, although a trend (P = 0.0687) for increased susceptibility was observed with the H1 haplotype (Table 2). In contrast, in vaccinated monkeys, the H2 and H6 haplotypes were associated with increased susceptibility (P = 0.0199) and resistance (P = 0.0087) to infection, respectively (Table 2), while the trend for an association of the H1 haplotype with increased susceptibility observed in the controls disappeared after vaccination (P = 0.5821).
The effects of the challenge dose and the IB haplotype on the likelihood of being aviremic were confirmed in a logistic regression model (the stepwise selection method was chosen to select the best-fit model, using maximum likelihood estimates) for controls and vaccinees. Specifically, in vaccinated animals the influence of the challenge dose (for 10 versus 15 to 20 MID50, odds ratio = 16.2, 95% confidence interval [CI] = 4.0 to 65.6; P < 0.0001) and the H6 haplotype (odds ratio =7.2; 95% CI = 1.3 to 39.2; P = 0.0215) on the aviremic status remained significant, whereas the association of H2 with increased susceptibility to infection was lost.
Acute infection.
The challenge dose and the IB haplotype had no significant impact on the peak viral load in control and Tat-vaccinated macaques. Notably, while CD4 T-cell loss in the controls was not affected by the challenge dose, it was significantly (P = 0.0099) contained in vaccinated monkeys inoculated with 15 MID50. This was confirmed in a multivariate model (analysis of variance), which included the challenge dose and haplotype as explicative factors (P = 0.0284).
Chronic phase.
Of the 71 viremic monkeys analyzed, 37 were controls and 34 were vaccinated. Of the 37 controls, 21 (57%) remained viremic and 16 (43%) became controllers (see Table S1 in the supplemental material). Of the 34 vaccinated monkeys, 17 remained viremic and 17 became controllers. Univariate analysis did not reveal significant effects of the challenge dose or IB haplotype on the viremic status (controller or viremic) in either controls or vaccinees (see Table S1 in the supplemental material), although a trend (P = 0.0570) to remain viremic was observed for vaccinees carrying H2, the IB haplotype associated with increased susceptibility to infection. A multivariate analysis (exact logistic regression model, backward-selection method) aimed at evaluating the effects of the MID50 and IB haplotype on the likelihood that viremic macaques became controllers confirmed the lack of significant associations in control monkeys, while in vaccinated macaques it strengthened the association of H2 haplotype with the viremic status (odds ratio = 0.1; 95% CI = 0.0 to 0.7; P = 0.0125) and disclosed a previously unnoticed association of the H5 haplotype with the viremic status (odds ratio = 0.1; 95% CI = 0.0 to 0.7; P = 0.0237).
The challenge dose did not significantly influence the viral load set point in control or vaccinated macaques, although there was a trend (P = 0.0560) for a lower set point in controls challenged with 15 MID50 (see Table S2 in the supplemental material). In controls, the H1 haplotype was associated with a higher viral load set point (P = 0.0280) (see Table S2 in the supplemental material) and greater CD4 T-cell loss (P = 0.0343) (Table 3). In vaccinees, trends for a higher (H2, P = 0.0942; H5, P = 0.0804) and a lower (H6, P = 0.0715) viral load set point were noticed (see Table S2 in the supplemental material). Further, CD4 T-cell loss was significantly contained in vaccinees challenged with 15 MID50 (P = 0.0067) or carrying H6 (P < 0.0001) (Table 3).
TABLE 3.
CD4 T-cell count changesa in viremic control or vaccinated monkeys, stratified by challenge dose or class IB haplotype
| Dose or haplotype | Controls |
Vaccinated |
||||
|---|---|---|---|---|---|---|
| Total no. of monkeys (n = 37) | Mean ± SE CD4 T-cell count change | P valueb | Total no. of monkeys (n= 34) | Mean ± SE CD4 T-cell count change | P valueb | |
| MID50s | ||||||
| 10 | 7 | −1,386 ± 227 | 0.8395 | 6 | −1,348 ± 357 | 0.7896 |
| 15 | 22 | −1,223 ± 128 | 0.3723 | 23 | −807 ± 96 | 0.0067 |
| 20 | 8 | −1,453 ± 231 | 5 | −1,465 ± 179 | ||
| Haplotypes | ||||||
| H1 | 12 | −1,604 ± 206 | 14 | −1,144 ± 191 | ||
| Non-H1 | 25 | −1,159 ± 100 | 0.0343 | 20 | −898 ± 111 | 0.2445 |
| H2 | 14 | −1,406 ± 154 | 10 | −1,178 ± 241 | ||
| Non-H2 | 23 | −1,241 ± 130 | 0.4305 | 24 | −925 ± 105 | 0.2684 |
| H3 | 13 | −1,280 ± 180 | 12 | −815 ± 127 | ||
| Non-H3 | 24 | −1,316 ± 121 | 0.8656 | 22 | −1,100 ± 140 | 0.1882 |
| H4 | 11 | −1,100 ± 188 | 14 | −994 ± 135 | ||
| Non-H4 | 26 | −1,389 ± 115 | 0.1872 | 20 | −1,003 ± 150 | 0.9641 |
| H5 | 4 | −1,266 ± 244 | 4 | −1,419 ± 363 | ||
| Non-H5 | 33 | −1,308 ± 109 | 0.8977 | 30 | −943 ± 104 | 0.1372 |
| H6 | 5 | −954 ± 194 | 4 | −535 ± 20 | ||
| Non-H6 | 32 | −1,358 ± 109 | 0.1676 | 30 | −1,061 ± 111 | <0.0001 |
Changes for the chronic phase were defined as the difference from the average value of multiple prechallenge determinations.
Student's t test.
A multivariate model (analysis of variance) that included vaccination, challenge dose, and haplotype as explicative factors was then applied to CD4 T-cell counts. Animals challenged with 10 or 15 MID50 were cumulated, since the results for these two groups did not significantly differ (P = 0.2649). This analysis showed that CD4 loss was influenced by both vaccination (P = 0.0391) and challenge dose (10 to 15 versus 20 MID50, P = 0.0168) (Fig. 1B). In addition, the association of the H1 haplotype with severe CD4 T-cell loss was confirmed (P = 0.0369) (Fig. 1C).
Thus, both the infectious dose and the MHC IB haplotype are important when the vaccine effects in monkey studies are evaluated. However, the impact of the challenge dose on infection susceptibility and severity appears to be negligible in control animals, in substantial agreement with our former data (4). Similarly, the influence of class IB haplotypes in controls was limited to H1 in acute infection (increased susceptibility, P = 0.0687) and chronic infection (viral load set point, P = 0.0280; CD4 loss, P = 0.0343).
In contrast, both the infectious dose and IB haplotype significantly affected the protection offered by the vaccine. In particular, vaccination significantly reduced infection acquisition at 10 MID50 but not at 15 or 20 MID50 (P < 0.0001), underscoring the importance of using in vaccine efficacy studies challenge doses not exceedingly higher than those responsible for transmission in humans (22).
MHC-encoded host factors, including immune response genes, contribute importantly to susceptibility to human immunodeficiency virus (HIV)/SIV infection and progression (1, 7, 19), confounding the interpretation of efficacy afforded by vaccine candidates (13). The associations in rhesus monkeys of Mamu-A*01, Mamu-B*08, and Mamu B*17 with resistance to SIV represent good examples of this influence (15, 21, 24, 25). The results of our studies with cynomolgus macaques suggest a different scenario. In fact, while the impact of IB haplotypes on infection susceptibility/resistance was very limited in the controls, in the vaccinees, haplotypes H2 and H6 were associated with increased susceptibility (P = 0.0199) and resistance (P = 0.0087), respectively, to becoming viremic (Table 2), a finding confirmed for haplotype H6 in a multivariate analysis.
Once they were infected, vaccinees did not differ from controls with respect to peak viremia, regardless of the challenge dose or IB haplotype, whereas CD4 T-cell loss was contained during acute infection (P = 0.0099) in vaccinees challenged with 15 MID50. In contrast, a relevant influence of these factors was evident in chronic infection. In fact, a significant containment of CD4 T-cell loss was apparent in vaccinees but not in control animals challenged with 15 MID50 (P = 0.0067) (Table 3), the most-represented challenge dose group (n = 45), confirming the preservation of CD4 T-cell counts observed during acute infection with 15 MID50 (P = 0.0099). Further, vaccinated macaques challenged with 10 to 15 MID50 exhibited significantly milder CD4 T-cell losses than those challenged with 20 MID50 (P = 0.0168) (Fig. 1B). A protective role for vaccination was also suggested by comparing CD4 T-cell losses in the controls and vaccinees, regardless of the challenge dose (P = 0.0391) (Fig. 1 B).
With regard to the impact of the IB haplotype, both haplotypes H2 and H5 were significantly associated in multivariate analysis with the viremic status in chronic infection, suggesting poor containment. The association of haplotype H1 with increased susceptibility to infection and a worse chronic infection observed in the controls was not found in vaccinated macaques, suggesting a protective effect of vaccination. Taken together, these associations suggest that IB haplotypes do not influence per se infection susceptibility or progression but, rather, affect responsiveness to vaccination and immunity to the virus.
Analysis of the immune responses in macaques with known IB haplotypes is warranted to confirm this hypothesis and provide relevant information for the development of monkey models suitable to evaluate both pathogenesis and vaccine efficacy. Notably, on the basis of these preclinical data, Tat has been tested in phase I preventative and therapeutic trials (2, 9-11, 16) and is being evaluated in a phase II therapeutic trial (http://www.hiv1tat-vaccines.info/).
Supplementary Material
Acknowledgments
We thank P. Sergiampietri and I. Ronci for editorial assistance; Franco Corrias, P. Pupino-Carbonelli, E. Iale, F. Incitti, N. Verrone, A. Marini, A. Avitabile, M. Chiodi, and M. Azzetti for hematoclinical analysis of cynomolgus monkey samples and for the handling of the animal facility; P. Cocco, D. Diamanti, and F. Costa for technical support; Roberto Belli for fluorescence-activated cell sorter analysis; and Silvia Baroncelli and Donatella Negri for assistance with data collection.
This work was supported in part by the Italian AIDS National Program from the Ministry of Health (to B.E.) and the Italian Concerted Action on Vaccine (ICAV); MHC analyses were supported by U.S. National Institute of Allergy and Infectious Diseases contract HHSN266200400088C/N01-A1-30061.
Footnotes
Published ahead of print on 16 June 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Anastassopoulou, C. G., and L. G. Kostrikis. 2003. The impact of human allelic variation on HIV-1 disease. Curr. HIV Res. 1:185-203. [DOI] [PubMed] [Google Scholar]
- 2.Bellino, S., V. Francavilla, O. Longo, A. Tripiciano, G. Paniccia, A. Arancio, V. Fiorelli, A. Scoglio, B. Collacchi, M. Campagna, A. Lazzarin, G. Tambussi, C. T. Din, R. Visintini, P. Narciso, A. Antinori, G. D'Offizi, M. Giulianelli, M. Carta, A. Di Carlo, G. Palamara, M. Giuliani, M. E. Laguardia, P. Monini, M. Magnani, F. Ensoli, and B. Ensoli. 2009. Parallel conduction of the phase I preventive and therapeutic trials based on the Tat vaccine candidate. Rev. Recent Clin. Trials 4:195-204. [DOI] [PubMed] [Google Scholar]
- 3.Borsetti, A., S. Baroncelli, M. T. Maggiorella, S. Bellino, S. Moretti, L. Sernicola, R. Belli, B. Ridolfi, S. Farcomeni, D. R. Negri, A. Cafaro, B. Ensoli, and F. Titti. 2008. Viral outcome of simian-human immunodeficiency virus SHIV-89.6P adapted to cynomolgus monkeys. Arch. Virol. 153:463-472. [DOI] [PubMed] [Google Scholar]
- 4.Cafaro, A., A. Caputo, C. Fracasso, M. T. Maggiorella, D. Goletti, S. Baroncelli, M. Pace, L. Sernicola, M. L. Koanga-Mogtomo, M. Betti, A. Borsetti, R. Belli, L. Akerblom, F. Corrias, S. Butto, J. Heeney, P. Verani, F. Titti, and B. Ensoli. 1999. Control of SHIV-89.6P-infection of cynomolgus monkeys by HIV-1 Tat protein vaccine. Nat. Med. 5:643-650. [DOI] [PubMed] [Google Scholar]
- 5.Cafaro, A., A. Caputo, M. T. Maggiorella, S. Baroncelli, C. Fracasso, M. Pace, A. Borsetti, L. Sernicola, D. R. Negri, P. ten Haaft, M. Betti, Z. Michelini, I. Macchia, E. Fanales-Belasio, R. Belli, F. Corrias, S. Butto, P. Verani, F. Titti, and B. Ensoli. 2000. SHIV89.6P pathogenicity in cynomolgus monkeys and control of viral replication and disease onset by human immunodeficiency virus type 1 Tat vaccine. J. Med. Primatol. 29:193-208. [DOI] [PubMed] [Google Scholar]
- 6.Cafaro, A., F. Titti, C. Fracasso, M. T. Maggiorella, S. Baroncelli, A. Caputo, D. Goletti, A. Borsetti, M. Pace, E. Fanales-Belasio, B. Ridolfi, D. R. Negri, L. Sernicola, R. Belli, F. Corrias, I. Macchia, P. Leone, Z. Michelini, P. ten Haaft, S. Butto, P. Verani, and B. Ensoli. 2001. Vaccination with DNA containing tat coding sequences and unmethylated CpG motifs protects cynomolgus monkeys upon infection with simian/human immunodeficiency virus (SHIV89.6P). Vaccine 19:2862-2877. [DOI] [PubMed] [Google Scholar]
- 7.den Uyl, D., I. E. van der Horst-Bruinsma, and M. van Agtmael. 2004. Progression of HIV to AIDS: a protective role for HLA-B27? AIDS Rev. 6:89-96. [PubMed] [Google Scholar]
- 8.Ensoli, B., L. Buonaguro, G. Barillari, V. Fiorelli, R. Gendelman, R. A. Morgan, P. Wingfield, and R. C. Gallo. 1993. Release, uptake, and effects of extracellular human immunodeficiency virus type 1 Tat protein on cell growth and viral transactivation. J. Virol. 67:277-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ensoli, B., V. Fiorelli, F. Ensoli, A. Cafaro, F. Titti, S. Butto, P. Monini, M. Magnani, A. Caputo, and E. Garaci. 2006. Candidate HIV-1 Tat vaccine development: from basic science to clinical trials. AIDS 20:2245-2261. [DOI] [PubMed] [Google Scholar]
- 10.Ensoli, B., V. Fiorelli, F. Ensoli, A. Lazzarin, R. Visintini, P. Narciso, A. Di Carlo, P. Monini, M. Magnani, and E. Garaci. 2008. The therapeutic phase I trial of the recombinant native HIV-1 Tat protein. AIDS 22:2207-2209. [DOI] [PubMed] [Google Scholar]
- 11.Ensoli, B., V. Fiorelli, F. Ensoli, A. Lazzarin, R. Visintini, P. Narciso, A. Di Carlo, A. Tripiciano, O. Longo, S. Bellino, V. Francavilla, G. Paniccia, A. Arancio, A. Scoglio, B. Collacchi, M. J. Ruiz Alvarez, G. Tambussi, D. C. Tassan, G. Palamara, A. Latini, A. Antinori, G. D'Offizi, M. Giuliani, M. Giulianelli, M. Carta, P. Monini, M. Magnani, and E. Garaci. 2009. The preventive phase I trial with the HIV-1 Tat-based vaccine. Vaccine 28:371-378. [DOI] [PubMed] [Google Scholar]
- 12.Florese, R. H., R. W. Wiseman, D. Venzon, J. A. Karl, T. Demberg, K. Larsen, L. Flanary, V. S. Kalyanaraman, R. Pal, F. Titti, L. J. Patterson, M. J. Heath, D. H. O'Connor, A. Cafaro, B. Ensoli, and M. Robert-Guroff. 2008. Comparative study of Tat vaccine regimens in Mauritian cynomolgus and Indian rhesus macaques: influence of Mauritian MHC haplotypes on susceptibility/resistance to SHIV(89.6P) infection. Vaccine 26:3312-3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaur, G., and N. Mehra. 2009. Genetic determinants of HIV-1 infection and progression to AIDS: susceptibility to HIV infection. Tissue Antigens 73:289-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krebs, K. C., Z. Jin, R. Rudersdorf, A. L. Hughes, and D. H. O'Connor. 2005. Unusually high frequency MHC class I alleles in Mauritian origin cynomolgus macaques. J. Immunol. 175:5230-5239. [DOI] [PubMed] [Google Scholar]
- 15.Loffredo, J. T., J. Maxwell, Y. Qi, C. E. Glidden, G. J. Borchardt, T. Soma, A. T. Bean, D. R. Beal, N. A. Wilson, W. M. Rehrauer, J. D. Lifson, M. Carrington, and D. I. Watkins. 2007. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J. Virol. 81:8827-8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Longo, O., A. Tripiciano, V. Fiorelli, S. Bellino, A. Scoglio, B. Collacchi, M. J. Alvarez, V. Francavilla, A. Arancio, G. Paniccia, A. Lazzarin, G. Tambussi, C. T. Din, R. Visintini, P. Narciso, A. Antinori, G. D'Offizi, M. Giulianelli, M. Carta, A. Di Carlo, G. Palamara, M. Giuliani, M. E. Laguardia, P. Monini, M. Magnani, F. Ensoli, and B. Ensoli. 2009. Phase I therapeutic trial of the HIV-1 Tat protein and long term follow-up. Vaccine 27:3306-3312. [DOI] [PubMed] [Google Scholar]
- 17.Maggiorella, M. T., S. Baroncelli, Z. Michelini, E. Fanales-Belasio, S. Moretti, L. Sernicola, A. Cara, D. R. Negri, S. Butto, V. Fiorelli, A. Tripiciano, A. Scoglio, A. Caputo, A. Borsetti, B. Ridolfi, R. Bona, P. ten Haaft, I. Macchia, P. Leone, M. R. Pavone-Cossut, F. Nappi, M. Ciccozzi, J. Heeney, F. Titti, A. Cafaro, and B. Ensoli. 2004. Long-term protection against SHIV89.6P replication in HIV-1 Tat vaccinated cynomolgus monkeys. Vaccine 22:3258-3269. [DOI] [PubMed] [Google Scholar]
- 18.Mee, E. T., N. Berry, C. Ham, U. Sauermann, M. T. Maggiorella, F. Martinon, E. J. Verschoor, J. L. Heeney, R. Le Grand, F. Titti, N. Almond, and N. J. Rose. 2009. Mhc haplotype H6 is associated with sustained control of SIVmac251 infection in Mauritian cynomolgus macaques. Immunogenetics 61:327-339. [DOI] [PubMed] [Google Scholar]
- 19.Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors 1. Proc. Natl. Acad. Sci. U. S. A. 97:2709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Connor, S. L., A. J. Blasky, C. J. Pendley, E. A. Becker, R. W. Wiseman, J. A. Karl, A. L. Hughes, and D. H. O'Connor. 2007. Comprehensive characterization of MHC class II haplotypes in Mauritian cynomolgus macaques. Immunogenetics 59:449-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pal, R., D. Venzon, N. L. Letvin, S. Santra, D. C. Montefiori, N. R. Miller, E. Tryniszewska, M. G. Lewis, T. C. VanCott, V. Hirsch, R. Woodward, A. Gibson, M. Grace, E. Dobratz, P. D. Markham, Z. Hel, J. Nacsa, M. Klein, J. Tartaglia, and G. Franchini. 2002. ALVAC-SIV-gag-pol-env-based vaccination and macaque major histocompatibility complex class I (A*01) delay simian immunodeficiency virus SIVmac-induced immunodeficiency. J. Virol. 76:292-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pilcher, C. D., H. C. Tien, J. J. Eron, Jr., P. L. Vernazza, S. Y. Leu, P. W. Stewart, L. E. Goh, and M. S. Cohen. 2004. Brief but efficient: acute HIV infection and the sexual transmission of HIV. J. Infect. Dis. 189:1785-1792. [DOI] [PubMed] [Google Scholar]
- 23.Romano, J. W., R. N. Shurtliff, E. Dobratz, A. Gibson, K. Hickman, P. D. Markham, and R. Pal. 2000. Quantitative evaluation of simian immunodeficiency virus infection using NASBA technology. J. Virol. Methods 86:61-70. [DOI] [PubMed] [Google Scholar]
- 24.Seaman, M. S., S. Santra, M. H. Newberg, V. Philippon, K. Manson, L. Xu, R. S. Gelman, D. Panicali, J. R. Mascola, G. J. Nabel, and N. L. Letvin. 2005. Vaccine-elicited memory cytotoxic T lymphocytes contribute to Mamu-A*01-associated control of simian/human immunodeficiency virus 89.6P replication in rhesus monkeys. J. Virol. 79:4580-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valentine, L. E., J. T. Loffredo, A. T. Bean, E. J. Leon, C. E. MacNair, D. R. Beal, S. M. Piaskowski, Y. C. Klimentidis, S. M. Lank, R. W. Wiseman, J. T. Weinfurter, G. E. May, E. G. Rakasz, N. A. Wilson, T. C. Friedrich, D. H. O'Connor, D. B. Allison, and D. I. Watkins. 2009. Infection with “escaped” virus variants impairs control of simian immunodeficiency virus SIVmac239 replication in Mamu-B*08-positive macaques. J. Virol. 83:11514-11527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Voltan R., A. Castaldello, E. Brocca-Cofano, G. Altavilla, A. Caputo, M. Laus, K. Sparnacci, B. Ensoli, S. Spaccasassi, M. Ballestri, and L. Tondelli. 2007. Preparation and characterization of innovative protein-coated poly(methylmethacrylate) core-shell nanoparticles for vaccine purposes. Pharm. Res. 24:1870-1882. [DOI] [PubMed] [Google Scholar]
- 26.Wiseman, R. W., J. A. Wojcechowskyj, J. M. Greene, A. J. Blasky, T. Gopon, T. Soma, T. C. Friedrich, S. L. O'Connor, and D. H. O'Connor. 2007. Simian immunodeficiency virus SIVmac239 infection of major histocompatibility complex-identical cynomolgus macaques from Mauritius. J. Virol. 81:349-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao, J., R. Voltan, B. Peng, A. vis-Warren, V. S. Kalyanaraman, W. G. Alvord, K. Aldrich, D. Bernasconi, S. Butto, A. Cafaro, B. Ensoli, and M. Robert-Guroff. 2005. Enhanced cellular immunity to SIV Gag following coadministration of adenoviruses encoding wild-type or mutant HIV Tat and SIV Gag. Virology 342:1-12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.