Abstract
In vitro infection of cells with the betaherpesvirus human cytomegalovirus (HCMV) stimulates an innate immune response characterized by phosphorylation of the transcription factor interferon regulatory factor 3 (IRF3) and subsequent expression of IRF3-dependent genes. While previous work suggests that HCMV envelope glycoprotein B is responsible for initiating this reaction, the signaling pathways stimulated by virus infection that lead to IRF3 phosphorylation have largely been uncharacterized. Recently, we identified Z DNA binding protein 1 (ZBP1), a sensor of cytoplasmic DNA, as an essential protein for this response. We now describe a human fibroblast cell line exhibiting a recessive defect that results in the absence of activation of IRF3 following treatment with HCMV but not Sendai virus or double-stranded RNA. In addition, we show that while exposure of these cells to soluble HCMV glycoprotein B is capable of triggering IRF3-dependent gene transcription, transfection of the cells with double-stranded DNA is not. Furthermore, we show that overexpression of ZBP1 in these cells reestablishes their ability to secrete interferon in response to HCMV and that multiple ZBP1 transcriptional variants exist in both wild-type and mutant cells. These results have two major implications for the understanding of innate immune stimulation by HCMV. First, they demonstrate that HCMV glycoprotein B is not the essential molecular pattern that induces an IRF3-dependent innate immune response. Second, IRF3-terminal signaling triggered by HCMV particles closely resembles that which is activated by cytoplasmic double-stranded DNA.
Human cytomegalovirus (HCMV) is a ubiquitous member of the betaherpesvirus family. The virus is an opportunistic and chronic pathogen exhibiting lifelong persistence despite eliciting strong and continual innate and adaptive antiviral immune responses. While infection is usually asymptomatic in healthy adults, it is a significant cause of morbidity and mortality during immunosuppression (33, 41). HCMV is also a major cause of birth defects when acute infection occurs during pregnancy (2). Chronic infection of the vasculature has also been linked strongly with the development of cardiovascular diseases such as atherosclerosis, restenosis, and transplant vascular sclerosis (reviewed in reference 62).
Infection of host cells by HCMV leads to rapid expression of type I interferons (IFNs), IFN-stimulated genes (ISGs), and proinflammatory cytokines (4, 8, 20, 38, 60, 72, 73), a response contributing to establishment of an antiviral state in infected and surrounding tissues. Type I IFNs include IFN-β as well as multiple subtypes of IFN-α (see reference 54). IFN-α/β induces the expression of so-called ISGs through activation of signaling pathways downstream of the type I IFN receptor. These includes Janus kinase (JAK)- and tyrosine kinase (Tyk)-dependent phosphorylation of signal transducer and activator of transcription 1 (STAT1) and STAT2. STAT1 and STAT2 associate with IFN regulatory factor 9 (IRF9) and accumulate in the nucleus, where the complex leads to ISG transcription. ISG-encoded proteins subsequently act to impair intracellular molecular activities required for virus replication. How HCMV replication proceeds in the presence of this antiviral response is poorly understood.
Induction of IFN-β transcription in response to exposure to microbial pathogens is an increasingly well-characterized cellular process (see references 25, 26, and 43). Cellular detection of pathogen-associated molecular patterns (PAMPs) leads to nuclear accumulation and formation on the IFN-β promoter region of an enhanceosome complex containing IRF3, nuclear factor κB (NF-κB), activating transcription factor 2 (ATF2), CREB-binding protein (CBP), and p300 (30, 34, 67, 71). While IFN-β expression has been shown to occur in the absence of NF-κB (65), IRF3 is necessary for HCMV-triggered IFN responses in human fibroblasts (15, 56). It is also important to note that the expression of a subset of ISGs, including viperin (5, 8, 19) and ISG56 (8, 15, 19, 45), can occur in an IRF3-dependent manner in the absence of IFN-mediated signaling. Moreover, IRF3 itself can induce the generation of an antiviral cellular state when IFN is not present (6, 8, 10, 15, 40, 72, 73), which likely plays a prominent role in the innate immune response to HCMV (40) since the virus can interfere with JAK/STAT-mediated gene expression (42), but no HCMV-encoded IRF3-inhibitory phenotypes have been identified.
IRF3 is a constitutively expressed protein normally moving freely between the cytoplasm and the nucleus. However, PAMP-initiated phosphorylation of C-terminal serine and threonine residues results in IRF3 homodimerization, association with CBP and p300, and nuclear accumulation (18, 30, 57, 59, 71). Phosphorylation of IRF3 occurs via TANK binding kinase 1 and IκB kinase ɛ (18, 30, 57, 59, 71). Phosphorylation signals originate from cell surface, endosomal, or cytoplasmic pattern recognition receptor (PRR) proteins that detect specific PAMPs. IRF3-terminal PRRs include the Toll-like receptors (TLRs) 3 and 4, the cytoplasmic RNA helicases retinoic acid-inducible gene I (RIG-I) (70) and melanoma differentiation-associated gene 5 (MDA5) (69), and the double-stranded DNA (dsDNA) sensors Z DNA binding protein 1 (ZBP1) (63) and RNA polymerase III (POL3) (1, 11). TLR3 and TLR4 react with extracellular dsRNA and lipopolysaccharide (LPS), respectively. RIG-I and MDA5 helicases recognize cytoplasmic poly(I:C) and virus-associated dsRNA and generate downstream signaling via N-terminal caspase recruitment domains (CARDs) (13, 21, 46, 50, 51, 69). The CARD-containing molecule IFN-β promoter stimulator 1 (IPS1; also called MAVS, VISA, and cardif [27, 35, 58, 68, 70]) acts as an adaptor molecule essential to RIG-I- and MDA5-initiated IRF3 phosphorylation. Cytoplasmic B-form dsDNA is also known to trigger IRF3 activation following detection by ZBP1 or POL3. ZBP1 was first shown to be involved in IFN-β induction by both cytosolic DNA and herpes simplex virus type 1 (HSV-1) (63, 66), yet the components of the ZBP1-dependent signaling pathway are mostly uncharacterized. Alternatively, POL3 reacts to dsDNA by using it as a template for synthesis of dsRNA, which then reacts with RIG-I to activate IRF3 and IFN-β expression via IPS1 (1, 11).
Activation of IRF3 has been shown to occur in response to infection with multiple herpesviruses, including HCMV (4, 14, 15, 20, 24, 37, 38, 40, 47), an ICP0-null mutant of the alphaherpesvirus HSV-1 (12, 31, 36, 47), and the gammaherpesvirus Epstein-Barr virus (EBV) (52, 53). Virus-associated IRF3-activating ligands (i.e., PAMPs) and receptors have been described in some of these cases. For example, small, EBV-encoded RNAs have been shown to trigger IRF3 activation via RIG-I and IPS1 (53). Interestingly, repetitive regions of the genomic DNAs of two other gammaherpesviruses, murine herpesvirus 68 (MHV68) and human herpesvirus 8 (HHV8), have been shown to be sufficient for induction of type I IFN, thus indicating another possible PAMP in this virus family (55). Other studies have also pointed to a role for alphaherpesvirus-associated DNA in the stimulation of IRF3-dependent IFN expression. For example, both dsDNA sensors have been implicated as important for IFN-β synthesis in response to infection with HSV-1, in separate studies (11, 63). Tsitoura et al. also described IRF3-dependent innate immune activation by HSV-based amplicon DNA vectors in human fibroblasts (64). Moreover, a recent publication from our laboratory demonstrated an essential role for the dsDNA sensor ZBP1 in HCMV-triggered IRF3 activation (14).
Our recent paper showing that ZBP1 is essential for IRF3 activation and ISG induction by HCMV (14) contrasts with earlier work indicating that the HCMV UL55-encoded envelope glycoprotein B (gB) triggers this response (4, 6, 60). Stimulation of IRF3-dependent antiviral responses in human fibroblasts by a soluble form of gB has been demonstrated repeatedly (4, 6, 60). In fact, a great deal of similarity is evident between patterns of host cell gene expression triggered by HCMV gB, HCMV particles, and IFN-β (60). Based on these observations, it was hypothesized that the contact between HCMV gB and an unknown cellular receptor prior to viral entry represents the initiating stimulus of HCMV-mediated innate immune activation (4). Additional work has identified glycoproteins of other herpesviruses as capable of triggering an IRF3-dependent IFN response. For example, HSV glycoprotein D can induce IFN-α secretion from peripheral blood mononuclear cells (PBMCs) (3), and Reske et al. have shown elicitation of IFN synthesis from dendritic cells by HSV glycoprotein complexes (48). A soluble form of glycoprotein gpK8.1 from the gammaherpesvirus HHV8 induces IRF3 activation and an antiviral state in human fibroblasts and endothelial cells (44). Thus, glycoprotein-dependent ISG induction is assumed to be one of the mechanisms of innate immune activation by herpesviruses. Unfortunately, showing that glycoproteins are the sole herpesvirus-associated, IRF3-stimulating PAMP has historically been difficult because these molecules are essential for viral entry, thereby making deletion mutants intractable.
Here we describe the identification of a defective fibroblast cell line in which IRF3-dependent activation occurs normally following exposure to transfected poly(I:C) and infection with an RNA virus but is absent in response to treatment with HCMV. We investigated the phenotypic basis for this trait and further examined the cells' innate immune response to dsDNA and HCMV gB in order to explore similarities in the IRF3 activation process between these stimuli. Our results show obvious overlaps in the patterns of IRF3-dependent ISG expression triggered by HCMV particles and cytoplasmic dsDNA, and these patterns clearly differ from those triggered by HCMV gB. These observations rule out gB as a critical innate immune-activating PAMP during HCMV infection and highlight the crucial importance of a cytoplasmic DNA-dependent pathway.
MATERIALS AND METHODS
Reagents and antibodies.
The dsRNA mimic poly(I:C) was obtained from Amersham Biosciences and resuspended in Millipure water. Hygromycin B was obtained from Invivogen and used at 300 μg/ml of cell culture medium. Puromycin was obtained from Clontech and used at 2 μg/ml of cell culture medium. Lipofectamine-LTX (LF-LTX) transfection reagent was obtained from Invitrogen and used according to the manufacturer's instructions. HiPerfect transfection reagent was obtained from Qiagen. Human recombinant IFN-β was obtained from PBL. Recombinant HCMV glycoprotein B was obtained from DevaTal. ONE-Glo lysis/luciferin reagent was obtained from Promega. Polyethylene glycol 1500 (PEG 1500) was obtained from Roche. Antibodies against the following antigens were used: glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Santa Cruz), IRF3 (BD Biosciences [for indirect immunofluorescence assay {IFA}]) and Santa Cruz [for immunoblotting]), phospho-IRF3 (Millipore), hemagglutinin (HA) (Santa Cruz), ZBP1 (a kind gift from Stefan Rothenburg), pp71 (a kind gift from Tom Shenk), ISG56 (a kind gift from Ganes Sen), viperin (a kind gift from Peter Cresswell), and HCMV immediate-early (IE) protein (a kind gift from Jay Nelson).
Virus and cell culture.
Human foreskin fibroblasts stably transfected with the human telomerase gene to extend passage life (THF cells) were described previously (7). A monoclonal THF-derived cell line stably expressing an HA-tagged version of the HCMV pp71 protein was described previously (9). Cells were propagated in Dulbecco's minimal essential medium (DMEM) containing 10% fetal calf serum (FCS) and antibiotics at 37°C in 5% CO2. HCMV strain AD169 was obtained from ATCC and propagated, purified, and titrated as described previously (14). Inactivation of HCMV particles by use of UV irradiation was performed as described previously (14, 15). Unless otherwise indicated, HCMV treatments were performed in duplicate at a multiplicity of infection (MOI) of 3 for 6 h. Sendai virus (SeV) was obtained from Charles River Laboratories and exposed to cells in duplicate at 160 HA units/ml of cell culture medium. HCMV glycoprotein B was used at 20 μg/ml of cell culture medium. Cell transfection was performed in 24-well dishes by adding 2 μl LF-LTX per 1 μg poly(I:C) or dsDNA in 0.1 ml DMEM and letting the plates sit at room temperature for 30 min. Cells were washed with phosphate-buffered saline (PBS), and 0.5 ml DMEM was added, followed by addition of LF-LTX-poly(I:C) or LF-LTX-dsDNA.
RNA interference.
Cells were plated at 30 to 40% confluence in 35-mm dishes the day before transfection with small interfering RNA (siRNA). Five microliters of siRNA (20 μM stock) was mixed with 10 μl HiPerfect in 95 μl Opti-MEM (Gibco) and added to cells containing 2.3 ml Opti-MEM. Cells were transfected twice, 8 h apart, and incubated for 16 h, and Opti-MEM was replaced with DMEM with 10% FCS. Cells were allowed to expand for 3 to 4 days to near confluence and were transfected once more 16 h before treatment. siRNA sequences were as follows: nonspecific (NS), 5′-GGACGUAGAAGAGGGUGUAGAG-3′; and UL82, 5′-GGACCUGCGUACCAACAUA-3′.
Generation of stable cell lines.
Stable cell lines were constructed using retroviral expression vectors. UL82-HF cells stably overexpressing human ZBP1 (UL82-HF-ZBP1 cells) were constructed by subcloning full-length ZBP1 from a pcDNA3.1 expression vector (described in reference 16) into the pRetro-Easy2 retroviral vector (Applied Biological Materials). THF cells stably expressing green fluorescent protein (THF-GFP cells) were constructed by subcloning GFP from pEGFP-C1 (Clontech) into pRetro-Easy2. IRF3 was stably degraded in THF and UL82-HF cells by stably expressing the NPro open reading frame from bovine viral diarrhea virus, as described previously (17). Reconstruction of THF cells stably expressing UL82 was performed as described previously (9). Replication-defective recombinant retrovirus was produced by transfecting the retroviral vector into Phoenix A or 293T cells (for expression of NPro from the pdl-NPro lentiviral vector; a kind gift from Roger Everett) by use of LF-LTX and harvesting the supernatant after 48 h (28). Supernatant was centrifuged (3,000 × g for 10 min) and filtered through two 0.45-μm filters to remove cell debris. Subconfluent target cells were exposed to retrovirus for 16 h in the presence of 5 μg ml−1 Polybrene. After cells reached confluence, they were split into DMEM plus 10% FCS containing 1 μg ml−1 puromycin. Transduced cells were passaged in the presence of increasing puromycin concentrations (to 3 μg ml−1) until cultures were fully resistant.
Type I interferon quantification assays.
Secretion of type I IFN by target cells was quantified as described previously (14). Briefly, confluent target cells in 24-well dishes were treated as indicated. At 6 h posttreatment, cells were washed three times with 1× PBS, and 500 μl DMEM plus 10% FCS was added for 16 h. The medium was harvested and added to THF cells stably transfected with an expression cassette containing firefly luciferase under the control of the IFN-dependent response element (termed THF-ISRE cells) and grown to confluence in a 96-well dish (50 μl medium was added to each of four wells). At 6 h post-medium transfer, 50 μl ONE-Glo lysis/luciferin reagent was added to each well, and luminescence was measured on a Veritas luminometer (Turner Biosystems).
RNA isolation, semiquantitative RT-PCR, and sequencing.
Total RNA was isolated using a Mini RNA isolation II kit according to the manufacturer's protocol (Zymo Research) and was quantified by UV spectrometry. RNA samples were treated with DNase by use of a DNA-Free RNA kit according to the manufacturer's protocol (Zymo Research). Single-stranded cDNA for use as a PCR template was made from total RNA by use of random hexamers to prime first-strand synthesis by Superscript III reverse transcriptase (RT; Invitrogen), as described in the manufacturer's protocol. Comparison of mRNA expression between samples (e.g., infected versus untreated) was performed using SYBR green-based semiquantitative real-time RT-PCR (qPCR) on an Applied Biosystems sequence detection system according to the ΔΔCT method of Livak and Schmittgen (32). GAPDH was used as a housekeeping gene to establish a baseline against which target genes were compared between samples (described in reference 14). Sequencing of ZBP1 mRNA transcripts was performed following overnight treatment of cells with 1,000 U IFN-β. Following RNA isolation and DNase treatment as described above, oligo(dT) priming was used for first-strand cDNA synthesis. PCR was performed using primers hDLM1f and hDLM3r as described previously (49). PCR products were cloned into pCR4-TOPO (Invitrogen) according to the manufacturer's instructions and were sequenced as described previously (49). ZBP1 transcript-specific qPCR was performed using the following primers: transcript E 5′ primer, CGCTTCCCAGACAGCACAACAGCACG; transcript E 3′ primer, CCAGACCCTGCGACTCCTCC; transcript F 5′ primer, CCTCTTGCTCTCTAAGCCAG; and transcript F 3′ primer, GTTGAGGAATCACCTGGTGCC.
IFA.
IFA was used to examine the subcellular localization of IRF3. Cells were grown on coverslips in 24-well plates and treated as described in the text. At room temperature, cells were washed twice with PBS, fixed for 30 min in 3.7% formaldehyde, washed, and quenched for 10 min using 50 mM NH4Cl. Cells were permeabilized with 0.1% Triton X-100 for 7 min and washed three times with PBS containing 2% bovine serum albumin (BSA). Cells were incubated with primary antibody in PBS containing 2% BSA at 37°C for 1 h, washed three times in PBS containing 2% BSA (10 min for each wash), and incubated with fluorescently labeled secondary antibody diluted 1:1,000 in PBS containing 2% BSA for 1 h. Cells were washed twice in PBS containing 2% BSA (10 min for each wash) and once in PBS. Secondary antibodies were as follows: goat anti-rabbit 555 (Invitrogen), goat anti-mouse 555 (Invitrogen), goat anti-mouse 488 (Invitrogen), goat anti-mouse 594 (Invitrogen), goat anti-rabbit 647 (Invitrogen), and donkey anti-mouse 647 (Invitrogen). Coverslips were mounted on a microscope slide with Vectashield mounting medium (Vector Laboratories, Burlingame, CA) containing 4′,6-diamidino-2-phenylindole (DAPI). Cell fusion assays were performed as follows. THF-GFP and UL82-HF cells (30,000 each) were coplated on coverslips in 24-well dishes at 16 h pretreatment. Cells were exposed to PEG for 1 min, after which cells were washed three times with warm DMEM plus 10% FCS. Cells were then treated as described in the text.
Immunoblotting.
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) immunoblots were performed as follows. Following trypsinization and cell pelleting at 2,000 × g for 10 min, whole-cell lysates were harvested in 2% SDS lysis buffer (50 mM Tris-HCl, 20% glycerol). Lysates were electrophoresed in 10% polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Millipore) by semidry transfer at 400 mA for 1.5 h. Blots were blocked at room temperature for 2 h, using 10% nonfat milk in 1× PBS containing 0.1% Tween 20. Blots were exposed to primary antibody in 5% nonfat milk in 1× PBS containing 0.1% Tween 20 for 16 h at 4°C. Blots were then washed in 1× PBS containing 0.1% Tween 20 for 20, 15, and 5 min, followed by deionized water for 5 min. One-hour exposures to horseradish peroxidase (HRP)-conjugated secondary antibodies and subsequent washes were performed as described for primary antibodies. Antibody was visualized using enhanced chemiluminescence (Pierce).
RESULTS
A stable human fibroblast cell line fails to secrete IFN-β or to synthesize ISG proteins following exposure to human cytomegalovirus.
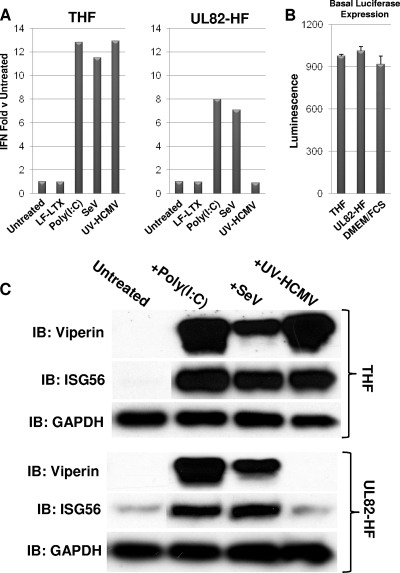
A monoclonal human foreskin fibroblast cell line (hereafter called UL82-HF) stably expressing both the catalytic subunit of human telomerase and an HA-tagged version of the HCMV open reading frame UL82, which encodes the viral tegument protein pp71, was described previously (9). Exposure of human fibroblasts (either primary cells or those stably expressing telomerase) to innate immune-stimulatory factors, such as transfected poly(I:C), single-stranded RNA (ssRNA) viruses, and HCMV particles, rapidly leads to direct expression of IFN-β (8, 15, 60, 72, 73). We examined whether these stimuli were capable of doing so in UL82-HF cells, originally under the suspicion that pp71 might exhibit innate immune-inhibitory activity. We exposed both UL82-HF cells and parental cells that stably express telomerase only (THF cells) to transfected poly(I:C), SeV, or HCMV rendered transcriptionally inactive by exposure to UV irradiation (UV-HCMV). UV inactivation was performed in order to neutralize all known or uncharacterized inhibitory phenotypes exhibited by live virus that are directed at innate immune activation. As shown in Fig. 1 A, type I IFN secretion was induced relative to that in untreated cells in THF cells following exposure to poly(I:C), SeV, and UV-HCMV. However, while exposure of UL82-HF cells to poly(I:C) and SeV resulted in IFN secretion, none was observed following treatment with UV-HCMV. It is worth noting that IFN-dependent luciferase expression by reporter cells was essentially indistinguishable following exposure to sterile DMEM-FCS, medium from untreated THF cells, and medium from untreated UL82-HF cells (Fig. 1B), thus ruling out differences in basal IFN expression between cell types impacting the observed ISG/IFN changes between stimulated and untreated cells. As shown in Fig. 1A, poly(I:C)- and SeV-triggered IFN secretion relative to that in untreated cells was less dramatic in UL82-HF cells than in THF cells. Whether this represents a diminished ability to respond to these stimuli is currently unknown, but the components of the signaling pathway known to be downstream of these stimuli that were examined here appeared to function normally. Nevertheless, IFN secretion following HCMV exposure was undetectable in UL82-HF cells, and thus these cells exhibit a specific defect in their ability to generate IFN in response to this stimulus but not in response to cytoplasmic dsRNA or an RNA virus.
FIG. 1.
Synthesis of IFN-β, ISG56, and viperin in THF and UL82-HF cells following exposure to poly(I:C), SeV, and UV-HCMV. (A) Expression of type I IFN-dependent luciferase from THF-ISRE cells exposed to medium collected at 24 h posttreatment from either THF or UL82-HF cells treated with LF-LTX, transfected poly(I:C), SeV, or UV-HCMV as described in the text. Values presented are normalized to untreated cells (set to 1) and are representative of duplicate experiments. (B) Samples (50 μl) of media from THF or UL82-HF cells grown in 9 wells of a 24-well dish were transferred to 4 wells each of THF-ISRE reporter cells grown to confluence in 96-well plates. IFN-dependent luciferase expression was measured as described in the text. Luciferase was also measured in THF-ISRE cells exposed only to fresh DMEM-FCS. (C) Immunoblots showing synthesis of ISG56, viperin, and GAPDH in untreated THF and UL82-HF cells or in THF and UL82-HF cells following 6 h of exposure to transfected poly(I:C), SeV, or UV-HCMV.
In addition to IFN-β, innate immune stimuli can trigger the direct (IFN-independent) expression of other antiviral molecules (including ISGs), such as ISG56 and viperin (5, 8, 10, 15, 19, 45). We therefore next asked whether synthesis of other antiviral proteins occurred in response to UV-HCMV or if the absence of HCMV-triggered IFN secretion in UL82-HF cells represents an effect targeted to that protein. Since type I IFN is not induced in response to HCMV in these cells, any observed synthesis of ISG proteins following exposure to the virus must necessarily be the result of IFN-independent signaling. As illustrated in Fig. 1B, exposure of THF and UL82-HF cells to transfected poly(I:C) and to SeV led to synthesis of viperin and ISG56 proteins. However, while UV-HCMV stimulated their translation in THF cells, no protein was observed in UL82-HF cells following treatment with UV-HCMV. We therefore concluded that UL82-HF cells synthesize type I IFN and antiviral proteins in response to poly(I:C) and SeV but not in response to HCMV.
Human cytomegalovirus-associated accumulation of IFN-β and ISG mRNAs does not occur in UL82-HF cells.
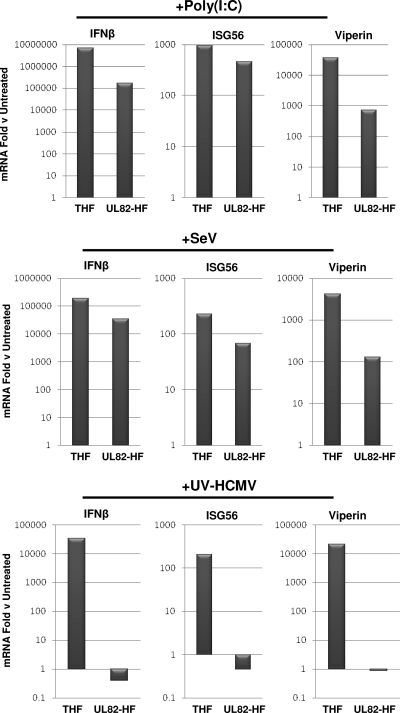
We next decided to investigate if the absence of IFN and antiviral proteins in UL82-HF cells following HCMV exposure was due to differences in corresponding mRNA abundance or to some other effect (e.g., translational impairment, protein degradation, etc.). We therefore used qPCR to compare mRNA abundances in THF and UL82-HF cells treated with innate immune stimuli relative to those in untreated control cells. As shown in Fig. 2, treatment with poly(I:C) and SeV led to large amounts of IFN-β, ISG56, and viperin mRNAs relative to those in untreated cells for both cell lines. While mRNA fold changes for these genes relative to the levels in untreated cells were noticeably lower in UL82-HF cells than in THF cells, we attributed this largely to higher basal mRNA expression levels of these genes in untreated UL82-HF cells than in untreated THF cells, as indicated by lower qPCR cycle threshold values (not shown), which translate into lower overall inductive fold changes. This is possibly reflected in higher background levels of ISG56 protein, as detected by immunoblotting, in UL82-HF versus THF cells (Fig. 1B). In contrast to the cases of poly(I:C) and SeV, exposure of UL82-HF cells to UV-HCMV did not lead to accumulation of IFN-β, ISG56, and viperin mRNAs, despite accumulation of these mRNAs in parental THF cells. We thus concluded that ISG and IFN-β mRNA accumulation in response to HCMV [but not poly(I:C) or SeV] does not occur in UL82-HF cells.
FIG. 2.
IFN-β, ISG56, and viperin mRNA accumulation in THF and UL82-HF cells following exposure to poly(I:C), SeV, and UV-HCMV. Values presented are fold changes of mRNA levels in treated versus untreated cells at 6 h posttreatment, as determined by semiquantitative RT-PCR as described in the text. Data are representative of duplicate experiments.
IRF3 phosphorylation and nuclear localization occur following exposure of UL82-HF cells to poly(I:C) and SeV but not to HCMV.
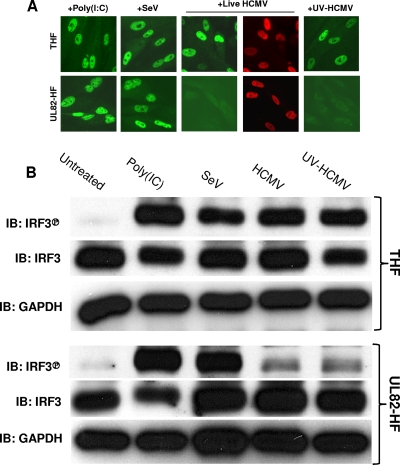
Since HCMV-triggered accumulation of IFN-β, ISG56, and viperin mRNAs and proteins was not detected in UL82-HF cells, we asked if this absence correlated with a lack of HCMV-triggered activation of IRF3. IRF3 activation is characterized by phosphorylation of C-terminal serine residues followed by its nuclear accumulation. As shown in Fig. 3 A, exposure of both THF and UL82-HF cells to poly(I:C) or SeV resulted in obvious nuclear accumulation of IRF3 protein, as detected by IFA. However, while treatment of THF cells with live HCMV or UV-HCMV also resulted in IRF3 nuclear accumulation, these stimuli failed to induce this response in UL82-HF cells.
FIG. 3.
IRF3 activation in THF and UL82-HF cells following exposure to poly(I:C), SeV, HCMV, and UV-HCMV. (A) IFA showing subcellular localization of IRF3 (green) and HCMV UL123-encoded IE protein (red) in THF and UL82-HF cells following 6 h of treatment with transfected poly(I:C), SeV, live HCMV, or UV-HCMV as described in the text. (B) Immunoblots showing the presence of total IRF3 and IRF3 phosphorylated on Ser398, as well as GAPDH, in untreated THF and UL82-HF cells or in THF and UL82-HF cells following 6 h of exposure to poly(I:C), SeV, live HCMV, or UV-HCMV.
We next examined if the lack of HCMV-triggered IRF3 nuclear accumulation in UL82-HF cells was due to an absence of IRF3 phosphorylation. To address this, we used immunoblotting with an antibody that reacts with IRF3 phosphorylated on Ser398. As shown in Fig. 3B, IRF3 was highly phosphorylated in both THF and UL82-HF cells following exposure to poly(I:C) and SeV. However, while exposure of THF cells to live HCMV or UV-HCMV also resulted in strong IRF3 phosphorylation, these stimuli failed to trigger a similar effect in UL82-HF cells. We therefore concluded that relative to parental cells, UL82-HF cells exhibit a defect in the innate response to HCMV infection, but not to transfected poly(I:C) or SeV infection, that leads to IRF3 Ser398 phosphorylation and nuclear accumulation.
The absence of HCMV-triggered IRF3 phosphorylation in UL82-HF cells is the result of a recessive defect.
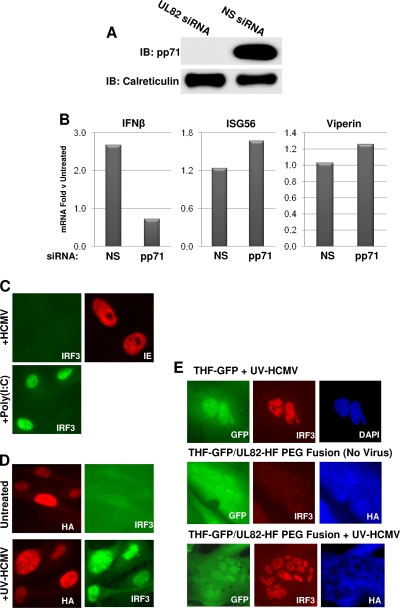
As described above, UL82-HF cells stably express the HCMV protein pp71. We therefore asked whether the lack of HCMV-stimulated IRF3 activation and accumulation of IRF3-dependent mRNAs observed in these cells was due to the presence of pp71. To address this, we used RNA interference to knock down expression of the UL82 transgene. As shown in Fig. 4 A, transfection of siRNA directed against the UL82 transcript reduced pp71 protein expression in UL82-HF cells to a level undetectable by immunoblotting. However, as shown in Fig. 4B, pp71 knockdown failed to restore UV-HCMV-mediated accumulation of IFN-β, ISG56, or viperin mRNA to levels approaching those seen in THF cells. Furthermore, IRF3 nuclear accumulation in response to UV-HCMV following pp71 knockdown also did not occur (Fig. 4C). To further address this, we reconstructed a THF cell line stably expressing pp71 by use of the original retroviral vector (9) and examined HCMV-triggered IRF3 nuclear accumulation in these cells. As shown in Fig. 4D, IRF3 nuclear accumulation occurred in THF cells following exposure to UV-HCMV even during stable expression of pp71. This further indicates that pp71 is not directly responsible for the lack of HCMV-triggered IRF3 activation observed in UL82-HF cells.
FIG. 4.
HCMV-triggered IRF3-dependent activity in UL82-HF cells following knockdown of UL82 and in THF and UL82-HF cell fusions. (A) Immunoblots showing siRNA-mediated knockdown of stably expressed pp71 in UL82-HF cells. (B) Accumulation of IFN-β, ISG56, and viperin mRNAs in UV-HCMV-exposed UL82-HF cells relative to those in unexposed cells following transfection with UL82-directed or nonspecific (NS) siRNA, as determined by semiquantitative qPCR. (C) Indirect IFA showing subcellular localization of IRF3 (green) and HCMV IE protein (red) in UL82-HF cells exposed to HCMV or poly(I:C) following transfection with UL82-directed siRNA. (D) Indirect IFA showing subcellular localization of IRF3 (green) and HA (red) in THF cells that were reconstructed to stably express HA-tagged pp71. Cells were left untreated or exposed to UV-HCMV. (E) Indirect IFA showing stably expressed GFP (green), subcellular localization of IRF3 (red), and DAPI staining (blue) or pp71-HA expression (blue) in THF-GFP and UL82-HF cells in the presence or absence of UV-HCMV and PEG, as described in the text.
While the absence of HCMV-mediated IRF3 activation in UL82-HF cells was not caused directly by pp71, it was possible that pp71 exerted an IRF3-targeted inhibitory effect via a specific alteration to the cells that remained (residually) after the protein's removal. In this case, the effects of pp71 on UL82-HF cells would still be manifested as a dominant and active inhibitory phenotype that targets only the HCMV-specific pathway. We therefore next asked whether this trait represents a defect in UL82-HF cells that can be rescued through complementation or, rather, is a dominant inhibitory phenotype. To address this possibility, we employed PEG-mediated cell fusion. Using retrovirus-mediated transduction, we constructed a THF cell line that stably expresses GFP (THF-GFP). We then cocultured UL82-HF and THF-GFP cells in the presence of PEG to promote fusion between the different cell types. If the lack of HCMV-mediated IRF3 activation in UL82-HF cells was the result of a dominant trait exhibited by the cells, then fused THF-GFP and UL82-HF cells should also exhibit this phenotype. However, if this was due to a recessive defect in UL82-HF cells, then THF-GFP cells should be able to “repair” the deficiency in fused cells. Fusion between THF-GFP and UL82-HF cells can be detected by staining of GFP-positive multinucleated cellular masses with anti-HA antibody, which detects the HA-tagged pp71 protein in UL82-HF cells. As shown in Fig. 4E, IRF3 nuclear accumulation occurred in THF-GFP cells following exposure to UV-HCMV, just as in the parental THF cells. In the absence of UV-HCMV, IRF3 nuclear accumulation did not occur in multinucleated HA- and GFP-positive PEG fusions (Fig. 4E). However, when exposed to UV-HCMV, HA- and GFP-positive fusions displayed obvious IRF3 nuclear accumulation. These results indicate that the absence of HCMV-stimulated IRF3 activation in UL82-HF cells is a recessive defect that is capable of being reestablished by the components of parental cells. Since this defect appears to be specific to HCMV-triggered, IRF3-terminal signaling, UL82-HF cells represent a unique and powerful tool for detailed characterization of this response and the related pathway(s).
HCMV glycoprotein B triggers expression of IFN-β and ISGs in UL82-HF cells.
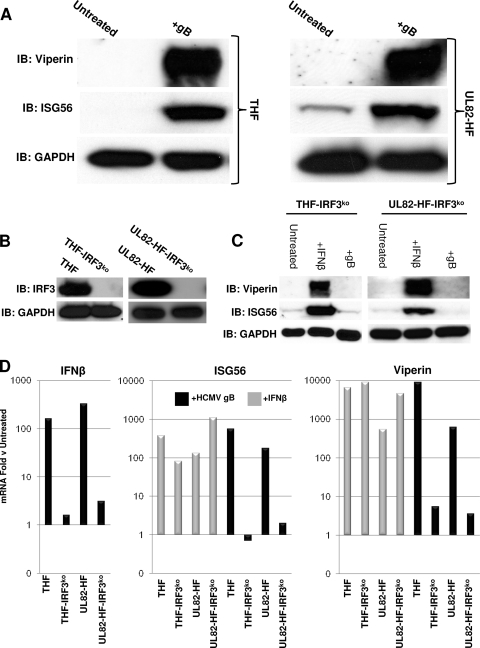
Previous work has demonstrated activation of the IRF3-dependent IFN and antiviral response in human fibroblasts by a soluble form of the HCMV UL55-encoded envelope gB (4, 6, 60). It has been hypothesized that gB acts as the herpesvirus-associated molecular pattern that initiates innate immune activation during virus-cell contact (4, 6, 60). However, it was not possible to directly test this hypothesis because gB is essential for HCMV fusion and entry (22) and because viral entry is required for ISG induction (24, 38). The HCMV-selective defect in the UL82-HF cell line thus presented a unique opportunity to determine whether ISG induction by gB and HCMV particles occurs via similar mechanisms. We therefore examined whether exposure to HCMV gB induced IRF3-dependent accumulation of mRNAs and corresponding proteins in UL82-HF cells. As shown in Fig. 5 A, treatment of both THF and UL82-HF cells with 20 μg ml−1 gB resulted in synthesis of both ISG56 and viperin proteins. Oddly, treatment of neither cell type with gB resulted in IRF3 Ser398 phosphorylation (data not shown). Since it is possible that gB treatment leads to activation of IRF3 via phosphorylation of different residues, we decided to ask whether the ISG induction observed in THF and UL82-HF cells following gB treatment was dependent on IRF3. To address this, we constructed THF and UL82-HF cell lines in which IRF3 is stably degraded by overexpression of the bovine viral diarrhea virus NPro protein, as described by Everett et al. (17). As shown in Fig. 5B, IRF3 was undetectable by immunoblotting in THF-IRF3ko and UL82-HF-IRF3ko cells. Figure 5C illustrates that ISG56 and viperin proteins were detected following treatment of both THF-IRF3ko and UL82-HF-IRF3ko cells with IFN-β, yet these proteins were induced only by gB in IRF3-positive cells (Fig. 5A). Furthermore, as shown in Fig. 5D, exposure of IRF3-positive THF and UL82-HF cells to gB resulted in strong accumulations of IFN-β, ISG56, and viperin mRNAs, but this did not occur in the absence of IRF3. Based on these observations, we concluded that HCMV glycoprotein B triggers an IRF3-dependent innate immune response in both THF and UL82-HF cells. These data also confirm that gB induces accumulation of IRF3-dependent IFN-β and ISGs, but they rule out the possibility that IRF3-dependent ISG induction by gB occurs via the same signal transduction pathway as that of HCMV particles.
FIG. 5.
IRF3-dependent mRNA accumulation in THF and UL82-HF cells following treatment with HCMV gB. (A) Immunoblots showing synthesis of viperin and ISG56 following exposure of THF and UL82-HF cells to 20 μg ml−1 HCMV gB. (B) Immunoblots showing NPro-mediated degradation of IRF3 from stably transfected THF and UL82-HF cells. (C) Immunoblots showing synthesis of viperin and ISG56 in THF-IRF3ko and UL82-HF-IRF3ko cells either left untreated or exposed to IFN-β or HCMV gB. (D) Accumulation of IFN-β, ISG56, and viperin mRNAs following exposure of THF, THF-IRF3ko, UL82-HF, and UL82-HF-IRF3ko cells to 20 μg ml−1 HCMV gB (IFN-β, ISG56, and viperin) or IFN-β (ISG56 and viperin). Values presented are fold changes relative to levels in untreated cells.
dsDNA does not trigger IRF3-dependent gene expression or IRF3 activation in UL82-HF cells.
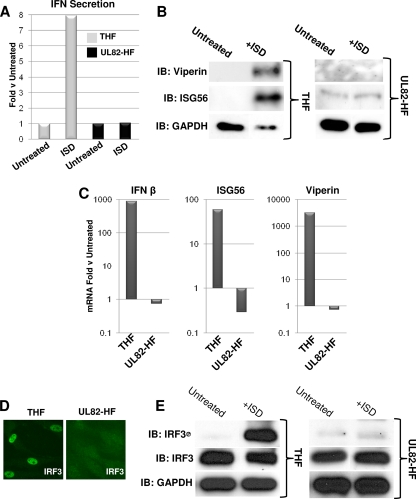
Recent work from our laboratory (14) indicates that HCMV triggers IRF3 activation in human fibroblasts via a pathway that requires ZBP1, a cytoplasmic sensor of dsDNA. An IRF3-terminal pathway can be activated in fibroblasts by cytoplasmic AT-rich dsDNA (23, 61). We therefore asked whether the IRF3-terminal signaling pathway activated by cytoplasmic IFN-stimulatory dsDNA (ISD) (61) was also inactive in UL82-HF cells. As shown in Fig. 6 A, transfection of ISD into THF cells induced secretion of type I IFN. However, we did not detect ISD-triggered IFN secretion in UL82-HF cells. In addition, while transfection of ISD into THF cells led to synthesis of ISG56 and viperin proteins, protein synthesis was not detected in UL82-HF cells following ISD transfection (Fig. 6B). We next examined ISD-induced accumulation of the corresponding mRNAs as described above. As shown in Fig. 6C, transfection of ISD led to accumulations of IFN-β, ISG56, and viperin mRNAs in THF cells relative to those in untreated cells, but no such induction was seen in UL82-HF cells. We next investigated the activation status of IRF3 following ISD transfection by examining the protein's nuclear accumulation and Ser398 phosphorylation. As shown in Fig. 6D, transfection of ISD led to IRF3 nuclear accumulation in THF cells but not in UL82-HF cells. Likewise, ISD transfection induced IRF3 phosphorylation in THF cells but not in UL82-HF cells (Fig. 6E). These observations led us to conclude that UL82-HF cells exhibit a defect in the IRF3-terminal signaling pathway(s) activated by exposure to HCMV particles and ISD. Whether this defect is based on the actual detection of the stimuli or a shared downstream signaling component is not known.
FIG. 6.
ISD does not activate IRF3-dependent gene expression in UL82-HF cells. (A) Expression of type I IFN-dependent luciferase from THF-ISRE cells exposed to medium collected at 24 h posttreatment from THF or UL82-HF cells either transfected with ISD or left untreated. Values presented are normalized to untreated cells (set to 1). (B) Immunoblots showing synthesis of viperin and ISG56 following 6 h of transfection of 5 μg ml−1 ISD into THF and UL82-HF cells. (C) Accumulation of IFN-β, ISG56, and viperin mRNAs following transfection of THF and UL82-HF cells with 5 μg ml−1 ISD. Values presented are fold changes relative to the levels in untreated cells. (D) IFA showing subcellular localization of IRF3 in THF and UL82-HF cells transfected with 5 μg ml−1 ISD for 6 h. (E) Immunoblots showing IRF3 Ser398 phosphorylation status in THF and UL82-HF cells transfected with 5 μg ml−1 ISD for 6 h.
Stable overexpression of ZBP1 in UL82-HF cells reestablishes HCMV-triggered IFN secretion.
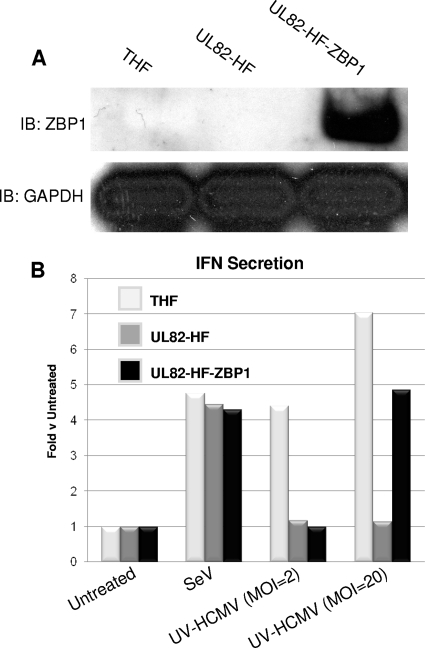
As discussed in our recently published work, ZBP1 is both essential for and capable of enhancing HCMV-induced IRF3-dependent cellular responses, including type I IFN secretion (14). Given that ZBP1 is both a sensor of dsDNA and required for HCMV-triggered IRF3 activation and that UL82-HF cells are nonresponsive to both stimuli, we asked whether the defect in these cells could involve and perhaps be reestablished by the presence of ZBP1. To address this, we constructed a UL82-HF cell line that stably overexpresses full-length ZBP1 (UL82-HF-ZBP1), as shown in Fig. 7 A. We next quantified IFN secretion following exposure of THF, UL82-HF, and UL82-HF-ZBP1 cells to SeV or UV-HCMV at two different MOIs (2 and 20). As shown in Fig. 7B, SeV induced similar levels of IFN secretion from all three cell types. Treatment with UV-HCMV at an MOI of 2 resulted in IFN secretion from THF cells but not from UL82-HF or UL82-HF-ZBP1 cells. However, while UV-HCMV at an MOI of 20 did not induce IFN secretion from UL82-HF cells, substantial IFN secretion was detected in THF and UL82-HF-ZBP1 cells. These results suggest that the defect in UL82-HF cells is likely related to ZBP1 or the ZBP1-dependent signal transduction pathway.
FIG. 7.
Stable overexpression of ZBP1 reestablishes the ability of UL82-HF cells to secrete type I IFN in response to HCMV. (A) Immunoblots showing expression of ZBP1 in THF, UL82-HF, and UL82-HF-ZBP1 cells. (B) Expression of type I IFN-dependent luciferase from THF-ISRE cells exposed to medium collected at 24 h posttreatment from THF, UL82-HF, or UL82-HF-ZBP1 cells left untreated, exposed to SeV, or exposed to UV-HCMV at an MOI of 2 PFU/cell or 20 PFU/cell. Values presented are normalized to those for untreated cells (set to 1).
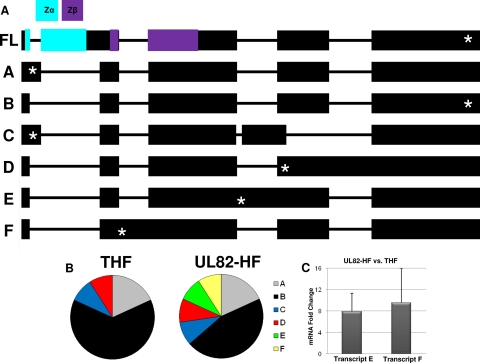
To determine if differences in ZBP1 mRNA primary sequence exist between THF and UL82-HF cells, we amplified and cloned random ZBP1 mRNA transcripts from both cell types and sequenced 11 clones for each. In all, six transcript variants were detected (Fig. 8). Of these, four variants were shared between the cell types and two additional variants (labeled E and F) were found only in UL82-HF cells (Fig. 8). The ZBP1 gene contains 10 exons, including those encoding two DNA binding domains (Zα and Zβ), and undergoes complex regulation that involves abundant differential splicing (49). It is thus conceivable that differences in the ZBP1 transcript profile between THF and UL82-HF cells could contribute to the phenotypic disparity between the cell types. Interestingly, transcripts containing an intact Zα DNA binding domain (49) were not detected in either cell type (Fig. 8). Transcript F encoded a premature stop codon in the Zβ DNA binding domain, which initially suggested the possibility that this variant interferes with functional ZBP1 signaling in UL82-HF cells. To determine if basal levels of transcript E or F differ between UL82-HF and THF cells, we performed qPCR on four independently isolated RNA samples from each cell type, using primers specific to 5′ regions identified as unique relative to other sequenced transcripts. Levels of both transcripts were higher in UL82-HF cells than in THF cells (7.94-fold ± 3.36 SEM and 9.56-fold ± 6.44 SEM for transcripts E and F, respectively) (Fig. 8). These results indicate that differences in ZBP1 transcript variants between UL82-HF and THF cells are detectable and could be responsible for the observed phenotypic differences between the cell types.
FIG. 8.
ZBP1 transcript variants identified in THF and UL82-HF cells. (A) Sequence maps and labels (A to F and FL [full length]) for ZBP1 mRNA transcript variants identified in THF and UL82-HF cells. Approximate locations of the Zα and Zβ DNA binding domains are color coded as indicated. Asterisks indicate approximate locations of initial predicted stop codons. (B) Cell type-specific distribution of transcript variants found in THF and UL82-HF cells. (C) Accumulation of ZBP1 transcripts E and F in UL82-HF cells versus THF cells, expressed as mean fold changes ± SEM, as determined by qPCR.
DISCUSSION
An altered human fibroblast cell line (UL82-HF) that exhibits a specific defect in the ability to activate IRF3-dependent gene expression following exposure to HCMV, but not SeV or poly(I:C), was able to respond to glycoprotein B but not to cytoplasmic immunostimulatory dsDNA. These data, along with previously published work, point toward dsDNA as being important for HCMV-triggered IRF3 activation. Our previous publication demonstrated the necessity of ZBP1, a sensor of cytoplasmic dsDNA, for activation of IRF3-dependent activity following exposure to HCMV and the unimportance of IPS1 to this response (14). Other work has identified herpesviral genomic DNA as important for the activation of IRF3-dependent responses (55, 64), and we have observed IRF3 activation in THF cells following transfection with HCMV DNA (unpublished data). Previous work has shown that virus replication is not needed for IRF3 and innate immune activation by HCMV (8, 14, 15, 39, 60, 72, 73), thereby ruling out newly synthesized viral molecules such as proteins or dsRNA. Thus, taken together, these results strongly suggest that HCMV activates IRF3 via double-stranded DNA.
In contrast, our results do not support viral glycoprotein B as the HCMV PAMP, as suggested previously (4, 6, 60), yet it is important to acknowledge that the immunostimulatory ability of virion-associated gB could vary between cell types. Similar to previous studies, we observed IRF3-dependent ISG induction following exposure of cells to a soluble form of gB. Other studies further showed that viral attachment is insufficient for HCMV-triggered, IRF3-dependent innate immune activation, whereas viral entry is required. For instance, Netterwald et al. failed to observe ISG induction during AD169 infection in the presence of CFI02, a synthetic virus fusion inhibitor targeting gB, yet a CFI02-resistant HCMV strain was capable of stimulating ISG transcription (38). Juckem et al. further showed that HCMV did not induce IFN-β when viral entry was inhibited by depletion of cholesterol-rich microdomains (24). They further described how using an inhibitor of virus fusion and entry blocked induction of IFN responses (presumably IRF3-dependent responses) but not synthesis of NF-κB-dependent inflammatory cytokines, such as interleukin-6 (IL-6). Taken together, these results indicate that viral attachment is sufficient to activate NF-κB but that gB-mediated viral entry is required for IRF3 activation. However, since gB is essential for viral entry, it was not possible to determine whether gB or the fusion process itself was the ultimate trigger of IRF3. The ability of gB to activate IRF3 in UL82-HF cells now strongly suggests that IFN activation by soluble gB does not mimic HCMV-dependent IFN induction. The implications and molecular mechanism of gB-mediated ISG induction remain to be determined.
We currently do not know the exact molecular basis of the defect in UL82-HF cells. The obvious conclusion that the stably expressed viral tegument protein pp71 is responsible was refuted by multiple lines of evidence. First, siRNA-mediated knockdown of pp71 expression did not reconstitute HCMV-triggered IRF3-dependent responses. Second, a separately constructed THF cell line stably expressing pp71 did not exhibit this deficiency. Third, this trait represents a recessive defect rather than a dominant (active) block to IRF3 activation, since HCMV-triggered IRF3 nuclear localization was “repaired” following cellular fusion with parental cells. Many fundamental questions remain regarding the cellular receptors and pathways through which HCMV-stimulated innate immune activation occurs, and these cells are thus a unique tool for answering many related inquiries. For example, a complementation approach could be used to precisely identify components of the HCMV- and ISD-specific signaling pathways leading to IRF3 activation, as described here for ZBP1.
A precedent exists for characterization of a cell line with a defect in innate immune reactivity to a specific stimulus. Leaman et al. described the creation through mutagenesis of a U4C cell line that was nonresponsive to poly(I:C) (29). They attributed this to extremely high rates of IRF3 degradation in mutant cells as well as a defect in poly(I:C)-triggered NF-κB activation (29, 45). In the case of UL82-HF cells, we can rule out differing levels of IRF3 protein or a generalized defect in IRF3 activation, since both poly(I:C) and SeV stimulated phosphorylation of the molecule.
While it is intriguing that ZBP1 is capable of reestablishing the HCMV-mediated type I IFN response in UL82-HF cells, the fact that it does so only during high-MOI exposure is a curious finding requiring additional exploration. It is also worth noting that overexpression in UL82-HF cells of STING, another molecule essential to HCMV-triggered IRF3 activation (14), had no restorative effect in these cells (data not shown). In this context, the complementing effect of ZBP1 appears to be specific to the protein. We further examined differences between THF and UL82-HF cells with respect to ZBP1 by randomly sequencing transcript variants present in the two cell types. Of 22 total mRNAs sequenced between the cell types, we observed six variants of ZBP1, two of which were found only in UL82-HF cells. Regulation of ZBP1 is highly complex, as demonstrated by the work of Rothenburg et al. (49), in which alternative splicing in various tissues was demonstrated to the extent that the potential for production of over 2,000 different splice variants was calculated. While the complete frequency spectrum of ZBP1 transcripts present in THF and UL82-HF cells cannot be inferred based on these data, qPCR analysis detected increased basal levels of two variants in UL82-HF cells. The identification of multiple ZBP1 transcript variants and the quantitative differences in expression levels of specific transcripts between cell types allow the possibility that either (i) THF cells express ZBP1 variants that function with respect to HCMV-induced innate signaling and lack sufficiency in UL82-HF cells or (ii) UL82-HF cells express ZBP1 mRNAs that encode forms of the protein that exhibit dominant negative effects. In support of the former alternative, fusion of THF and UL82-HF cells, as well as overexpression of full-length ZBP1 in UL82-HF cells, was able to restore HCMV-triggered, IRF3-terminal signaling. In support of the latter alternative, the two ZBP1 transcript variants identified as exhibiting increased basal expression in UL82-HF cells encoded premature stop codons, which could give rise to truncated proteins (in the case of variant F, this stop codon prevents full expression of the Zβ DNA binding domain). ZBP1 is known to dimerize following interaction with dsDNA (66), and thus it is possible that an interaction between functional protein and a specific truncated version of the protein disrupts normal signaling and that this disruption is diminished in the presence of overabundant full-length ZBP1. Distinguishing between these and other possible explanations for the phenotypic differences between the cell types will require additional exploration of the diversity of ZBP1 transcripts present in the two cell types along with a detailed characterization of the molecular and biochemical effects of distinct transcript variants and variant profiles.
In conclusion, we describe a fibroblast cell line defective in its IRF3 activation response to HCMV particles and cytoplasmic dsDNA but not to HCMV gB, RNA virus infection, or cytoplasmic dsRNA. Our results indicate that the activation of IRF3 and IRF3-dependent transcription occurring in response to HCMV infection is therefore not triggered by gB, as has previously been hypothesized. Two overarching questions remain regarding HCMV-triggered innate immune responses. First, what is the HCMV-associated PAMP responsible for stimulating this reaction? Our results indicate that gB is not essential to this response and that HCMV-mediated IRF3 activation closely resembles that induced by dsDNA. It is also possible that a combination of molecules and cellular processes is required. Second, which cellular signaling pathways lead from HCMV infection to IRF3 activation? While a separate cytoplasmic dsDNA-sensing apparatus involving POL3 and RIG-I has been described (1, 11), our previous results show that this pathway is not relevant to innate sensing of HCMV but that the ZBP1-dependent pathway is (14). Here we reinforced that conclusion by demonstrating that overexpressed ZBP1 is capable of reestablishing IFN secretion in UL82-HF cells. However, individual components of the ZBP1-dependent pathway remain largely uncharacterized. Future work will focus on further characterizing defects in the HCMV-triggered, ZBP1-dependent pathway in these cells by use of a complementation approach.
Acknowledgments
We thank Stefan Rothenburg (NIH) for the ZBP1 antibody. We thank Tom Shenk (Princeton University) for the pp71 antibody. We thank Ganes Sen (Cleveland Clinic) for the ISG56 antibody. We thank Peter Cresswell (Yale University School of Medicine) for the viperin antibody. We thank Jay Nelson (Oregon Health and Science University) for the HCMV IE antibody. We thank Roger Everett (University of Glasgow) for the NPro lentivirus.
This work was supported by American Heart Association grant 0730325N (V.R.D.), by a grant from the Medical Research Foundation of Oregon (V.R.D.), and by NIH grant R01AI070890 (K.J.F.).
Footnotes
Published ahead of print on 23 June 2010.
REFERENCES
- 1.Ablasser, A., F. Bauernfeind, G. Hartmann, E. Latz, K. A. Fitzgerald, and V. Hornung. 2009. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 10:1065-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alford, C. A., S. Stagno, R. F. Pass, and W. J. Britt. 1990. Congenital and perinatal cutomegalovirus infection. Rev. Infect. Dis. 12:745-753. [DOI] [PubMed] [Google Scholar]
- 3.Ankel, H., D. F. Westra, S. Welling-Wester, and P. Lebon. 1998. Induction of interferon-alpha by glycoprotein D of herpes simplex virus: a possible role of chemokine receptors. Virology 251:317-326. [DOI] [PubMed] [Google Scholar]
- 4.Boehme, K. W., J. Singh, S. T. Perry, and T. Compton. 2004. Human cytomegalovirus elicits a coordinated cellular antiviral response via envelope glycoprotein B. J. Virol. 78:1202-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boudinot, P., S. Riffault, S. Salhi, C. Carrat, C. Sedlik, N. Mahmoudi, B. Charley, and A. Benmansour. 2000. Vesicular stomatitis virus and pseudorabies virus induce a vig1/cig5 homologue in mouse dendritic cells via different pathways. J. Gen. Virol. 81:2675-2682. [DOI] [PubMed] [Google Scholar]
- 6.Boyle, K. A., R. L. Pietropaolo, and T. Compton. 1999. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol. Cell. Biol. 19:3607-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bresnahan, W. A., G. E. Hultman, and T. Shenk. 2000. Replication of wild-type and mutant human cytomegalovirus in life-extended human diploid fibroblasts. J. Virol. 74:10816-10818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Browne, E. P., B. Wing, D. Coleman, and T. Shenk. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319-12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantrell, S. R., and W. A. Bresnahan. 2005. Interaction between the human cytomegalovirus UL82 gene product (pp71) and hDaxx regulates immediate-early gene expression and viral replication. J. Virol. 79:7792-7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chew, T., R. Noyce, S. E. Collins, M. H. Hancock, and K. L. Mossman. 2009. Characterization of the interferon regulatory factor 3-mediated antiviral response in a cell line deficient for IFN production. Mol. Immunol. 46:393-399. [DOI] [PubMed] [Google Scholar]
- 11.Chiu, Y. H., J. B. Macmillan, and Z. J. Chen. 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138:576-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins, S. E., R. S. Noyce, and K. L. Mossman. 2004. Innate cellular response to virus particle entry requires IRF3 but not virus replication. J. Virol. 78:1706-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui, S., K. Eisenacher, A. Kirchhofer, K. Brzozka, A. Lammens, K. Lammens, T. Fujita, K. K. Conzelmann, A. Krug, and K. P. Hopfner. 2008. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell 29:169-179. [DOI] [PubMed] [Google Scholar]
- 14.DeFilippis, V. R., D. Alvarado, T. Sali, S. Rothenburg, and K. Fruh. 2010. Human cytomegalovirus induces the interferon response via the DNA sensor ZBP1. J. Virol. 84:585-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeFilippis, V. R., B. Robinson, T. M. Keck, S. G. Hansen, J. A. Nelson, and K. J. Fruh. 2006. Interferon regulatory factor 3 is necessary for induction of antiviral genes during human cytomegalovirus infection. J. Virol. 80:1032-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deigendesch, N., F. Koch-Nolte, and S. Rothenburg. 2006. ZBP1 subcellular localization and association with stress granules is controlled by its Z-DNA binding domains. Nucleic Acids Res. 34:5007-5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everett, R. D., D. F. Young, R. E. Randall, and A. Orr. 2008. STAT-1- and IRF-3-dependent pathways are not essential for repression of ICP0-null mutant herpes simplex virus type 1 in human fibroblasts. J. Virol. 82:8871-8881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 19.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76:5532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gravel, S. P., and M. J. Servant. 2005. Roles of an IkappaB kinase-related pathway in human cytomegalovirus-infected vascular smooth muscle cells: a molecular link in pathogen-induced proatherosclerotic conditions. J. Biol. Chem. 280:7477-7486. [DOI] [PubMed] [Google Scholar]
- 21.Hornung, V., J. Ellegast, S. Kim, K. Brzozka, A. Jung, H. Kato, H. Poeck, S. Akira, K. K. Conzelmann, M. Schlee, S. Endres, and G. Hartmann. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994-997. [DOI] [PubMed] [Google Scholar]
- 22.Isaacson, M. K., and T. Compton. 2009. Human cytomegalovirus glycoprotein B is required for virus entry and cell-to-cell spread but not for virion attachment, assembly, or egress. J. Virol. 83:3891-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishii, K. J., C. Coban, H. Kato, K. Takahashi, Y. Torii, F. Takeshita, H. Ludwig, G. Sutter, K. Suzuki, H. Hemmi, S. Sato, M. Yamamoto, S. Uematsu, T. Kawai, O. Takeuchi, and S. Akira. 2006. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat. Immunol. 7:40-48. [DOI] [PubMed] [Google Scholar]
- 24.Juckem, L. K., K. W. Boehme, A. L. Feire, and T. Compton. 2008. Differential initiation of innate immune responses induced by human cytomegalovirus entry into fibroblast cells. J. Immunol. 180:4965-4977. [DOI] [PubMed] [Google Scholar]
- 25.Kawai, T., and S. Akira. 2007. Antiviral signaling through pattern recognition receptors. J. Biochem. 141:137-145. [DOI] [PubMed] [Google Scholar]
- 26.Kawai, T., and S. Akira. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131-137. [DOI] [PubMed] [Google Scholar]
- 27.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981-988. [DOI] [PubMed] [Google Scholar]
- 28.Kinsella, T. M., and G. P. Nolan. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 7:1405-1413. [DOI] [PubMed] [Google Scholar]
- 29.Leaman, D. W., A. Salvekar, R. Patel, G. C. Sen, and G. R. Stark. 1998. A mutant cell line defective in response to double-stranded RNA and in regulating basal expression of interferon-stimulated genes. Proc. Natl. Acad. Sci. U. S. A. 95:9442-9447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, R., R. S. Noyce, S. E. Collins, R. D. Everett, and K. L. Mossman. 2004. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J. Virol. 78:1675-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 33.Ljungman, P. 1996. Cytomegalovirus infections in transplant patients. Scand. J. Infect. Dis. 100(Suppl.):59-63. [PubMed] [Google Scholar]
- 34.Merika, M., A. J. Williams, G. Chen, T. Collins, and D. Thanos. 1998. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol. Cell 1:277-287. [DOI] [PubMed] [Google Scholar]
- 35.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 36.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Navarro, L., K. Mowen, S. Rodems, B. Weaver, N. Reich, D. Spector, and M. David. 1998. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol. Cell. Biol. 18:3796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Netterwald, J. R., T. R. Jones, W. J. Britt, S. J. Yang, I. P. McCrone, and H. Zhu. 2004. Postattachment events associated with viral entry are necessary for induction of interferon-stimulated genes by human cytomegalovirus. J. Virol. 78:6688-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noyce, R. S., S. E. Collins, and K. L. Mossman. 2006. Identification of a novel pathway essential for the immediate-early, interferon-independent antiviral response to enveloped virions. J. Virol. 80:226-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paladino, P., D. T. Cummings, R. S. Noyce, and K. L. Mossman. 2006. The IFN-independent response to virus particle entry provides a first line of antiviral defense that is independent of TLRs and retinoic acid-inducible gene I. J. Immunol. 177:8008-8016. [DOI] [PubMed] [Google Scholar]
- 41.Pass, R. F. 2001. Cytomegalovirus, p. 2675-2705. In P. M. Howley, D. M. Knipe, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 42.Paulus, C., S. Krauss, and M. Nevels. 2006. A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling. Proc. Natl. Acad. Sci. U. S. A. 103:3840-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perry, A. K., G. Chen, D. Zheng, H. Tang, and G. Cheng. 2005. The host type I interferon response to viral and bacterial infections. Cell. Res. 15:407-422. [DOI] [PubMed] [Google Scholar]
- 44.Perry, S. T., and T. Compton. 2006. Kaposi's sarcoma-associated herpesvirus virions inhibit interferon responses induced by envelope glycoprotein gpK8.1. J. Virol. 80:11105-11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peters, K. L., H. L. Smith, G. R. Stark, and G. C. Sen. 2002. IRF-3-dependent, NFkappa B- and JNK-independent activation of the 561 and IFN-beta genes in response to double-stranded RNA. Proc. Natl. Acad. Sci. U. S. A. 99:6322-6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pichlmair, A., O. Schulz, C. P. Tan, T. I. Naslund, P. Liljestrom, F. Weber, and C. Reis e Sousa. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997-1001. [DOI] [PubMed] [Google Scholar]
- 47.Preston, C. M., A. N. Harman, and M. J. Nicholl. 2001. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J. Virol. 75:8909-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reske, A., G. Pollara, C. Krummenacher, D. R. Katz, and B. M. Chain. 2008. Glycoprotein-dependent and TLR2-independent innate immune recognition of herpes simplex virus-1 by dendritic cells. J. Immunol. 180:7525-7536. [DOI] [PubMed] [Google Scholar]
- 49.Rothenburg, S., T. Schwartz, F. Koch-Nolte, and F. Haag. 2002. Complex regulation of the human gene for the Z-DNA binding protein DLM-1. Nucleic Acids Res. 30:993-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saito, T., and M. Gale, Jr. 2008. Differential recognition of double-stranded RNA by RIG-I-like receptors in antiviral immunity. J. Exp. Med. 205:1523-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saito, T., D. M. Owen, F. Jiang, J. Marcotrigiano, and M. Gale, Jr. 2008. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Samanta, M., D. Iwakiri, T. Kanda, T. Imaizumi, and K. Takada. 2006. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. EMBO J. 25:4207-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Samanta, M., D. Iwakiri, and K. Takada. 2008. Epstein-Barr virus-encoded small RNA induces IL-10 through RIG-I-mediated IRF-3 signaling. Oncogene 27:4150-4160. [DOI] [PubMed] [Google Scholar]
- 54.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanchez, D. J., D. Miranda, Jr., V. Arumugaswami, S. Hwang, A. E. Singer, A. Senaati, A. Shahangian, M. J. Song, R. Sun, and G. Cheng. 2008. A repetitive region of gammaherpesvirus genomic DNA is a ligand for induction of type I interferon. J. Virol. 82:2208-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 57.Servant, M. J., B. ten Oever, C. LePage, L. Conti, S. Gessani, I. Julkunen, R. Lin, and J. Hiscott. 2001. Identification of distinct signaling pathways leading to the phosphorylation of interferon regulatory factor 3. J. Biol. Chem. 276:355-363. [DOI] [PubMed] [Google Scholar]
- 58.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669-682. [DOI] [PubMed] [Google Scholar]
- 59.Sharma, S., B. R. tenOever, N. Grandvaux, G. P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 60.Simmen, K. A., J. Singh, B. G. Luukkonen, M. Lopper, A. Bittner, N. E. Miller, M. R. Jackson, T. Compton, and K. Fruh. 2001. Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B. Proc. Natl. Acad. Sci. U. S. A. 98:7140-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stetson, D. B., and R. Medzhitov. 2006. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24:93-103. [DOI] [PubMed] [Google Scholar]
- 62.Streblow, D. N., S. L. Orloff, and J. A. Nelson. 2001. Do pathogens accelerate atherosclerosis? J. Nutr. 131:2798S-2804S. [DOI] [PubMed] [Google Scholar]
- 63.Takaoka, A., Z. Wang, M. K. Choi, H. Yanai, H. Negishi, T. Ban, Y. Lu, M. Miyagishi, T. Kodama, K. Honda, Y. Ohba, and T. Taniguchi. 2007. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448:501-505. [DOI] [PubMed] [Google Scholar]
- 64.Tsitoura, E., J. Thomas, D. Cuchet, K. Thoinet, P. Mavromara, and A. L. Epstein. 2009. Infection with herpes simplex type 1-based amplicon vectors results in an IRF3/7-dependent, TLR-independent activation of the innate antiviral response in primary human fibroblasts. J. Gen. Virol. 90:2209-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, X., S. Hussain, E. J. Wang, X. Wang, M. O. Li, A. Garcia-Sastre, and A. A. Beg. 2007. Lack of essential role of NF-kappa B p50, RelA, and cRel subunits in virus-induced type 1 IFN expression. J. Immunol. 178:6770-6776. [DOI] [PubMed] [Google Scholar]
- 66.Wang, Z., M. K. Choi, T. Ban, H. Yanai, H. Negishi, Y. Lu, T. Tamura, A. Takaoka, K. Nishikura, and T. Taniguchi. 2008. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc. Natl. Acad. Sci. U. S. A. 105:5477-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 68.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727-740. [DOI] [PubMed] [Google Scholar]
- 69.Yoneyama, M., M. Kikuchi, K. Matsumoto, T. Imaizumi, M. Miyagishi, K. Taira, E. Foy, Y. M. Loo, M. Gale, Jr., S. Akira, S. Yonehara, A. Kato, and T. Fujita. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851-2858. [DOI] [PubMed] [Google Scholar]
- 70.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 71.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. U. S. A. 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu, H., J. P. Cong, and T. Shenk. 1997. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. U. S. A. 94:13985-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]