Abstract
The PB2 subunit of the influenza virus RNA polymerase is a major virulence determinant of influenza viruses. However, the molecular mechanisms involved remain unknown. It was previously shown that the PB2 protein, in addition to its nuclear localization, also accumulates in the mitochondria. Here, we demonstrate that the PB2 protein interacts with the mitochondrial antiviral signaling protein, MAVS (also known as IPS-1, VISA, or Cardif), and inhibits MAVS-mediated beta interferon (IFN-β) expression. In addition, we show that PB2 proteins of influenza viruses differ in their abilities to associate with the mitochondria. In particular, the PB2 proteins of seasonal human influenza viruses localize to the mitochondria while PB2 proteins of avian influenza viruses are nonmitochondrial. This difference in localization is caused by a single amino acid polymorphism in the PB2 mitochondrial targeting signal. In order to address the functional significance of the mitochondrial localization of the PB2 protein in vivo, we have generated two recombinant human influenza viruses encoding either mitochondrial or nonmitochondrial PB2 proteins. We found that the difference in the mitochondrial localization of the PB2 proteins does not affect the growth of these viruses in cell culture. However, the virus encoding the nonmitochondrial PB2 protein induces higher levels of IFN-β and, in an animal model, is attenuated compared to the isogenic virus encoding a mitochondrial PB2. Overall this study implicates the PB2 protein in the regulation of host antiviral innate immune pathways and suggests an important role for the mitochondrial association of the PB2 protein in determining virulence.
Influenza A viruses have a natural reservoir in aquatic birds, with only certain subtypes (H1N1, H2N2, and H3N2) causing sustained infection in the human population. However, in Hong Kong in 1997, 18 humans were infected with an H5N1 avian influenza virus, and 6 died (5, 48). This subtype reemerged in humans in Southeast Asia in 2003 and since then has infected over 450 individuals in 15 countries in Asia, Europe, and Africa (http://www.who.int/). A major cause for concern is the high case fatality rate associated with these viruses (∼60%), which is thought to be due to the virus's ability to replicate in extrapulmonary sites as well as its ability to induce an exaggerated cytokine response, referred to as hypercytokinemia (6).
Influenza viruses encode three polymerase subunits, the PB2, PB1, and PA proteins that form the trimeric viral RNA-dependent RNA polymerase (8). The RNA polymerase is responsible for the transcription and replication of the viral RNA genome in the nucleus of infected cells. The PB2 protein specifically plays a role in generating 5′-capped RNA fragments from cellular pre-mRNA molecules that are used as primers for viral transcription (8, 15). Interestingly, the PB2 protein has been found to affect host range as well as virulence of influenza viruses (1, 18, 24, 32, 46, 47). In particular, a single gene reassortant virus encoding the PB2 gene from an avian influenza virus, A/mallard/New York/78 (H2N2), in the background of the remaining genes from a human influenza virus, A/Los Angeles/2/87 (H3N2), replicated efficiently in embryonated eggs and primary chicken cells but not in Madin-Darby canine kidney (MDCK) cells (47). However, if position 627 of the PB2 protein was mutated from a glutamic acid to lysine, the virus acquired the ability to replicate efficiently in MDCK cells. Furthermore, it has been noted that an E627K mutation in the PB2 protein of H5N1 influenza A viruses increased viral replication in mice (13, 18, 19, 26, 29, 46).
Although the role of the PB2 gene in determining host range and virulence is well documented, the exact molecular mechanisms involved remain unknown. It has been suggested that the host range restriction of avian influenza viruses in mammalian cells could be due to the inability of the avian influenza virus polymerase proteins and nucleoprotein to form a ribonucleoprotein (RNP) complex and successfully replicate the viral genome in mammalian cells (24, 31, 32). In particular, the generation of avian/mammalian heterokaryons suggested that a mammalian cellular factor(s) affects RNP complex formation, specifically by altering PB2-NP interactions. However, the identity of this cellular factor(s) remains unknown. While these studies suggest a potential mechanism to explain how the PB2 gene affects the host range of influenza viruses, they do not entirely explain how it affects virulence.
Although most of the PB2 protein in infected cells localizes to the nucleus, where it forms a complex with the PB1 and PA polymerase subunits, PB2 protein was also detected in the mitochondria (2, 54). The mitochondrial localization signal of the PB2 protein has been mapped to the N-terminal region, and it has been proposed that the mitochondrial PB2 protein could contribute to the preservation of mitochondrial function during influenza virus infection by stabilizing the mitochondrial membrane potential. However, the functional significance of the mitochondrial localization of the PB2 protein for viral infection has not been elucidated in detail. Mitochondria play an essential function in cellular respiration (17), and they are involved in the regulation of apoptosis (14) and the induction of beta interferon (IFN-β) in response to viral infection. In particular, the mitochondrial antiviral signaling protein, MAVS (also known as IPS-1, VISA, or Cardif), which is located on the outer membrane of the mitochondria and acts downstream of the viral RNA sensors RIG-I and Mda5, plays a crucial role in inducing IFN-β expression (21, 33, 43, 55).
In this study, we investigated the functional significance of the mitochondrial association of the PB2 protein. We found that the PB2 protein interacts with MAVS and that overexpression of the PB2 protein results in the inhibition of IFN-β expression. Surprisingly, we also found that PB2 proteins from H5N1 influenza A viruses, in contrast with seasonal human influenza viruses, do not associate with mitochondria. The lack of mitochondrial association of H5N1 influenza virus PB2 proteins is caused by a single amino acid polymorphism in the N-terminal mitochondrial localization signal. A recombinant human influenza virus encoding a nonmitochondrial mutant PB2 protein induced higher levels of IFN-β expression in cell culture and was less virulent in an animal model than the wild-type (WT) virus encoding a mitochondrial PB2 protein. These findings demonstrate the importance of the mitochondrial PB2 protein in regulating innate immune responses.
MATERIALS AND METHODS
Plasmids.
Plasmids expressing the PB2, PB1, PA, and NP influenza virus proteins (pcDNA-PB2, pcDNA-PB1, pcDNA-PA, and pcDNA-NP, respectively) from A/WSN/33 (H1N1) were described previously (10). The plasmids expressing the C-terminal green fluorescent protein (GFP)-tagged PB1 and PB2 proteins (pcDNA-PB1-GFP and pcDNA-PB2-GFP, respectively) were also described previously (12). Tandem affinity purification (TAP)-tagged PB2 expression vectors were created as follows. Viral RNA was isolated from A/Ann Arbor/6/60 ca (H2N2; ca indicates cold-adapted virus), followed by reverse transcription (RT) of the PB2 gene segment. The PB2 gene segment cDNAs from A/Hong Kong/483/97 (H5N1) and A/Hong Kong/486/97 (H5N1) were amplified from vectors pBD-483-PB2 and pBD-486-PB2, respectively (3). Amplified PB2 cDNAs were inserted into the pcDNA-PB2-TAP plasmid to replace the PB2 open reading frame (9). The A/turkey/England/50-92/91 (H5N1) PB2-TAP vector was kindly provided by R. Harvey (University of Oxford). Site-directed mutagenesis of the protein expression vector pcDNA-PB2-TAP and the viral RNA (vRNA) expression plasmid pPOLI-PB2-RT (11) was performed using Phusion HF polymerase. Plasmids for the rescue of A/WSN/33 (H1N1) were described previously (11).
A luciferase reporter plasmid under the control of the IFN-β promoter (pIFN-luc) was kindly provided by A. García-Sastre (Mt. Sinai School of Medicine, New York, NY). To control for cell numbers and transfection efficiency, a Renilla control plasmid (pGL4.75[hRluc/CMV]) (Promega) was used. pFlag-MAVS (21) (kindly provided by S. Akira, Osaka University, Japan) was used to express exogenous MAVS.
Immunofluorescence.
Monolayers of Vero or DF-1 cells (kindly provided by M. Iqbal, Institute for Animal Health, Compton, United Kingdom) were seeded onto 13-mm glass coverslips in 24-well dishes and transfected with 1 μg of expression plasmids using 2.5 μl of Lipofectamine 2000 (Invitrogen). Cells were incubated for 24 h and stained with 250 nM MitoTracker Red CMXRos (Molecular Probes) in minimal essential medium (MEM) for 30 to 45 min. Cells were fixed in 4% electron microscopy (EM) grade paraformaldehyde (Electron Microscopy Sciences), followed by permeabilization in 0.5% Triton X-100 in phosphate-buffered saline (PBS). PB2 protein was detected by using a polyclonal rabbit anti-PB2 antibody (2) and a Cy2-conjugated AffiniPure donkey anti-rabbit secondary antibody (Jackson ImmunoResearch). Images were taken using a Zeiss laser scanning microscope (LSM). Overlays and brightness of individual images were manipulated using LSM Image Browser (Zeiss) or Adobe Photoshop.
Alternatively, 50 to 70% confluent monolayers of Vero cells on glass coverslips in a 24-well dish were infected at a multiplicity of infection (MOI) of 1. Cells were incubated at 37°C for 8 h. Following staining of mitochondria with 100 nM MitoTracker Red CMXRos for 30 min, cells were fixed, permeabilized, and stained with PB2-specific antibody as above. Images were taken using a Zeiss Axioplan microscope with a ×63 (numerical aperture, 1.25) oil immersion objective and processed in Photoshop.
Immunoprecipitation and Western blotting.
Approximately 2 × 107 293T cells were plated onto a 15-cm dish and transfected with 20 μg of expression plasmids using 77 μl of FuGENE HD, followed by incubation at 37°C. Forty-eight hours posttransfection, cells were collected, washed in ice-cold PBS, and lysed in 2.4 ml of Tris lysis buffer (50 mM Tris-HCl, pH 8.0, 25% glycerol, 0.5% Igepal CA-630, 200 mM NaCl, 1 mM dithiothreitol [DTT], 1 tablet/10 ml Complete Mini EDTA-free protease inhibitor cocktail [Roche]) on ice for 45 min. Clarified cell lysate (150 μl) was used for immunoprecipitations with 5 μl of rabbit anti-OctA-Probe (Flag) (Santa Cruz) or rabbit anti-MAVS (Abcam) and 6 mg of protein A-Sepharose (Sigma Aldrich). Beads were washed with Tris wash buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% Igepal CA-630) and resuspended in 50 μl of SDS-PAGE protein loading dye. Cell lysates and immunoprecipitates were analyzed by SDS-PAGE, and proteins were transferred to nitrocellulose. Membranes were incubated with mouse anti-OctA-Probe (Flag) or mouse anti-VISA (Abnova) primary antibodies, followed by a goat anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibody (Dako). Signals were detected using an Immobilon Western Chemiluminescent HRP Substrate kit (Millipore) and Hyperfilm (Amersham).
IFN assays.
Semiconfluent monolayers of 293T cells in six-well plates were transfected with 100 ng of pIFN-luc, pGL4.75(hRluc/CMV), and pFlag-MAVS and 2 μg of influenza virus protein expression plasmids using 5 μl of FuGENE HD. Cells were incubated for 24 to 48 h, lysed in 250 μl of passive lysis buffer (PLB; Promega); luciferase and Renilla activities were assessed by using a Dual-Luciferase Assay kit (Promega). To measure IFN-β mRNA expression during influenza virus infection, A549 cells were infected at an MOI of 0.3. Twelve hours postinfection, total cellular RNA was isolated using TRIzol reagent (Invitrogen), followed by reverse transcription of mRNA into cDNA using SuperScript II (Invitrogen) and a T20 primer. Quantitative PCR (qPCR) was performed with IFN-β forward and reverse primers (25) using a QuantiTect SYBR green PCR kit (Qiagen) on a Corbett Rotor-Gene RG-3000 cycler. Data were analyzed using comparative analysis software (Rotor-Gene 6). Infections were performed in triplicate twice using two independently rescued viruses of the same genetic background, and these two sets were subsequently averaged. IFN-β expression in the lungs of infected mice was measured using a VeriKine Mouse IFN-β enzyme-linked immunosorbent assay (ELISA) kit (PBL Interferon Source) according to the manufacturer's instructions. An IFN-β standard curve was created which was used to calculate the IFN-β concentrations in the lung samples.
Virus rescue and growth curves.
Recombinant influenza A/WSN/33 viruses were rescued in 293T cells using a plasmid-based rescue system (11). Amplification of viruses was performed on MDBK cells, and the identities of the viruses were confirmed by sequencing the PB2 gene. To determine the growth kinetics of the rescued viruses, confluent monolayers of MDBK and Vero cells were infected at an MOI of 0.001 and incubated at 37°C. At 4, 8, 24, 48, and 72 h postinfection, 150 μl of the cellular supernatant was harvested and stored at −80°C until further analysis. The 50% tissue culture infective dose (TCID50) was determined in MDCK cells.
Viral RNA assay.
The accumulation of PB2 and NA gene-specific vRNA, mRNA, and cRNA in A549 cells infected with the WSN or WSN N9D virus was analyzed by primer extension as described previously (10, 39, 53) using the total RNA preparations obtained for the IFN assay.
Purification of TAP-tagged recombinant viral RNA polymerase.
Purification of TAP-tagged recombinant viral RNA polymerase was performed as described previously (7, 12) with modifications. Approximately 2.5 × 106 293T cells were plated onto a 6-cm dish and transfected with 7.5 μg of plasmid DNA (pcDNA-PA-TAP, pcDNA-PB1, pcDNA-PB2 WT, or pcDNA-PB2 N9D) using 7.5 μl of TurboFect (Fermentas), followed by incubation at 37°C for 48 h. The cells were collected, washed once in PBS, and lysed in 530 μl of HEPES lysis buffer (50 mM HEPES, pH 8.0, 25% glycerol, 0.5% Igepal CA-630, 0.2 M NaCl, 1 mM β-mercaptoethanol, 1 tablet/10 ml mini-EDTA free protease inhibitor cocktail) on ice for 60 min. Lysates were clarified by centrifuging at 13,000 rpm for 10 min at 4°C, followed by the binding of 500 μl of lysate to 100 μl of IgG-Sepharose at 4°C for 3 to 4 h in a total volume of 2.5 ml. After samples were washed in IgG wash buffer (10 mM HEPES, 10% glycerol, 0.1% Igepal CA-630, 150 mM NaCl, 0.1 mM phenylmethylsulfonyl fluoride [PMSF]), the purified proteins were cleaved from the beads by incubation with 2 μl of AcTEV protease (Invitrogen) in 100 μl of cleavage buffer (IgG wash buffer, 1 mM DTT) at 4°C for 2 h. Finally, 50 μl of 100% glycerol was added to the purified polymerase. Polymerase proteins were analyzed by SDS-PAGE and visualized using a SilverXpress silver stain kit (Invitrogen).
In vitro polymerase activity assays.
In vitro activity of the purified polymerases was determined as described previously (10) with modifications. Briefly, 1.5 μl of TAP-purified polymerase was added to 1.5 μl of ApG activity buffer (10 mM MgCl2, 2 mM DTT, 1 mM ApG, 1.87 U/μl rRNasin RNase inhibitor [Promega], 0.3 μM [α-32P]GTP [3,000 Ci/mmol; Amersham], 1.33 μM promoter RNA [equal amounts of 3′-end vRNA {5′-GGCCUGCUUUUGCU-3′} and 5′-end vRNA {5′-AGUAGAAACAAGGCC-3′}; Dharmacon], 2 mM ATP, 1 mM CTP, 1 mM UTP) or 1 μl of globin mRNA activity buffer (10 mM MgCl2, 2 mM DTT, 36 ng/μl globin mRNA [Sigma], 1.87 U/μl rRNasin RNase inhibitor [Promega], 0.3 μM [α-32P]GTP [3,000 Ci/mmol; Amersham], 1.33 μM promoter RNA, 2 mM ATP, 1 mM CTP, 1 mM UTP) and incubated at 30°C for 2 h (ApG activity) or overnight (globin mRNA activity). Samples were mixed with 3 μl of formamide loading dye (90% formamide, 10 mM EDTA, bromophenol blue, cyanoxylene) and then heated at 95°C for 5 min. Transcription products were analyzed on a 6 M urea-25% PAGE gel at 1,500 V for 3 to 4 h. To visualize products, Kodak BioMax MS films were exposed to the gel with BioMax intensifying screens. Transcription product signals were measured using a Fuji phosphorimager and analyzed using AIDA densitometry software.
Mouse infections.
Eight-week-old female B6 (C57B/6NTac [Taconic]) mice were infected with WSN or WSN N9D virus following the NIH Animal Care and Use Committee Guidelines. Briefly, mice were lightly anesthetized using 4% isoflurane in an induction chamber attached to a precision vaporizer (Viking Medical). Following anesthesia, viruses diluted to the appropriate titer in 1× Leibovitz's L-15 medium were administered in a 50-μl volume intranasally. To measure the virulence in terms of 50% mouse lethal dose (MLD50), groups of five mice were infected with dilutions of virus ranging from 101 to 105 TCID50s/50 μl. Following infection, mice were weighed daily and were euthanized if they lost >25% of their original body weight. The MLD50 of the viruses was calculated as described previously (30). To determine the viral growth kinetics in vivo, groups of four mice were infected with 104 TCID50s/50 μl intranasally. Lungs were harvested at 1, 3, and 5 days postinfection (dpi), weighed, homogenized to generate 10%, wt/vol, suspensions in 1× Leibovitz's L-15 medium, and stored at −80°C until titration. Samples were titrated on MDCK monolayers as described elsewhere (30).
RESULTS
PB2 proteins of different influenza A virus subtypes differ in their ability to localize to the mitochondria.
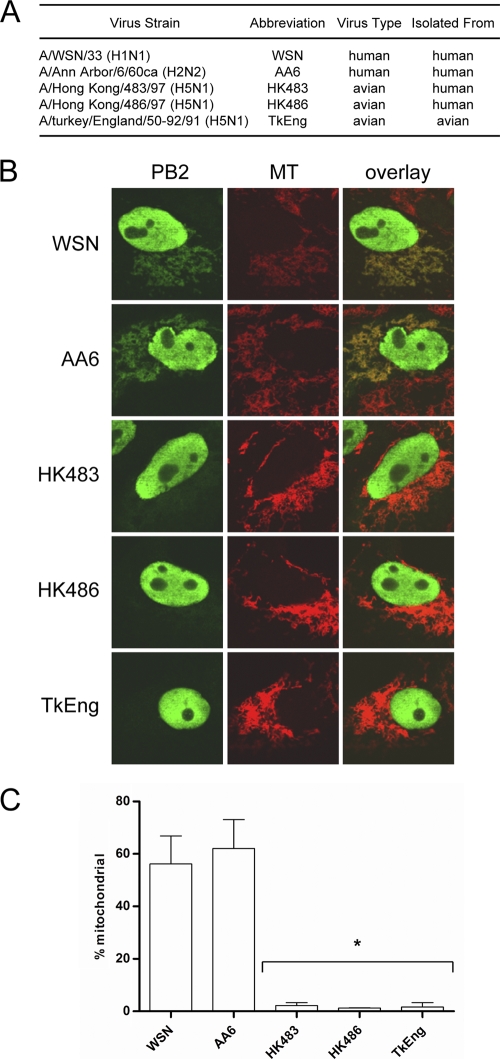
In order to assess the intracellular distribution of PB2 proteins from different influenza virus subtypes, we transfected Vero cells with plasmids expressing TAP-tagged PB2 proteins of H1N1, H2N2, and H5N1 influenza viruses (Fig. 1A). At 24 h posttransfection the mitochondria were stained with MitoTracker Red, and PB2 proteins were detected via indirect immunofluorescence. The PB2 proteins from the H1N1 and H2N2 viruses displayed significant levels of mitochondrial signal, as expected (2). In contrast, PB2 proteins from all three H5N1 viruses tested showed no detectable mitochondrial association (Fig. 1B). To quantitatively measure mitochondrial localization, we scored cells showing PB2 protein expression as positive or negative for mitochondrial PB2 signals (Fig. 1C). The PB2 proteins from the H1N1 and H2N2 viruses were found in the mitochondria in 56% and 62% of cells expressing the PB2 protein, respectively. In contrast, the PB2 proteins from the H5N1 viruses, HK483, HK486, and TkEng, were found in the mitochondria of only 2%, 1%, and 2% of PB2-expressing cells, respectively. Similar results were observed in chicken fibroblasts (DF-1), showing that the difference in distribution of the PB2 proteins of different influenza virus subtypes was not host cell specific (data not shown).
FIG. 1.
PB2 proteins of different influenza virus subtypes display different localization patterns in Vero cells. (A) List of viruses used to examine PB2 localization (ca, cold adapted). (B) Images of cellular distribution of PB2 proteins in transfected Vero cells. Mitochondria (MT) were stained with MitoTracker Red. (C) PB2-expressing cells were scored for mitochondrial localization of PB2. Columns represent the percentage of PB2-expressing cells containing mitochondrial PB2 signal. Bars represent standard errors of the means based on three independent experiments (n = 61 to 107 cells/experiment). *, P < 0.0075, based on an unpaired, two-tailed Student's t test.
Difference in mitochondrial localization is due to an amino acid polymorphism at position 9 of the PB2 protein.
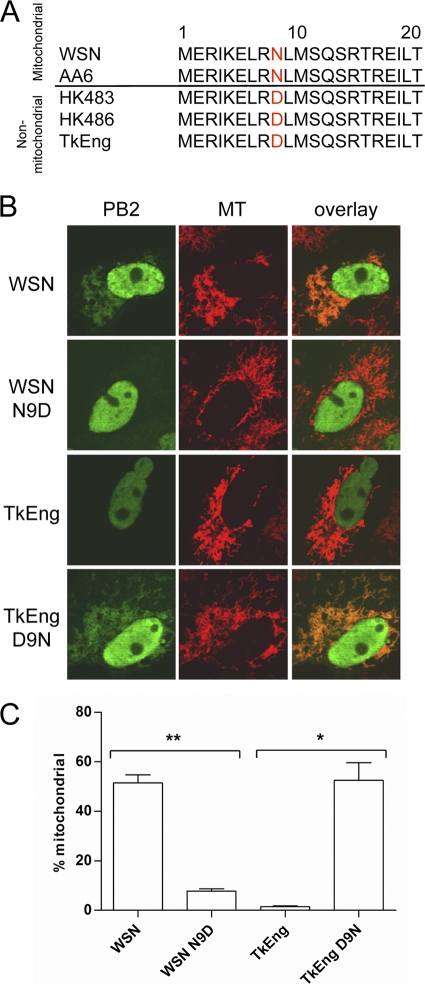
The mitochondrial localization signal of the PB2 protein has been shown to be located within the N-terminal region (2, 54). To determine whether there were any differences in the amino acid sequence of the mitochondrial localization signals of the five PB2 proteins, we aligned the N-terminal 21 amino acids of each PB2 protein (Fig. 2A). We found that the three PB2 proteins that did not localize to the mitochondria all encoded an aspartic acid at position 9, whereas the two mitochondrial proteins encoded an asparagine. To determine whether this polymorphism was responsible for the difference in mitochondrial localization, we mutated position 9 of the PB2 proteins of the WSN and TkEng viruses and assessed their localization in Vero cells. We found that the N9D mutation in the WSN PB2 protein reduced mitochondrial association from 52% to 8% (Fig. 2B and C). In contrast, the D9N mutation in the TkEng PB2 protein increased mitochondrial association from 1.5% to 52.5%. Similar results were observed in avian DF-1 cells (data not shown). These results demonstrate that the polymorphism at position 9 is responsible for the different localization patterns of PB2 proteins of H5N1 viruses compared to the H1N1 and H2N2 viruses. It should be noted that a previous study reported mitochondrial association of the PB2 protein from the TkEng virus; however, quantitative analysis was not performed (2). Here, we have demonstrated that only about 1.5 to 2% of the TkEng PB2-expressing cells show mitochondrial PB2. At this point, it is unclear why a minority of cells expressing PB2 with an aspartic acid at position 9 still show mitochondrial localization of PB2 or, in fact, why not all cells expressing PB2 with an asparagine at position 9 show mitochondrial PB2. We can only speculate that this may be due to cell variability. In particular, nuclear and mitochondrial transport of PB2 depends on host factors, and the relative amounts of these factors might differ in individual cells.
FIG. 2.
Polymorphism at position 9 is responsible for the differential localization of the PB2 protein. (A) Alignment of the mitochondrial localization signals of influenza A virus PB2 proteins. (B) Images of cellular distribution of wild-type and mutant PB2 proteins in transfected Vero cells. (C) PB2-expressing cells were scored for mitochondrial localization of PB2. Columns represent the percentage of PB2-expressing cells containing mitochondrial PB2 signal. Bars represent standard errors of the means based on three independent experiments (n = 63 to 110 cells/experiment). **, P < 0.0002; *, P < 0.021 (based on an unpaired, two-tailed Student's t test).
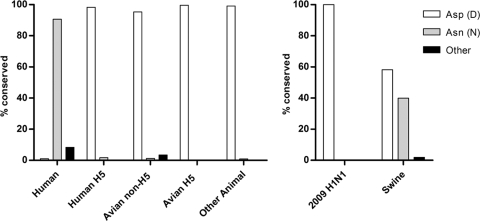
The two mitochondrial PB2 proteins were derived from H1N1 and H2N2 viruses while all three nonmitochondrial PB2 proteins originated from H5N1 viruses, suggesting that mitochondrial localization may be subtype specific. To investigate this further, we aligned PB2 protein sequences from the NCBI influenza resource database (http://www.ncbi.nlm.nih.gov/genomes/FLU/). Influenza A virus isolates from avian species as well as from other animals, including dogs, cats, and horses, regardless of subtype, predominantly encoded an aspartic acid at position 9 (non-H5N1, 95.3%; H5N1, 99.6%) (Fig. 3A). H5N1 viruses isolated from humans also encoded an aspartic acid (98.3%). In contrast, seasonal H1N1, H2N2, and H3N2 human influenza viruses encoded predominantly an asparagine (91%). Interestingly, 39.9% and 58% of swine influenza virus isolates encoded an asparagine or aspartic acid, respectively (Fig. 3B), whereas 100% of human isolates of the 2009 H1N1 pandemic virus PB2 protein encoded an aspartic acid. Overall, these data demonstrate that the mitochondrial localization of the PB2 protein is not subtype specific. Rather, it is related to the origin of influenza viruses, with nonmitochondrial aspartic acid predominating in all isolates except seasonal human influenza viruses, suggesting that mitochondrial association may be important in adaptation to a human host.
FIG. 3.
Alignment of PB2 amino acid sequences of influenza A viruses reveals host-specific conservation of position 9. PB2 protein sequences from the NCBI influenza virus database were grouped based on host and hemagglutinin subtype, followed by analysis of the amino acid at position 9. Human (excluding H5 isolates and swine-origin 2009 H1N1), n = 2686; human H5, n = 117; avian non-H5, n = 1215; avian H5, n = 788; other animal, n = 112; human swine-origin 2009 H1N1, n = 57; swine, n = 263.
PB2 protein interacts with the mitochondrial antiviral signaling protein, MAVS.
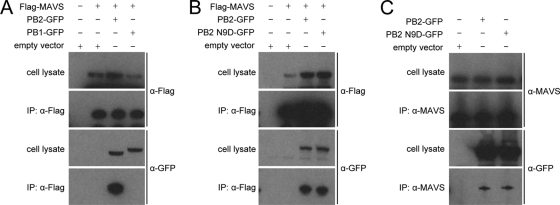
All three subunits of the influenza virus RNA polymerase complex are required for polymerase function (8), yet neither the PB1 nor the PA protein is known to localize to the mitochondria (12). We therefore hypothesized that the mitochondrial PB2 protein might be performing a function independent of its role in transcription and replication. Mitochondria are involved in innate immune responses via the MAVS protein (21, 33, 43, 55), and therefore we investigated whether the mitochondrial PB2 protein could affect MAVS function and, consequently, the expression of IFN-β. To determine whether the mitochondrial PB2 protein interacts with the MAVS protein, we performed an immunoprecipitation assay using 293T cells cotransfected with a Flag-MAVS plasmid and either a PB2-GFP or a PB1-GFP expression plasmid as a negative control. The PB2-GFP protein was coimmunoprecipitated with Flag-tagged MAVS, but the PB1-GFP protein was not (Fig. 4A), demonstrating that this interaction is specific to the PB2 protein and not its GFP tag. We also performed immunoprecipitations with the PB2 N9D mutant and found that the PB2-GFP protein with the N9D mutation was also coimmunoprecipitated with Flag-MAVS (Fig. 4B). No PB2-GFP, wild-type or mutant, was immunoprecipitated in the absence of an antibody (results not shown). Both the wild-type and N9D mutant PB2 proteins could also be coimmunoprecipitated with endogenous MAVS (Fig. 4C). Taken together, these results show that the PB2 protein specifically interacts with MAVS. In addition, the MAVS interaction domain of the PB2 protein is unaffected by mutation at position 9 in the mitochondrial localization signal as both the mitochondrial and nonmitochondrial PB2 proteins can interact with MAVS in vitro.
FIG. 4.
Immunoprecipitation reveals interaction between Flag-MAVS and PB2-GFP. (A) Lysates from 293T cells transfected with Flag-MAVS and pcDNA-PB2-GFP or pcDNA-PB1-GFP and immunoprecipitates (IP) were analyzed by Western blotting using anti-Flag and anti-GFP antibodies. Immunoprecipitations of MAVS were performed with anti-Flag antibody. (B) Lysates from 293T cells transfected with Flag-MAVS and pcDNA-PB2-GFP or pcDNA-PB2 N9D-GFP and immunoprecipitates were analyzed by Western blotting using anti-Flag and anti-GFP antibodies. Immunoprecipitations of MAVS were performed with anti-Flag antibody. (C) Lysates from 293T cells transfected with pcDNA-PB2-GFP or pcDNA-PB2 N9D-GFP and immunoprecipitates were analyzed by Western blotting using anti-MAVS and anti-GFP antibodies. Immunoprecipitations of MAVS were performed with anti-MAVS antibody. α, anti.
PB2 protein affects levels of type I interferon in transfected cells.
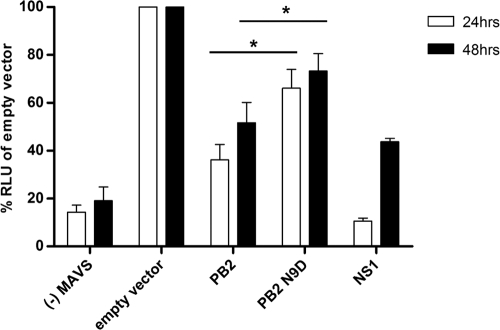
To determine whether the PB2 protein affects IFN-β expression, we performed an IFN-β promoter-driven luciferase reporter gene assay (Fig. 5). We overexpressed MAVS to induce the IFN-β promoter (43). The expression of the influenza virus NS1 protein, a potent inhibitor of IFN-β expression (used as a positive control), caused a significant decrease in luciferase reporter activity at both 24 and 48 h posttransfection (11% and 44%, respectively, compared to the negative control). Expression of mitochondrial PB2 protein also resulted in a significant reduction in luciferase activity at both 24 and 48 h posttransfection (36% and 52%, respectively). However, when the nonmitochondrial PB2 protein was expressed, the effect on luciferase reporter activity was significantly (P < 0.01) less than that of the mitochondrial PB2 protein. These results suggest that the PB2 protein could be involved in regulating innate immune responses by inhibiting IFN-β expression and that the mitochondrial association of the PB2 protein is an important factor in determining the effect on IFN-β expression.
FIG. 5.
PB2 inhibits MAVS-induced IFN-β production. 293T cells were transfected with a luciferase reporter plasmid under the control of an IFN-β promoter as well as a Renilla control plasmid, Flag-MAVS expression plasmid, and plasmids expressing either wild-type or N9D mutant PB2 or NS1 protein from A/WSN/33 or an empty vector. Twenty-four and 48 h later, cells were lysed, and both luciferase and Renilla activities were measured. Renilla-adjusted luciferase activity (RLU) in the presence of overexpressed Flag-MAVS but in the absence of viral proteins (empty vector) was set to 100%. Activity in the presence of viral proteins PB2, PB2 N9D, or NS1 was expressed as a percentage of that of the empty vector control. Only low levels of activity were detected in the absence (−) of Flag-MAVS. Bars represent standard errors of the means, based on between four and seven experiments. *, P < 0.01 based on a paired, two-tailed student's t test.
A recombinant influenza A virus encoding a nonmitochondrial PB2 protein induces higher expression of IFN-β in vitro.
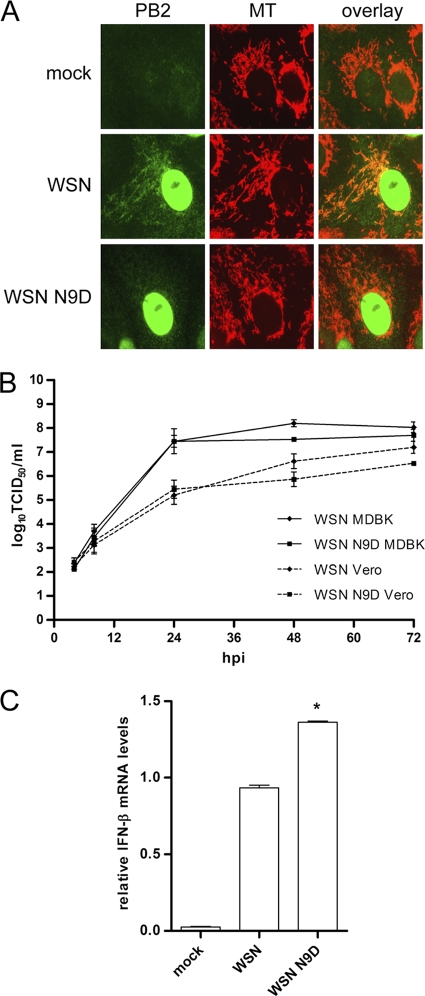
To assess the effect of the mitochondrial localization of the PB2 protein on virus growth and the induction of IFN-β during infection in vitro, recombinant A/WSN/33 (H1N1) influenza viruses encoding either a wild-type PB2 protein (WSN) or a PB2 protein with an N9D mutation (WSN N9D) were generated by reverse genetics. We then analyzed the localization of the PB2 protein by indirect immunofluorescence in Vero cells infected with the WSN or WSN N9D virus. As expected, the wild-type PB2 protein was detected in the mitochondria as well as the nucleus, whereas the PB2 protein in WSN N9D virus-infected cells was detected in only the nucleus (Fig. 6A). We also analyzed the growth properties of the two viruses in cell culture. We found no significant difference in the growth kinetics of the WSN and WSN N9D viruses in both MDBK and Vero cells (Fig. 6B), suggesting that mitochondrial localization of the PB2 protein does not affect viral replication in vitro.
FIG. 6.
Wild-type and PB2 N9D mutant influenza viruses have similar growth kinetics in vitro but induce different levels of IFN-β mRNA. (A) Vero cells were infected with either WSN or WSN N9D virus at an MOI of 1. At 8 h postinfection, the cells were stained with MitoTracker Red (MT), and PB2 was visualized by indirect immunofluorescence. (B) MDBK or Vero cells were infected with either WSN or WSN N9D virus at an MOI of 0.001 and incubated at 37°C for 72 h. Aliquots of cell supernatants were collected at 4, 8, 24, 48, and 72 h postinfection (hpi), followed by analysis of TCID50 using MDCK cells. Data points represent an average of three independent infections. (C) A549 cells were infected with either WSN or WSN N9D virus at an MOI of 0.3. Twelve hours postinfection, total RNA was isolated, and RT-qPCR was performed to measure IFN-β mRNA levels. Infections were performed in triplicate twice using two independently rescued viruses of the same genetic background, and results for these two sets were subsequently averaged. Error bars represents standard errors of the means. *, P < 0.02, based on a paired, two-tailed Student's t test.
To assess IFN-β expression in response to virus infection, A549 cells were infected with the WSN or WSN N9D viruses, and IFN-β mRNA was quantified by RT-qPCR at 12 h postinfection. Both viruses induced significant levels of IFN-β mRNA compared to the negative control (mock infection). However, we found that IFN-β mRNA levels were about 1.5-fold higher (P < 0.02) in cells infected with the WSN N9D virus than in cells infected with the WSN virus (Fig. 6C). Thus, the expression of the nonmitochondrial PB2 protein in the context of viral infection results in increased levels of IFN-β expression. These results are in agreement with the observed reduction in IFN-β expression in transfected cells expressing the mitochondrial PB2 protein (Fig. 5) and suggest a role for the PB2 protein in the regulation of type I interferon expression.
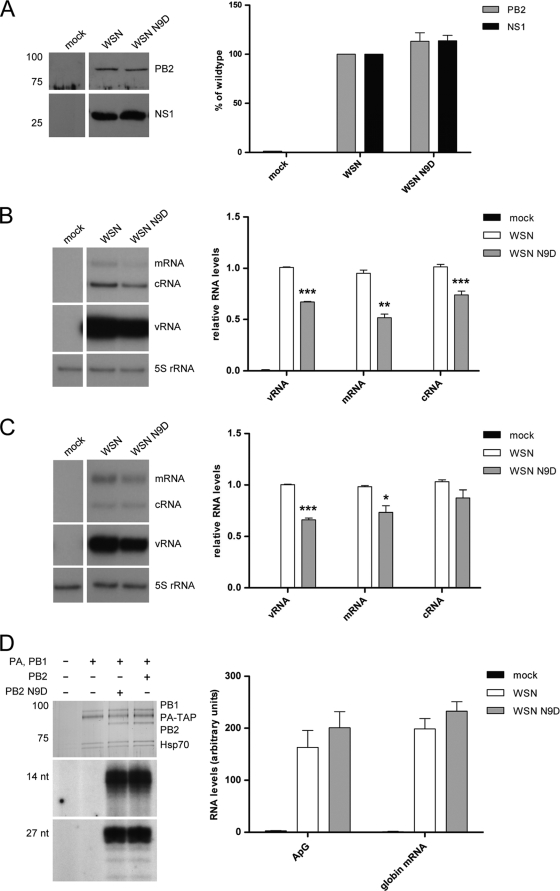
To address the possibility that the difference in IFN-β production was due to differences in viral protein or RNA expression, the viral PB2 and NS1 protein levels and mRNA, cRNA, and vRNA levels in infected cells were determined by Western blotting (Fig. 7A) and primer extension analysis (Fig. 7B and C), respectively. The NS1 and PB2 proteins of the two viruses were expressed at similar levels (Fig. 7A). Interestingly, the PB2 and neuraminidase vRNA, mRNA, and cRNA levels were reduced in the WSN N9D virus-infected cells compared to levels in the WSN virus-infected cells, although in the case of the neuraminidase cRNA this difference was not statistically significant (Fig. 7B and C). These results suggested that the N9D mutation might have affected polymerase function. To investigate this further, we expressed TAP-tagged WSN and WSN N9D polymerase complexes in transfected cells, purified them using IgG chromatography (Fig. 7D), and tested their activity in ApG-primed and capped RNA-primed transcription. In vitro transcription assays revealed that the wild-type and PB2 N9D polymerase complexes had similar activities, suggesting that the N9D mutation is unlikely to directly affect polymerase function (Fig. 7D and E).
FIG. 7.
Accumulation of viral proteins and RNAs in A549 cells infected with wild-type or PB2 N9D mutant influenza virus and the effect of the N9D mutation on RNA polymerase activity. (A) Western blotting and quantitative analysis of PB2 and NS1 in lysates from A549 cells infected with WSN or WSN N9D at an MOI of 0.3 for 12 h. (B and C) Primer extension analysis of the three viral RNA species of the PB2 (B) and neuraminidase (C) genes in A549 cells infected with WSN or WSN N9D at an MOI of 0.3 for 12 h. Bars represent standard errors of the means based on four independent experiments. ***, P ≤ 0.0004; **, P < 0.005; *, P < 0.03 (based on a paired, two-tailed Student's t test). (D) 293T cells were transfected with plasmids expressing PA-TAP and PB1 and either wild-type PB2 or N9D PB2 from A/WSN/33. Cells were lysed at 48 h posttransfection, and proteins were purified by IgG-Sepharose column chromatography. Purified proteins were analyzed by SDS-PAGE and stained by silver (top panel). In vitro transcription assays were performed with ApG dinucleotide primer (middle panel) or in the presence of globin mRNA as a donor of capped-RNA primer (bottom panel). Transcription products were analyzed by PAGE and visualized by autoradiography. Quantitation of transcription products was done by phosphorimager analysis of experiments performed in triplicate.
Thus, it remains unclear why PB2 and neuraminidase RNA levels were reduced in cells infected with WSN N9D compared to cells infected with the WSN virus. However, the finding that increased IFN-β expression was detected in the presence of a reduced amount of vRNA, the inducer of the RIG-I pathway in influenza virus-infected cells (38), is in agreement with our hypothesis that the difference in IFN-β levels induced by the two viruses is due to the difference in the mitochondrial localization of their PB2 proteins and not due to differences in vRNA levels.
In vivo analysis of recombinant influenza viruses encoding mitochondrial or nonmitochondrial PB2 protein.
The results described in the previous section demonstrated that an influenza virus encoding a nonmitochondrial PB2 protein induced higher levels of IFN-β expression than a virus encoding a mitochondrial PB2 protein. However, this difference in IFN-β expression did not affect the growth kinetics of the two viruses in cell culture. To assess the effect of the mitochondrial localization of the PB2 protein on the pathogenicity and replication of the viruses in vivo, we infected C57/B6 (B6) mice intranasally and monitored them daily for weight loss and mortality. Mice infected with the WSN virus displayed a higher mortality rate and more weight loss than those infected with the WSN N9D virus (Fig. 8A and B), demonstrating that the WSN virus was more virulent for mice. The MLD50 of the WSN virus was approximately 3-fold lower (103.4 versus 103.8 TCID50s) than that of the WSN N9D virus. In addition, the WSN virus induced death more rapidly; at all doses, the WSN virus resulted in death earlier than the WSN N9D virus even when the dose of virus significantly exceeded the MLD50 (104 to 105 TCID50s) (Fig. 8C). Thus, the WSN N9D virus was attenuated in the B6 mice even though its growth was not attenuated in cell culture.
FIG. 8.
WSN N9D virus expressing nonmitochondrial PB2 is attenuated in vivo. (A) Groups of five C57/B6 mice were infected intranasally with 104 TCID50s/50 μl of either WSN or WSN N9D virus. Animals were weighed daily and were euthanized when they lost more than 25% of their weight; percent survival is plotted on the y axis. (B) Weights of infected mice were measured daily and recorded as percentages of initial weight. Mean percent weights on each day are shown. (C) Groups of five C57/B6 mice were infected intranasally with the indicated dose of either WSN or WSN N9D virus. The table shows the mean time to death (MTD) in days and percent survival at each dose (%). (D and E) Groups of four C57/B6 mice were infected intranasally with 104 TCID50s of the WSN or WSN N9D virus, and the lungs were harvested at 1, 3, and 5 dpi. Viral titers (log10 TCID50/g of tissue) were determined on Vero 76 cells (D). The dashed line represents the limit of detection (101.5 TCID50s/g). Columns represent mean viral titers. Error bars represent standard errors of the means. *, P < 0.05, based on a Mann-Whitney test. (E) IFN-β was measured using a mouse IFN-β ELISA kit. Columns represent mean observed concentrations of cytokines from four mice. Error bars represent standard errors of the means. **, P < 0.01; ***, P < 0.001 (two-way analysis of variance with a Bonferroni posttest correction).
To determine whether in vivo replication of the two viruses may have been responsible for the observed differences in virulence, the growth kinetics of the two viruses were determined in the lungs of B6 mice. At 1 dpi, both the WSN and WSN N9D viruses reached titers of approximately 106 TCID50s/g. However, by 3 dpi, the WSN virus reached significantly higher titers than the WSN N9D virus (P < 0.05), and this difference was maintained at 5 dpi (Fig. 8D). An IFN-β ELISA was performed to determine whether this decrease in pulmonary viral titer was due to an increase in IFN-β expression. At 1 dpi no statistically significant difference in IFN-β expression was observed (Fig. 8E). However, at both 3 and 5 dpi, IFN-β expression in the lungs of mice infected with the WSN N9D virus was lower, suggesting that the attenuation of the WSN N9D virus is not due to an increase in IFN-β expression. The data suggest that the inability of the PB2 protein to associate with the mitochondria results in an attenuation of virulence associated with a lower pulmonary viral load.
DISCUSSION
In this report, we examined the functional significance of the association of the PB2 subunit of the influenza virus RNA polymerase complex with mitochondria. We demonstrated that the PB2 protein interacts with MAVS and is able to inhibit IFN-β expression. Moreover, we found that a virus encoding a nonmitochondrial PB2 protein is restricted in replication and is less virulent in mice than a virus encoding a mitochondrial PB2 protein. These findings suggest a molecular mechanism by which the PB2 protein might affect virulence.
A comparison of the intracellular localization of PB2 proteins originating from various influenza virus subtypes indicated that only PB2 proteins of seasonal human influenza viruses associate with mitochondria. In contrast, the PB2 proteins of avian influenza viruses, including avian viruses of the H5N1 subtype, do not associate with mitochondria. We determined that this difference was caused by a single amino acid polymorphism at amino acid residue 9 within the previously characterized mitochondrial targeting signal of the PB2 protein, with an asparagine residue resulting in mitochondrial localization and an aspartic acid leading to nonmitochondrial localization. Comparison of PB2 protein sequences revealed that, of the three pandemic influenza viruses of the 20th century, only the 1918 “Spanish” influenza virus encodes an aspartic acid at position 9, while the 1957 “Asian” influenza and 1968 “Hong Kong” influenza pandemic viruses encode an asparagine. This correlates with the genetic characterization of these viruses that shows that the 1918 virus was generated by direct transmission from an avian host to humans, while the 1957 and 1968 viruses were produced by reassortment between previously circulating human and avian influenza viruses (42, 50). As the PB2 segment of the 1957 and 1968 pandemic viruses originates from the 1918 virus, at some point after 1918 a mutation from aspartic acid to asparagine at position 9 must have occurred. The first human influenza virus isolates originating from the early 1930s already encoded an asparagine. Thus, the presence of an asparagine at position 9 in subsequent seasonal influenza viruses could reflect a founder effect. However, the fact that asparagine has been maintained in most of the PB2 proteins of seasonal H1N1, H2N2, and H3N2 viruses since 1930, with only very few isolates containing a residue other than asparagine (less than 10%) (Fig. 3), highlights the importance of this residue in the adaptation of influenza viruses to humans. Moreover, this position has been previously identified as a “signature” amino acid that distinguishes human from avian influenza viruses (34, 35). Interestingly, one human H5N1 isolate, A/Hong Kong/482/97, and one human H9N2 isolate, A/Hong Kong/1073/99, were reported to encode an asparagine at position 9 instead of aspartic acid. In addition, another human H5N1 isolate, A/Hong Kong/483/97, was reported to encode an asparagine (45) although, according to another report (20), it encodes an aspartic acid, suggesting that this virus may have existed as a quasispecies within the human host it was isolated from. Taken together, these results provide further support for the role of this residue in the adaptation of avian influenza viruses to humans.
We also examined the amino acid conservation at position 9 of the PB2 proteins of swine influenza isolates and the 2009 H1N1 pandemic viruses. Swine influenza isolates contain either asparagine or aspartic acid (39.9% and 58%), consistent with the idea that pigs can be infected by both avian and human influenza viruses. However, all isolates of the 2009 pandemic H1N1 virus have an aspartic acid, in agreement with the phylogenetic analysis suggesting that the PB2 segment in this reassortant virus is of avian origin (36). It is intriguing that no swine-origin pandemic 2009 H1N1 influenza viruses have been isolated yet with a mutation at this position. It should also be noted that the PB2 protein of the pandemic 2009 H1N1 viruses contains a glutamic acid at position 627 that is normally associated with avian influenza viruses (36). These observations suggest that the ability of these viruses to successfully replicate and transmit in humans is specified by other genetic determinants, eliminating the requirements for an asparagine at position 9 and a lysine at position 627.
How can a single amino acid change in the N-terminal region have such a dramatic effect on the mitochondrial association of the PB2 protein? Mitochondrial localization signals of proteins tend to be amphipathic alpha helices rich in leucines, serines, and arginines (51). The N terminus of the PB2 protein was also predicted to be an amphipathic alpha helix containing leucine, serine, and arginine residues (2), and recently, a crystal structure of the N terminus of the PB2 protein has confirmed its alpha-helical nature (49). To address the possibility that the aspartic acid at position 9 might disrupt the alpha helix, thus blocking mitochondrial localization, we performed a JPred secondary structure prediction (http://www.compbio.dundee.ac.uk/∼www-jpred/) for the N-terminal 20 amino acids of the PB2 protein. An N9D mutation did not significantly alter the predicted alpha-helical structure (data not shown), suggesting that the inability of the PB2 protein with an aspartic acid at position 9 to localize to the mitochondria is not due to a change in secondary structure. We speculate that the change in charge caused by the replacement of a neutral with a negatively charged amino acid might alter the interaction of the PB2 protein with components of the mitochondrial import complex that requires an amphipathic alpha helix with positively charged residues to mediate mitochondrial import (37, 41).
In order to address the functional significance of the mitochondrial association of the PB2 protein, we investigated the involvement of the PB2 protein in the regulation of host innate immune responses. Influenza virus infection results in IFN-β expression triggered by the binding of 5′ triphosphorylated full-length genomic RNA to RIG-I (38). MAVS, located in the outer membrane of mitochondria, has been shown to play an important role in this pathway by acting downstream of RIG-I. We found that the PB2 protein interacts with MAVS and that expression of the PB2 protein results in the inhibition of MAVS-mediated IFN-β expression, suggesting that the PB2 protein could inhibit the activity of MAVS. Our results are consistent with a recent study which proposed that the influenza virus RNA polymerase may play a previously unappreciated role in modulating interferon production (44). In particular, overexpression of PB1, PB2, and NP, individually or in combination, was sufficient to inhibit cellular interferon responses to either vRNA transfection or viral infection. Our study suggests a possible molecular mechanism for the previously observed inhibition of interferon production by the PB2 protein.
Several other viruses also express proteins that inhibit IFN-β expression via MAVS. For example, the hepatitis C virus (HCV) NS3/4A protein was shown to cleave the mitochondrial transmembrane domain of MAVS, thus releasing the protein into the cytoplasm and rendering it inactive (28). Influenza virus PB2 protein appears to affect MAVS function by a different mechanism as we detected no degradation of MAVS by Western blot analysis or release of MAVS by indirect immunofluorescence of cells expressing Flag-MAVS and PB2 protein (data not shown). We speculate that the PB2 protein, by binding to MAVS, could potentially block interactions between MAVS and other proteins of the antiviral signaling pathway, thus reducing IFN-β expression. Further studies, e.g., mapping the interaction domains of the PB2 protein and MAVS, constructing PB2 mutants deficient in MAVS binding, and constructing MAVS mutants lacking the C-terminal mitochondrial targeting sequence (43), will be required to elucidate the exact molecular mechanisms by which the PB2 protein inhibits MAVS function. Furthermore, studies of the exact location of the PB2 protein and MAVS in the mitochondria, e.g., by using electron microscopy, may help to address the question of whether PB2 and MAVS localize to the same or different mitochondrial compartments. It should be noted that the nonmitochondrial PB2 protein was also found to interact with MAVS in vitro and inhibited IFN-β expression in a PB2 overexpression assay, although to a lesser extent than the mitochondrial PB2 protein. However, considering that MAVS is preferentially localized to the mitochondria and that its mitochondrial localization has been shown to be important for its function in IFN-β expression (43), we speculate that in vivo only the mitochondrial PB2 protein is likely to significantly affect MAVS function. Thus, the inhibitory interaction between the PB2 protein and MAVS must take place at the mitochondria.
In order to address the importance of the mitochondrial association of the PB2 protein in the context of viral infection in vivo, we generated a recombinant A/WSN/33 influenza virus with an N9D mutation that renders its PB2 protein nonmitochondrial. Consistent with the observation that overexpression of mitochondrial PB2 protein results in lower levels of IFN-β than overexpression of nonmitochondrial PB2 protein, we found that the mutant WSN virus with the nonmitochondrial PB2 protein induced higher levels of IFN-β in A549 cells than the wild-type virus encoding the mitochondrial PB2 protein. Surprisingly, IFN-β expression did not affect viral growth kinetics. Both the WSN and WSN N9D viruses replicated to similar titers in IFN-β-competent MDBK cells as well as in IFN-β-incompetent Vero cells, suggesting that mitochondrial PB2 protein is not required for virus replication in cell culture.
Interestingly, the accumulation of viral RNAs in cells infected with the WSN N9D virus was reduced, albeit less than 2-fold, compared to levels in the cells infected with WSN wild-type virus. A similar reduction in all three types of viral RNAs was observed in a cell-based recombinant ribonucleoprotein reconstitution assay performed in 293T cells in which the neuraminidase gene served as a reporter gene (data not shown). Therefore, although an effect on polymerase activity could not be detected in transcription assays in vitro using purified polymerase preparations (Fig. 7D), we cannot completely exclude the possibility that the N9D change has an effect on polymerase function. The possibility remains that polymerase function is affected within the cell; position 9 might alter the association of the polymerase with a host factor(s) involved in regulating polymerase function. Whatever the mechanism, the differences observed in viral RNA levels do not seem to be sufficient to affect viral growth in cell culture.
In contrast to cell culture, the mutant virus was attenuated in an animal model, as manifested by an increase in the MLD50 of the virus, delayed time to death, and a decrease in virus titers in the lungs of mice. We suggest that the reduction in viral titers could be caused by the lack of mitochondrial PB2 protein although we cannot completely exclude the possibility that the N9D mutation had some effect on the accumulation on viral RNAs that might have affected replication in vivo. Interestingly, in contrast to the in vitro data, the lungs of mice infected with the WSN N9D virus expressed lower levels of IFN-β than mice infected with the WSN wild-type virus at both 3 and 5 dpi. These results suggest that the mitochondrial PB2 protein does not inhibit IFN-β expression in vivo. However, the decreased IFN-β expression in the lungs of mice infected with WSN N9D may be the result of reduced viral replication. At both 3 and 5 dpi, the titer of WSN N9D is approximately 10-fold lower than that of the WSN virus, and the amounts of IFN-β are reduced by half in the lungs of mice infected with WSN N9D. Therefore, we speculate that if the viral titers were identical, the levels of IFN-β in WSN N9D-infected mice would be higher than those in WSN-infected mice. In agreement with our observations, it was reported recently that a D9N mutation in the PB2 protein of an avian H5N1 virus resulted in increased virulence in mice (22). Although these authors have not analyzed the effect of this mutation on the mitochondrial association of the PB2 protein, our study would suggest that the D9N mutation changed the localization from nonmitochondrial to mitochondrial. Thus, the increase in virulence could have been caused by a more efficient inhibition of MAVS-mediated IFN-β expression due to the presence of the PB2 protein in mitochondria. Taken together, these data suggest that the mitochondrial association of the PB2 protein affects the virulence of influenza viruses without affecting their growth in cell culture, possibly via an effect on MAVS and, consequently, on IFN-β expression that affects the overall immune response and virulence.
The identification of the PB2 protein as an interferon antagonist suggests that influenza viruses use multiple strategies to evade antiviral host responses. The viral NS1 protein functions as an interferon antagonist by inhibiting IFN-β expression and signaling at multiple steps (16, 38). More recently, the viral RNA polymerase complex has been implicated in the inhibition of antiviral host responses by inducing the degradation of the large subunit of host RNA polymerase II (40, 52). Further support for the involvement of the RNA polymerase in the regulation of innate immune pathways was provided by the discovery of multiple physical associations between the RNA polymerase subunits and proteins of these pathways (44). Thus, influenza virus targets multiple components of diverse signaling pathways, indicating that viral regulation of cellular processes is extremely complex and involves multiple viral and host genes. The observation that blocking mitochondrial localization of the PB2 protein by mutagenesis results in higher levels of IFN-β expression in both a transient transfection system and in the context of viral infection argues in favor of an important role of the mitochondrial PB2 protein in the outcome of these complex regulatory processes.
It should be noted that MAVS was also found to play a role in the induction of apoptosis (27). In fact, it was shown previously that viruses with mutations in the PB2 protein inhibiting mitochondrial localization induced higher mitochondrial membrane potential loss than the wild-type virus encoding a mitochondrial PB2 protein (2). The loss of mitochondrial membrane potential, a key indicator of the induction of apoptosis (23), suggests that the PB2 protein may have a dual function within the mitochondria, being involved in the regulation of apoptotic pathways as well. Another influenza virus protein, PB1-F2, is known to associate with mitochondria and regulate apoptosis (4). It remains to be determined whether there is any interplay between PB1-F2 and PB2 in the regulation of apoptosis.
In summary, we have demonstrated that the PB2 subunit of the influenza virus RNA polymerase complex interacts with MAVS and inhibits MAVS-mediated IFN-β expression. A recombinant influenza virus encoding a nonmitochondrial PB2 protein was found to induce higher levels of IFN-β expression in cell culture than an isogenic virus encoding a mitochondrial PB2 and was attenuated in an animal model in comparison. These findings highlight the importance of the mitochondrial association of the PB2 protein in the viral life cycle and suggest that the PB2 protein is involved in the regulation of antiviral innate immune pathways. We also demonstrated that the PB2 proteins of H5N1 influenza viruses do not localize to the mitochondria, suggesting a possible mechanism by which H5N1 influenza viruses induce hypercytokinemia in humans, a factor that might be crucial in determining their virulence. Identification of the viral and cellular factors that determine the virulence of influenza viruses and understanding the molecular mechanisms of virus-host interactions are critical for developing control strategies for influenza viruses.
Acknowledgments
We thank S. Akira, A. García-Sastre, M. Iqbal, W. Barclay, and R. Harvey for materials and G. G. Brownlee for helpful discussions and critical reading of the manuscript. We extend our thanks to the Comparative Medicine Branch (NIH) for their help with animal care and husbandry.
This study was supported by grants from the Wellcome Trust (084349), the MRC (G0700848), and the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, NIH. K.M.G. was funded by the International Biomedical Research Alliance and OxCam Graduate Partnerships Program (NIH).
Footnotes
Published ahead of print on 10 June 2010.
REFERENCES
- 1.Almond, J. W. 1977. A single gene determines the host range of influenza virus. Nature 270:617-618. [DOI] [PubMed] [Google Scholar]
- 2.Carr, S. M., E. Carnero, A. Garcia-Sastre, G. G. Brownlee, and E. Fodor. 2006. Characterization of a mitochondrial-targeting signal in the PB2 protein of influenza viruses. Virology 344:492-508. [DOI] [PubMed] [Google Scholar]
- 3.Chen, H., R. A. Bright, K. Subbarao, C. Smith, N. J. Cox, J. M. Katz, and Y. Matsuoka. 2007. Polygenic virulence factors involved in pathogenesis of 1997 Hong Kong H5N1 influenza viruses in mice. Virus Res. 128:159-163. [DOI] [PubMed] [Google Scholar]
- 4.Chen, W., P. A. Calvo, D. Malide, J. Gibbs, U. Schubert, I. Bacik, S. Basta, R. O'Neill, J. Schickli, P. Palese, P. Henklein, J. R. Bennink, and J. W. Yewdell. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 7:1306-1312. [DOI] [PubMed] [Google Scholar]
- 5.Claas, E. C., A. D. Osterhaus, R. van Beek, J. C. De Jong, G. F. Rimmelzwaan, D. A. Senne, S. Krauss, K. F. Shortridge, and R. G. Webster. 1998. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351:472-477. [DOI] [PubMed] [Google Scholar]
- 6.de Jong, M. D., C. P. Simmons, T. T. Thanh, V. M. Hien, G. J. Smith, T. N. Chau, D. M. Hoang, N. V. Chau, T. H. Khanh, V. C. Dong, P. T. Qui, B. V. Cam, Q. Ha do, Y. Guan, J. S. Peiris, N. T. Chinh, T. T. Hien, and J. Farrar. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 12:1203-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng, T., J. Sharps, E. Fodor, and G. G. Brownlee. 2005. In vitro assembly of PB2 with a PB1-PA dimer supports a new model of assembly of influenza A virus polymerase subunits into a functional trimeric complex. J. Virol. 79:8669-8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelhardt, O. G., and E. Fodor. 2006. Functional association between viral and cellular transcription during influenza virus infection. Rev. Med. Virol. 16:329-345. [DOI] [PubMed] [Google Scholar]
- 9.Engelhardt, O. G., M. Smith, and E. Fodor. 2005. Association of the influenza A virus RNA-dependent RNA polymerase with cellular RNA polymerase II. J. Virol. 79:5812-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fodor, E., M. Crow, L. J. Mingay, T. Deng, J. Sharps, P. Fechter, and G. G. Brownlee. 2002. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76:8989-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fodor, E., L. Devenish, O. G. Engelhardt, P. Palese, G. G. Brownlee, and A. Garcia-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fodor, E., and M. Smith. 2004. The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex. J. Virol. 78:9144-9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fornek, J. L., L. Gillim-Ross, C. Santos, V. Carter, J. M. Ward, L. I. Cheng, S. Proll, M. G. Katze, and K. Subbarao. 2009. A single-amino acid-substitution in a polymerase protein of an H5N1 influenza virus is associated with systemic infection and impaired T-cell activation in mice. J. Virol. 83:11102-11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green, D. R., and J. C. Reed. 1998. Mitochondria and apoptosis. Science 281:1309-1312. [DOI] [PubMed] [Google Scholar]
- 15.Guilligay, D., F. Tarendeau, P. Resa-Infante, R. Coloma, T. Crepin, P. Sehr, J. Lewis, R. W. Ruigrok, J. Ortin, D. J. Hart, and S. Cusack. 2008. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat. Struct. Mol. Biol. 15:500-506. [DOI] [PubMed] [Google Scholar]
- 16.Hale, B. G., R. E. Randall, J. Ortin, and D. Jackson. 2008. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89:2359-2376. [DOI] [PubMed] [Google Scholar]
- 17.Hatefi, Y. 1985. The mitochondrial electron transport and oxidative phosphorylation system. Annu. Rev. Biochem. 54:1015-1069. [DOI] [PubMed] [Google Scholar]
- 18.Hatta, M., P. Gao, P. Halfmann, and Y. Kawaoka. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840-1842. [DOI] [PubMed] [Google Scholar]
- 19.Hatta, M., Y. Hatta, J. H. Kim, S. Watanabe, K. Shinya, T. Nguyen, P. S. Lien, Q. M. Le, and Y. Kawaoka. 2007. Growth of H5N1 influenza A viruses in the upper respiratory tracts of mice. PLoS Pathog. 3:1374-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiromoto, Y., Y. Yamazaki, T. Fukushima, T. Saito, S. E. Lindstrom, K. Omoe, R. Nerome, W. Lim, S. Sugita, and K. Nerome. 2000. Evolutionary characterization of the six internal genes of H5N1 human influenza A virus. J. Gen. Virol. 81:1293-1303. [DOI] [PubMed] [Google Scholar]
- 21.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981-988. [DOI] [PubMed] [Google Scholar]
- 22.Kim, J. H., M. Hatta, S. Watanabe, G. Neumann, T. Watanabe, and Y. Kawaoka. 2010. Role of host-specific amino acids in the pathogenicity of avian H5N1 influenza viruses in mice. J. Gen. Virol. 91:1284-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, R., M. Emi, K. Tanabe, S. Murakami, Y. Uchida, and K. Arihiro. 2006. Regulation and interplay of apoptotic and non-apoptotic cell death. J. Pathol. 208:319-326. [DOI] [PubMed] [Google Scholar]
- 24.Labadie, K., E. Dos Santos Afonso, M. A. Rameix-Welti, S. van der Werf, and N. Naffakh. 2007. Host-range determinants on the PB2 protein of influenza A viruses control the interaction between the viral polymerase and nucleoprotein in human cells. Virology 362:271-282. [DOI] [PubMed] [Google Scholar]
- 25.Law, H. K., C. Y. Cheung, H. Y. Ng, S. F. Sia, Y. O. Chan, W. Luk, J. M. Nicholls, J. S. Peiris, and Y. L. Lau. 2005. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood 106:2366-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le, Q. M., Y. Sakai-Tagawa, M. Ozawa, M. Ito, and Y. Kawaoka. 2009. Selection of H5N1 influenza virus PB2 during replication in humans. J. Virol. 83:5278-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lei, Y., C. B. Moore, R. M. Liesman, B. P. O'Connor, D. T. Bergstralh, Z. J. Chen, R. J. Pickles, and J. P. Ting. 2009. MAVS-mediated apoptosis and its inhibition by viral proteins. PLoS One 4:e5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, X. D., L. Sun, R. B. Seth, G. Pineda, and Z. J. Chen. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. U. S. A. 102:17717-17722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu, X., T. M. Tumpey, T. Morken, S. R. Zaki, N. J. Cox, and J. M. Katz. 1999. A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J. Virol. 73:5903-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuoka, Y., E. W. Lamirande, and K. Subbarao. 2009. The mouse model for influenza. Curr. Protoc. Microbiol. Chapter 15, unit 15G 3. doi: 10.1002/9780471729259.mc15g03s13. [DOI] [PubMed]
- 31.Mehle, A., and J. A. Doudna. 2009. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc. Natl. Acad. Sci. U. S. A. 106:21312-21316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehle, A., and J. A. Doudna. 2008. An inhibitory activity in human cells restricts the function of an avian-like influenza virus polymerase. Cell Host Microbe 4:111-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 34.Miotto, O., A. Heiny, T. W. Tan, J. T. August, and V. Brusic. 2008. Identification of human-to-human transmissibility factors in PB2 proteins of influenza A by large-scale mutual information analysis. BMC Bioinformatics 9(Suppl. 1):S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naffakh, N., P. Massin, N. Escriou, B. Crescenzo-Chaigne, and S. van der Werf. 2000. Genetic analysis of the compatibility between polymerase proteins from human and avian strains of influenza A viruses. J. Gen. Virol. 81:1283-1291. [DOI] [PubMed] [Google Scholar]
- 36.Neumann, G., T. Noda, and Y. Kawaoka. 2009. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459:931-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neupert, W., and J. M. Herrmann. 2007. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76:723-749. [DOI] [PubMed] [Google Scholar]
- 38.Rehwinkel, J., C. P. Tan, D. Goubau, O. Schulz, A. Pichlmair, K. Bier, N. Robb, F. Vreede, W. Barclay, E. Fodor, and C. Reis e Sousa. 2010. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140:397-408. [DOI] [PubMed] [Google Scholar]
- 39.Robb, N. C., M. Smith, F. T. Vreede, and E. Fodor. 2009. NS2/NEP protein regulates transcription and replication of the influenza virus RNA genome. J. Gen. Virol. 90:1398-1407. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez, A., A. Perez-Gonzalez, M. J. Hossain, L. M. Chen, T. Rolling, P. Perez-Brena, R. Donis, G. Kochs, and A. Nieto. 2009. Attenuated strains of influenza A viruses do not induce degradation of RNA polymerase II. J. Virol. 83:11166-11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roise, D., and G. Schatz. 1988. Mitochondrial presequences. J. Biol. Chem. 263:4509-4511. [PubMed] [Google Scholar]
- 42.Scholtissek, C., W. Rohde, V. Von Hoyningen, and R. Rott. 1978. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology 87:13-20. [DOI] [PubMed] [Google Scholar]
- 43.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122:669-682. [DOI] [PubMed] [Google Scholar]
- 44.Shapira, S. D., I. Gat-Viks, B. O. Shum, A. Dricot, M. M. de Grace, L. Wu, P. B. Gupta, T. Hao, S. J. Silver, D. E. Root, D. E. Hill, A. Regev, and N. Hacohen. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139:1255-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaw, M., L. Cooper, X. Xu, W. Thompson, S. Krauss, Y. Guan, N. Zhou, A. Klimov, N. Cox, R. Webster, W. Lim, K. Shortridge, and K. Subbarao. 2002. Molecular changes associated with the transmission of avian influenza a H5N1 and H9N2 viruses to humans. J. Med. Virol. 66:107-114. [DOI] [PubMed] [Google Scholar]
- 46.Shinya, K., S. Hamm, M. Hatta, H. Ito, T. Ito, and Y. Kawaoka. 2004. PB2 amino acid at position 627 affects replicative efficiency, but not cell tropism, of Hong Kong H5N1 influenza A viruses in mice. Virology 320:258-266. [DOI] [PubMed] [Google Scholar]
- 47.Subbarao, E. K., W. London, and B. R. Murphy. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 67:1761-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subbarao, K., A. Klimov, J. Katz, H. Regnery, W. Lim, H. Hall, M. Perdue, D. Swayne, C. Bender, J. Huang, M. Hemphill, T. Rowe, M. Shaw, X. Xu, K. Fukuda, and N. Cox. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279:393-396. [DOI] [PubMed] [Google Scholar]
- 49.Sugiyama, K., E. Obayashi, A. Kawaguchi, Y. Suzuki, J. R. Tame, K. Nagata, and S. Y. Park. 2009. Structural insight into the essential PB1-PB2 subunit contact of the influenza virus RNA polymerase. EMBO J. 28:1803-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taubenberger, J. K., A. H. Reid, R. M. Lourens, R. Wang, G. Jin, and T. G. Fanning. 2005. Characterization of the 1918 influenza virus polymerase genes. Nature 437:889-893. [DOI] [PubMed] [Google Scholar]
- 51.von Heijne, G. 1986. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 5:1335-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vreede, F. T., A. Y. Chan, J. Sharps, and E. Fodor. 2010. Mechanisms and functional implications of the degradation of host RNA polymerase II in influenza virus infected cells. Virology 396:125-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vreede, F. T., T. E. Jung, and G. G. Brownlee. 2004. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J. Virol. 78:9568-9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woodfin, B. M., and A. L. Kazim. 1993. Interaction of the amino-terminus of an influenza virus protein with mitochondria. Arch. Biochem. Biophys. 306:427-430. [DOI] [PubMed] [Google Scholar]
- 55.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727-740. [DOI] [PubMed] [Google Scholar]