Abstract
HIV-1 employs the cellular nuclear import machinery to actively transport its preintegration complex (PIC) into the nucleus for integration of the viral DNA. Several viral karyophilic proteins and cellular import factors have been suggested to contribute to HIV-1 PIC nuclear import and replication. However, how HIV interacts with different cellular machineries to ensure efficient nuclear import of its preintegration complex in dividing and nondividing cells is still not fully understood. In this study, we have investigated different importin α (Impα) family members for their impacts on HIV-1 replication, and we demonstrate that short hairpin RNA (shRNA)-mediated Impα3 knockdown (KD) significantly impaired HIV infection in HeLa cells, CD4+ C8166 T cells, and primary macrophages. Moreover, quantitative real-time PCR analysis revealed that Impα3-KD resulted in significantly reduced levels of viral 2-long-terminal repeat (2-LTR) circles but had no effect on HIV reverse transcription. All of these data indicate an important role for Impα3 in HIV nuclear import. In an attempt to understand how Impα3 participates in HIV nuclear import and replication, we first demonstrated that the HIV-1 karyophilic protein integrase (IN) was able to interact with Impα3 both in a 293T cell expression system and in HIV-infected CD4+ C8166 T cells. Deletion analysis suggested that a region (amino acids [aa] 250 to 270) in the C-terminal domain of IN is involved in this viral-cellular protein interaction. Overall, this study demonstrates for the first time that Impα3 is an HIV integrase-interacting cofactor that is required for efficient HIV-1 nuclear import and replication in both dividing and nondividing cells.
HIV-1 replicates productively in nondividing cells, such as monocytes (49, 61, 74), macrophages (23, 37, 59, 65, 71), dendritic cells (47, 64), and resting CD4+ T lymphocytes (86), through its ability to undergo active nuclear import by hijacking the host nuclear import machinery. Moreover, active nuclear import is not only required for nondividing-cell infection but also plays a role in the infection of proliferating cells (35). This ability of HIV-1 to enter the nucleus at interphase may contribute significantly to the very high replication rate observed in infected individuals (30, 70, 73) and is one of the crucial steps in HIV-1 replication, which plays a leading role in the establishment of infection and AIDS pathogenesis.
The viral double-stranded DNA (dsDNA), which associates with viral and cellular proteins, forms a high-molecular-mass nucleoprotein complex called the preintegration complex (PIC) in the cytosol of an infected cell (15, 51). This large complex has to actively enter the nucleus through the intact nuclear membrane in order to be integrated. At the molecular level, the active nuclear import ability of HIV-1 is attributed to the karyophilic properties of viral PICs. It is known that several viral nucleophilic proteins, including integrase (IN), matrix (MA), and Vpr, are associated with this nucleoprotein complex and play significant roles in HIV-1 nuclear import (8, 20, 22, 29, 53, 72). Moreover, a unique DNA structure in the viral cDNA, known as the central DNA flap, has also been implicated in this viral replication step (3, 17, 70, 84, 85). Interestingly, HIV-1 IN and the central DNA flap collectively contribute to HIV-1 nuclear import not only in nondividing cells but also in dividing cells. On the other hand, even though Vpr and MA have been shown to be involved in PIC nuclear import (29, 72, 81), later studies have questioned the significance of the MA or Vpr protein in this step: a virus with a complete deletion of the MA nuclear localization signal (NLS) can still support HIV-1 replication (58), and HIV-1 without Vpr was able to replicate efficiently in susceptible cells (20). Hence, in contrast to IN and the DNA flap, it is quite possible that MA and Vpr may act only as accessory factors in PIC nuclear import (56).
IN is a key enzymatic protein of 32 kDa produced by proteolytic cleavage of the Pol polyprotein and is incorporated into progeny viruses during viral assembly. The presence of IN was initially identified as an absolute requirement for genomic integration of viral cDNA. Later studies have demonstrated the involvement of IN at various stages of HIV replication, including nuclear import. However, the precise molecular mechanism by which HIV-1 IN contributes to PIC nuclear import is still not fully understood. In particular, it remains to be determined which host nuclear import pathway(s) is employed by HIV IN to ensure active HIV nuclear translocation. To date, at least three cellular nuclear import factors, including importin α1 (Impα1), Imp7, and transportin-SR2 (TRN-SR2), have been suggested to interact with HIV-1 IN and are involved in viral nuclear import (2, 10, 16, 20). Imp7, a member of the Impβ family, was initially identified as one of the receptors that mediates the nuclear import of ribosomal proteins and the glucocorticoid receptor (18, 31). Recently, our lab and Fassati et al. have demonstrated, by using cell-based coimmunoprecipitation and in vitro pulldown assays, respectively, that Imp7 is able to interact with HIV-1 IN (2, 16). However, the exact function of this host protein in HIV-1 nuclear import is still controversial (2, 16, 83, 87). TRN-SR2 is an Impβ family member and shuttles the serine/arginine (SR)-rich pre-mRNA splicing factors from the cytoplasm into the nucleus (34, 44, 45). TRN-SR2 has recently been implicated in HIV-1 nuclear import by two functional genomic screening studies (7, 42). Interestingly, using a yeast two-hybrid system and in vitro pulldown assays, Christ et al. showed that HIV IN interacts with TRN-SR2 and revealed that the knockdown (KD) of TRN-SR2 affected the formation of 2-long-terminal-repeat (2-LTR) circles and the nuclear translocation of fluorescently labeled PIC (10). However, the latest study of Krishnan et al. indicated that the HIV-1 capsid (CA), not IN, determines TRN-SR2 dependency during HIV-1 replication (43).
While the contribution of either the IN-Imp7 or the IN-TRN-SR2 interaction to HIV-1 nuclear import and viral replication remains unclear and controversial, another well-characterized Impα/Impβ-mediated nuclear import pathway (reviewed in references 24 and 46) was also suggested to be utilized by HIV-1 for efficient nuclear import of viral DNA. In fact, an earlier study by Gallay and coworkers suggested for the first time that HIV-1 may infect nondividing cells through the recognition of IN by the host importin/karyopherin pathway (20). Their study showed that HIV-1 was able to interact with the cellular import adaptor Impα1/Rch1 (a 32-amino-acid [aa] N-terminally truncated form of Impα1) in an in vitro pulldown assay and found that two IN mutants, the K186Q and Q214L Q216L mutants, were unable to bind to Rch1. This observation was further confirmed by another in vitro binding study by Hearps and Jans (27). However, the functional significance of this Impα1-mediated nuclear import pathway and the impact of the IN-Impα1 interaction on HIV-1 replication still remain to be elucidated. Intriguingly, it is known that the Impα family contains six isoforms in human cells: Impα1/Rch1 (13), Impα3/Qip1 (38, 62), Impα4 (38), Impα5 (12, 75, 76), Impα6 (38), and Impα7 (41). These six isoforms are grouped into three subfamilies (41). Sequence analysis revealed that different subfamilies have about 50% sequence identity. Within a subfamily, the identity is at least 80%. Some in vitro studies have indicated that various isoforms can recognize the same NLS-bearing proteins, but each isoform has a different binding efficiency (40, 41, 52, 77). However, the question of whether Impα isoforms can substitute for one another in vivo is still controversial, and several Impα isoforms have been shown to have specific substrates during nuclear import (5, 40, 50, 57, 63, 77). Importantly, some studies have provided evidence for a differentiation-associated alteration of Impα isoform gene expression in human cells. During neural differentiation of embryonic stem cells, Impα1 expression was drastically reduced to nearly undetectable levels, whereas high levels of Impα3 and Impα5 expression were induced (82). A similar observation was also made when human leukemia HL-60 cells were induced to differentiate into monocytes/macrophages (67) and when rat pancreatic AR42J cells were stimulated for differentiation toward a neuroendocrine phenotype (39). This appears to be a common phenomenon during cell differentiation. Since HIV-1 is capable of infecting both proliferating CD4+ T cells and differentiated macrophages, it is interesting to investigate how different Impα isoforms can contribute to HIV infection.
In the present study, we have investigated the contributions of different Impα isoforms to HIV-1 replication, and we demonstrate that Impα3 is required for efficient HIV infection of various susceptible cells, including primary macrophages. Quantitative PCR analysis revealed that Impα3-KD resulted in a significantly reduced level of viral 2-LTR circles, suggesting a role for Impα3 in HIV nuclear import. Moreover, we demonstrate that HIV-1 IN is able to interact with Impα3 and that a region (aa 250 to 270) in the C-terminal domain (CTD) of IN is involved in this viral-cellular protein interaction. All of these results provide evidence that Impα3 may be hijacked by HIV-1 IN for efficient HIV nuclear import and replication in both dividing and nondividing cells, including macrophages.
MATERIALS AND METHODS
Construction of different expression plasmids and HIV proviruses.
The cDNA encoding Homo sapiens importin α3 (karyopherin α4) was PCR amplified from pCMV6 Entry Impα3-myc (OriGene Technologies) using primers 5′-ATAGGATCCGTCGACGCGGACAACGAGAAACTGG-3′ (forward) and 5′-CTGCGGATCCAGCGGCCGCGTACGCGT-3′ (reverse), which introduced a BamHI and a NotI restriction site, respectively. The amplified fragment was subcloned into the SvCMVin-T7 vector at the 3′ end of a T7 tag by using BamHI and NotI restriction sites and was named T7-Impα3. To construct pAcGFP-IN, a cDNA fragment encoding full-length HIV IN was digested from yellow fluorescent protein-labeled IN (YFP-IN) (2) using the BglII/BamHI enzymes and was cloned into the pAcGFP1-C vector (Clontech) at the 3′ end of green fluorescent protein (GFP). To construct IN deletion mutants pAcGFP-IN1-212, pAcGFP-IN1-250, pAcGFP-IN1-270, andpAcGFP-IN50-288, the corresponding coding fragments were PCR amplified from AcGFP-IN by using specific primers and were subcloned into the pAcGFP-C expression vector at the 3′ end of GFP. The MA-YFP expression plasmid has been described previously (2), and Vpr-YFP was constructed by inserting an HxBru Vpr cDNA (no ATG) into the pCMV-YFP-N1 vector. The Moloney murine leukemia virus (MMLV)-based vector pFB-Luc was purchased from Stratagene. The HIV-1 proviral clone pNL4.3-Nef+/GFP+ (pNL4.3-GFP) and the HIV-1 provirus pNL-BruΔBgl/Luc have been described previously (2, 4). To generate the pNL-BruΔBgl/Luc/R− provirus, the ApaI-SalI region in pNL-BruΔBgl/Luc was replaced by the same fragment from a previous HIV provirus, HxBru-R− (80).
Antibodies and chemicals.
The antibodies and chemicals used in this study are as follows. The rabbit anti-GFP polyclonal antibody, the mouse anti-T7 monoclonal antibody, and the mouse anti-Impα3 antibody were obtained from Molecular Probes, Novagen, and Abcam, Inc., respectively. The mouse anti-HIV p24 and anti-CD4 monoclonal antibodies used in this study have been described previously (1, 80). The rabbit antibody against hemagglutinin (HA) was obtained from Sigma. The horseradish peroxidase (HRP)-conjugated anti-GFP antibody and anti-HA were purchased from Miltenyi Biotec. The HRP-conjugated donkey anti-rabbit IgG and sheep anti-mouse IgG were purchased from Amersham Biosciences. The rabbit anti-IN antibodies (catalog no. 757) and the purified recombinant HIV-1 NL4.3 IN protein (catalog no. 9420) were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. The Western blot detection ECL kit was purchased from Perkin-Elmer Life Science (Boston, MA). NP-40 Alternative and puromycin were obtained from Calbiochem.
Cell culture, transfection, establishment of Impα KD cell lines, and cell proliferation assay.
Human 293T cells, HeLa cells, and MLV packaging phoenix cells were maintained in continuous culture using Dulbecco's modified Eagle medium (DMEM) supplemented with 1% penicillin-streptomycin and 10% fetal calf serum (FCS). CD4+ C8166 T lymphocytes were maintained in RPMI 1640 medium containing 1% penicillin-streptomycin and 10% FBS. Peripheral blood mononuclear cells (PBMC) were isolated from the blood of healthy adult volunteers by sedimentation on a Ficoll (Lymphoprep; Axis-Shield) gradient, plated at the desired density in 12-well plates, and grown in DMEM containing 10% fetal bovine serum (FBS) and 10 ng/ml macrophage colony-stimulating factor (M-CSF; R&D Systems) for 1 week, until they differentiated into mature macrophages. The DNA transfection experiments were carried out in 293T cells or HeLa cells by a standard calcium phosphate DNA precipitation method or the Lipofectamine 2000 transfection protocol. At 48 h posttransfection, the cells were harvested and subjected to further experiments.
To stably knock down different Impα isoforms, pLKO.1 lentiviral vectors harboring short hairpin RNA (shRNA) targeting Impα1, Impα3, Impα5, and Impα7 were obtained from Open Biosystems. The hairpin consists of a 21-base stem and a 6-base loop. The sense oligonucleotide sequence targeting Impα1, Impα3, Impα5, and Impα7 are as follows: 5′-CTACCTCTGAAGGCTACACTT-3′, 5′-GCCCTCTCTTACCTTACTGAT-3′, 5′-GCAGTTATTCAAGCGGAGAAA-3′, and 5′-GCTGCCATGTTCGATAGTCTT-3′. First, vesicular stomatitis virus glycoprotein G (VSV-G)-pseudotyped lentiviral particles (LVPs) harboring each shRNA were produced in 293T cells by cotransfecting pLKO1-shRNA, an HIV packaging plasmid (pCMVΔR8.2), and a VSV-G expression plasmid. The pLKO.1 vector plasmid expressing scrambled shRNA (ScRNA) (obtained from Open Biosystems) was used to produce control vector particles. At 48 h posttransfection, the LVPs from supernatants were pelleted by ultracentrifugation (32,000 rpm for 1.5 h) and were used to transduce HeLa and C8166 T cells. After 48 h posttransduction, cells were cultured with fresh medium containing puromycin (0.5 to 2 μg/ml). After selection for 6 to 7 days, the knockdown levels of each Impα isoform were evaluated by Western blotting using corresponding antibodies, and meanwhile the cell lines were prepared for different analyses, including HIV infection experiments. To produce Impα3-KD and ScRNA-treated monocyte-derived macrophages (MDMs), the MDMs were treated with M-CSF for 1 week in 12-well plates and were then treated with LVPs harboring ScRNA or Impα3 shRNA twice in a 24-h interval. Both transduced and nontransduced MDMs were analyzed for their morphology and KD efficiency at day 4 posttransduction, as well as for HIV infection.
The 4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate (WST-1) assay (Roche) was used to measure the proliferation of Impα1-, Impα3-, Impα5-, and ScRNA-treated C8166 cell lines. Briefly, after different shRNA-transduced C8166 cell lines were generated for 6 to 7 days, each cell line was cultured at a density of 15 × 103 cells/well in a 96-well format and was maintained at 37°C. On different culture days, WST-1 was added to the culture at 10 μl/well and was incubated at 37°C for 4 h. After a thorough shaking for 1 min, the absorbance at 490 nm was recorded using a microplate (enzyme-linked immunosorbent assay [ELISA]) reader. To examine the expression of the CD4 receptor on the surfaces of C8166 T cells transduced with different shRNAs, equal amounts of cells were incubated with a monoclonal anti-CD4 antibody (OKT4) at 4°C, followed by the addition of a fluorescein isothiocyanate (FITC)-conjugated anti-mouse antibody. After three washes with phosphate-buffered saline (PBS), cell populations were analyzed by a fluorescence-activated cell sorter (FACS) (FACSCalibur; Becton Dickinson).
In vitro binding assay and cell-based coimmunoprecipitation (co-IP) experiments.
To test the HIV IN-Impα3 interaction by an in vitro binding assay, glutathione S-transferase (GST) and Impα3-GST fusion proteins expressed in Escherichia coli JM101 were purified as described previously (19). Equal amounts of purified GST and Impα3-GST were incubated with AcGFP- or AcGFP-IN-transfected 293T cell lysates. Then 100 μl of glutathione-Sepharose 4B beads (Amersham Biosciences) was added to the mixture and incubated for 2 h at 4°C. Beads were then washed five times with 0.25% NP-40 lysis buffer. Bound proteins were eluted with sodium dodecyl sulfate (SDS)-gel loading buffer, resolved by 12.5% SDS-polyacrylamide gel electrophoresis (PAGE), and detected by Western blotting using mouse anti-GFP antibodies.
To detect the interaction between Impα3 and HIV IN, Vpr, MA, or different IN deletion mutants in mammalian cells, 293T cells were cotransfected with T7-Impα3 and the AcGFP-INwt/mut, MA-YFP, or Vpr-YFP expression plasmid. At 48 h posttransfection, the cells were harvested and washed with PBS. Then 90% of the cells were lysed in NP-40 lysis buffer (199 medium containing 0.25% NP-40 Alternative and a protease inhibitor cocktail [Roche]) on ice for 30 min, and the lysates were clarified by centrifugation (13,000 rpm for 30 min at 4°C). After centrifugation, the clarified supernatant was subjected to immunoprecipitation using a rabbit anti-GFP antibody. The immunoprecipitates were then resolved by 10% SDS-PAGE, and the T7-Impα3 that was pulled down together with GFP-IN was analyzed by Western blotting using mouse anti-T7 antibodies. In addition, the presence of GFP-INwt/mut in the immunoprecipitates was detected by an anti-GFP antibody. To detect the expression of T7-Impα3 and/or GFP-IN wt/mut, 10% of the cells were lysed using Tris buffer containing 0.5% NP-40 and were subjected to Western blotting using mouse anti-T7 or anti-GFP antibodies, respectively.
To detect the binding of HIV-1 IN to endogenous Impα3 in HIV-1-infected CD4+ C8166 T cells, 10 × 106 CD4+ C8166T cells were infected with an HxBru or HxBru-IN-HA virus (2). After 72 h of infection, cells were lysed in NP-40 lysis buffer and were immunoprecipitated with an anti-HA antibody. Then the immunoprecipitates were resolved by 10% SDS-PAGE, followed by Western blotting using rabbit anti-Impα3 and anti-HA antibodies. Meanwhile, the intracellular p24 protein was checked by loading 1/30 of the cell lysate onto an 12% SDS-PAGE gel, followed by Western blotting using a mouse anti-p24 antibody.
Virus production and infection.
To study the effect of Impα knockdown on HIV replication, VSV-G-pseudotyped single-cycle-replicating pNL-Bru-ΔBgl/Luc/R+ and pNL-Bru-ΔBgl/Lucp24/R− viruses and HIV-1 pNL4.3-GFP were produced in 293T cells as described previously (1, 4). The virus titer was quantified with an HIV-1 p24 antigen capture assay kit (purchased from the NCI—Frederick AIDS Vaccine Program). A total of 0.5 × 106 Impα1-, Impα3-, or Impα5-KD or ScRNA-treated HeLa or C8166 T cells were infected with equal amounts of VSV-G-pseudotyped pNL-Bru-ΔBgl/Luc/R+ virus (5 to 10 ng p24). After 2 h of incubation, cells were washed twice and were then cultured in complete RPMI medium at 37°C for 48 h. Then 1 × 106 cells from each sample were collected and lysed in 50 μl of luciferase (Luc) lysis buffer (Promega), and 10 μl of the cell lysate was subjected to the Luc assay by using a POLARstar OPTIMA microplate reader (BMG Labtech, Germany). Luc activity was expressed as relative luciferase units (RLU). To test the effect of Impα3-KD on HIV infection in macrophages, the macrophages were first transduced by LVPs expressing an Impα3 shRNA or ScRNA. After 4 days of transduction, cells were infected with a VSV-G-pseudotyped pNL-BruΔBgl/Luc/R+ or pNL-BruΔBgl/Luc/R− virus (30 ng of p24/sample). After overnight infection, the cells were washed, cultured in complete DMEM, and harvested at different time intervals. All cells were lysed in Luc lysis buffer, and equal amounts of cell lysates (adjusted by protein concentration) were subjected to the Luc assay.
To monitor HIV-1 replication kinetics in Impα3-KD C8166 cells, pNL4.3-GFP+ viruses (multiplicity of infection [MOI], 0.02) were used to infect C8166 T cells, and at different time points after infection, HIV replication levels were monitored by measuring the HIV-1 Gag antigen present in the supernatant using an HIV-1 p24gag ELISA. In addition, at day 4 postinfection, the GFP-positive HIV-1 infected cells were fixed with 4% paraformaldehyde in PBS and were observed by fluorescence microscopy. Meanwhile, the levels of HIV p24gag protein in the infected cells were analyzed by Western blotting with an anti-p24 antibody.
For the production of VSV-G-pseudotyped MLV vector particles, an MLV-based retroviral plasmid (pFB-Luc; purchased from Stratagene) containing the luciferase gene and a VSV-G plasmid were cotransfected into the MLV packaging phoenix cell line. After 48 h, supernatants containing MLV particles were filtered through a 0.45-μm-pore size filter and were used to infect CD4+ C8166 T cells. At 48 and 72 h postinfection, the level of MLV infection was monitored by measurement of Luc activity.
Real-time quantitative PCR (RQ-PCR) analysis.
Stable C8166 cell lines with Impα3 knockdown or ScRNA were infected with the pNL4.3-GFP virus stock, which was produced from C8166 T cells infected with the same virus strain. pNL4.3-GFP viruses were used to infect C8166 T cells, followed by PCR analysis, in order to avoid carryover plasmid DNA contamination. Heat-inactivated virus (pretreated at 65°C for 30 min) was used as a negative control for infection. After infection, cells were incubated for 2 h at 37°C, washed twice, and cultured in complete RPMI medium. To restrict viral replication to a single cycle, azidothymidine (AZT) was added to the cell cultures after 12 h of infection. At 12 and 24 h postinfection, 1 × 106 infected cells were harvested, and DNA was isolated using a QIAamp blood DNA minikit (Qiagen). The total levels of HIV-1 DNA, 2-LTR circles, and integrated DNA were quantified in an Mx3000P real-time PCR system (Stratagene, CA) with the following protocols. The quantitative PCR for total HIV-1 DNA was carried out using primers targeting the Gag region outside the HIV genomic sequence of PLKO.1 shRNA (Open Biosystems). The reaction was carried out in a 20-μl final volume, consisting of 1× FastStart DNA Master SYBR green I (Roche Diagnostics, Germany) and 0.2 μM (each) sense (TD-Gag Fr [5′-ATCAAGCAGCCATGCAAATG-3′]) and antisense (TD-Gag-Rv [5′-CTGAAGGGTACTAGTAGTTCC-3′]) primers. The 2-LTR circle DNA was quantified using primers MH535 (5′-AACTAGGGAACCCACTGCTTAAG-3′ and MH536 (5′-TCCACAGATCAAGGATATCTTGTC-3′) and the 2LTR probe (3′-6-carboxyfluorescein [FAM]-ACACTACTTGAAGCACTCAAGGCAAGCTTT-6-carboxytetramethylrhodamine [TAMRA]-5′), targeting LTR-LTR junctions, as described previously (9). The integrated viral DNA was quantified by an Alu-LTR-nested PCR procedure as described previously (68), with minor modifications. Briefly, the first round of PCR was carried out using primers targeting Alu (Alu-Fr [5′-TCCCAGCTACTCGGGAGGCTGAGG-3′]) and the Gag region (Int-Gag [5′-GTCCAGAATGCTGGTAGGGCTATACA-3′]) outside the HIV genomic sequence of the PLKO.1-ShRNA vector. The second round of PCR was carried out using primers TD-Gag Fr and TD-Gag Rv as described above. A first-round PCR without Taq DNA polymerase was followed by a second round of PCR to rule out background amplification from unintegrated viral DNA. Total DNA, 2-LTR circle DNA, and integrated DNA were expressed as copy numbers per cell, with the DNA template normalized by β-globin gene amplification using primers Bglo1 (forward) (5′-CAACTTCATCACGTTCACC-3′) and glob2 (reverse) (5′-GAAGAGCCAAGGACAGGTAC-3′).
RESULTS
Inhibitory effects of shRNA-mediated knockdown of Impα1, Impα3, Impα5, and Impα7 on VSV-G-pseudotyped HIV-1 infection in HeLa cells.
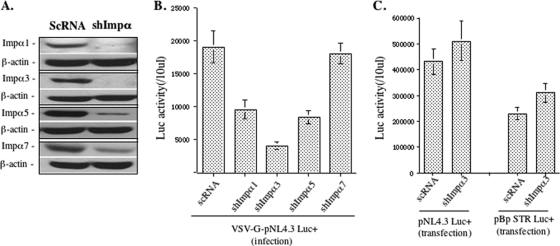
In this study, we first investigated the effects of different Impα isoforms on HIV-1 infection. A shRNA-lentiviral vector system (pLKO1 lentiviral vector) was used to knock down Impα1, Impα3, Impα5, or Impα7 in HeLa cells, and four stable Impα knockdown (Impα-KD) cell lines were generated. Briefly, lentiviral vector particles (LVPs) containing Impα1, Impα3, Impα5, or Impα7 shRNA produced from 293T cells, as described in Materials and Methods, were used to transduce 0.2 × 106 HeLa cells. In parallel, LVPs containing a scrambled shRNA (ScRNA) were used as a control. After 48 h, transduced cells were cultured in DMEM containing puromycin (2 μg/ml) for 6 days. Then the knockdown of each Impα was verified by immunoblotting with the corresponding antibodies. It is worth noting that we could not compare the relative expression levels of distinct Impα isoforms due to the different affinities of antibodies. However, the results clearly showed that approximately 90 to 95% of Impα1, Impα3, and Impα5 was knocked down, and for Impα7, the protein level was reduced to about 30% compared to the level in ScRNA-transduced cells (Fig. 1A).
FIG. 1.
shRNA-mediated knockdown of Impα1, Impα3, Impα5, and Impα7 in HeLa cells inhibited infection with VSV-G-pseudotyped HIV-1. (A) HeLa cells were transduced with lentiviral vectors harboring Impα1, Impα3, Impα5, or Impα7 shRNA or a scrambled shRNA (ScRNA) and were selected with puromycin (2 μg/ml) for 7 days. Cells were subsequently analyzed for Impα1, Impα3, Impα5, or Impα7 knockdown (KD) by Western blotting using the corresponding antibodies. β-Actin is included as an internal control. The results shown are representative of three different experiments. (B) A total of 0.2 × 106 HeLa cells with KD of Impα1, Impα3, Impα5, or Impα7, or transduced with a vector harboring ScRNA, were challenged by VSV-G-pseudotyped, luciferase (Luc)-expressing HIV-1 (pNL-BruΔBgl/Luc) (10 ng virus-associated p24 antigen). The same amount of cells was collected after 48 h of infection and analyzed for Luc activity. (C) HeLa cells with Impα3-KD or ScRNA were transfected with the pNL-BruΔBgl/Luc provirus or an MMLV vector carrying a luciferase gene plasmid (pBp-STR Luc+). Forty-eight hours later, the same amount of cells was collected and analyzed for Luc activity. Data shown are means and standard errors and are representative of the results for duplicate samples from two independent experiments.
We then tested a VSV-G-mediated single-cycle HIV infection in each Impα-KD or ScRNA cell line by infecting with equal amounts of VSV-G-pseudotyped, firefly luciferase-expressing HIV-1 (pNL-BruΔBgl/Luc) (10 ng of HIV p24 antigen). After 48 h, the HIV-1 infection level was monitored by measuring the luciferase (Luc) activity from an equal amount of infected cells (Fig. 1B). Compared with ScRNA cells, we observed about 50% inhibition of HIV-1 infection in Impα1- or Impα5-KD cells, and a 3.5-fold reduction of infection in Impα3-KD cells. In contrast, no decrease in Luc activity was found in Impα7-KD cells (Fig. 1B). Since the luc gene was used to replace the nef gene in the virus, these findings suggest that Impα1, Impα3, and Impα5, especially Impα3, may be involved in the steps before or at early gene expression during the HIV replication cycle.
Since Impα3-KD was shown to have a significant inhibitory effect on HIV infection, we next tested whether Impα3-KD-mediated inhibition was due to its effect on HIV transcription and gene expression. Impα3-KD and ScRNA-treated HeLa cells were transfected with pNL-BruΔBgl/Luc proviral DNA, and the Luc activity in the transfected cells was measured at 48 h posttransfection. Interestingly, Impα3-KD cells displayed a Luc activity level similar to that of control cells, suggesting that Impα3-KD did not affect HIV gene expression, at least for Nef (Fig. 1C). Meanwhile, an MMLV vector plasmid (pBpSTR-Luc+) in which Luc expression is driven by a cytomegalovirus (CMV) promoter was also transfected into Impα3-KD HeLa cells. Consistently, Luc activity was not reduced in Impα3-KD cells (Fig. 1C). Taken together, these results provide evidence to suggest that knockdown of Impα3 in HIV-1 target cells might specifically impair early steps during HIV-1 replication, possibly at reverse transcription, nuclear import, and/or integration.
Impα3-KD in CD4+ C8166 T cells significantly inhibited HIV-1 infection.
The next question we asked was whether knockdown of various Impα isoforms in HIV-susceptible cells would have similar effects on HIV infection. Impα1, Impα3, or Impα5 shRNAs were introduced into CD4+ C8166 T cells by use of the LVPs described above. After 1 week of selection with puromycin, Western blotting showed that the expression of Impα1, Impα3, and Impα5 was reduced as much as 90% or more (Fig. 2A). Moreover, specific knockdown of any one of these isoforms did not affect the expression of the other Impα isoforms. It was noticed that the efficient knockdown of Impα lasted only for about 3 to 4 weeks. Subsequently, the expression of the targeted Impα isoform would be increased even in the presence of puromycin (data not shown). Thus, after puromycin selection for 1 week, each of the cell lines was prepared for different analyses. First, the effect of each Impα-KD on C8166 T-cell growth was monitored by using a WST-1 assay (Fig. 2B). The results showed that the Impα5-KD C8166 T-cell line grew at a rate similar to that of ScRNA-transduced cells. However, the growth of C8166 T cells with Impα1-KD or Impα3-KD was moderately inhibited, even though the knockdown of these importin isoforms was not lethal to the cells.
FIG. 2.
Effects of Impα1-, Impα3-, and Impα5-KD on the infection of CD4+ C8166 cells with VSV-G-pseudotyped HIV-1. (A) C8166 cells were transduced with lentiviral vectors harboring either Impα1, Impα3, or Impα5 shRNA or a scrambled shRNA and were selected with puromycin (0.5 μg/ml) for 7 days. Cells were subsequently analyzed for Impα1, Impα3, or Impα5 expression by Western blotting using the corresponding antibodies. β-Actin was included as an internal control. The results shown are representative of three different experiments. (B) A WST assay was performed to determine the growth of cell populations with Impα1-, Impα3-, or Impα5-KD, or with ScRNA, at different time points, as indicated. (C) A total of 0.5 × 106 C8166 T cells with Impα1-, Impα3-, or Impα5-KD, or with ScRNA, were challenged with VSV-G-pseudotyped, Luc-expressing HIV-1 (pNL-Bru-Luc+/E−) (10 ng virus-associated p24 antigen). After 48 h of infection, equal amounts of cells were collected and were analyzed for Luc activity. (D) C8166 T cells with Impα3-KD or with ScRNA were infected with a VSV-G-pseudotyped MLV vector containing the luc gene, and the Luc activity of infected cells was checked at 48 and 72 h postinfection. Data shown are means and standard errors and are representative of the results for duplicate samples from a typical experiment, which were confirmed in two other independent experiments.
To determine the contribution of Impα1, Impα3, or Impα5 to HIV-1 infection in C8166 T cells, we first infected the same amount of Impα-KD C8166 T cells with VSV-G-pseudotyped, luciferase-expressing HIV-1, as described above. The data showed that knockdown of Impα3 inhibited HIV-1 replication 4-fold, while the shRNA-mediated silencing of Impα1 or Impα5 reduced Luc activity only about 50 to 60% from that in shRNA-transduced cells (Fig. 2C). These results are consistent with our observation for HeLa cells with KD of Impα (Fig. 1B). To further test whether such inhibitory effects were only specific for HIV, we also tested a VSV-G-pseudotyped MLV vector infection in Impα3-KD and ScRNA-transduced C8166 T-cell lines. Since a luc gene was inserted into the MLV vector, MLV infection was also monitored by the measurement of Luc activity at 48 and 72 h postinfection (Fig. 2D). The results showed that the MLV infection in dividing Impα3-KD C8166 T cells was reduced approximately 40 to 50% from that in ScRNA-transduced cells. All of these studies indicate that Impα3-KD in CD4+ T cells had a profound inhibitory effect on HIV-1 infection and a modest inhibitory effect on MLV infection in Impα3 KD cells. It is possible that the modest impairment of MLV infection is at least partially due to slower cell proliferation, since the MLV PIC requires cell division.
To further investigate HIV-1 replication and spread in Impα3-KD C8166 T cells, we infected 0.5 × 106 Impα3-KD or ScRNA-transduced C8166 cells with equal amounts of wild-type HIV-1 (pNL4.3-GFP+) (MOI, 0.02). At different time intervals, the HIV replication kinetics were monitored by measuring HIV p24gag production in the supernatant with an HIV p24 ELISA. The results showed that HIV-1 replication progressed quickly in ScRNA-transduced cells and that viral replication peaked at day 4, at which time cells were rapidly killed by HIV-induced cytopathic effects and syncytium formation. In contrast, in Impα3-KD cells, viral infection was significantly attenuated, and no viral p24gag was detected in the first 3 days. At days 4 and 5, only low levels of p24 were detected by ELISA (Fig. 3A) and by Western blotting (Fig. 3B, right). Similar results were obtained by fluorescence microscopy (Fig. 3B, left). At day 6 postinfection, viral replication started to increase. To exclude the possibility that Impα3-KD-mediated attenuation of HIV infection was the result of altered expression of the HIV-1 receptor on the cell surface, we measured the levels of the CD4 receptor on Impα3-KD and ScRNA-transduced C8166 cells by anti-CD4 staining and FACS analysis. No difference was observed between these two transduced cell lines (Fig. 3C), indicating that Impα3-KD had no effect on the expression of the CD4 receptor on the cell surface. Taken together, our results indicate that Impα3-KD in CD4+ C8166 T cells efficiently inhibited HIV-1 replication and spread.
FIG. 3.
Impα3 knockdown significantly inhibited the infection of CD4+ C8166 T cells with wild-type HIV-1. (A) Inhibitory effect of Impα3-KD on HIV-1 replication and progression in CD4+ C8166 T cells. A total of 0.5 × 106 Impα3-KD or ScRNA-transduced C8166 cells were infected with HIV-1 pNL4.3-GFP at an MOI of 0.02. At various days postinfection (x axis), the supernatant was collected, and HIV p24gag levels were measured in order to monitor virus replication. (B) After 4 days of infection, the infected (GFP-positive) Impα3-KD and ScRNA-transduced C8166 cells were visualized by fluorescence microscopy (left), and the viral protein p24gag was detected by Western blotting using an anti-p24 antibody (right). (C) Levels of expression of the CD4 receptor on the surfaces of Impα3-KD, ScRNA-transduced, or normal (Mock) C8166 T cells were observed by anti-CD4 staining and flow cytometry analysis.
shRNA-mediated Impα3 depletion resulted in reduced HIV-1 2-LTR circle formation without affecting reverse transcription.
To pinpoint the step(s) at which HIV-1 replication was affected in Impα3-KD cells, we examined HIV-1 reverse transcription, 2-LTR circles, and integrated-DNA levels by RQ-PCR analysis. First, C8166 T cells with Impα3-KD or ScRNA were infected with the pNL4.3-GFP+ virus. After 2 h of infection, the cells were washed and cultured in RPMI medium. To restrict viral replication to a single cycle, AZT was added to the culture after 12 h of infection. At 12 and 24 h postinfection, cells were harvested and DNA was extracted by using a QIAamp DNA blood minikit to quantify late reverse transcripts, 2-LTR circles, and the integrated provirus, as described in Materials and Methods. The results showed that similar levels of viral cDNA synthesis were detected in Impα3-KD and ScRNA-transduced C8166 T cells (Fig. 4A). However, a 4- to 5-fold reduction in the level of 2-LTR circles was steadily detected in HIV-infected Impα3-KD cells at both 12 and 24 h postinfection (Fig. 4B). Meanwhile, the integrated proviral DNA in HIV-1-infected Impα3-KD and ScRNA-transduced C8166 T cells was analyzed. At 12 h postinfection, no integrated proviral DNA could be detected in either Impα3-KD or ScRNA-transduced cell samples, but at 24 h postinfection, the level of integrated proviral DNA observed in Impα3-KD cells was approximately 7-fold lower than that in ScRNA-transduced cells (Fig. 4C). These significant decreases in the levels of both 2-LTR circles and integrated proviral DNA were well correlated with the attenuated HIV replication in Impα3-KD C8166 cells (Fig. 3). All of these data suggest that HIV-1 replication was affected at a step(s) after reverse transcription, which included HIV DNA nuclear import.
FIG. 4.
Effects of Impα3-KD on HIV reverse transcription, the formation of 2-LTR DNA, and the level of integrated provirus. Impα3-KD and ScRNA-transduced C8166 T cells were infected with HIV-1 pNL4.3-GFP for 2 h, washed twice, and cultured in RPMI medium. At distinct time points after infection (12 and 24 h), DNA was extracted from infected cells, and HIV-1 late reverse transcription products (A), HIV-1 2-LTR circles (B), and the integrated DNA level (C) were analyzed by RQ-PCR using the corresponding primers as described in Materials and Methods. Data shown are means and standard errors and are representative of the results for duplicate samples from a typical experiment, which were confirmed in two other independent experiments.
Interaction between Impα3 and HIV-1 IN.
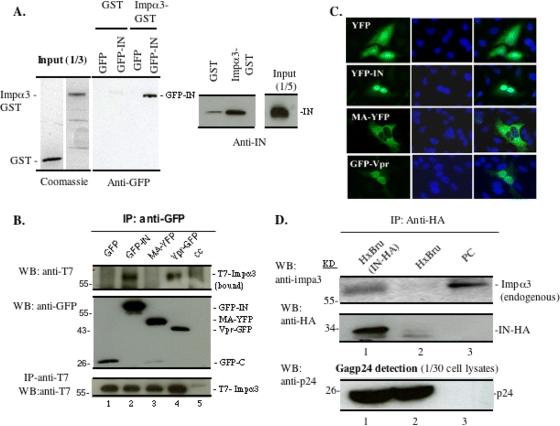
The results reported above suggest that Impα3 is required for HIV nuclear import. However, how Impα3 is involved in this HIV replication step is still an open question. It is possible that during the early stages of viral replication, HIV may be able to recruit this cellular importin by using its nucleophilic proteins to facilitate PIC nuclear import. Since HIV IN plays an important role during HIV nuclear import (1, 2, 6, 10, 16, 20), we first used an in vitro pulldown assay to test for an interaction between HIV IN and Impα3. The result showed that recombinant Impα3-GST, but not GST alone, bound to GFP-IN that was overexpressed in 293T cells (Fig. 5 A). To test whether HIV IN could bind directly to Impα3 in vitro, similar amounts of purified GST and GST-Impα3 were incubated with purified recombinant HIV-1 IN in 0.25% NP-40 lysis buffer for 4 h at 4°C, followed by an additional 1-h incubation with glutathione-Sepharose 4B beads. Then the bound protein complex was eluted out with 100 mM glutathione and was loaded onto a 12.5% SDS-PAGE gel, followed by Western blot analysis with anti-IN antibodies. The results showed that the purified HIV-1 IN was able to interact with GST-Impα3 but not with GST alone (Fig. 5A, right). Thus, IN may bind to Impα3 through a direct protein-protein interaction. Then we checked the interaction between Impα3 and IN by a cell-based coimmunoprecipitation (co-IP) assay. In parallel, the interaction between Impα3 and other HIV-1 karyophilic proteins, MA and Vpr, was also tested. Briefly, a T7-tagged human Impα3 (CMV-T7-Impα3) expresser was cotransfected with a plasmid expressing GFP, GFP-IN, MA-YFP, or Vpr-YFP into 293T cells, as indicated in Fig. 5B. After 48 h, cells were lysed with NP-40 buffer (0.25% NP-40 in 199 medium). The lysates were subsequently subjected to immunoprecipitation (IP) using a rabbit anti-GFP antibody, and the coprecipitated Impα3 was detected by Western blotting with a mouse anti-T7 antibody (Fig. 5B, top). The results showed that T7-Impα3 could be detected in the GFP-IN and Vpr-YFP IP samples, but not in the GFP or the MA-YFP IP sample. These data indicate that both HIV-1 IN and Vpr, but not GFP or MA-YFP, were able to specifically interact with Impα3 in 293T cells. Also, we investigated the intracellular localizations of these fusion proteins in HeLa cells and showed that while GFP-IN and Vpr-YFP were localized predominately in the cell nucleus, MA-YFP was located outside the nucleus (Fig. 5C), confirming a previous observation (14).
FIG. 5.
Impα3 interacts with HIV-1 integrase (IN) in vitro and in cotransfected 293T cells and HIV-infected C8166 T cells. (A) (Left) Cell lysates from 293T cells expressing GFP or GFP-IN were incubated with either GST alone or a recombinant GST-Impα3 fusion protein. After GST pulldown, the bound proteins were analyzed by Western blotting (WB) with an anti-GFP antibody. (Right) Equal amounts of GST and GST-Impα3 were incubated with purified recombinant HIV-1 IN followed by GST pulldown and were analyzed on an SDS-PAGE gel by Western blotting with rabbit anti-IN antibodies. (B) Impα3 interacts with HIV-1 IN and Vpr in 293T cells. A full-length human T7-Impα3 expression plasmid was cotransfected with a plasmid expressing GFP, GFP-IN, MA-YFP, or Vpr-YFP into 293T cells. (Top) After 48 h of transfection, 90% of the transfected cells were lysed in 0.25% NP-40 buffer and were immunoprecipitated with a rabbit anti-GFP antibody. The immunoprecipitates were resolved using 10% SDS-PAGE, and the bound T7-Impα3 was detected by WB with an anti-T7 antibody. (Center) The presence of GFP-INwt/mut in immunoprecipitates was detected by an anti-GFP antibody. (Bottom) To check protein expression, the remaining 10% of the transfected cells were lysed with 0.5% NP-40, run directly on a 10% SDS-PAGE gel, and probed with anti-T7 antibodies. (C) HeLa cells were transfected with a GFP, GFP-IN, MA-YFP, or Vpr-YFP plasmid. After 48 h, cells were fixed and labeled with an anti-GFP antibody, and the nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Cells were then analyzed by fluorescence microscopy (with a 40× objective lens). (D) Impα3 interacts with HIV-1 IN in virus-infected C8166 T cells. Approximately 10 × 106 C8166 T cells were infected with an HxBru or HxBru-IN-HA virus. (Top) Seventy-two hours after infection, cells were lysed and immunoprecipitated with a rabbit anti-HA antibody, and the bound endogenous Impα3 was detected by Western blotting using rabbit anti-Impα3. The normal C8166 cell lysate was loaded as a positive control (PC). (Center) The immunoprecipitated HA-IN was also detected by WB using a mouse anti-HA antibody. (Bottom) Lysates from the infected C8166 cells were directly loaded onto an SDS-PAGE gel to detect HIV p24gag protein by WB using an anti-p24 antibody.
To further test whether the interaction between IN and endogenous Impα3 occurs during HIV-1 infection, we infected 10 × 106 C8166 T cells with either an HxBru or an HxBru-IN-HA virus (2), and at 72 h after infection, cells were lysed and immunoprecipitated with an anti-HA antibody. Then immunoprecipitates were resolved by 10% SDS-PAGE followed by Western blotting using rabbit anti-Impα3 and anti-HA antibodies, respectively. These results revealed that endogenous Impα3 was coprecipitated by IN-HA in the HxBru-IN-HA-infected sample but not in the HxBru-infected sample (Fig. 5D, top and center, compare lanes 1 and 2). To confirm that similar levels of infection occurred in the two infected cultures, the HIV p24gag level in each sample was examined, and the levels in the two infected samples were found to be similar (Fig. 5D, bottom, compare lanes 2 and 3). These results clearly demonstrated the interaction between Impα3 and viral IN in HIV-1-infected T cells.
The C-terminal region of HIV-1 IN is involved in its interaction with Impα3.
We next tried to delineate the region(s) of IN required for its interaction with Impα3. First, we cotransfected either wild-type GFP-IN or the N- or C-terminal domain deletion mutant (GFP-IN50-288 or GFP-IN1-212) with T7-Impα3 in 293T cells and tested their binding abilities by a co-IP assay. Interestingly, the results revealed that GFP-IN50-288 bound to T7-Impα3 as efficiently as did GFP-IN (Fig. 6A, lanes 2 and 4). However, the C-terminal deletion mutant GFP-IN1-212 was unable to bind to T7-Impα3 (Fig. 6A, lane 3), suggesting that the C-terminal domain (CTD) of IN may be required for Impα3 binding.
FIG. 6.
Requirement of the C-terminal domain of HIV-1 IN for IN-Impα3 interaction. (A) Plasmids encoding either GFP, GFP-IN, the C-terminal deletion mutant GFP-IN1-212, or the N-terminal deletion mutant GFP-IN50-288 were each cotransfected with the T7-Impα3 expression plasmid into 293T cells, and the interaction of each form of IN with Impα3 was analyzed by anti-GFP immunoprecipitation followed by Western blotting (WB) with an anti-T7 antibody. (Top) Bound T7-Impα3 was detected by IP with an anti-GFP antibody followed by WB using an anti-T7 antibody. (Center) The immunoprecipitated GFP, wild-type GFP-IN, and GFP-IN deletion mutants were detected by WB with an anti-GFP antibody. (Bottom) T7-Impα3 expression was detected with an anti-T7 antibody. (B) Plasmids expressing wild-type GFP-IN or the different IN C-terminal deletion mutants, including GFP-IN1-212, GFP-IN1-250, and GFP-IN1-270, were each cotransfected with a T7-Impα3 expression plasmid into 293T cells, and the Impα3 binding of each IN mutant was analyzed by the co-IP assay as described in Materials and Methods. (Top) Bound T7-Impα3 was detected by IP with anti-GFP followed by WB using anti-T7 antibodies. (Center) The immunoprecipitated GFP, wild-type GFP-IN, and GFP-IN deletion mutants were detected by WB with an anti-GFP antibody. (Bottom) Total T7-Impα3 expression in the transfected cells was examined by Western blotting using an anti-T7 antibody. (C). Intracellular localizations of different GFP-IN mutants. HeLa cells were transfected with plasmids expressing GFP, GFP-IN, GFP-IN1-212, GFP-IN1-250, or GFP-IN1-270. After 48 h, cells were fixed and labeled with an anti-GFP antibody, and the nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Cells were then analyzed by fluorescence microscopy (with a 40× objective lens).
To further clarify the region(s) in the CTD of IN that may contribute to the binding to Impα3, we constructed another two C-terminal deletion mutants, GFP-IN1-250 and GFP-IN1-270, and tested their binding to T7-Impα3. As shown in Fig. 6B, the GFP-IN1-212 and GFP-IN1-250 mutants were unable to bind efficiently to Impα3. However, the GFP-IN1-270 mutant restored a strong binding ability. Meanwhile, we examined the intracellular localization of each GFP-IN mutant by transfecting each of them into HeLa cells. After 48 h of transfection, cells were fixed, and the intracellular localization of each GFP-IN mutant was visualized by an anti-GFP antibody followed by an FITC-conjugated anti-rabbit antibody. The results revealed that while GFP-IN1-212 and GFP-IN1-250 were localized outside the nucleus, GFP-IN1-270 was located primarily in the nuclei of transfected cells (Fig. 6C). All of these results suggest that a region encompassing aa 251 to 270 in the C-terminal domain of IN is required both for Impα3 binding and for nuclear localization.
Importance of Impα3 for HIV-1 infection in primary macrophages.
Productive infection of nondividing cells, such as macrophages, depends on the active nuclear import of the HIV-1 PIC. Since the results described above provide evidence for the requirement of Impα3 for HIV nuclear import, it is important to investigate whether Impα3 is also necessary for the infection of primary macrophages by HIV-1. First, peripheral blood mononuclear cells (PBMC) were isolated from healthy donors and cultivated in DMEM supplemented with 10% FBS for 2 h, and nonadherent cells were removed. The adherent cells were cultured for 1 week in the presence of macrophage colony-stimulating factor (M-CSF) (10 ng/ml) to allow differentiation into monocyte-derived macrophages (MDMs) (Fig. 7A). Then the MDMs were transduced with equal amounts of lentiviral vectors containing either scrambled shRNA or Impα3 shRNA. After 4 days of transduction, both transduced and nontransduced MDMs were analyzed for their morphology (Fig. 7A) and the efficiency of Impα3-KD. The results showed that at 4 days after the introduction of the Impα3 shRNA, the Impα3 expression level in macrophages was suppressed by 75 to 80% (Fig. 7A, top).
FIG. 7.
Inhibitory effect of Impα3-KD on HIV-1 infection in human primary macrophages. (A) MDMs from different donors were transduced with equal amounts of lentiviral vectors carrying ScRNA or Impα3 shRNA. (Top) At day 4 posttransduction, endogenous Impα3 expression in Impα3 shRNA- and ScRNA-treated macrophages was detected by Western blotting using an anti-Impα3 antibody. β-Actin was included as an internal control. (Bottom) Nontransduced (a), ScRNA-transduced (b), and Impα3 shRNA-transduced (c) MDMs were observed under a microscope with a 20× objective lens. (B and C) Impα3 shRNA- or ScRNA-transduced MDMs from donor 1 or donor 2 were infected with VSV-G-pseudotyped pNL-Bru-Luc+/R+ HIV-1. Equal amounts of cells were collected and analyzed for Luc activity at various days postinfection (B) or at day 7 (C). (D) Impα3 shRNA- or ScRNA-transduced MDMs from donor 3 were infected with a VSV-G-pseudotyped R+ or R− pNL-Bru-Luc+ virus. After 7 days of infection, equal amounts of cells were collected and analyzed for Luc activity.
Then Impα3-KD and ScRNA-transduced macrophages from donors 1 and 2 were infected with VSV-G-pseudotyped pNL-BruΔBgl/Luc/R+ HIV-1 (30 ng virus-associated p24 antigen), and at different time points (3, 5, 7, and 9 days for donor 1; day 7 for donor 2) after infection, macrophages were lysed and subjected to Luc assays to analyze viral replication (Fig. 7B and C). In ScRNA-transduced macrophages, the Luc activity reached a peak at day 7 after infection. In contrast, HIV could not establish a productive infection in Impα3-KD MDMs (Fig. 7B). At the peak time of virus infection, the Luc activity detected in Impα3-KD MDMs was more than 10-fold lower than that in ScRNA-transduced macrophages. A similar difference was also seen for donor 2 (Fig. 7C), clearly indicating that Impα3 is required for productive HIV infection in macrophages.
Since HIV Vpr was also able to bind to Impα3, the possible role of their interaction was also tested. The MDMs were infected with equal amounts of VSV-G-pseudotyped Vpr+ or Vpr− pNL-BruΔBgl/Luc viruses, and Luc activities were measured after 7 days of infection. In agreement with previous studies, our results showed a requirement of Vpr for productive infection in macrophages, since the Luc activity produced by Vpr− virus infection was 4- to 5-fold lower than that produced by Vpr+ virus in ScRNA-transduced macrophages (Fig. 7D, compare bar 3 with bar 1). However, we could not see a correlation between Vpr and Impα3. The results from Fig. 7D showed that while Luc activity induced by Vpr+ virus infection was 7.6-fold lower in Impα3-KD macrophages than in ScRNA-transduced macrophages (Fig. 7D, compare bar 2 with bar 1), the Luc activity induced by Vpr− virus infection in Impα3-KD macrophages also showed a 6.6-fold decrease (Fig. 7D, compare bar 4 with bar 3). These data suggest that the attenuated HIV infection in Impα3-KD macrophages was Vpr independent. Even though there were differences in HIV-1 susceptibility from one donor to another, we consistently observed a significant impairment of HIV infection in Impα3-KD macrophages derived from each donor, and these results were consistent with the observations in HeLa and C8166 T-cell lines. These findings strongly support the notion that Impα3 is an important cellular importin that is involved in HIV-1 replication in macrophages.
DISCUSSION
HIV-1 employs the cellular nuclear import machinery to actively transport its PIC into the nucleus for its integration. Several viral karyophilic proteins and different cellular import proteins have been suggested to contribute to HIV-1 PIC nuclear import and replication. In this study, by using a shRNA-mediated knockdown (KD) technique, we have demonstrated that Impα3 contributes significantly to HIV-1 nuclear import and replication in CD4+ T cells. Moreover, our data also showed that Impα3 is required for HIV infection in primary macrophages. In an attempt to unravel the mechanism underlying its contribution, we demonstrated that Impα3 was bound by IN during HIV-1 infection in CD4+ C8166 T cells, and the deletion analysis suggested that a region (aa 250 to 270) in the C-terminal domain of IN was involved in this viral-cellular protein interaction. All of these studies provide evidence that Impα3, one member of the cellular Impα family, plays an important role in HIV-1 nuclear import and replication by interacting with HIV IN.
The Impα/Impβ-mediated nuclear import pathway is a well-characterized pathway for numerous macromolecules (reviewed in references 24 to 26 and 46). The Impα family contains six isoforms in human cells and acts as an adaptor by binding both the import substrate and importin β, which is essential for the classical nuclear import pathway in living cells. Accumulated evidence indicates that HIV-1 proteins, including MA, Vpr, and IN, are able to interact with Impα1/Rch1 (an 32-amino-acid N-terminally truncated form of Impα1) in in vitro binding assays (20, 21, 56, 72), even though the contribution of these interactions to HIV-1 replication remains to be elucidated.
In this study, we first investigated the functional roles of different isoforms of the importin α family, including Impα1, Impα3, Impα5, and Impα7, in HIV-1 replication by specific knockdown of each isoform in human cells. By using the pLKO1 lentiviral vector encoding small hairpin RNAs of Impα1, Impα3, Impα5, and Impα7, we were able to efficiently knock down their expression in HeLa cells or C8166 cells and to generate stable cell lines by using puromycin selection. Interestingly, the VSV-G-mediated single-cycle HIV infection potential in various Impα-KD stable cell lines was significantly varied. An approximately 4-fold reduction of HIV infection was observed in Impα3-KD HeLa cells and C8166 T cells, while only about a 0.8- to 1.2-fold decrease of HIV infection was seen in Impα1- and Impα5-KD cells (Fig. 1 and 2). However, knockdown of the expression of Impα7 in HeLa cells had no effect on HIV-1 replication (Fig. 1B). Since a previous study had revealed that downregulation of Impα1, Impα3, Impα5, or Impα7 by small interfering RNA (siRNA) affected HeLa cell proliferation to different extents (57), we checked the proliferation of Impα1-, Impα3-,Imp α5-, and Impα7-KD C8166 T cells by using a WST assay. We observed that the proliferation of both Impα1- and Impα3-KD cells was modestly inhibited (30%) but that Impα5 and Impα7 downregulation did not show an effect on cell growth (Fig. 2B; data not shown for Impα7). Similar results were obtained by using a trypan blue exclusion assay for ScRNA-transduced and Impα3-KD HeLa cells (data not shown). The reason for Impα1- and Impα3-KD-mediated proliferation inhibition is unknown. It is possible that some proteins that are important for cellular proliferation could not efficiently enter into the nucleus due to the specific import receptor knockdown. However, the significant Impα3-KD-induced attenuation of HIV-1 infection appears to be specific to the virus, since Impα1 induced a similar degree of proliferation inhibition but inhibited HIV-1 infection only modestly. Moreover, we did not detect any effect of Impα3 knockdown on the expression of an HIV-1 gene transfected into Impα3-KD and ScRNA-transduced HeLa cells (Fig. 1C), suggesting that knockdown of Impα3 does not affect the cellular environment required for HIV-1 gene expression. All of these data indicate that Impα3 played an important role in the early stages of HIV-1 replication in dividing T cells. Moreover, since knockdown of Impα1 or Impα5 also resulted in ca. 50% reduction of viral infection, we could not exclude the possibility that these Impα isoforms also contribute to efficient HIV-1 replication. Interestingly, some previous studies have shown that the expression levels of each importin α isoform may differ in different cell and tissue types and may depend on the state of cellular metabolism and differentiation (33, 52, 69). In this regard, we could not exclude the possibility that HIV-1 IN may employ various Impα isoforms for nuclear import of the virus in different susceptible cell types and under different conditions.
The importance of Impα3 in HIV-1 replication in dividing and nondividing cells was further confirmed by infecting Impα3-KD CD4+ C8166 T cells with wild-type HIV-1 (pNL4.3-GFP). Viral replication was drastically attenuated, and at day 4 postinfection, Impα3-KD led to a 10- to 15-fold reduction in virus replication (Fig. 3). Moreover, in primary macrophages, Impα3-KD also significantly inhibited HIV infection (Fig. 7). To further test at which step HIV infection was affected, we performed real-time PCR analysis to examine HIV total-DNA synthesis, 2-LTR circles, and integrated-DNA levels in HIV-1-infected Impα3-KD and ScRNA-transduced C8166 T cells. Impα3-KD resulted in drastic reductions in the levels of 2-LTR circles and integrated DNA (Fig. 4B and C). In contrast to HIV infection, infection with the VSV-G-pseudotyped MLV vector was reduced only 40 to 50% in Impα3-KD C8166 T cells (Fig. 2D). Since MLV replication is dependent on cell mitosis, the modestly impaired MLV infection could be at least partially due to the slower proliferation of Impα3-KD C8166 T cells, as shown in Fig. 2B. Thus, our study strongly indicates that Impα3-KD in CD4+ T cells had profound inhibitory effects on HIV-1 infection by specifically blocking viral nuclear import.
We next asked how HIV-1 hijacks Impα3 for efficient nuclear import and replication. It is known that several HIV karyophilic proteins, including IN, MA, and Vpr, are associated with this nucleoprotein complex and participate in HIV-1 nuclear import (8, 20, 22, 29, 53, 72). Therefore, we examined the possible interaction between Impα3 and these three HIV karyophilic proteins. Interestingly, we identified Impα3 as a novel partner of HIV-1 IN by an in vitro GST pulldown assay and by in vivo co-IP assays in virus-infected C8166 T cells. Moreover, our deletion analysis revealed that GFP-IN1-212 and GFP-IN1-250 lost the ability to bind to Impα3, but GFP-IN1-270 was able to bind to this cellular protein. These results provided convincing evidence for the specificity of the IN-Impα3 interaction and suggested that a C-terminal region encompassing residues 250 to 270 in HIV IN may be involved in this viral-cellular protein interaction. Consistently, our localization study also showed that the Impα3-binding-deficient mutants GFP-IN1-212 and GFP-IN1-250 were excluded from the nucleus, while the GFP-IN1-270 mutant, like wild-type GFP-IN, was localized predominantly in the nucleus (Fig. 6C). It is worth noting that the role of the C-terminal domain of IN in assisting HIV-1 cDNA nuclear import has been observed in several studies. Gallay et al. first reported that wild-type IN recognizes Impα1 and suggested that a bipartite nuclear localization signal (NLS) exists at the catalytic core (186KRK188) and the C-terminal domains (215KELQKQITK219) (20). Our previous study also showed that the K215A K219A and K240A K244E CTD mutants affected the efficiency of HIV-1 cDNA nuclear import (1). In the current study, our deletion analysis suggested that a region from aa 251 to 270 in the C-terminal domain of IN may contribute to the interaction with Impα3 and to the nuclear localization of IN. All this information supports the notion that the C-terminal domain of IN plays an important role in IN and HIV PIC nuclear import by interacting with cellular proteins. Interestingly, our previous study also revealed that an IN C-terminal mutant (INKKRK) in which three lysines (K) at positions 240, 244, and 264 and an arginine (R) at position 263 were changed to alanines lost the ability to bind to Imp7 (2). Since two of these amino acids are located within the region from aa 251 to 270 in the C-terminal domain of IN, this raises the possibility that a similar region in the C-terminal domain of IN may be involved in the interaction of IN with two cellular importins. Therefore, how HIV IN coordinates with these two importins during HIV replication is another interesting question requiring further investigation. It appears that Impα3 could be the major importin recruited during HIV nuclear import, since previous studies, including ours, showed that HIV replication was not significantly affected when HeLa cells with an Imp7 siRNA were infected with the wild-type virus (2, 87). However, it is still possible that the presence of Imp7 may facilitate HIV nuclear import (83). At this point, a more-detailed substitution mutation analysis is under way to precisely define the critical interaction interfaces between IN and Impα3.
In addition to HIV IN, another karyophilic protein, Vpr, was also shown to bind to Impα3 (Fig. 5B). Consistent with this, a recent study also indicates that HIV Vpr is able to bind to three Impα isoforms (Impα1, Impα3, and Impα5) in an in vitro binding assay (54). However, the evidence provided by our study did not support the notion that the Vpr-Impα3 interaction plays a major role in HIV replication. First, Impα3-KD significantly inhibited HIV nuclear import and replication in dividing HeLa and C8166 T-cell lines, while Vpr was shown to be dispensable for HIV infection in these susceptible dividing cell lines. Second, in Impα3-KD macrophages, similar degrees of decrease in Vpr− virus infection (6-fold reduced Luc activity) and Vpr+ virus infection (7-fold decrease) were observed (Fig. 7C). These observations suggest that the impaired HIV replication in the Impα3-KD dividing T-cell line and macrophage is Vpr independent. However, in agreement with previous observations (11, 29, 66, 72), our results also showed that in the absence of Vpr, viral replication in both normal and Impα3-KD macrophages was significantly attenuated (Fig. 7C, compare bar 3 to bar 1), indicating that Vpr facilitates HIV-1 replication in macrophages, but possibly through a different mechanism. Indeed, a number of studies have suggested that Vpr is capable of binding to nuclear import factors, including Impα1 and hCG1, and facilitates HIV nuclear import and infection in macrophages (11, 28, 32, 48, 54, 55, 72). In addition to HIV-1 karyophilic proteins, recent studies have also demonstrated that the HIV-1 capsid (CA) protein plays an important role in HIV-1 infection in growth-arrested cells (78, 79) and is a determinant of the transportin-3 requirement for HIV-1 replication (43). These studies suggest that a substantial shedding of components of PIC (uncoating) and/or a significant conformational change in PIC is required for efficient HIV nuclear import. It is possible that an uncoating process mediated by CA is necessary in order for HIV-1 karyophilic proteins to be accessible to recruiting cellular cofactors required for HIV-1 nuclear import. It is also possible that viral uncoating and nuclear import of karyophilic proteins could be independent but could both be required for efficient HIV PIC nuclear import. We believe that these are very interesting questions to be addressed in future studies.
It is known that HIV-1 nuclear import is essential for productive infection not only in nondividing but also in dividing cells. The roles of HIV IN and the central DNA flap in HIV-1 nuclear import in both types of susceptible cells have been documented (1, 6, 84). The contribution of IN to HIV-1 nuclear import in dividing cells was also suggested by testing the interaction between IN and Imp7 (2) and between IN and TRN-SR2 (10). Consistent with this, other studies by Katz et al. (35, 36) and Rivière et al. (60) indicate that efficient HIV-1 and Rous sarcoma virus (RSV) nuclear entry can occur independently of mitotic nuclear disassembly in cycling cells. In the present study, our results showed that, in addition to inhibiting HIV infection in macrophages, Impα3-KD also affected HIV-1 2-LTR DNA levels and viral replication in dividing C8166 T cells. Thus, we concluded that Impα3 is required for efficient HIV-1 PIC nuclear transport and viral replication in both dividing and nondividing cells. Further investigation into the molecular mechanism of IN-Impα3 interaction and its contribution to HIV replication in different natural virus target cells will facilitate a better understanding of the HIV-1 nuclear import process and will benefit the development of novel anti-HIV strategies.
Acknowledgments
We thank Leslie Slaney for technical support. We are also grateful to D. Grandgenett and R. Craigie for providing their anti-IN antiserum and recombinant IN protein, which were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
Z. Ao is the recipient of a postdoctoral fellowship from the CIHR International Infectious Disease & Global Health Training Program, and K. Danappa Jayappa and Y. Zheng are recipients of Manitoba Health Research Council/Manitoba Institute of Child Health (MHRC/MICH) scholarships. X. Yao is the recipient of a Basic Science Career Development research award from the Manitoba Medical Service Foundation. This work was supported by a CIHR grant (HOP-81180) and a Leaders Opportunity Fund award from the Canadian Foundation of Innovation (CFI) to X. Yao.
Footnotes
Published ahead of print on 16 June 2010.
REFERENCES
- 1.Ao, Z., K. R. Fowke, E. A. Cohen, and X. Yao. 2005. Contribution of the C-terminal tri-lysine regions of human immunodeficiency virus type 1 integrase for efficient reverse transcription and viral DNA nuclear import. Retrovirology 2:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ao, Z., G. Huang, H. Yao, Z. Xu, M. Labine, A. W. Cochrane, and X. Yao. 2007. Interaction of human immunodeficiency virus type 1 integrase with cellular nuclear import receptor importin 7 and its impact on viral replication. J. Biol. Chem. 282:13456-13467. [DOI] [PubMed] [Google Scholar]
- 3.Ao, Z., X. Yao, and E. A. Cohen. 2004. Assessment of the role of the central DNA flap in human immunodeficiency virus type 1 replication by using a single-cycle replication system. J. Virol. 78:3170-3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ao, Z., Z. Yu, L. Wang, Y. Zheng, and X. Yao. 2008. Vpr14-88-Apobec3G fusion protein is efficiently incorporated into Vif-positive HIV-1 particles and inhibits viral infection. PLoS One 3:e1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aratani, S., T. Oishi, H. Fujita, M. Nakazawa, R. Fujii, N. Imamoto, Y. Yoneda, A. Fukamizu, and T. Nakajima. 2006. The nuclear import of RNA helicase A is mediated by importin-α3. Biochem. Biophys. Res. Commun. 340:125-133. [DOI] [PubMed] [Google Scholar]
- 6.Bouyac-Bertoia, M., J. D. Dvorin, R. A. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. [DOI] [PubMed] [Google Scholar]
- 7.Brass, A. L., D. M. Dykxhoorn, Y. Benita, N. Yan, A. Engelman, R. J. Xavier, J. Lieberman, and S. J. Elledge. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921-926. [DOI] [PubMed] [Google Scholar]
- 8.Bukrinsky, M., S. Haggerty, M. P. Dempsey, N. Sharova, A. Adzhubel, L. Spitz, P. Lewis, D. Goldfarb, M. Emerman, and M. Stevenson. 1993. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 365:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butler, S. L., E. P. Johnson, and F. D. Bushman. 2002. Human immunodeficiency virus cDNA metabolism: notable stability of two-long terminal repeat circles. J. Virol. 76:3739-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christ, F., W. Thys, J. De Rijck, R. Gijsbers, A. Albanese, D. Arosio, S. Emiliani, J. C. Rain, R. Benarous, A. Cereseto, and Z. Debyser. 2008. Transportin-SR2 imports HIV into the nucleus. Curr. Biol. 18:1192-1202. [DOI] [PubMed] [Google Scholar]
- 11.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 12.Cortes, P., Z. S. Ye, and D. Baltimore. 1994. RAG-1 interacts with the repeated amino acid motif of the human homologue of the yeast protein SRP1. Proc. Natl. Acad. Sci. U. S. A. 91:7633-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuomo, C. A., S. A. Kirch, J. Gyuris, R. Brent, and M. A. Oettinger. 1994. Rch1, a protein that specifically interacts with the RAG-1 recombination-activating protein. Proc. Natl. Acad. Sci. U. S. A. 91:6156-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Depienne, C., P. Roques, C. Creminon, L. Fritsch, R. Casseron, D. Dormont, C. Dargemont, and S. Benichou. 2000. Cellular distribution and karyophilic properties of matrix, integrase, and Vpr proteins from the human and simian immunodeficiency viruses. Exp. Cell Res. 260:387-395. [DOI] [PubMed] [Google Scholar]
- 15.Fassati, A., and S. P. Goff. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 75:3626-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fassati, A., D. Gorlich, I. Harrison, L. Zaytseva, and J. M. Mingot. 2003. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J. 22:3675-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Follenzi, A., L. E. Ailles, S. Bakovic, M. Geuna, and L. Naldini. 2000. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 25:217-222. [DOI] [PubMed] [Google Scholar]
- 18.Freedman, N. D., and K. R. Yamamoto. 2004. Importin 7 and importin alpha/importin beta are nuclear import receptors for the glucocorticoid receptor. Mol. Biol. Cell 15:2276-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedrich, B., C. Quensel, T. Sommer, E. Hartmann, and M. Kohler. 2006. Nuclear localization signal and protein context both mediate importin alpha specificity of nuclear import substrates. Mol. Cell. Biol. 26:8697-8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. U. S. A. 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallay, P., V. Stitt, C. Mundy, M. Oettinger, and D. Trono. 1996. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J. Virol. 70:1027-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallay, P., S. Swingler, C. Aiken, and D. Trono. 1995. HIV-1 infection of nondividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell 80:379-388. [DOI] [PubMed] [Google Scholar]
- 23.Gartner, S., P. Markovits, D. M. Markovitz, M. H. Kaplan, R. C. Gallo, and M. Popovic. 1986. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science 233:215-219. [DOI] [PubMed] [Google Scholar]
- 24.Goldfarb, D. S., A. H. Corbett, D. A. Mason, M. T. Harreman, and S. A. Adam. 2004. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 14:505-514. [DOI] [PubMed] [Google Scholar]
- 25.Görlich, D., S. Kostka, R. Kraft, C. Dingwall, R. A. Laskey, E. Hartmann, and S. Prehn. 1995. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr. Biol. 5:383-392. [DOI] [PubMed] [Google Scholar]
- 26.Görlich, D., and R. A. Laskey. 1995. Roles of importin in nuclear protein import. Cold Spring Harbor Symp. Quant. Biol. 60:695-699. [DOI] [PubMed] [Google Scholar]
- 27.Hearps, A. C., and D. A. Jans. 2006. HIV-1 integrase is capable of targeting DNA to the nucleus via an importin alpha/beta-dependent mechanism. Biochem. J. 398:475-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hehl, E. A., P. Joshi, G. V. Kalpana, and V. R. Prasad. 2004. Interaction between human immunodeficiency virus type 1 reverse transcriptase and integrase proteins. J. Virol. 78:5056-5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heinzinger, N. K., M. I. Bukinsky, S. A. Haggerty, A. M. Ragland, V. Kewalramani, M. A. Lee, H. E. Gendelman, L. Ratner, M. Stevenson, and M. Emerman. 1994. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc. Natl. Acad. Sci. U. S. A. 91:7311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123-126. [DOI] [PubMed] [Google Scholar]
- 31.Jäkel, S., and D. Gorlich. 1998. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 17:4491-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamata, M., Y. Nitahara-Kasahara, Y. Miyamoto, Y. Yoneda, and Y. Aida. 2005. Importin-alpha promotes passage through the nuclear pore complex of human immunodeficiency virus type 1 Vpr. J. Virol. 79:3557-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamei, Y., S. Yuba, T. Nakayama, and Y. Yoneda. 1999. Three distinct classes of the alpha-subunit of the nuclear pore-targeting complex (importin-alpha) are differentially expressed in adult mouse tissues. J. Histochem. Cytochem. 47:363-372. [DOI] [PubMed] [Google Scholar]
- 34.Kataoka, N., J. L. Bachorik, and G. Dreyfuss. 1999. Transportin-SR, a nuclear import receptor for SR proteins. J. Cell Biol. 145:1145-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katz, R. A., J. G. Greger, P. Boimel, and A. M. Skalka. 2003. Human immunodeficiency virus type 1 DNA nuclear import and integration are mitosis independent in cycling cells. J. Virol. 77:13412-13417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katz, R. A., J. G. Greger, K. Darby, P. Boimel, G. F. Rall, and A. M. Skalka. 2002. Transduction of interphase cells by avian sarcoma virus. J. Virol. 76:5422-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kesson, A. M., W. R. Fear, F. Kazazi, J. M. Mathijs, J. Chang, N. J. King, and A. L. Cunningham. 1993. Human immunodeficiency virus type 1 infection of human placental macrophages in vitro. J. Infect. Dis. 168:571-579. [DOI] [PubMed] [Google Scholar]
- 38.Köhler, M., S. Ansieau, S. Prehn, A. Leutz, H. Haller, and E. Hartmann. 1997. Cloning of two novel human importin-alpha subunits and analysis of the expression pattern of the importin-alpha protein family. FEBS Lett. 417:104-108. [DOI] [PubMed] [Google Scholar]
- 39.Köhler, M., A. Fiebeler, M. Hartwig, S. Thiel, S. Prehn, R. Kettritz, F. C. Luft, and E. Hartmann. 2002. Differential expression of classical nuclear transport factors during cellular proliferation and differentiation. Cell. Physiol. Biochem. 12:335-344. [DOI] [PubMed] [Google Scholar]
- 40.Köhler, M., D. Gorlich, E. Hartmann, and J. Franke. 2001. Adenoviral E1A protein nuclear import is preferentially mediated by importin α3 in vitro. Virology 289:186-191. [DOI] [PubMed] [Google Scholar]
- 41.Köhler, M., C. Speck, M. Christiansen, F. R. Bischoff, S. Prehn, H. Haller, D. Gorlich, and E. Hartmann. 1999. Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Mol. Cell. Biol. 19:7782-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.König, R., Y. Zhou, D. Elleder, T. L. Diamond, G. M. Bonamy, J. T. Irelan, C. Y. Chiang, B. P. Tu, P. D. De Jesus, C. E. Lilley, S. Seidel, A. M. Opaluch, J. S. Caldwell, M. D. Weitzman, K. L. Kuhen, S. Bandyopadhyay, T. Ideker, A. P. Orth, L. J. Miraglia, F. D. Bushman, J. A. Young, and S. K. Chanda. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135:49-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krishnan, L., K. A. Matreyek, I. Oztop, K. Lee, C. H. Tipper, X. Li, M. J. Dar, V. N. Kewalramani, and A. Engelman. 2010. The requirement for cellular transportin 3 (TNPO3 or TRN-SR2) during infection maps to human immunodeficiency virus type 1 capsid and not integrase. J. Virol. 84:397-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lai, M. C., R. I. Lin, S. Y. Huang, C. W. Tsai, and W. Y. Tarn. 2000. A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J. Biol. Chem. 275:7950-7957. [DOI] [PubMed] [Google Scholar]
- 45.Lai, M. C., R. I. Lin, and W. Y. Tarn. 2001. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. U. S. A. 98:10154-10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lange, A., R. E. Mills, C. J. Lange, M. Stewart, S. E. Devine, and A. H. Corbett. 2007. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 282:5101-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langhoff, E., E. F. Terwilliger, H. J. Bos, K. H. Kalland, M. C. Poznansky, O. M. Bacon, and W. A. Haseltine. 1991. Replication of human immunodeficiency virus type 1 in primary dendritic cell cultures. Proc. Natl. Acad. Sci. U. S. A. 88:7998-8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le Rouzic, E., A. Mousnier, C. Rustum, F. Stutz, E. Hallberg, C. Dargemont, and S. Benichou. 2002. Docking of HIV-1 Vpr to the nuclear envelope is mediated by the interaction with the nucleoporin hCG1. J. Biol. Chem. 277:45091-45098. [DOI] [PubMed] [Google Scholar]
- 49.Lewin, S. R., J. Kirihara, S. Sonza, L. Irving, J. Mills, and S. M. Crowe. 1998. HIV-1 DNA and mRNA concentrations are similar in peripheral blood monocytes and alveolar macrophages in HIV-1-infected individuals. AIDS 12:719-727. [DOI] [PubMed] [Google Scholar]
- 50.Melen, K., R. Fagerlund, J. Franke, M. Kohler, L. Kinnunen, and I. Julkunen. 2003. Importin alpha nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J. Biol. Chem. 278:28193-28200. [DOI] [PubMed] [Google Scholar]
- 51.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyamoto, Y., N. Imamoto, T. Sekimoto, T. Tachibana, T. Seki, S. Tada, T. Enomoto, and Y. Yoneda. 1997. Differential modes of nuclear localization signal (NLS) recognition by three distinct classes of NLS receptors. J. Biol. Chem. 272:26375-26381. [DOI] [PubMed] [Google Scholar]
- 53.Nie, Z., D. Bergeron, R. A. Subbramanian, X. J. Yao, F. Checroune, N. Rougeau, and E. A. Cohen. 1998. The putative alpha helix 2 of human immunodeficiency virus type 1 Vpr contains a determinant which is responsible for the nuclear translocation of proviral DNA in growth-arrested cells. J. Virol. 72:4104-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nitahara-Kasahara, Y., M. Kamata, T. Yamamoto, X. Zhang, Y. Miyamoto, K. Muneta, S. Iijima, Y. Yoneda, Y. Tsunetsugu-Yokota, and Y. Aida. 2007. Novel nuclear import of Vpr promoted by importin alpha is crucial for human immunodeficiency virus type 1 replication in macrophages. J. Virol. 81:5284-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Popov, S., M. Rexach, L. Ratner, G. Blobel, and M. Bukrinsky. 1998. Viral protein R regulates docking of the HIV-1 preintegration complex to the nuclear pore complex. J. Biol. Chem. 273:13347-13352. [DOI] [PubMed] [Google Scholar]
- 56.Popov, S., M. Rexach, G. Zybarth, N. Reiling, M. A. Lee, L. Ratner, C. M. Lane, M. S. Moore, G. Blobel, and M. Bukrinsky. 1998. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 17:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quensel, C., B. Friedrich, T. Sommer, E. Hartmann, and M. Kohler. 2004. In vivo analysis of importin alpha proteins reveals cellular proliferation inhibition and substrate specificity. Mol. Cell. Biol. 24:10246-10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reil, H., A. A. Bukovsky, H. R. Gelderblom, and H. G. Gottlinger. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rich, E. A., I. S. Chen, J. A. Zack, M. L. Leonard, and W. A. O'Brien. 1992. Increased susceptibility of differentiated mononuclear phagocytes to productive infection with human immunodeficiency virus-1 (HIV-1). J. Clin. Invest. 89:176-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rivière, L., J. L. Darlix, and A. Cimarelli. 2010. Analysis of the viral elements required in the nuclear import of HIV-1 DNA. J. Virol. 84:729-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schuitemaker, H., N. A. Kootstra, M. H. Koppelman, S. M. Bruisten, H. G. Huisman, M. Tersmette, and F. Miedema. 1992. Proliferation-dependent HIV-1 infection of monocytes occurs during differentiation into macrophages. J. Clin. Invest. 89:1154-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seki, T., S. Tada, T. Katada, and T. Enomoto. 1997. Cloning of a cDNA encoding a novel importin-alpha homologue, Qip1: discrimination of Qip1 and Rch1 from hSrp1 by their ability to interact with DNA helicase Q1/RecQL. Biochem. Biophys. Res. Commun. 234:48-53. [DOI] [PubMed] [Google Scholar]
- 63.Sekimoto, T., N. Imamoto, K. Nakajima, T. Hirano, and Y. Yoneda. 1997. Extracellular signal-dependent nuclear import of Stat 1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 16:7067-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shen, R., L. E. Smythies, R. H. Clements, L. Novak, and P. D. Smith. 2010. Dendritic cells transmit HIV-1 through human small intestinal mucosa. J. Leukoc. Biol. 87:663-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith, P. D., G. Meng, G. M. Shaw, and L. Li. 1997. Infection of gastrointestinal tract macrophages by HIV-1. J. Leukoc. Biol. 62:72-77. [DOI] [PubMed] [Google Scholar]
- 66.Subbramanian, R. A., A. Kessous-Elbaz, R. Lodge, J. Forget, X. J. Yao, D. Bergeron, and E. A. Cohen. 1998. Human immunodeficiency virus type 1 Vpr is a positive regulator of viral transcription and infectivity in primary human macrophages. J. Exp. Med. 187:1103-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suzuki, T., Y. Ishigami, N. Okada, A. Kaneko, R. Fukutomi, and M. Isemura. 2008. Differentiation-associated alteration in gene expression of importins and exportins in human leukemia HL-60 cells. Biomed. Res. 29:141-145. [DOI] [PubMed] [Google Scholar]
- 68.Suzuki, Y., N. Misawa, C. Sato, H. Ebina, T. Masuda, N. Yamamoto, and Y. Koyanagi. 2003. Quantitative analysis of human immunodeficiency virus type 1 DNA dynamics by real-time PCR: integration efficiency in stimulated and unstimulated peripheral blood mononuclear cells. Virus Genes 27:177-188. [DOI] [PubMed] [Google Scholar]
- 69.Tsuji, L., T. Takumi, N. Imamoto, and Y. Yoneda. 1997. Identification of novel homologues of mouse importin alpha, the alpha subunit of the nuclear pore-targeting complex, and their tissue-specific expression. FEBS Lett. 416:30-34. [DOI] [PubMed] [Google Scholar]
- 70.Van Maele, B., J. De Rijck, E. De Clercq, and Z. Debyser. 2003. Impact of the central polypurine tract on the kinetics of human immunodeficiency virus type 1 vector transduction. J. Virol. 77:4685-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van't Wout, A. B., N. A. Kootstra, G. A. Mulder-Kampinga, N. Albrecht-van Lent, H. J. Scherpbier, J. Veenstra, K. Boer, R. A. Coutinho, F. Miedema, and H. Schuitemaker. 1994. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J. Clin. Invest. 94:2060-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vodicka, M. A., D. M. Koepp, P. A. Silver, and M. Emerman. 1998. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 12:175-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. H. Hahn, M. S. Saag, and G. M. Shaw. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373:117-122. [DOI] [PubMed] [Google Scholar]
- 74.Weinberg, J. B., T. J. Matthews, B. R. Cullen, and M. H. Malim. 1991. Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J. Exp. Med. 174:1477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weis, K., I. W. Mattaj, and A. I. Lamond. 1995. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science 268:1049-1053. [DOI] [PubMed] [Google Scholar]
- 76.Weis, K., U. Ryder, and A. I. Lamond. 1996. The conserved amino-terminal domain of hSRP1 alpha is essential for nuclear protein import. EMBO J. 15:1818-1825. [PMC free article] [PubMed] [Google Scholar]
- 77.Welch, K., J. Franke, M. Kohler, and I. G. Macara. 1999. RanBP3 contains an unusual nuclear localization signal that is imported preferentially by importin-α3. Mol. Cell. Biol. 19:8400-8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamashita, M., and M. Emerman. 2004. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J. Virol. 78:5670-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yamashita, M., and M. Emerman. 2006. Retroviral infection of non-dividing cells: old and new perspectives. Virology 344:88-93. [DOI] [PubMed] [Google Scholar]
- 80.Yao, X. J., J. Friborg, F. Checroune, S. Gratton, F. Boisvert, R. P. Sekaly, and E. A. Cohen. 1995. Degradation of CD4 induced by human immunodeficiency virus type 1 Vpu protein: a predicted alpha-helix structure in the proximal cytoplasmic region of CD4 contributes to Vpu sensitivity. Virology 209:615-623. [DOI] [PubMed] [Google Scholar]
- 81.Yao, X. J., R. A. Subbramanian, N. Rougeau, F. Boisvert, D. Bergeron, and E. A. Cohen. 1995. Mutagenic analysis of human immunodeficiency virus type 1 Vpr: role of a predicted N-terminal alpha-helical structure in Vpr nuclear localization and virion incorporation. J. Virol. 69:7032-7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yasuhara, N., N. Shibazaki, S. Tanaka, M. Nagai, Y. Kamikawa, S. Oe, M. Asally, Y. Kamachi, H. Kondoh, and Y. Yoneda. 2007. Triggering neural differentiation of ES cells by subtype switching of importin-alpha. Nat. Cell Biol. 9:72-79. [DOI] [PubMed] [Google Scholar]
- 83.Zaitseva, L., P. Cherepanov, L. Leyens, S. J. Wilson, J. Rasaiyaah, and A. Fassati. 2009. HIV-1 exploits importin 7 to maximize nuclear import of its DNA genome. Retrovirology 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101:173-185. [DOI] [PubMed] [Google Scholar]
- 85.Zennou, V., C. Serguera, C. Sarkis, P. Colin, E. Perret, J. Mallet, and P. Charneau. 2001. The HIV-1 DNA flap stimulates HIV vector-mediated cell transduction in the brain. Nat. Biotechnol. 19:446-450. [DOI] [PubMed] [Google Scholar]
- 86.Zhang, Z., T. Schuler, M. Zupancic, S. Wietgrefe, K. A. Staskus, K. A. Reimann, T. A. Reinhart, M. Rogan, W. Cavert, C. J. Miller, R. S. Veazey, D. Notermans, S. Little, S. A. Danner, D. D. Richman, D. Havlir, J. Wong, H. L. Jordan, T. W. Schacker, P. Racz, K. Tenner-Racz, N. L. Letvin, S. Wolinsky, and A. T. Haase. 1999. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science 286:1353-1357. [DOI] [PubMed] [Google Scholar]
- 87.Zielske, S. P., and M. Stevenson. 2005. Importin 7 may be dispensible for human immunodeficiency virus type 1 and simian immunodeficiency virus infection of primary macrophages. J. Virol. 79:11541-11546. [DOI] [PMC free article] [PubMed] [Google Scholar]