Abstract
Herpes simplex virus type 1 (HSV-1) glycoprotein K (gK) and the UL20 protein (UL20p) are strictly required for virus-induced cell fusion, and mutations within either the gK or UL20 gene cause extensive cell fusion (syncytium formation). We have shown that gK forms a functional protein complex with UL20p, which is required for all gK and UL20p-associated functions in the HSV-1 life cycle. Recently, we showed that the amino-terminal 82 amino acids (aa) of gK (gKa) were required for the expression of the syncytial phenotype of the mutant virus gBΔ28 lacking the carboxyl-terminal 28 amino acids of gB (V. N. Chouljenko, A. V. Iyer, S. Chowdhury, D. V. Chouljenko, and K. G. Kousoulas, J. Virol. 83:12301-12313, 2009). This work suggested that the amino terminus of gK may directly or indirectly interact with gB and/or other viral glycoproteins. Two-way coimmunoprecipitation experiments revealed that UL20p interacted with gB in infected cells. Furthermore, the gKa peptide was coimmunoprecipitated with gB but not gD. Three recombinant baculoviruses were constructed, expressing the amino-terminal 82 aa of gKa together with either the extracellular portion of gB (30 to 748 aa), gD (1 to 340 aa), or gH (1 to 792 aa), respectively. Coimmunoprecipitation experiments revealed that gKa physically interacted with the extracellular portions of gB and gH but not gD. Three additional recombinant baculoviruses expressing gKa and truncated gBs encompassing aa 30 to 154, 30 to 364, and 30 to 500 were constructed. Coimmunoprecipitation experiments showed that gKa physically interacted with all three truncated gBs. Computer-assisted prediction of possible gKa binding sites on gB suggested that gKa may interact predominantly with gB domain I (E. E. Heldwein, H. Lou, F. C. Bender, G. H. Cohen, R. J. Eisenberg, and S. C. Harrison, Science 313:217-220, 2006). These results imply that the gK/UL20p protein complex modulates the fusogenic properties of gB and gH via direct physical interactions.
Herpes simplex virus type 1 (HSV-1) can enter into cells via the fusion of its viral envelope with cellular membranes. Also, the virus can spread from infected to uninfected cells by causing virus-induced cell fusion, allowing virions to enter into uninfected cells without being exposed to extracellular spaces. These membrane fusion phenomena are known to be mediated by viral glycoproteins and other viral proteins (reviewed in reference 36). Although wild-type viruses cause a limited amount of virus-induced cell fusion, certain mutations cause extensive virus-induced cell-to-cell fusion (syncytial, or syn, mutations). These syncytial mutations are located predominantly within the UL20 gene (5, 27, 28); the UL24 gene (25, 38); the UL27 gene, encoding glycoprotein gB (7, 15, 18, 32); and the UL53 gene, coding for gK (6, 11, 24, 34, 35, 37).
The presence of syncytial mutations within different viral genes, as well as other accumulating evidence, suggests that virus-induced cell fusion is mediated by the concerted action and interactions of the viral glycoproteins gD, gB, and gH/gL as well as gK and the membrane protein UL20p. Specifically, recent studies have shown that gD interacts with both gB and gH/gL (1, 2, 21). However, gB and gH/gL can also interact with each other even in the absence of gD (3). In this membrane fusion model, the binding of gD to its cognate receptors, including nectin-1, herpesvirus entry mediator (HVEM), and other receptors (8, 19, 30, 39-42), is thought to trigger sequential conformational changes in gH/gL and gB causing the fusion of the viral envelope with cellular membranes during virus entry as well as fusion among cellular membranes (22, 23). The transient coexpression of gB, gD, and gH/gL causes cell-to-cell fusion (31, 43), suggesting that these four viral glycoproteins are necessary and sufficient for membrane fusion. However, this transient fusion system does not accurately depict virus-induced cell fusion. Specifically, viral glycoprotein K (gK) and the UL20 membrane protein (UL20p) have been shown to be strictly required for virus-induced cell fusion (10, 27, 29). Moreover, syncytial mutations within gK (6, 11, 24, 34, 35, 37) or UL20 (5, 27, 28) promote extensive virus-induced cell fusion, and viruses lacking gK enter more slowly than the wild-type virus into susceptible cells (17). In contrast, the transient coexpression of gK carrying a syncytial mutation with gB, gD, and gH/gL did not enhance cell fusion, while the coexpression of wild-type gK with gB, gD, and gH/gL was reported previously to inhibit cell fusion in certain cell lines (4). To date, there is no direct evidence that either gK or UL20p interacts with gB, gD, gH, or gL.
The X-ray structure of the ectodomain of HSV-1 gB has been determined and was predicted to assume at least two major conformations, one of which may be necessary for the fusogenic properties of gB (23). Single-amino-acid changes within the carboxyl terminus of gB located intracellularly as well as the deletion of the terminal 28 amino acids (aa) of gB cause extensive virus-induced cell fusion, presumably because they alter the extracellular conformation of gB (15, 31, 43). We have previously shown that HSV-1 gK and UL20p functionally and physically interact and that these interactions are absolutely necessary for their coordinate intracellular transport, cell surface expression, and functions in the HSV-1 life cycle (13, 16). In contrast to gB, syncytial mutations in gK map predominantly within extracellular domains of gK and particularly within the amino-terminal portion of gK (domain I) (12), while syncytial mutations of UL20 are located within the amino terminus of UL20p shown to be located intracellularly (27).
Recently, we showed that the a peptide composed of the amino-terminal 82 amino acids of gK (gKa) can complement in trans for gB-mediated cell fusion caused by the deletion of the carboxyl-terminal 28 amino acids of gB, suggesting that the gKa peptide interacted with gB or other viral glycoproteins involved in virus-induced cell fusion (10). In this work, we demonstrate that UL20p and the amino terminus of gKa physically interact with gB in infected cells, while the gKa peptide is also capable of binding to the extracellular portion of gH, suggesting that gK/UL20p modulates virus-induced cell fusion via direct interactions with gB and gH.
MATERIALS AND METHODS
Cell culture.
African green monkey kidney (Vero) and human embryonic kidney (HEK-293T) cells were obtained from the American Type Culture Collection (Rockville, MD). Cells were maintained in Dulbecco's modified Eagle's medium (Gibco-BRL, Grand Island, NY) supplemented with 10% fetal calf serum and antibiotics. Spodoptera frugiperda (Sf9; Invitrogen, Carlsbad, CA) insect cells were maintained in monolayer and/or suspension cultures at 27°C using serum-free Sf-900 II culture medium (SFM) with antibiotics (Pen Strep at a 1:200 dilution; Gibco). Cells were maintained in Dulbecco's modified Eagle's medium (Gibco-BRL, Grand Island, NY) supplemented with 10% fetal calf serum and antibiotics.
Construction of recombinant baculoviruses.
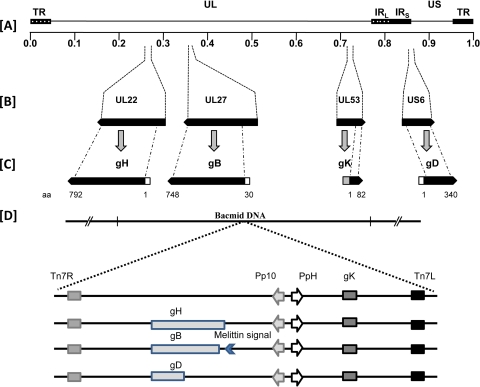
Recombinant baculoviruses were generated by using the Bac-to-Bac baculovirus expression system (Invitrogen). To create all the viral constructs, the pFastbac Dual vector was used, which contains two multiple-cloning sites to allow the expression of two heterologous genes, one controlled by the polyhedrin (PH) promoter and the other controlled by the p10 promoter. Fail Safe DNA polymerase (Epicentre Biotechnologies, Inc., Madison, WI) was used for all DNA amplifications. The synthetic oligonucleotides that were used for the PCR-based construction of different recombinant DNAs are shown in Table 1. Forward primer gK-Flag-EcoRI-F and reverse primer gK-Flag-BamHI-R were used to clone the first 82 amino acids of gKa into transient expression vector p3XFLAG-CMV14 (Sigma-Aldrich Biotechnology, St. Louis, MO) in frame with the carboxyl-terminal 3×FLAG epitope, as previously described (10). This plasmid was used as a PCR template to generate a DNA fragment encoding the 82 amino acids of gK in frame with the 3×FLAG epitope and with a 9-aa histidine tag using forward primer gK-Flag-EcoRI-F and reverse primer gK-Flag-9His-HindIII-R. EcoRI and HindIII restriction sites were utilized to clone the gKa gene cassette into the pFast Dual vector (Invitrogen, Inc.) under the control of the PPH promoter, resulting in the transfer vector pBD-gKa. DNA sequences encoding predicted ectodomains of gB (amino acids 30 to 748), gH (amino acids 1 to 792), and gD (amino acids 1 to 340) were first cloned into the pcDNA 3.1/V5/His (Invitrogen) expression vector by using primer pairs gB-F/gB-aa 748-R, gH-F/gH-aa792-R, and gD-F/gD-aa 340-R, respectively. The presence of the V5 epitope at the carboxyl end of the each protein was confirmed by Western immunoblotting using transiently transfected 293T cells (not shown). DNA fragments encoding the above-mentioned regions were PCR amplified by using primer pairs gB-Mel-NheI-F/pcDNA-KpnI-R, gD-NheI-F/pcDNA-KpnI-R, gH-NsiI-F/pcDNA-SphI-R, and the respective plasmids as templates. Plasmid pBD-gKa was used as the backbone for the construction of the transfer vectors pgKa/gB748, pgKa/gH792, pgKa/gD340, placing each gene cassette under the control of the baculovirus Pp10 promoter. NsiI and SphI restriction sites were used to clone the PCR-amplified gH gene cassette into plasmid vector pBD-gKa. The PCR products encoding the gB and gD gene cassettes were ligated into NheI and KpnI restriction sites of pBD-gKa. In the case of gB, the signal sequence of gB was replaced with the signal sequence of the melittin gene to facilitate enhanced gB expression in insect cells. Additional plasmids expressing gB truncations of 500, 364, and 154 amino acids were created as described above. These truncated portions of gB were ligated into the transfer vector pBD-gKa under the control of the Pp10 promoter using NheI and KpnI restriction sites.
TABLE 1.
Synthetic oligonucleotides used to construct recombinant baculoviruses
| Primer | Sequencea |
|---|---|
| gK-Flag-EcoRI-F | 5′-TGATCAGAATTCGCCATGCTCGCCGTCCGTTCC-3′ |
| gK-Flag-BamHI-R | 5′-TGATCAGGATCCGCAGATATGGGCGTGGTTGCG-3′ |
| gK-Flag-9His-HindIII-R | 5′-ATATATAAGCTTCTAGTGATGGTGATGGTGATGGTGATGATGACCGGTACGCTTGTCATCGTCATCCTTGTAGTC-3′ |
| gB-F | 5′-ACCATGCGCCAGGGCGCCCCCGC-3′ |
| gB-aa 748-R | 5′-CGCGCGCCCCAGGTCGCCCATC-3′ |
| gB-aa 500-R | 5′-GGAGGAGGTGGTCTTGATGCG-3′ |
| gB-aa 364-R | 5′-GCAGACCGACGGGCGCTTTGGCAC-3′ |
| gB-aa 154-R | 5′-GATGTTCTCCTTGAAGACCACCGC-3′ |
| gB-Mel-NheI-F | 5′-TATATAGCTAGCatgaaattcttagtcaacgttgcccttgtttttatggtcgtatacatttct tacatctatgccGCGGCTCCGAGTTCCCCCGGCACG-3′ |
| gH-F | 5′-ACCATGGGGAATGGTTTATGGTTC-3′ |
| gH-aa 792-R | 5′-GGCTAGCAAATGAATGACGGTTC-3′ |
| gH-NsiI-F | 5′-TATATAATGCATATGGGGAATGGTTTATGGTTC-3′ |
| gD-F | 5′-ACCATGGGaGGtGCTGCCGCCAGG-3′ |
| gD-aa 340-R | 5′-CATGTTGTTCGGGGTGGCCGG-3′ |
| gD-NheI-F | 5′-TATATAGCTAGCATGGGaGGtGCTGCCGCCAGG-3′ |
| pcDNA-KpnI-R | 5′-TATATAGGTACCCACAGTCGAGGCTGATCAGCG-3′ |
| pcDNA-SphI-R | 5′-TATATAGCATGCCACAGTCGAGGCTGATCAGCG-3′ |
Nucleotides in boldface type indicate restriction sites. Nucleotides in lowercase and italics code for the melittin sequence. Single lowercase letters depict nucleotide changes in gD inserted to facilitate primer synthesis without altering the amino acid sequence.
The recombinant pFastBac constructs were individually transformed into Escherichia coli DH10Bac cells (Invitrogen) to generate the corresponding recombinant bacmids. Sf9 insect cells (American Type Culture Collection, Rockville, MD) were transfected with recombinant bacmid DNA with Cellfectin II (Invitrogen), and the recombinant baculovirus vectors were amplified by repeated passages. The recombinant viruses produced are as follows: VgKa, VgKa/gB748, VgKa/gB500, VgKa/gB364, VgKa/gB154, gKa/gH792, and VgKa/gD340.
Immunoprecipitation assays.
Sf9 cell lysates were prepared by harvesting the cells at 72 h postinfection (hpi). The cells collected from one T75 flask were washed with phosphate-buffered saline (PBS) and lysed for 30 min on ice with 1 ml of NP-40 lysis buffer (Invitrogen) supplemented with phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich) and a cocktail of protease inhibitors (Roche Diagnostics, Indianapolis, IN). Immunoprecipitation reactions were performed by using Dynabeads protein G (Invitrogen) according to the manufacturer's instructions. Briefly, 50 μl of Dynabeads was incubated for 10 min at room temperature with either 2 μl of anti-V5 antibody (Invitrogen) or 1 μl of anti-FLAG antibody (Sigma-Aldrich) for each reaction. After one washing step using the magnetic rack, 1 ml of clarified infected cell lysates was incubated for 15 min with the protein G antibody complexes. After immunoprecipitation and three washing steps, the eluted samples were prepared for sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western immunoblot analysis by the addition of sample buffer containing β-mercaptoethanol.
Transfections and Western immunoblot analysis.
Subconfluent Vero cells in six-well plates were transfected with 5 μg of plasmid gKa or control plasmid containing the gB gene sequence cloned in a reverse orientation using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's directions. Twenty-four hours posttransfection, cells were infected with the ΔgK31-68 virus at a multiplicity of infection (MOI) of 5, and infected cells were collected at 24 hpi. For the gB/UL20p interaction experiments, Vero cells were infected with either the V3032 or V3024 virus at an MOI of 5, and cell were collected at 24 hpi. All cells were washed with PBS and lysed on ice for 30 min in NP-40 lysis buffer supplemented with a cocktail of protease inhibitors (Invitrogen Life Technologies, Carlsbad, CA). Western immunoblot analysis was carried out essentially as described previously (10). Briefly, samples were electrophoretically separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Tris-HEPES-SDS gradient 4 to 20% gels; Thermo-Scientific, Inc., Rockford, IL). Following electrophoresis, two identical gels were transferred onto a nitrocellulose membrane under a constant current. Membranes were blocked in Tris-buffered saline containing 0.1% Tween 20 (TBST) plus 5% nonfat milk. Membranes were probed with either primary monoclonal anti-V5 antibody (1:2,000; Invitrogen) or monoclonal anti-FLAG antibody (1:4,000; Sigma-Aldrich), anti-gB monoclonal antibody (1:6,000; Virusys), or anti-gD monoclonal antibody (1:40,000; Virusys). Goat anti-mouse secondary antibody conjugated with horseradish peroxidase (HRP) (1:10,000; Abcam, Inc., Cambridge, MA) and ECL (GE Healthcare, Piscataway, NJ) substrate were used for detection purposes except for the immunoblots shown in Fig. 1 and 2. Results shown in Fig. 1 and 2 were produced by using Immobilon-Western chemiluminescence-HRP substrate (Millipore Corp., Billerica, MA). For results shown in Fig. 1A, the secondary antibody used was goat polyclonal antibody to the mouse IgG F(c)-F(ab)2 fragment conjugated with HRP (1:5,000; Abcam). For results shown in Fig. 1B and C and Fig. 2, the secondary antibody used was goat polyclonal antibody to the mouse IgG-F(ab)2-F(ab)2 fragment conjugated with HRP (1:5,000; Abcam). Clean-Blot IP detection reagent (HRP, dilution 1:200; Thermo-Scientific) and Immobilon-Western chemiluminescence-HRP substrate (Millipore Corp., Billerica, MA) were used to produce the immunoblots shown in Fig. 5B to assist in a better visualization of the gB364 truncation that overlapped with the IgG heavy chain. Certain samples used for Western immunoblot analysis were treated with endoglycosidase H (Endo H; New England BioLabs) according to the manufacturer's instructions.
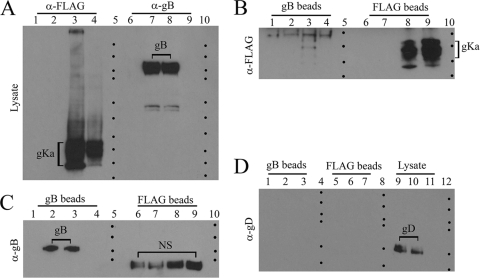
FIG. 1.
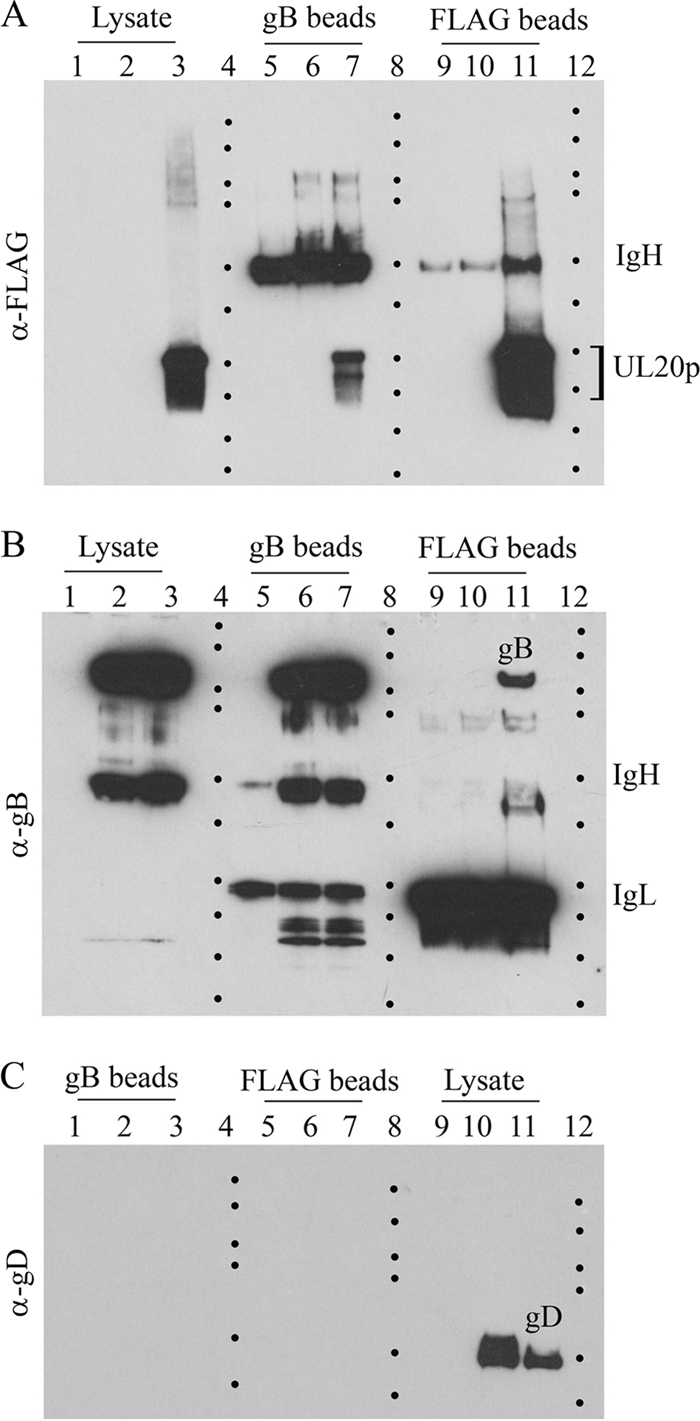
UL20p interacts with gB in virus-infected cells. Vero cell monolayers were infected with either the V3032 virus (having only gK tagged with the ProtC epitope) or the V3024 virus (having UL20p tagged with the 3×FLAG epitope and gK tagged with the ProtC epitope) or were mock infected. Cellular extracts were prepared at 24 hpi and in the presence of gB or UL20p and were visualized with Western immunoblots before or after immunoprecipitation with either anti-gB (α-gB) or anti-UL20p antibody. (A) Immunoblots probed with the anti-UL20p antibody (anti-3×FLAG). Lane 1, antigen from mock-infected cells; lane 2, V3032-infected cell extracts; lane 3, V3024-infected cell extracts; lane 5, mock-infected cell extracts immunoprecipitated with gB (gB beads) and then reacted with anti-UL20p antibody; lane 6, V3032-infected cell extracts immunoprecipitated with gB and reacted with anti-UL20p antibody; lane 7, V3024-infected cell extracts immunoprecipitated with anti-gB and reacted with the anti-UL20p antibody. Lanes 9, 10, and 11 are the same as lanes 5, 6, and 7, respectively, except that the initial immunoprecipitation was performed with anti-UL20p antibody and immunoblots were probed with anti-UL20p antibody. Lanes 4, 8, and 12 demarcate the relative positions of molecular mass markers (10 to 250 kDa; Bio-Rad). (B) Exact same arrangement of reactivity patterns shown above (A) except that the immunoblot was probed with anti-gB antibody.(C) Immunoblot probed with anti-gD antibody. Lanes 9, 10, and 11 are lysates from mock-infected cells, V3032-infected cells, and V3024-infected cells probed with the anti-gD antibody, respectively. Lanes 1, 2, and 3 are the same samples used for lanes 9, 10, and 11, respectively, except that the samples were first immunoprecipitated with anti-gB antibody prior to probing with the anti-gD antibody. Similarly, samples in lanes 5, 6, and 7 are the same samples shown in lanes 9, 10, and 11, respectively, except that the samples were first immunoprecipitated with anti-FLAG antibody and subsequently probed with anti-gD antibody.
FIG. 2.
The gKa peptide interacts with gB. Vero cell monolayers were mock transfected, transfected with a plasmid expressing the gKa peptide under the control of the HCMV-IE promoter, or a control plasmid containing the gB coding sequence in a reverse orientation. Twenty-four hours posttransfection, cells were infected with HSV-1(F) gKΔ31-68 virus, and after 24 hpi, cellular extracts were prepared and examined for the presence of gB or gKa before and after immunoprecipitation with either anti-gB or anti-FLAG (anti-gKa). (A) Lanes 1 to 4 show immunoblots reacted with anti-gKa antibody. Lane 1, mock-transfected cell extracts; lane 2, cellular extracts transfected with the control plasmid and subsequently infected with gKΔ31-68 virus; lane 3, cellular extracts transfected with the gKa plasmid and subsequently infected with gKΔ31-68; lane 4, gKa-transfected cellular extracts. Lanes 6 to 9 represent the same samples as those in lanes 1 to 4, respectively, except that the immunoblot was probed with anti-gB antibody. Lanes 5 and 10 demarcate the relative positions of the molecular mass markers (10 to 250 kDa). (B) The sample arrangement in of immunoblot is the same as that described above (A) except that all cellular extracts were first immunoprecipitated with either anti-gB (lanes 1 to 4) or anti-gKa (lanes 6 to 9) antibody and the immunoblot was probed with anti-gKa antibody. Lanes 5 and 10 demarcate the relative positions of the molecular mass markers (10 to 25 kDa). (C) The sample arrangement of the immunoblot is the same as that described above (B) except that the immunoblot was probed with anti-gB antibody. Lanes 5 and 10 demarcate the relative positions of the molecular mass markers (75 to 250 kDa). (D) Immunoblot probed with anti-gD antibody. Lane 1, cellular extracts transfected with control plasmid and subsequently infected with the gKΔ31-68 virus; lane 2, cellular extracts transfected with the gKa plasmid and subsequently infected with gKΔ31-68; lane 3, gKa-transfected cellular extracts. Samples represented in lanes 1, 2, and 3 were immunoprecipitated with anti-gB antibody and probed with anti-gD. Lanes 5 to 7 are the same as lanes 1 to 3, respectively, with the exception that samples were immunoprecipitated with the anti-FLAG antibody and probed with anti-gD antibody. Similarly, lanes 9 to 11 are lanes 1 to 3, respectively, probed with anti-gD antibody without any prior immunoprecipitation. Lanes 4, 8, and 12 demarcate the relative positions of the molecular mass markers (37 to 250 kDa).
FIG. 5.
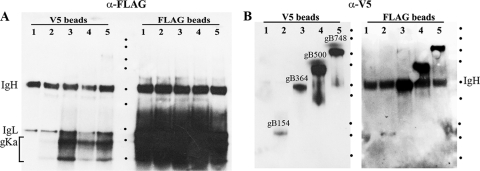
Coimmunoprecipitation experiments. Sf9 cellular extracts obtained after infection with each of the recombinant baculoviruses expressing gKa in combination with either gB, gD, or gH were used in coimmunoprecipitation experiments using the anti-FLAG antibody to precipitate gKa and anti-V5 antibody immobilized on magnetic beads to immunoprecipitate either gB, gD, or gH. The presence of gKa, gB, gD, and gH was detected in Western immunoblots by using anti-FLAG antibody immobilized on magnetic beads (A, B, and C) or anti-V5 antibody (D and E). B and E are lighter exposures of immunoblots shown in A and D, respectively. C shows replicate samples shown in A treated with Endo H. Lane 1, uninfected Sf9 cell lysates; lane 2, lysates from Sf9 cells infected with a baculovirus expressing gKa alone; lane 3, lysates containing gKa plus gD; lane 4, lysates containing gKa plus gH; lane 5, lysates containing gKa plus gB. Heavy and light chains of immunoglobulin are represented by IgH and IgL, respectively. Dots demarcate relative positions of the molecular mass ladder (250, 150, 100, 75, 50, 37, 25, 20, 15, and 10 kDa).
Structure prediction.
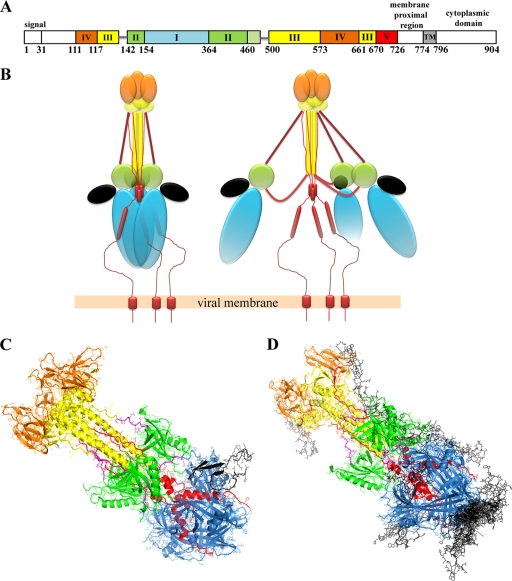
The PDB crystal structure file for gB (PDB accession number 2GUM) was downloaded from the RSCB Protein Data Bank (http://www.rcsb.org/pdb/home/home.do). The secondary structure of the 82-aa gKa was predicted by using the Protein Homology/Analogy Recognition Engine (PHYRE) algorithm (http://www.sbg.bio.ic.ac.uk/phyre/html/index.html) (26). The PDB output file with the highest estimation precision (20%) was used as the input into ZDOCK, version 3.0.1 (http://zdock.bu.edu/), along with the gB PDB file. ZDOCK utilizes pairwise shape complementarity, desolvation, and electrostatics to generate 2,000 possible protein-protein docking combinations (9). The 2,000 ZDOCK output files were further ranked by using ZRANK (33), which ranks ZDOCK outputs based on optimized energy function. The highest-ranking complex from the ZDOCK output is shown in Fig. 6C. The 15 top-ranking complexes juxtaposed on the gB structure are shown in Fig. 6D (the corresponding ZRANK scores are not shown).
FIG. 6.
Baculovirus expression of gB truncations with gKa. Additional recombinant baculoviruses coexpressing gB500, gB364, and gB154 with gKa were constructed. The anti-FLAG antibody (A) or the anti-V5 antibody (B) was used to detect the expression of gKa and gB truncations in Western immunoblots. Lane 1, lysates containing gK alone; lane 2, lysates containing gKa plus gB154; lane 3, lysates containing gKa plus gB364; lane 4, lysates containing gKa plus gB500; lane 5, lysates containing gKa plus gB748. Dots demarcate the relative positions of the molecular mass ladder (250, 150, 100, 75, 50, 37, 25, 20, and 15 kDa).
RESULTS
UL20p interacts with gB in HSV-1-infected cells.
We have shown previously that gK forms a functional complex with UL20p, which is absolutely necessary for their coordinate intracellular transport, cell surface expression, and functions of both gK and UL20p in virion assembly and virus-induced cell fusion (13, 14, 16, 28). The recombinant virus V3024 has gK and UL20p tagged with ProtC and 3×FLAG epitope tags inserted in frame at the amino terminus of UL20p and gK domain III, respectively (13). The anti-FLAG (anti-UL20p) antibody did not react with antigen obtained from mock- or V3032 virus-infected cells (the V3032 virus has only gK tagged with the ProtC epitope tag) but strongly reacted with UL20p obtained from V3024-infected cells, migrating with the expected molecular mass of 25 to 27 kDa (Fig. 1A, lanes 1, 2, and 3, respectively). The anti-FLAG (anti-UL20p) antibody did not react with anti-FLAG (anti-UL20p) immunoprecipitates from mock- or V3032-infected cells, while it reacted with UL20p expressed in V3024-infected cells (Fig. 1A, lanes, 9, 10, and 11, respectively). Similarly, the anti-FLAG (anti-UL20p) antibody did not react with anti-gB immunoprecipitates from mock- or V3032-infected cells but reacted with UL20p present in V3024-infected cell extracts (Fig. 1A, lanes 5, 6, and 7, respectively).
Reverse immunoprecipitation experiments were also performed to test whether gB could immunoprecipitate UL20p. The anti-gB antibody did not react with mock-infected cell extracts but readily detected gB in V3032- and V3024-infected cell extracts, migrating with the expected molecular mass of approximately 120 kDa (Fig. 1B, lanes 1, 2, and 3, respectively). The anti-gB antibody did not react with anti-gB immunoprecipitates from mock-infected cells but readily detected the presence of full-copy gB in anti-gB immunoprecipitates from V3032- and V3024-infected cells (Fig. 1B, lanes 5, 6, and 7, respectively). The anti-gB antibody did not react with anti-FLAG (anti-UL20p) immunoprecipitates from either mock- or V3032-infected cells, while it detected the presence of full-copy gB in anti-FLAG (anti-UL20p) immunoprecipitates from V3024-infected cells (Fig. 1B, lanes 9, 10, and 11, respectively). The anti-gD antibody did not react with lysates from mock-infected cells but readily detected gD expressed in V3032- and V3024-infected cell extracts (Fig. 1C, lanes 9, 10, and 11, respectively). The anti-gD antibody did not react with either anti-gB or anti-FLAG immunoprecipitates from mock-, V3032-, or V3024-infected cells (Fig. 1C, lanes 1, 2, 3, 5, 6, and 7, respectively).
The gKa peptide interacts with gB in HSV-1-infected cells.
We investigated whether the gKa peptide physically interacts with gB in infected cells. Vero cells were transfected with a plasmid expressing the gKa peptide under the control of the human cytomegalovirus (HCMV) immediate-early (IE) promoter or a control plasmid containing the gB coding sequence in a reverse orientation (predicted not to be able to encode any protein). The gKa peptide contains a 3×FLAG epitope tag inserted in frame at its carboxyl terminus. Twenty-four hours posttransfection, cells were infected with HSV-1(F) gKΔ31-68 virus, which lacks aa 31 to 68 in the amino terminus of gK (10). The anti-FLAG (anti-gKa) antibody did not react with Vero cell extracts derived from untransfected Vero cells or cells transfected with the control plasmid, while it reacted strongly with the gKa protein species from cells transfected with the gKa-expressing plasmid and subsequently infected with the gKΔ31-68 virus or transfected with the gKa-expressing plasmid alone (Fig. 2A, lanes 1, 2, 3, and 4, respectively). The anti-gB antibody did not react with cellular extracts obtained from Vero cells or gKa-transfected plasmid alone (Fig. 2A, lanes 6 and 9, respectively). However, gB was readily detected in cells transfected with either the control or gKa plasmids and infected with the gKΔ31-68 virus (Fig. 2A, lanes 7 and 8, respectively). The anti-FLAG (anti-gKa) antibody detected gKa in anti-gB immunoprecipitates obtained from cells transfected with the gKa-expressing plasmid and subsequently infected with the gKΔ31-68 virus (Fig. 2B, lane 3). There was no gKa detected in untreated cell extracts, cell extracts transfected with the control plasmid and infected with the gKΔ31-68 virus, or cell extracts transfected with the gKa plasmid alone (Fig. 2B, lanes 1, 2, and 4, respectively). The anti-FLAG (anti-gKa) antibody did not react with anti-gKa immunoprecipitates from mock-infected cell extracts or from cells transfected with a control plasmid and subsequently infected with the gKΔ31-68 virus (Fig. 2B, lanes 6 and 7, respectively). In contrast, the anti-FLAG (anti-gKa) antibody readily detected gKa protein species in anti-FLAG (anti-gKa) immunoprecipitates from gKa-transfected and subsequently gKΔ31-68 virus-infected cell extracts or from cell extracts transfected with gKa alone (Fig. 2B, lanes 8 and 9, respectively).
Reverse pulldown experiments were also performed to test whether the gKa peptide was able to immunoprecipitate full-length gB. The anti-gB antibody did not react with anti-gB immunoprecipitates from untreated cell extracts or from cell extracts transfected with the gKa plasmid alone (Fig. 2C, lanes 1 and 4, respectively). The anti-gB antibody readily detected full-copy gB in gB immunoprecipitates from control or gKa plasmid-transfected cells, which were subsequently infected with the gKΔ31-68 virus (Fig. 2C, lanes 2 and 3, respectively). Anti-FLAG (anti-gKa) immunoprecipitates probed with anti-gB antibody failed to clearly detect the presence of gB, although higher exposures of X-ray films indicated small amounts of gB present only in cell extracts transfected with gKa and subsequently infected with the gKΔ31-68 virus (not shown). In contrast, the anti-gD antibody did not react with any protein species present in anti-gB or anti-FLAG (anti-gKa) immunoprecipitates, while it was readily detected in whole lysates from cells infected with the gKΔ31-68 virus but not in cell extracts transfected with gKa alone (Fig. 2D, lanes 1, 2, and 3 for anti-gB immunoprecipitates; lanes 5, 6, and 7 for anti-FLAG; and lanes 9, 10, and 11 for whole lysates).
Expression of gB, gH, gD, and the gKa peptide in insect (Sf9) cells.
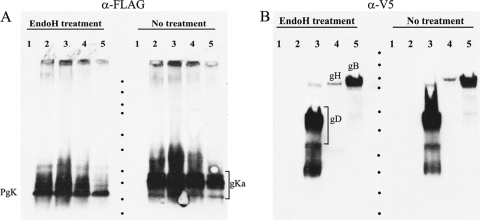
To further substantiate potential interactions between gKa and gB in a more robust expression system, recombinant baculoviruses VgKa, VgKa/gB748, VgKa/gH792, and VgKa/gD340 were constructed to simultaneously express the gKa peptide in combination with gB, gH, and gD, respectively (see Materials and Methods) (Fig. 3). All viral glycoproteins were efficiently detected in Western immunoblots of infected Sf9 cellular extracts using anti-V5 monoclonal antibody to detect gB, gD, or gH, since all three constructs contained the V5 epitope tag inserted in frame at the carboxyl terminus of each truncated gB, gD, or gH glycoprotein. The truncated viral glycoproteins gB and gD exhibited apparent molecular masses of approximately 95 to 100 kDa and 45 to 50 kDa, respectively. The truncated gH (1 to 792 aa) had an apparent molecular mass of approximately 100 kDa. Both gB748 and gD340 were efficiently expressed and detected, while gH792 appeared to be produced less efficiently than gB748 or gD340. Endoglycosidase H (Endo H) treatment of Sf9 lysates slightly reduced the apparent molecular masses of gB and gH, while a reduction in the molecular mass of gD was not apparent (Fig. 4B). The anti-FLAG (anti-gKa) antibody was used to detect the expression of the gKa peptide (Fig. 4A). The gKa peptide was efficiently expressed, migrating in SDS gels with an apparent molecular mass of approximately 15 to 22 kDa. Additional gKa-related protein species with larger apparent molecular masses were also detected, presumably representing multimeric forms or slower-migrating gKa-related species. Endo H treatment of Sf9 cellular extracts containing gKa reduced the apparent molecular mass of gK to a range of faster-migrating protein species, with the fastest gK-derived species migrating with an apparent molecular mass of 15 kDa (Fig. 4A).
FIG. 3.
Construction of recombinant baculoviruses. (A and B) The approximate locations of the herpesviral genes encoding the viral glycoproteins gH, gB, gK, and gD in the prototypical arrangement of the HSV-1 viral genome are shown. (C) The portions of gH, gB, gK, and gD expressed by the baculoviruses are gH at aa 1 to 792, gB at aa 30 to 748, gK at aa 1 to 82, and gD at aa 1 to 340. (D) Schematic of the gene cassettes expressing truncated gH792, gB748, gKa, and gD340 inserted in recombinant baculovirus genomes. The signal sequence of gB (aa 1 to 29) was replaced with the melittin signal sequence. The truncated gH, gD, and gK glycoproteins contain their native signal sequences. UL, unique long; US, unique short; TR, terminal repeat; IR, inverted repeat. Tn7R and Tn7L are transposon sites in the baculovirus bacmid. Pp10 and PPH are baculovirus promoters for the p10 and polyhedrin genes, respectively.
FIG. 4.
Expression and characterization of gB748, gH792, gD340, and gKa by recombinant baculoviruses. Sf9 cellular extracts obtained after infection with recombinant baculoviruses expressing gKa, gKa plus gB, gKa plus gH, and gKa plus gD were separated by SDS-PAGE, and the expression of gKa was detected by using anti-FLAG antibody (A), while the expression of gB, gD, and gH was detected by using anti-V5 antibody (B) in Western immunoblots. Replicate samples were also treated with Endo H. PgK denotes the putative precursor to gKa. Lane 1, uninfected Sf9 cell lysates; lane 2, lysates from Sf9 cells infected with a baculovirus expressing gKa alone; lane 3, lysates containing gKa plus gD; lane 4, lysates containing gKa plus gH; lane 5, lysates containing gKa plus gB. The locations of the major gB, gD, gH, and gKa species are shown. Dots demarcate the relative positions of the molecular mass ladder (250, 150, 100, 75, 50, 37, 25, 20, 15, and 10 kDa).
Physical interactions between gKa and gB, gD, or gH.
To assess whether the gKa peptide interacted with either gB, gD, or gH, coimmunoprecipitation experiments were performed by using either anti-FLAG (anti-gKa) or anti-B, -gD, or -gH antibodies (anti-V5) in pairs, as described in Materials and Methods. The gKa peptide was efficiently detected in anti-FLAG (anti-gKa) immunoprecipitates from Sf9 cellular extracts containing gKa alone, gKa plus gD, gK plus gH, and gK plus gB probed with the anti-FLAG (anti-gKa) antibody (Fig. 5A). The gKa peptide was expressed at similar levels by all baculoviruses, which is easily visualized in a shorter film exposure of the same immunoblot (Fig. 5B). The gKa peptide was detected in anti-gB and anti-gH immunoprecipitates in insect cell extracts containing gKa plus gB and gKa plus gH but not in cell extracts expressing gKa plus gD or gKa alone (Fig. 3A). To better visualize gKa-associated protein species, cellular extracts were treated with Endo H after immunoprecipitation. The 15-kDa gKa was detected only in the presence of gB and gH coexpression (Fig. 5C). Similar experiments were performed to detect truncated gB, gD, and gH glycoproteins. The truncated gB, gD, and gH glycoproteins were detected in anti-V5 immunoprecipitates probed with the anti-V5 antibody (Fig. 5D). Similarly, gB and gH, but not gD, were detected in anti-FLAG (anti-gKa) immunoprecipitates obtained from cellular extracts containing gKa plus gB, gKa plus gH, and gKa plus gD probed with anti-V5 antibody (Fig. 3D). Because the truncated gD migrated with an apparent molecular mass similar to that of the IgG heavy chain, a lighter exposure of the same immunoblot shown in Fig. 3D was obtained to improve the visualization of gD only for the anti-V5 immunoprecipitates probed with the anti-V5 antibody (Fig. 5E).
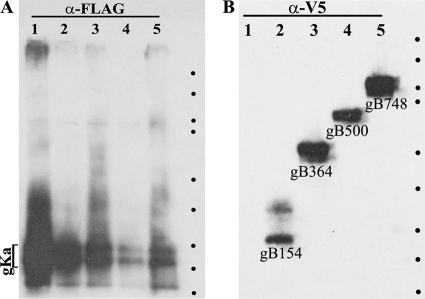
Mapping of gB domains that interact with the gKa peptide.
To delineate gB domains that interact with the gKa peptide, additional recombinant baculoviruses coexpressing gKa with truncated gB proteins of 500, 364, and 154 amino acids were produced as described above (see also Materials and Methods). The gKa peptide was readily detected in Western immunoblots of Sf9 cellular extracts infected with these recombinant baculoviruses using the anti-FLAG (anti-gKa) antibody (Fig. 6A). Similarly, the truncated gBs were detected in Western immunoblots using anti-V5 antibody migrating with approximate apparent molecular masses of 95 to 100 kDa (gB748), 60 to 65 kDa (gB500), 45 to 50 kDa (gB364), and 18 to 20 kDa (gB154). gB154 appeared to produce two protein species, with the 18- to 20-kDa species being more prominent than a 30-kDa protein species (Fig. 6B).
Anti-V5 immunoprecipitates of Sf9 cellular extracts containing either gKa plus gB748, gKa plus gB500, gKa plus gB364, or gKa plus gB154 probed with anti-V5 antibody revealed the presence of truncated gB proteins with apparent molecular masses of approximately 95 to 100 kDa, 60 to 65 kDa, 45 to 50 kDa, and 18 to 20 kDa, respectively. Conversely, anti-FLAG (anti-gKa) immunoprecipitates probed with the anti-V5 antibody clearly revealed the presence of gB748, gB500, and gB154. gB364 comigrated with the IgG heavy chain, but it was readily detected due to the substantial increase in band intensity over the IgG heavy-chain background (Fig. 7B). Similarly, the gKa peptide was present only in gB-containing anti-V5 immunoprecipitates probed with the anti-FLAG antibody but not in the sample that expressed gKa alone. All anti-FLAG (anti-gKa) immunoprecipitates contained large amounts of the gKa peptide detected with the anti-FLAG antibody (Fig. 7A).
FIG. 7.
gKa interacts with truncated gB proteins. Coimmunoprecipitation experiments were performed as explained in the legend of Fig. 3. gKa and gB were detected in Western immunoblots using anti-FLAG (A) or anti-V5 (B) antibody. Lane 1, lysates containing gK alone; lane 2, lysates containing gKa plus gB154; lane 3, lysates containing gKa plus gB364; lane 4, lysates containing gKa plus gB500; lane 5, lysates containing gKa plus gB748. Heavy and light chains of immunoglobulin are represented by IgH and IgL, respectively. Dots demarcate the relative positions of the molecular mass ladder (250, 150, 100, 75, 50, 37, 25, 20, and 15 kDa).
DISCUSSION
HSV-1 extensively utilizes membrane fusion for virus entry into cells as well as for spreading from infected to uninfected cells. These membrane fusion events are thought to be mediated by the concerted action of viral glycoproteins and other membrane proteins, among which the glycoproteins gB, gD, gH, and gL are key players. Although the viral glycoprotein gK and the membrane protein UL20p are known to be intimately involved in virus-induced cell fusion, their role in these membrane fusion phenomena remain undefined. Recently, we showed that a peptide composed of the 82 amino-terminal amino acids of gKa complemented virus-induced cell fusion caused by a syncytial mutation in the carboxyl terminus of gB, providing for the first time a functional association between gK and gB (10). In this work, we show that the gKa peptide interacts with both the extracellular portion of gH and the amino terminus of gB. Furthermore, UL20p, the functional partner of gK, interacts with full-length gB in virus-infected cells. This work provides for the first time experimental evidence that the gK/UL20p protein complex is part of the virus-induced fusion machine.
We have previously shown that gK and UL20p physically interact and that this interaction is absolutely essential for their coordinate intracellular transport, gK glycosylation, expression on cell surfaces, and functions in the virus life cycle (13, 14, 16, 27, 28). We chose to demonstrate physical interactions between UL20p and gB because we had previously constructed the recombinant virus V3024, which expresses a UL20p protein containing a 3×FLAG epitope inserted in frame at the amino terminus of UL20p. The anti-3×FLAG antibody is particularly strong in immunoprecipitating UL20p from infected cell extracts, which greatly facilitated these experiments. Two-way coimmunoprecipitation experiments clearly showed that UL20p physically interacted with gB but not gD. Furthermore, the immunoprecipitation of gB resulted in the coimmunoprecipitation of the gKa peptide, suggesting that the ability of the gKa peptide to complement the gK31-68-deleted virus for gB-mediated cell fusion (10) was due to direct interactions with gB. Attempts to immunoprecipitate wild-type gB with the gKa antibody failed, although a weak coimmunoprecipitation of gB was noted in multiple experiments (not shown). These data suggest that the gKa peptide did not interact sufficiently strongly to retain associations with the much larger wild-type gB under these immunoprecipitation conditions.
Interactions of gKa with gB were further substantiated by using baculovirus-expressed extracellular portions of gB as well as gH. It was previously shown that HSV-1 gH is stably expressed in insect cell cultures, migrating to cell surfaces in the absence of its interacting partner gL (20). The gKa peptide was efficiently expressed and N glycosylated in Sf9 cells. Endoglycosidase H digestion produced a peptide with an apparent molecular mass of 15 kDa, in agreement with the predicted gKa molecular mass. Two-way immunoprecipitation experiments established that the extracellular portions of gB and gH interacted with the gKa peptide in a specific manner, since this interaction was not observed with gD, which was abundantly expressed in insect cells. Apparently, gKa interacted with all truncated gB proteins, indicating that gB154 is the minimum gB domain that can physically interact with gKa. These results suggest that gKa most likely interacted with a linear domain of gB154, since it is unlikely that this small portion of gB assumes a conformational structure similar to that found for full-length gB. The lower levels of gH expression in Sf9 cells hampered somewhat the visualization of the reactive protein species. However, it is noted that pulldown experiments with anti-V5 (gH) antibody coimmunoprecipitated significant amounts of the gKa peptide. Similarly, the anti-FLAG (gKa) pulldown immunoprecipitated a significant portion of the expressed gH, suggesting a strong interaction between gKa and gH.
The postfusion X-ray structure of the HSV-1 gB737 extracellular portion has been produced to a 2.1-Å resolution, revealing a trimeric form resembling that of the VSV G fusogenic glycoprotein (23). The analysis of this structure showed the presence of individual domains, which are proximal to membrane sequences that may play direct roles in membrane fusion. This gB structure includes a triple-coiled-coil domain that is predicted to interact directly with membranes as well as other domains that may provide for gB conformations that have increased fusogenic potential (23). Based on the coimmunoprecipitation experiments with the gKa peptide, it can be deduced that gKa interacts with amino acid segments that are part of domains II, III, and IV included within the gB154 peptide but may also interact with multiple segments contained within domain I and part of domain II (gB364) (Fig. 6A). Based on the fact that gB364 was more strongly immunoprecipitated than gB154, it can be concluded that if multiple linear domains interact with gK, the linear amino acid subdomains within domain I (contained within gB364) interact with greater affinity than linear amino acid subdomains contained within domains II, III, and IV (contained within gB154). Additional work will be needed in the future to verify the specificity of the interaction between the amino terminus of gK and amino acid domains within the amino terminus of gB.
Previously, we showed that the gKa peptide functionally complemented gB-mediated cell fusion, suggesting that it may assume the proper three-dimensional structure required for binding and modulating gB's fusogenic properties. Therefore, in an attempt to relate the experimental observations produced by the coimmunoprecipitation experiments with the overall three-dimensional structure of gB, the three-dimensional structure of the gKa peptide spanning amino acids 26 to 82 was predicted by using the PHYRE algorithm (26) (Fig. 8). This program automatically excluded amino acids 1 to 26 as being noninformative with regard to structure prediction. Protein docking calculations using ZDOCK and the subsequent ranking of interacting complexes using ZRANK showed that gKa could interact with multiple sites on the gB molecule; however, most of these interactions involved gB domains I and II (Fig. 8D). The top-ranked gKa-gB complex depicted an interaction of gKa with gB domain I (Fig. 8C). This theoretical result supports the stronger binding observed with the gB364 truncation that contains all of gB domain I. Therefore, collectively, the experimental and prediction results suggest that there are two interactive sites between gKa and gB: one site is composed of amino acids within domain I, and the second site is composed of amino acids derived from either part of domain III or IV. Furthermore, because it is unlikely that these domains fold in a fashion similar to that found for the entire extracellular portion of gB, these gB domains interacting with gKa must be of a linear nature, composed of amino acids proximal to each other.
FIG. 8.
Schematic representation of potential interactions between gK and the three-dimensional X-ray structure of gB derived by protein docking algorithms. (A) Linear domain architecture of gB as previously shown by Heldwein et al. (23). (B) Schematic representation showing gKa interacting with gB domains I and II (23). (C) Schematic representation of the top-ranked gKa-gB complex produced by the ZDOCK and ZRANK algorithms. (D) Schematic representation of 15 top-ranked positions where gKa may interact with gB.
Based on the fact that the gKa peptide is located at the amino terminus of gK, which is predicted to protrude proximal to the extracellular surface of plasma membranes, we suggest that the gKa peptide interacts with the gB portion that is similarly located proximal to the extracellular plasma membrane surfaces and contained within domains I and II, as shown previously by Heldwein et al. (23) (Fig. 8B). The fact that gK is absolutely required for virus-induced cell fusion suggests that gK functions as a regulator of virus-induced cell fusion, causing the limited amounts of virus-induced cell fusion typically observed for wild-type-infected cells. In this hypothesis, syncytial mutations in gK enhance virus-induced cell fusion by physically tilting the regulatory balance of gK toward a more fusogenic gB and/or gH status. In this regard, previous results suggesting that gK may inhibit glycoprotein-mediated cell fusion in transient expression experiments (4) may suggest that additional virus-specified proteins are involved.
Collectively, data from this laboratory and many others are pointing to complex interactions among gD, gB, gH, gL, gK, and UL20 that control virus-induced cell fusion. The biological significance of this highly complicated virus-induced cell fusion machinery remains largely unresolved and will be the subject of future experiments.
Acknowledgments
This work was supported by grant AI43000 from the National Institute of Allergy and Infectious Diseases to K.G.K. We acknowledge financial support by the LSU School of Veterinary Medicine to BIOMMED.
We acknowledge Yong-Hwan Lee for helpful comments on the structure prediction analysis of gKa.
Footnotes
Published ahead of print on 23 June 2010.
REFERENCES
- 1.Atanasiu, D., J. C. Whitbeck, T. M. Cairns, B. Reilly, G. H. Cohen, and R. J. Eisenberg. 2007. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc. Natl. Acad. Sci. U. S. A. 104:18718-18723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avitabile, E., C. Forghieri, and G. Campadelli-Fiume. 2007. Complexes between herpes simplex virus glycoproteins gD, gB, and gH detected in cells by complementation of split enhanced green fluorescent protein. J. Virol. 81:11532-11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avitabile, E., C. Forghieri, and G. Campadelli-Fiume. 2009. Cross talk among the glycoproteins involved in herpes simplex virus entry and fusion: the interaction between gB and gH/gL does not necessarily require gD. J. Virol. 83:10752-10760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avitabile, E., G. Lombardi, and G. Campadelli-Fiume. 2003. Herpes simplex virus glycoprotein K, but not its syncytial allele, inhibits cell-cell fusion mediated by the four fusogenic glycoproteins, gD, gB, gH, and gL. J. Virol. 77:6836-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baines, J. D., P. L. Ward, G. Campadelli-Fiume, and B. Roizman. 1991. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J. Virol. 65:6414-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bond, V. C., and S. Person. 1984. Fine structure physical map locations of alterations that affect cell fusion in herpes simplex virus type 1. Virology 132:368-376. [DOI] [PubMed] [Google Scholar]
- 7.Bzik, D. J., B. A. Fox, N. A. DeLuca, and S. Person. 1984. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology 137:185-190. [DOI] [PubMed] [Google Scholar]
- 8.Campadelli-Fiume, G., F. Cocchi, L. Menotti, and M. Lopez. 2000. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev. Med. Virol. 10:305-319. [DOI] [PubMed] [Google Scholar]
- 9.Chen, R., L. Li, and Z. Weng. 2003. ZDOCK: an initial-stage protein-docking algorithm. Proteins 52:80-87. [DOI] [PubMed] [Google Scholar]
- 10.Chouljenko, V. N., A. V. Iyer, S. Chowdhury, D. V. Chouljenko, and K. G. Kousoulas. 2009. The amino terminus of herpes simplex virus type 1 glycoprotein K (gK) modulates gB-mediated virus-induced cell fusion and virion egress. J. Virol. 83:12301-12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Debroy, C., N. Pederson, and S. Person. 1985. Nucleotide sequence of a herpes simplex virus type 1 gene that causes cell fusion. Virology 145:36-48. [DOI] [PubMed] [Google Scholar]
- 12.Foster, T. P., X. Alvarez, and K. G. Kousoulas. 2003. Plasma membrane topology of syncytial domains of herpes simplex virus type 1 glycoprotein K (gK): the UL20 protein enables cell surface localization of gK but not gK-mediated cell-to-cell fusion. J. Virol. 77:499-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster, T. P., V. N. Chouljenko, and K. G. Kousoulas. 2008. Functional and physical interactions of the herpes simplex virus type 1 UL20 membrane protein with glycoprotein K. J. Virol. 82:6310-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster, T. P., J. M. Melancon, J. D. Baines, and K. G. Kousoulas. 2004. The herpes simplex virus type 1 UL20 protein modulates membrane fusion events during cytoplasmic virion morphogenesis and virus-induced cell fusion. J. Virol. 78:5347-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster, T. P., J. M. Melancon, and K. G. Kousoulas. 2001. An alpha-helical domain within the carboxyl terminus of herpes simplex virus type 1 (HSV-1) glycoprotein B (gB) is associated with cell fusion and resistance to heparin inhibition of cell fusion. Virology 287:18-29. [DOI] [PubMed] [Google Scholar]
- 16.Foster, T. P., J. M. Melancon, T. L. Olivier, and K. G. Kousoulas. 2004. Herpes simplex virus type 1 glycoprotein K and the UL20 protein are interdependent for intracellular trafficking and trans-Golgi network localization. J. Virol. 78:13262-13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster, T. P., G. V. Rybachuk, and K. G. Kousoulas. 2001. Glycoprotein K specified by herpes simplex virus type 1 is expressed on virions as a Golgi complex-dependent glycosylated species and functions in virion entry. J. Virol. 75:12431-12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gage, P. J., M. Levine, and J. C. Glorioso. 1993. Syncytium-inducing mutations localize to two discrete regions within the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B. J. Virol. 67:2191-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 20.Ghiasi, H., A. B. Nesburn, and S. L. Wechsler. 1991. Cell surface expression of herpes simplex virus type 1 glycoprotein H in recombinant baculovirus-infected cells. Virology 185:187-194. [DOI] [PubMed] [Google Scholar]
- 21.Gianni, T., M. Amasio, and G. Campadelli-Fiume. 2009. Herpes simplex virus gD forms distinct complexes with fusion executors gB and gH/gL in part through the C-terminal profusion domain. J. Biol. Chem. 284:17370-17382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hannah, B. P., E. E. Heldwein, F. C. Bender, G. H. Cohen, and R. J. Eisenberg. 2007. Mutational evidence of internal fusion loops in herpes simplex virus glycoprotein B. J. Virol. 81:4858-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heldwein, E. E., H. Lou, F. C. Bender, G. H. Cohen, R. J. Eisenberg, and S. C. Harrison. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217-220. [DOI] [PubMed] [Google Scholar]
- 24.Hutchinson, L., K. Goldsmith, D. Snoddy, H. Ghosh, F. L. Graham, and D. C. Johnson. 1992. Identification and characterization of a novel herpes simplex virus glycoprotein, gK, involved in cell fusion. J. Virol. 66:5603-5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobson, J. G., S. H. Chen, W. J. Cook, M. F. Kramer, and D. M. Coen. 1998. Importance of the herpes simplex virus UL24 gene for productive ganglionic infection in mice. Virology 242:161-169. [DOI] [PubMed] [Google Scholar]
- 26.Kelley, L. A., and M. J. Sternberg. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363-371. [DOI] [PubMed] [Google Scholar]
- 27.Melancon, J. M., T. P. Foster, and K. G. Kousoulas. 2004. Genetic analysis of the herpes simplex virus type 1 UL20 protein domains involved in cytoplasmic virion envelopment and virus-induced cell fusion. J. Virol. 78:7329-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melancon, J. M., P. A. Fulmer, and K. G. Kousoulas. 2007. The herpes simplex virus UL20 protein functions in glycoprotein K (gK) intracellular transport and virus-induced cell fusion are independent of UL20 functions in cytoplasmic virion envelopment. Virol. J. 4:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melancon, J. M., R. E. Luna, T. P. Foster, and K. G. Kousoulas. 2005. Herpes simplex virus type 1 gK is required for gB-mediated virus-induced cell fusion, while neither gB and gK nor gB and UL20p function redundantly in virion de-envelopment. J. Virol. 79:299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 31.Muggeridge, M. I. 2000. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J. Gen. Virol. 81:2017-2027. [DOI] [PubMed] [Google Scholar]
- 32.Pellett, P. E., K. G. Kousoulas, L. Pereira, and B. Roizman. 1985. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of monoclonal antibody-resistant mutants. J. Virol. 53:243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pierce, B., and Z. Weng. 2007. ZRANK: reranking protein docking predictions with an optimized energy function. Proteins 67:1078-1086. [DOI] [PubMed] [Google Scholar]
- 34.Pogue-Geile, K. L., G. T. Lee, S. K. Shapira, and P. G. Spear. 1984. Fine mapping of mutations in the fusion-inducing MP strain of herpes simplex virus type 1. Virology 136:100-109. [DOI] [PubMed] [Google Scholar]
- 35.Pogue-Geile, K. L., and P. G. Spear. 1987. The single base pair substitution responsible for the syn phenotype of herpes simplex virus type 1, strain MP. Virology 157:67-74. [DOI] [PubMed] [Google Scholar]
- 36.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 37.Ruyechan, W. T., L. S. Morse, D. M. Knipe, and B. Roizman. 1979. Molecular genetics of herpes simplex virus. II. Mapping of the major viral glycoproteins and of the genetic loci specifying the social behavior of infected cells. J. Virol. 29:677-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanders, P. G., N. M. Wilkie, and A. J. Davison. 1982. Thymidine kinase deletion mutants of herpes simplex virus type 1. J. Gen. Virol. 63:277-295. [DOI] [PubMed] [Google Scholar]
- 39.Satoh, T., J. Arii, T. Suenaga, J. Wang, A. Kogure, J. Uehori, N. Arase, I. Shiratori, S. Tanaka, Y. Kawaguchi, P. G. Spear, L. L. Lanier, and H. Arase. 2008. PILRalpha is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell 132:935-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shukla, D., J. Liu, P. Blaiklock, N. W. Shworak, X. Bai, J. D. Esko, G. H. Cohen, R. J. Eisenberg, R. D. Rosenberg, and P. G. Spear. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99:13-22. [DOI] [PubMed] [Google Scholar]
- 41.Spear, P. G., R. J. Eisenberg, and G. H. Cohen. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1-8. [DOI] [PubMed] [Google Scholar]
- 42.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]