Abstract
Recently, we reported the discovery and characterization of Tulane virus (TV), a novel rhesus calicivirus (CV) (T. Farkas, K. Sestak, C. Wei, and X. Jiang, J. Virol. 82:5408-5416, 2008). TV grows well in tissue culture, and it represents a new genus within Caliciviridae, with the proposed name of Recovirus. We also reported a high prevalence of CV antibodies in macaques of the Tulane National Primate Research Center (TNPRC) colony, including anti-norovirus (NoV), anti-sapovirus (SaV), and anti-TV (T. Farkas, J. Dufour, X. Jiang, and K. Sestak, J. Gen. Virol. 91:734-738, 2010). To broaden our knowledge about CV infections in captive nonhuman primates (NHP), 500 rhesus macaque stool samples collected from breeding colony TNPRC macaques were tested for CVs. Fifty-seven (11%) samples contained recovirus isolates. In addition, one NoV was detected. Phylogenetic analysis classified the recovirus isolates into two genogroups and at least four genetic types. The rhesus NoV isolate was closely related to GII human NoVs. TV-neutralizing antibodies were detected in 88% of serum samples obtained from primate caretakers. Binding and plaque reduction assays revealed the involvement of type A and B histo-blood group antigens (HBGA) in TV infection. Taken together, these findings indicate the zoonotic potential of primate CVs. The discovery of a genetically diverse and prevalent group of primate CVs and remarkable similarities between rhesus enteric CVs and human NoVs opens new possibilities for research involving in vitro and in vivo models of human NoV gastroenteritis.
Caliciviruses (CV) are important human and animal pathogens, causing a wide variety of diseases in their respective hosts. The family Caliciviridae consists of five established genera (Norovirus, Sapovirus, Lagovirus, Vesivirus, and Nebovirus). Recently, two new calicivirus genera have been proposed, represented by the Tulane virus (Recovirus) and the St. Valerien-like viruses (Valovirus) (11-13, 24, 36, 37, 39).
NoVs are recognized as the leading cause of epidemics of gastroenteritis (GE), causing 80 to 90% of nonbacterial GE outbreaks and more than 50% of all food-related GE outbreaks (7, 8, 29). They are also an important cause of sporadic GE in both children and adults. Based on phylogenetic analysis, NoVs are divided into five genogroups and more than 30 genetic clusters or genotypes (9, 46). This high genetic and, likely, antigenic variation, combined with the lack of a tissue culture or animal model, represent major obstacles for NoV research.
NoVs with close genetic and antigenic relatedness to human NoVs have been isolated from various animal species (6, 28, 33, 41). This not only provided opportunities for using some of these viruses as surrogates for human NoV research (44) but also raised the concern of the possible zoonotic nature of CV gastroenteritis.
Based on results of in vitro binding assays, volunteer challenge studies, and the analysis of NoV outbreaks, it was proposed that histo-blood group antigens (HBGA), including the ABO, Lewis, and secretor-type HBGAs, function as the NoV receptors (17, 19, 20, 27, 32). The involvement of other host factors in NoV replication and susceptibility to infection also has been implicated (14, 43).
Previously, we reported the isolation and characterization of a novel CV (Tulane virus; TV) from stool samples of juvenile rhesus macaques (11). TV represents a newly proposed genus (Recovirus) within Caliciviridae that phylogenetically shares a common origin with NoVs; however, TV can be grown in tissue culture (11). We also reported a high prevalence of anti-NoV, anti-SaV binding, and anti-TV-neutralizing (VN) antibodies in colony macaques, suggesting that CV infections are frequent in captive nonhuman primates (NHP) (10). The few NoV challenge studies conducted also suggest that NHPs are susceptible to NoV infection. Chimpanzees inoculated with the Norwalk virus developed seroresponses and virus shedding but without the manifestation of clinical disease (45). Subekti et al. reported the development of clinical illness characterized by diarrhea, dehydration, vomiting, and virus shedding in newborn pigtail macaques inoculated with the Toronto virus (40). In a study conducted by Rockx et al., one of the three rhesus macaques infected with Norwalk virus developed virus-specific IgM and IgG responses and shed the virus for 19 days postinoculation (38). To date, however, direct evidence of natural NoV or SaV infection in NHPs is missing. Moreover, the prevalence and genetic diversity of recoviruses have yet to be studied.
In this study, we undertook the molecular detection and genetic analysis of CVs circulating in colony macaques and examined the role of HBGAs in recovirus infection.
MATERIALS AND METHODS
Sample collection.
Stool, saliva, and serum samples were collected between April and July of 2008 from 500 randomly selected juvenile (≤3-year-old) rhesus macaques (Macaca mulatta) of Indian origin during the semiannual TNPRC breeding colony inventory. A rectal loop stool sample was collected from each animal, and ∼20% (wt/vol) stool suspensions were prepared in phosphate-buffered saline, pH 7.4 (PBS). Saliva samples were collected by cotton-tipped applicators that were immersed into 0.5 ml sterile PBS after sampling. Four ml of EDTA blood was obtained from each macaque, and serum was harvested. Samples were aliquoted and stored at −80°C.
All samples were obtained in accordance with the standards of the Association for Assessment and Accreditation of Laboratory Animal Care. Only animals negative for simian immunodeficiency virus, simian retrovirus, simian T-cell leukemia virus, and Cercopithecine herpesvirus (B virus) were used. Investigators adhered to the Guide for the Care and Use of Laboratory Animals prepared by the National Research Council. NHP samples were handled in compliance with biosafety level 2+ laboratory practices approved by the institutional biosafety committees of the participating institutions.
Saliva samples were collected from adult human volunteers at Cincinnati Children's Hospital Medical Center by following a protocol approved by the Institutional Review Board.
Virus stock.
In this study, to keep mutations at the lowest level, TV stock was prepared from plaque-purified TV at the third passage. Virus stock was aliquoted and stored at −80°C.
RNA extraction and RT-PCR.
Viral RNA was extracted by the QIAamp viral RNA mini kit on a QIAvac 24 plus vacuum manifold (Qiagen Inc., Valencia, CA) according to the manufacturer's instructions. Twenty-two stool samples along with negative (deionized water) and positive (cell culture-adapted TV of 104 PFU/ml) controls were extracted at a time. Extracted RNA was eluted into 30 μl buffer and stored at −80°C. CV-specific RNA was amplified from 3 μl of extracted RNA template using the AccessQuick reverse transcription-PCR (RT-PCR) system (Promega, Madison, WI), according to the manufacturer's instructions, with modified P289/P290 primers (9, 22) that target the RNA-dependent RNA polymerase region (RdRp). These primers produce 313-, 319-, and 331-bp products for TV, NoV, and SaV, respectively. RT-PCR conditions were as described previously (11).
DNA sequencing.
RT-PCR products were excised from agarose gels, recovered by a QIAquick gel extraction kit (Qiagen Inc., Valencia, CA), and cloned into pGEM-T vector (Promega, Madison, WI) according to the manufacturers' protocols. Positive clones were identified by PCR. Plasmid DNA was isolated from 2-ml cultures by a QIAprep spin miniprep kit (Qiagen Inc., Valencia, CA) according to the manufacturer's instructions and sequenced using M13 forward and reverse primers by the chain termination method on an ABI PRISM 3730 DNA analyzer (Applied Biosystems Inc., Foster City, CA). Each sample was sequenced in both directions from two independent clones.
Phylogenetic analysis.
Multiple-sequence alignments of partial RdRp nucleotide sequences were created using Omiga v2.0 software (Oxford Molecular Ltd., Oxford, United Kingdom). Dendrograms were constructed by the unweighted pair group method with arithmetic mean (UPGMA) and the neighbor-joining clustering methods of the Molecular Evolutionary Genetics Analysis (MEGA version 3.1) software with Jukes-Cantor distance calculations. The confidence values of the internal nodes were obtained by performing 1,025 bootstrap analyses. The following GenBank sequences were used in the analysis: Boxer (AF538679), Chiba (AB042808), Desert Storm (U04469), Gifu96 (AB045603), Hawaii (U07611), Hesse (AF093797), Hokkaido/83(AB231342), Lordsdale (X86557), Melksham (X81879), Mexico (U22498), Mex7076(AY579431), MOH (AF397156), Neustrelitz260 (AY772730), Norwalk (M87661), Pont de Roide 671 (AY682548), Southampton (L07418), Su4 (AB039777), Su25 (AB039780), SW918 (AB074893), SzUG1 (AB039774), Tulane/M33 (EU391643), VA207 (AY038599), Vietnam026 (AF504671), and Winchester (AJ277609). The GenBank accession numbers of partial RdRp sequences of the recovirus isolates and the rhesus NoV are HM035091 to HM035147 and HM035148, respectively.
VN assay.
A cytopathic effect (CPE)-based VN assay was performed with heat-inactivated (50°C, 30 min) serum samples and 100 50% tissue culture infectious doses (TCID50) of the prototype TV as described previously (10). All rhesus serum samples were tested in duplicate wells, with 2-fold dilutions starting from 1:10. In addition to rhesus serum samples, 100 archived human serum samples collected from TNPRC animal caretakers in the 1990s also were tested starting from a 1:2 dilution.
Confirmation of NoV in a rhesus macaque stool sample.
Based on the NoV sequence (FT244) obtained from the macaque stool sample with P289/P290, one forward (P121/131F, 5′-CCAGCTTGATGTAGGCGATT-3′) and two reverse (P121R, 5′-CTCAGCCATTGCACTCAAAG-3′, and P131R, 5′-GACCTGACACCTCAGCCATT-3′) strain-specific primers were designed. These primers produced a 121- and a 131-bp amplicon, respectively, within the RdRp region. To further corroborate the presence of NoV in the macaque stool and to eliminate the possibility of sample contamination at Cincinnati Children's Hospital Medical Center (CCHMC), two additional aliquots of the original sample were retested at the TNPRC using the strain-specific primers.
HBGA phenotyping.
The phenotyping of rhesus and human saliva samples was performed by enzyme-linked immunosorbent assay (ELISAs) using commercially available monoclonal antibodies (MAbs) specific to ABH and Lewis (Le) antigens as previously described (17). MAbs used in this study included anti-A (ABO1), anti-B (ABO2) (Diagast, Loos, France), anti-H type 1 (BG-4), anti-Lea (BG-5), anti-Leb (BG-6), anti-Lex (BG-7), and anti-Ley (BG-8) (Covance, Berkeley, CA). Rhesus saliva samples recovered from swabs were tested at a 1:25 dilution, and human saliva samples were tested at a 1:500 dilution. All samples were tested in duplicate wells.
HBGA binding.
Twenty-four human saliva samples (eight each of A, B, and O type) and type A and type B trisaccharides conjugated to bovine serum albumin (BSA) (Glycorex AB, Lund, Sweden) or to biotin-labeled polyacrylamide (PAA) (GlycoTech, Gaithersburg, MD) were used to evaluate the HBGA binding properties of the prototype TV, as described previously for NoV virus-like particles (VLPs) (17).
Briefly, ELISA plates (Corning Life Sciences, Lowell, MA) were coated with boiled human saliva samples at a 1:500 dilution, or with BSA-conjugated type A or B trisaccharide (1 μg/well), overnight at 4°C. To coat the biotin-labeled PAA-conjugated type A and B trisaccharides, ELISA plates were precoated with Neutravidin (1 μg/well) (Thermo Fisher Scientific Inc., Waltham, MA) for 3 h at room temperature, blocked with 5% skim milk-PBS, and incubated with the biotin-PAA type A or B trisaccharide (1 μg/well) overnight at 4°C. All plates were blocked with 5% skim milk-PBS, and 108 TCID50/well purified TV in 1% skim milk-PBS was added. Saliva- or synthetic oligosaccharide-coated wells without TV served as negative controls. Plates were incubated for 2 h at 37°C, and virus binding was detected by hyperimmune anti-TV mouse sera and horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG, IgA, and IgM (H+L) (Invitrogen, Carlsbad, CA). Samples were analyzed in at least two separate experiments. TV binding was established based on a P/N ratio of >2 and an optical density (OD) value equal to or more than the means of negative controls plus 2 standard deviations (SD).
Plaque reduction.
Saliva samples (n = 24) at a 1:100 dilution or type A and B synthetic oligosaccharides (0.1 to 10 μg/ml) were mixed with 50 to 100 PFU of TV and incubated for 1 h at 37°C. The following synthetic oligosaccharides were tested: BSA-conjugated type A and B trisaccharides (Glycorex AB, Lund, Sweden), PAA-conjugated type A and B trisaccharides (GlycoTech, Gaithersburg, MD), and A-Leb pentasaccharide and B-Leb pentasaccharide (Sigma-Aldrich, St. Louis, MO). Samples, including virus controls, were adsorbed to LLC-MK2 monolayers in 6-well tissue culture plates (Corning Life Sciences, Lowell, MA) for 1 h at 37°C. Plates were washed twice and overlaid with M199 culture medium containing 0.8% methyl cellulose. Plates were stained with crystal violet on day 4 postinfection, and plaques were counted. Each sample was tested in at least two wells in two separate experiments. Blocking efficiency was calculated by 1 − (PFU of sample/PFU of virus control) and expressed as a percentage.
Statistical analysis.
Statistical significance in binding and plaque reduction assays was calculated by a two-tailed t test with unequal variances, and P < 0.05 was considered significant.
RESULTS
Molecular detection of enteric CVs in colony rhesus macaques.
Fifty-eight (11.6%) of the 500 stool samples yielded CV-specific PCR product. All amplicons were cloned and sequenced, revealing 57 recovirus (268 bp) and 1 NoV sequence (274 bp). The recovirus sequences exhibited 61 to 90% nucleotide and 62 to 90% amino acid homology with the prototype TV (EU391643). The NoV sequence (FT244) revealed the highest nucleotide homology (94%) with human NoV isolates EU072307 and EU007752 from the GenBank depository.
Confirmation of NoV in rhesus macaque stool.
The amplification of the NoV-specific sequence from rhesus macaque stool sample was confirmed by repeated detection and sequencing at CCHMC. Since the CCHMC laboratory routinely handles human NoV samples, it was necessary to exclude the possibility of laboratory contamination. Therefore, aliquots of the original stool sample that were kept at the TNPRC were tested with NoV-specific primers, yielding sequences identical to those detected at CCHMC, thus confirming the authenticity of rhesus NoV strain FT244.
Phylogenetic analysis.
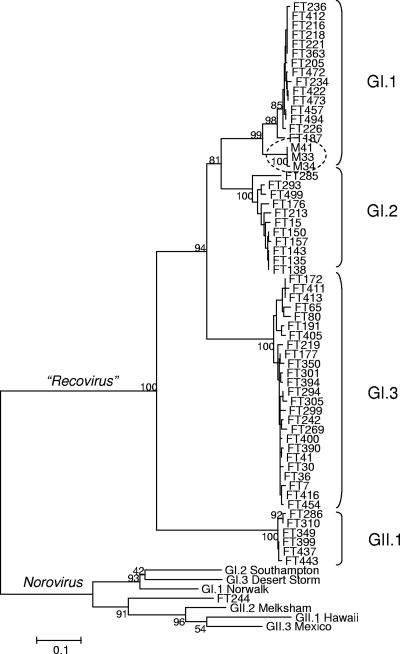
Phylogenetic analysis clearly divided the 57 recovirus isolates into four distinct groups, suggesting the existence of at least four recovirus genotypes within two genogroups (Fig. 1). Among the 57 isolates, 15 (26%), 11 (19%), 25 (44%), and 6 (11%) grouped to GI.1, GI.2, GI.3, and GII.1, respectively. The mean intergenogroup distances between the GI and GII recoviruses (0.536 to 0.613) were comparable to distances between the GI and the GII NoVs (0.534 to 0.695) (Table 1). Similarly, the mean distances between the recovirus genotypes within GI (0.251 to 0.359) were comparable to distances between the GI or GII NoV genotypes (0.289 to 0.352 and 0.251 to 0.346, respectively).
FIG. 1.
Rhesus enteric caliciviruses are genetically diverse and can be classified into four genetic types within the two genogroups. The dendrogram was constructed by the neighbor-joining clustering method of the MEGA (version 3.1) software with Jukes-Cantor distance calculations. The confidence values of the internal nodes were obtained by performing 1,025 bootstrap analyses. The prototype Tulane virus (M33) isolated in 2004 from the TNPRC rhesus colony is circled.
TABLE 1.
Phylogenetic distance scores between recovirus and norovirus genetic typesa
| Genogroup | Recovirus |
Norovirus |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| GI.1 | GI.2 | GI.3 | GII.1 | GI.1 | GI.2 | GI.3 | GII.1 | GII.2 | |
| GI.2 | 0.251 | ||||||||
| GI.3 | 0.359 | 0.321 | |||||||
| GII.1 | 0.536 | 0.540 | 0.613 | ||||||
| GI.1 | 1.182 | 1.080 | 1.034 | 1.100 | |||||
| GI.2 | 1.114 | 1.086 | 1.177 | 1.084 | 0.289 | ||||
| GI.3 | 1.210 | 1.049 | 1.097 | 1.205 | 0.340 | 0.352 | |||
| GII.1 | 1.458 | 1.390 | 1.202 | 1.264 | 0.598 | 0.598 | 0.573 | ||
| GII.2 | 1.210 | 1.091 | 1.122 | 1.254 | 0.658 | 0.623 | 0.589 | 0.251 | |
| GII.3 | 1.314 | 1.213 | 1.170 | 1.205 | 0.615 | 0.534 | 0.695 | 0.317 | 0.346 |
Between-group average phylogenetic distances were calculated based on nucleic acid sequence alignments of the corresponding partial RdRp region of recoviruses (268 bp) and noroviruses (274 bp). Group assignments of recovirus isolates were based on those shown in Fig. 1. Norovirus genogroups were represented by the Norwalk (G1.1), Southampton (GI.2), Desert Shield (GI.3), Hawaii (GII.1), Melksham (GII.2), and Mexico (GII.3) viruses. Numbers in bold are discussed in the text.
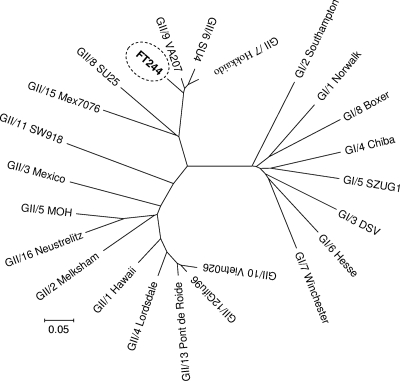
Phylogenetic analysis consistently placed the rhesus NoV isolate (FT244) in a branch rooting together with GII.6, GII.7, and GII.9 NoVs (Fig. 2). Multiple-sequence alignments and distance calculations revealed 90, 93, and 89% homology scores and distances of 0.088, 0.058, and 0.073 between FT244 and the GII.6, GII.7, and GII.9 NoV sequences used in the alignment, respectively.
FIG. 2.
Rhesus NoV isolate is closely related to human NoV isolates. The dendrogram, based on the alignment of 274-nucleotide RdRp sequences, was constructed by the neighbor-joining clustering method of the MEGA (version 3.1) software with Jukes-Cantor distance calculations. The rhesus NoV (FT244) isolate is circled.
Rhesus enteric CV infection and diarrhea.
Among the 500 breeding colony juvenile animals selected for this study, 101 (20%) exhibited clinical signs of diarrhea at any time within a 1-month period prior to sample collection. Eleven of the 57 (19%) recovirus-positive animals had a clinical history of watery diarrhea during the same period. Since diarrhea can be caused in NHPs by a variety of causes, and most of the animals in this study possessed TV-neutralizing antibodies, a controlled (experimental challenge) study with preselected seronegative animals is needed to corroborate or disprove the association between recovirus infection and clinical disease. Preparations for such an experiment are currently in progress.
TV-neutralizing antibodies and infection status.
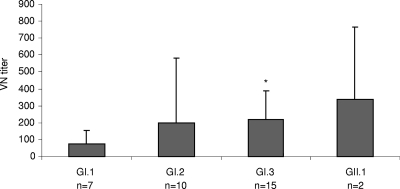
Serum samples collected at the time of the stool sample collection were available from 7 of the 15 GI.1, 10 of the 11 GI.2, 15 of the 25 GI.3, and 2 of the 6 GII.1 recovirus-infected animals. VN titers against the prototype TV (GI.1) ranged from ≤10 to 160 in GI.1, from 20 to ≥1,280 in GI.2, from 20 to 640 in GI.3, and from 40 to 640 in GII.1 recovirus-infected animals. The mean VN titer was lower in animals infected with GI.1 viruses than that in animals infected with other genotypes. Although statistically significant differences in VN titers were observed only between GI.1- and GI.3-infected animals (Fig. 3), such differences between genotypes may indicate homotypic antibody protection and the existence of serotypes.
FIG. 3.
Mean VN titers against the prototype TV (GI.1) in macaques with virus shedding. Animals were grouped by the genetic types of the recovirus isolates. VN titers were higher in animals infected with GI.2, GI.3, or GII.1 strains than in animals infected with GI.1 strains. Statistical significance was calculated between GI.1 (test virus) and the other groups using two-tailed t test with unequal variances. P < 0.05 was considered significant. Error bars represent SD. n, number of samples available for VN; *, P < 0.05.
TV-neutralizing antibodies in humans.
Overall, 88% of the 100 human serum samples tested neutralized the prototype TV at ≥1:2 dilution, indicating the potential for the zoonotic transmission of TVs. The distribution of end titers was 1:256 (2%), 1:128 (1%), 1:64 (0%), 1:32 (3%), 1:16 (15%), 1:8 (22%), 1:4 (22%), and 1:2 (23%).
HBGA phenotypes and infection status.
Saliva-based ABO phenotyping of the 500 animals revealed that these animals represented an almost-homotypic population. A total of 485 animals (97%) secreted the B antigen (type B), 2 animals (0.4%) secreted both A and B antigen (type AB), and 13 animals (2.6%) had no detectable level of A or B antigen in their saliva (type O). Eleven of the 13 O-type rhesus saliva contained the Lewis b (Leb) antigen, and all 13 were negative for the Lewis a (Lea) antigen, indicating that all, or perhaps all but two, of the animals in this study were secretor type. Among the 57 macaques that shed virus in their stools, 55 (96%) were type B and 2 (4%) were type O. One of the type O animals was infected with a GI.1 (FT218) and the other with a GI.3 (FT269) virus. To evaluate the possible association between Lewis types and infectious status, the 57 recovirus-positive animals, along with the 13 type O animals and 60 randomly selected VN antibody-negative animals, were typed for Lea, Leb, Lex, and Ley antigens. None of the animals expressed detectable levels of Lea antigen. The distribution of Leb, Lex, and Ley antigens was comparable among all animals regardless of infection status (infected versus VN negative). Furthermore, no difference in the distribution of Lewis antigens was found between animals infected by different genotypes (Table 2). Due to the homotypic distribution of HBGAs in the subset of 500 macaques tested, recovirus infection could not be linked to a particular HBGA type, although both type B and type O macaques were infected with recoviruses. Future studies of macaques possessing more-heterogeneous HBGA distribution are needed to determine a potential association between HBGA phenotypes and susceptibility to recovirus infection in a population-based study as described for NoVs (3, 23, 35).
TABLE 2.
Distribution of HBGAs in macaques assigned for this studya
| Genogroup (n) | No. (%) of HBGA: |
|||||||
|---|---|---|---|---|---|---|---|---|
| A | B | AB | O | Lea | Leb | Lex | Ley | |
| Colony (500) | 0 | 485 (97) | 2 (0.4) | 13 (2.6) | ND | ND | ND | ND |
| GI.1 infected (15) | 0 | 14 (93) | 0 | 1 (6) | 0 | 11 (73) | 7 (47) | 14 (93) |
| GI.2 infected (11) | 0 | 11 (100) | 0 | 0 | 0 | 7 (64) | 5 (45) | 11 (100 |
| GI.3 infected (25) | 0 | 24 (96) | 0 | 1 (4) | 0 | 20 (80) | 16 (64) | 24 (96) |
| GII.1 infected (6) | 0 | 6 (100) | 0 | 0 | 0 | 5 (83) | 3 (50) | 6 (100) |
| Total infected (57) | 0 | 55 (96) | 0 | 2 (4) | 0 | 43 (75) | 31 (54) | 55 (96) |
| VN negative (60) | 0 | 58 (97) | 0 | 2 (3) | 0 | 42 (70) | 31 (52) | 60 (100) |
All 500 animals were tested for ABO phenotypes. The recovirus-positive animals (n = 57) and 60 animals without detectable levels of VN antibodies against the prototype TV also were tested for Lea, Leb, Lex, and Ley. Saliva-based ELISA using specific MAbs was used. GI.1 to GI.3 and GII.1 indicate recovirus genotypes.
Saliva HBGA binding patterns of the prototype TV.
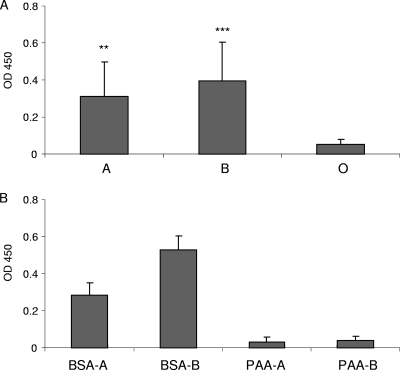
The prototype TV bound to six of the eight type A and seven of the eight type B saliva samples. None of the type O saliva samples exhibited binding (Fig. 4 A). No association of binding with Lewis antigens could be established in any of the three groups. The eight type O saliva samples represented all four Lewis types screened for in this study.
FIG. 4.
Saliva and synthetic oligosaccharide binding of the prototype TV. Eight type A, type B, and type O saliva samples (A) and BSA- or PAA-conjugated type A and type B trisaccharides (B) were tested, respectively. Mean OD values of separate experiments were calculated. Error bars represent SD. **, P < 0.01; ***, P < 0.0005.
Synthetic oligosaccharide binding of the prototype TV.
Both BSA- and PAA-conjugated type A and B synthetic oligosaccharides were recognized by the corresponding anti-A (ABO1) or anti-B (ABO2) (Diagast, Loos, France) MAb. OD values were comparable to those of saliva samples (OD of 1.5 to 2), suggesting that both conjugated oligosaccharides contained the corresponding sugar moieties and were efficiently coated onto the plates. Nevertheless, TV binding was observed only with BSA-conjugated type A and B trisaccharides but not with PAA-conjugated ones (Fig. 4B).
Saliva plaque reduction assays.
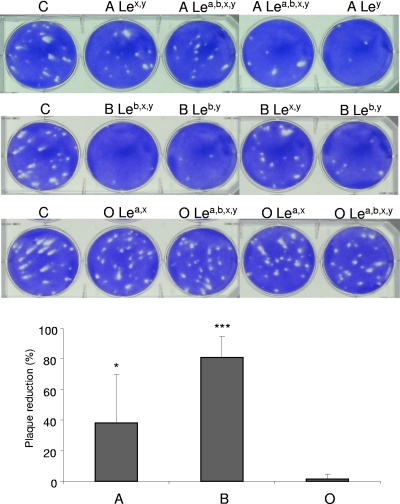
In agreement with the saliva binding assays, both type A and type B saliva samples caused significant plaque reduction, while type O samples had no effect (Fig. 5) regardless of the Lewis type. The type A, B, and O saliva samples had 7 to 79%, 50 to 90%, and 0 to 7% plaque reduction effect, respectively. Since variation among the virus control wells was ∼10%, reductions of ≤10% were not considered. Similarly to the binding assays, an association between plaque reduction and any of the Lewis types tested was not evident.
FIG. 5.
Saliva blocking of TV replication. Type A and type B, but not type O, saliva samples blocked TV replication. Eight type A, type B, and type O saliva samples were tested individually. C, virus control. ABO and Lewis types are indicated. Plaque reduction was calculated by 1 − (PFU of sample/PFU of virus control) and is expressed as a percentage. The means were calculated from the results of two independent tests with two different TV concentrations (∼50 and ∼100 PFU). Error bars represent SD. *, P < 0.05; ***, P < 0.0005.
Synthetic oligosaccharide plaque reduction assay.
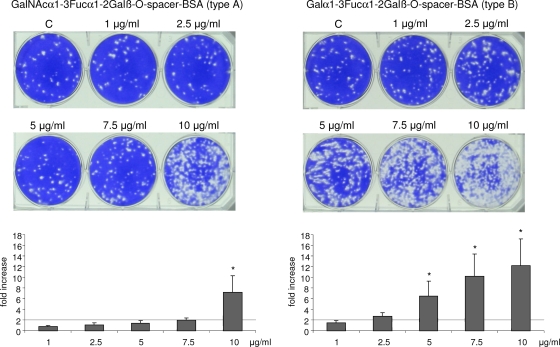
The pentasaccharide structures (ALeb and BLeb) and the PAA-conjugated trisaccharides exhibited no effect on the plaque numbers at any of the concentrations tested (0.1 to 20 μg/ml) (data not shown). The BSA-conjugated type A or B trisaccharides had no effect at concentrations of less than 1 μg/ml. However, a dose-dependent 2- to 12-fold increase in plaque formation was observed at higher concentrations, starting at 2.5 μg/ml with the BSA-conjugated type B trisaccharide and at 7.5 μg/ml with the type A trisaccharide, respectively (Fig. 6).
FIG. 6.
BSA-conjugated type A and type B trisaccharides increase TV plaque numbers in a concentration-dependent manner. C, virus control. Fold increase/decrease was calculated by PFU of sample/PFU of control. The mean was calculated from the results of two independent tests with two TV concentrations (∼50 and ∼100 PFU). Error bars represent SD. P values indicate statistical significance. *, P < 0.05.
DISCUSSION
Recently, we reported the high prevalence of NoV-, SaV-, and TV-specific antibodies in captive rhesus macaques (10). Although the existence of cross-reactive binding antibodies between human NoVs and the prototype TV suggested that anti-NoV antibodies detected in NHPs (10, 21) are a consequence of TV infections, there also is evidence suggesting that NHPs are susceptible to at least experimental NoV infection (38, 40, 45). In this study, we targeted the molecular detection of CVs in TNPRC rhesus macaques with a generic primer set (P289/P290) that has been used previously for the detection of a variety of CVs (11, 15, 22, 24, 33). A high prevalence of recoviruses was detected, with 57 (11.6%) of the 500 stool samples containing viruses closely related (62 to 90% nucleotide homology) to the prototype TV. According to phylogenetic analysis, the recovirus strains detected in this study can be grouped into four genetic types within two genogroups. Of the 57 isolates, 15 (26%), 11 (19%), 25 (44%), and 6 (11%) grouped to GI.1, GI.2, GI.3, and GII.1, respectively (Fig. 1). Such genetic diversity of recoviruses and circulation of genetically distinct strains in the TNPRC colony resembles the features of human NoVs. All the recoviruses detected in this study exhibited at least a 10% difference in nucleotides compared to those of the prototype TV, which was isolated from stool samples collected in 2004 from the same colony (11). The existence of homologous antibody protection and the existence of distinct serotypes that likely are associated with genotypes were suggested by the high prevalence (69%) of TV (GI.1)-neutralizing antibodies in the colony (herd immunity) and by the higher TV-neutralizing antibody titers in animals shedding viruses of heterologous (other than GI.1) genotypes (Fig. 3). Consequently, tissue culture adaptation of the different recovirus strains, identification of serotypes, assessment of protective level of VN antibodies, T- and B-cell epitope mapping, and relation of antigenic epitopes to carbohydrate binding sites will provide valuable information for NoV vaccine design and development.
Interestingly, in one rhesus macaque stool sample, a NoV was detected. The susceptibility of NHPs to experimental NoV infection was demonstrated previously (38, 40, 45). Nevertheless, this report is the first to describe the natural NoV infection of a NHP. Laboratory contamination was excluded by the repeated detection of the NoV from different aliquots of the original sample in two collaborating laboratories. Since stool samples were collected as individual rectal loops, contamination during sample collection could be excluded. The rhesus NoV isolate exhibited a 89 to 93% nucleotide homology with GII.6, GII.7, and GII.9 human NoVs (Fig. 2). Since the separation of these genotypes in the RdRp region analyzed is poor, capsid-based (ORF2) sequence analysis of strain FT244 will be necessary to determine its exact genotype. Nevertheless, data generated in this study clearly placed the rhesus NoV isolate in a close proximity with human NoV isolates, indicating the possibility of interspecies transmission. Despite the high prevalence of anti-NoV and anti-SaV antibodies in the TNPRC colony (10), only one NoV-infected animal was detected in this study, and no SaV was found. Whether the low detection rate of NoVs and SaVs compared to the high seroprevalence is due to the seasonal occurrence of these viruses in colony macaques, the unfitness of the primers, or other reasons remains to be elucidated. The possible zoonotic nature of enteric CV infections also is suggested by the high prevalence of TV-neutralizing antibodies in serum samples of animal caretakers and the involvement of HBGAs in TV infection. The end titers of human samples were lower than the end titers of NHP samples. However, these human samples were collected nearly 20 years ago, and antibody titers could be reduced during long-term storage. Parallel investigation of the epidemiology of enteric CVs in human and NHP populations therefore would be of interest.
Based on results obtained from in vitro binding assays and volunteer challenge studies, HBGAs have been identified as the putative cellular receptors for human NoVs (16, 17, 19, 20). The association between the HBGA binding abilities of NoVs and susceptibility to infection was established for some strains (27); however, for others, such a link was not found (26), indicating that different NoVs use different strategies to establish infection, and that factors other than HBGAs also might be involved. Since there is no effective tissue culture or animal model available to study this relationship, the potential involvement of HBGAs in recovirus infection was examined in this study. Despite the fact that NHPs and humans have four phenotypes of ABO blood groups (A, B, O, and AB), only humans and the anthropoid apes express the ABH antigens on the surface of red blood cells (1). Both New and Old World monkeys secrete ABH antigens in their saliva and possess anti-A and/or anti-B antibodies in their sera. Thus, ABO blood groups of rhesus macaques can be determined by the detection of A/B antigens in saliva or antibodies in serum. In this study, based on the reported involvement of ABO, Lewis, and host secretor status in NoV infection (16, 17, 20), saliva-based assays were used for the HBGA phenotyping of rhesus macaques. All commercially available MAbs used were confirmed to recognize the corresponding HBGA antigens except the anti-H type I (BG-4) antibody, which showed no reaction with any of the human saliva samples, even at a 1:10 dilution.
Saliva-based ABO phenotyping of the 500 animals used in this study revealed an almost-homotypic population, with 97% of the animals being type B. These results created difficulty in linking any particular ABO phenotype to susceptibility to recovirus infection or in excluding that possibility. Similarly, no association could be established for a particular Lewis phenotype. Among the 57 recovirus-positive macaques, 55 (96%) were type B and 2 (4%) were type O. The recovirus infection of type O animals also was indicated by the presence of VN serum antibodies. Thus, at least both type B and type O macaques are susceptible to recovirus infection. To assess the role of type A antigen and the possible differences among the different strains in population-based studies, macaques with a more-polymorphic distribution of the ABO types need to be tested.
Our findings are consistent with previous studies that reported group B as the major blood type (∼97%) found in rhesus macaques (25, 34). On the other hand, Malaivijitnond et al. described the polymorphism of the ABO blood group in rhesus macaques in Thailand, although variation among troops from different geographic locations was significant (30). Since ABO typing is not included in the routine assessment of research colony macaques, the identification of a colony with polymorphic HBGA distribution may pose a challenge.
In saliva binding assays, the prototype TV bound to both type A and B saliva but not to type O saliva (Fig. 4A). The type A and type B binding was confirmed by the binding of the prototype TV to BSA-conjugated type A and B trisaccharides (Fig. 4B). Interestingly, no binding to the PAA-conjugated type A or B trisaccharides was observed, even though the anti-A (ABO1) and anti-B (ABO2) MAbs recognized both of the corresponding BSA- and PAA-conjugated trisaccharides. Similar discrepancies have been noted previously for NoV VLP binding (17, 18). Huang et al. demonstrated that while specific MAbs recognize the HBGAs present in human milk or saliva regardless of the carrier molecule, the binding of NoV VLPs depends on both the specific carbohydrate structure and the carrier molecule (18). Marionneau et al. suggested that the optimal binding of NoVs to the carbohydrate ligands requires an optimal density of the ligand, and binding may disappear at low densities (31). Since PAA-conjugated trisaccharides with various carbohydrate-to-PAA ratios were not available for our study, this possibility remains to be elucidated.
Both previous observations with NoV VLPs and our experiments with TV indicate that the nature of carrier molecules plays a critical role in the binding of enteric CVs to carbohydrate structures. HBGA binding interfaces of different NoV strains have been characterized by cocrystallizing NoV P domain dimers with tri- and pentasaccharides (2, 4, 5). In our study, there was no demonstrable interaction between the prototype TV and unconjugated type A and B pentasaccharide structures or PAA-conjugated type A and B trisaccharides. This indicates that interaction between enteric CVs and HBGAs is more complex than is described in these structural studies, and future studies with more-complex HBGA structures are necessary.
Results of plaque reduction assays confirmed the involvement of the type A and B HBGAs in recovirus infection. The preincubation of the prototype TV with type A or B saliva samples resulted in a significant reduction of plaque numbers (Fig. 5). Type O saliva samples, the PAA-conjugated type A and B trisaccharides, and ALeb and BLeb pentasaccharides had no effect in plaque assays (data not shown). Contrary to our expectation, both BSA-conjugated type A and B trisaccharides increased the plaque numbers by 2- to 12-fold in a dose-dependent manner. This effect started at a lower concentration with the BSA-conjugated type B (2.5 μg/ml) than the type A (7.5 μg/ml) trisaccharide, indicating a higher-affinity interaction with the prototype TV.
These observations create several controversies. According to the ability of saliva samples to block TV infectivity in vitro, saliva should act as an antiviral during natural infection. It also is difficult to explain how, contrary to saliva samples, BSA-conjugated synthetic oligosaccharides increased TV infectivity. One explanation is the involvement of coreceptor-receptor interactions in TV infection. We hypothesize that attachment to the HBGA (coreceptor) is required to prime TV, possibly through conformational changes, to enable interaction with a yet-unknown cell surface molecule (receptor). In saliva, HBGA structures are attached to a wide variety of molecules, including high-molecular-weight mucins. It was shown that NoV VLPs bind to HBGA structures attached to high-molecular-weight carriers but not to HBGAs attached to low-molecular-weight carriers present in human milk and saliva (18). These large carriers of HBGAs, possibly due to steric hindrance, interfered with receptor binding when saliva was used in our TV plaque assays. On the other hand, the smaller BSA molecules had no interference with the receptor binding site. Assuming that TV receptor and coreceptor molecules need to be in physical proximity to support optimal attachment and entry, isolated receptor molecules far from the coreceptor will not be able to support virus attachment and entry. In the case of HBGA-primed TV virions, these isolated receptor molecules alone could be sufficient, which will lead to the increased efficiency of infection and plaque numbers.
The involvement of coreceptor/receptor interactions in enteric CV infections also is supported by previous studies showing that the cell surface expression of HBGAs alone does not confer susceptibility to NoV infection, even in cell types that are able to support the intracellular steps of NoV replication (14, 43).
Finally, the discrepancy with saliva acting as a natural antiviral in vitro could be explained by a yet-unknown mechanism (pH, digestive enzymes, etc.) that in the stomach or small intestine dissociates the HBGA-mucin complexes or otherwise digests the large carrier structures, thus exposing the receptor interaction sites.
In summary, this study describes the high prevalence of genetically diverse rhesus enteric CVs, in conjunction with evidence that suggests the zoonotic potential of enteric CV infections and the involvement of HBGAs in recovirus infection. Although more-detailed studies of recovirus HBGA interactions are necessary, including the characterization of the different isolates described in this study and in vivo studies to link HBGAs to susceptibility to recovirus infection, the data presented here, together with our previous reports (11, 42), collectively indicate the potential that recoviruses can be used to address research questions applicable to NoV gastroenteritis.
Acknowledgments
We thank Amanda Tardo and Diana Taft for their excellent technical assistance.
Grants from the National Institutes of Health-NCRR and NIAID R21RR024871 and P51RR000164-47 to K.S. were used to support this study. Partial salary support to T.F. was provided by NICHD grant HD013021 to A.M.
Footnotes
Published ahead of print on 16 June 2010.
REFERENCES
- 1.Apoil, P. A., F. Roubinet, S. Despiau, R. Mollicone, R. Oriol, and A. Blancher. 2000. Evolution of alpha 2-fucosyltransferase genes in primates: relation between an intronic Alu-Y element and red cell expression of ABH antigens. Mol. Biol. Evol. 17:337-351. [DOI] [PubMed] [Google Scholar]
- 2.Bu, W., A. Mamedova, M. Tan, M. Xia, X. Jiang, and R. S. Hegde. 2008. Structural basis for the receptor binding specificity of Norwalk virus. J. Virol. 82:5340-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bucardo, F., E. Kindberg, M. Paniagua, A. Grahn, G. Larson, M. Vildevall, and L. Svensson. 2009. Genetic susceptibility to symptomatic norovirus infection in Nicaragua. J. Med. Virol. 81:728-735. [DOI] [PubMed] [Google Scholar]
- 4.Cao, S., Z. Lou, M. Tan, Y. Chen, Y. Liu, Z. Zhang, X. C. Zhang, X. Jiang, X. Li, and Z. Rao. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 81:5949-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, J. M., A. M. Hutson, M. K. Estes, and B. V. Prasad. 2008. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc. Natl. Acad. Sci. U. S. A. 105:9175-9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dastjerdi, A. M., J. Green, C. I. Gallimore, D. W. Brown, and J. C. Bridger. 1999. The bovine Newbury agent-2 is genetically more closely related to human SRSVs than to animal caliciviruses. Virology 254:1-5. [DOI] [PubMed] [Google Scholar]
- 7.Fankhauser, R. L., S. S. Monroe, J. S. Noel, C. D. Humphrey, J. S. Bresee, U. D. Parashar, T. Ando, and R. I. Glass. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1-7. [DOI] [PubMed] [Google Scholar]
- 8.Fankhauser, R. L., J. S. Noel, S. S. Monroe, T. Ando, and R. I. Glass. 1998. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J. Infect. Dis. 178:1571-1578. [DOI] [PubMed] [Google Scholar]
- 9.Farkas, T., W. M. Zhong, Y. Jing, P. W. Huang, S. M. Espinosa, N. Martinez, A. L. Morrow, G. M. Ruiz-Palacios, L. K. Pickering, and X. Jiang. 2004. Genetic diversity among sapoviruses. Arch. Virol. 149:1309-1323. [DOI] [PubMed] [Google Scholar]
- 10.Farkas, T., J. Dufour, X. Jiang, and K. Sestak. 2010. Detection of norovirus-, sapovirus- and rhesus enteric calicivirus-specific antibodies in captive juvenile macaques. J. Gen. Virol. 91:734-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farkas, T., K. Sestak, C. Wei, and X. Jiang. 2008. Characterization of a rhesus monkey calicivirus representing a new genus of Caliciviridae. J. Virol. 82:5408-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green, K., R. Chanock, and A. Kapikian. 2001. Human caliciviruses, p. 841-874. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 13.Green, K. Y., T. Ando, M. S. Balayan, T. Berke, I. N. Clarke, M. K. Estes, D. O. Matson, S. Nakata, J. D. Neill, M. J. Studdert, and H. J. Thiel. 2000. Taxonomy of the caliciviruses. J. Infect. Dis. 181(Suppl. 2):S322-S330. [DOI] [PubMed] [Google Scholar]
- 14.Guix, S., M. Asanaka, K. Katayama, S. E. Crawford, F. H. Neill, R. L. Atmar, and M. K. Estes. 2007. Norwalk virus RNA is infectious in mammalian cells. J. Virol. 81:12238-12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo, M., J. F. Evermann, and L. J. Saif. 2001. Detection and molecular characterization of cultivable caliciviruses from clinically normal mink and enteric caliciviruses associated with diarrhea in mink. Arch. Virol. 146:479-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, P., T. Farkas, S. Marionneau, W. Zhong, N. Ruvoen-Clouet, A. L. Morrow, M. Altaye, L. K. Pickering, D. S. Newburg, J. LePendu, and X. Jiang. 2003. Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: identification of 4 distinct strain-specific patterns. J. Infect. Dis. 188:19-31. [DOI] [PubMed] [Google Scholar]
- 17.Huang, P., T. Farkas, W. Zhong, M. Tan, S. Thornton, A. L. Morrow, and X. Jiang. 2005. Norovirus and histo-blood group antigens: demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns. J. Virol. 79:6714-6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, P., A. L. Morrow, and X. Jiang. 2009. The carbohydrate moiety and high molecular weight carrier of histo-blood group antigens are both required for norovirus-receptor recognition. Glycoconj. J. 26:1085-1096. [DOI] [PubMed] [Google Scholar]
- 19.Hutson, A. M., R. L. Atmar, D. Y. Graham, and M. K. Estes. 2002. Norwalk virus infection and disease is associated with ABO histo-blood group type. J. Infect. Dis. 185:1335-1337. [DOI] [PubMed] [Google Scholar]
- 20.Hutson, A. M., R. L. Atmar, D. M. Marcus, and M. K. Estes. 2003. Norwalk virus-like particle hemagglutination by binding to histo-blood group antigens. J. Virol. 77:405-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, B., H. M. McClure, R. L. Fankhauser, S. S. Monroe, and R. I. Glass. 2004. Prevalence of rotavirus and norovirus antibodies in non-human primates. J. Med. Primatol. 33:30-33. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, X., P. W. Huang, W. M. Zhong, T. Farkas, D. W. Cubitt, and D. O. Matson. 1999. Design and evaluation of a primer pair that detects both Norwalk- and Sapporo-like caliciviruses by RT-PCR. J. Virol. Methods 83:145-154. [DOI] [PubMed] [Google Scholar]
- 23.Le Guyader, F. S., J. Krol, K. Ambert-Balay, N. Ruvoen-Clouet, B. Desaubliaux, S. Parnaudeau, J. C. Le Saux, A. Ponge, P. Pothier, R. L. Atmar, and J. Le Pendu. Comprehensive analysis of a norovirus-associated gastroenteritis outbreak, from the environment to the consumer. J. Clin. Microbiol. 48:915-920. [DOI] [PMC free article] [PubMed]
- 24.L'Homme, Y., R. Sansregret, E. Plante-Fortier, A. M. Lamontagne, M. Ouardani, G. Lacroix, and C. Simard. 2009. Genomic characterization of swine caliciviruses representing a new genus of Caliciviridae. Virus Genes 39:66-75. [DOI] [PubMed] [Google Scholar]
- 25.Lindén, S., J. Mahdavi, C. Semino-Mora, C. Olsen, I. Carlstedt, T. Boren, and A. Dubois. 2008. Role of ABO secretor status in mucosal innate immunity and H. pylori infection. PLoS Pathog. 4:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindesmith, L., C. Moe, J. Lependu, J. A. Frelinger, J. Treanor, and R. S. Baric. 2005. Cellular and humoral immunity following Snow Mountain virus challenge. J. Virol. 79:2900-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindesmith, L., C. Moe, S. Marionneau, N. Ruvoen, X. Jiang, L. Lindblad, P. Stewart, J. LePendu, and R. Baric. 2003. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 9:548-553. [DOI] [PubMed] [Google Scholar]
- 28.Liu, B. L., P. R. Lambden, H. Gunther, P. Otto, M. Elschner, and I. N. Clarke. 1999. Molecular characterization of a bovine enteric calicivirus: relationship to the Norwalk-like viruses. J. Virol. 73:819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopman, B. A., M. H. Reacher, Y. Van Duijnhoven, F. X. Hanon, D. Brown, and M. Koopmans. 2003. Viral gastroenteritis outbreaks in Europe, 1995-2000. Emerg. Infect. Dis. 9:90-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malaivijitnond, S., W. Sae-Low, and Y. Hamada. 2008. The human-ABO blood groups of free-ranging long-tailed macaques (Macaca fascicularis) and parapatric rhesus macaques (M. mulatta) in Thailand. J. Med. Primatol. 37:31-37. [DOI] [PubMed] [Google Scholar]
- 31.Marionneau, S., F. Airaud, N. V. Bovin, J. Le Pendu, and N. Ruvoen-Clouet. 2005. Influence of the combined ABO, FUT2, and FUT3 polymorphism on susceptibility to Norwalk virus attachment. J. Infect. Dis. 192:1071-1077. [DOI] [PubMed] [Google Scholar]
- 32.Marionneau, S., N. Ruvoen, B. Le Moullac-Vaidye, M. Clement, A. Cailleau-Thomas, G. Ruiz-Palacois, P. Huang, X. Jiang, and J. Le Pendu. 2002. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 122:1967-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martella, V., M. Campolo, E. Lorusso, P. Cavicchio, M. Camero, A. L. Bellacicco, N. Decaro, G. Elia, G. Greco, M. Corrente, C. Desario, S. Arista, K. Banyai, M. Koopmans, and C. Buonavoglia. 2007. Norovirus in captive lion cub (Panthera leo). Emerg. Infect. Dis. 13:1071-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moor-Jankowski, J., and W. W. Socha. 1978. Blood groups of macaques: a comparative study. J. Med. Primatol. 7:136-145. [DOI] [PubMed] [Google Scholar]
- 35.Nordgren, J., E. Kindberg, P. E. Lindgren, A. Matussek, and L. Svensson. Norovirus gastroenteritis outbreak with a secretor-independent susceptibility pattern, Sweden. Emerg. Infect. Dis. 16:81-87. [DOI] [PMC free article] [PubMed]
- 36.Oliver, S. L., E. Asobayire, A. M. Dastjerdi, and J. C. Bridger. 2006. Genomic characterization of the unclassified bovine enteric virus Newbury agent-1 (Newbury1) endorses a new genus in the family Caliciviridae. Virology 350:240-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliver, S. L., A. M. Dastjerdi, S. Wong, L. El-Attar, C. Gallimore, D. W. Brown, J. Green, and J. C. Bridger. 2003. Molecular characterization of bovine enteric caliciviruses: a distinct third genogroup of noroviruses (Norwalk-like viruses) unlikely to be of risk to humans. J. Virol. 77:2789-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rockx, B. H., W. M. Bogers, J. L. Heeney, G. van Amerongen, and M. P. Koopmans. 2005. Experimental norovirus infections in non-human primates. J. Med. Virol. 75:313-320. [DOI] [PubMed] [Google Scholar]
- 39.Smiley, J. R., K. O. Chang, J. Hayes, J. Vinje, and L. J. Saif. 2002. Characterization of an enteropathogenic bovine calicivirus representing a potentially new calicivirus genus. J. Virol. 76:10089-10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subekti, D. S., P. Tjaniadi, M. Lesmana, J. McArdle, D. Iskandriati, I. N. Budiarsa, P. Walujo, I. H. Suparto, I. Winoto, J. R. Campbell, K. R. Porter, D. Sajuthi, A. A. Ansari, and B. A. Oyofo. 2002. Experimental infection of Macaca nemestrina with a Toronto Norwalk-like virus of epidemic viral gastroenteritis. J. Med. Virol. 66:400-406. [DOI] [PubMed] [Google Scholar]
- 41.Sugieda, M., H. Nagaoka, Y. Kakishima, T. Ohshita, S. Nakamura, and S. Nakajima. 1998. Detection of Norwalk-like virus genes in the caecum contents of pigs. Arch. Virol. 143:1215-1221. [DOI] [PubMed] [Google Scholar]
- 42.Wei, C., T. Farkas, K. Sestak, and X. Jiang. 2008. Recovery of infectious virus by transfection of in vitro-generated RNA from Tulane calicivirus cDNA. J. Virol. 82:11429-11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White, L. J., J. M. Ball, M. E. Hardy, T. N. Tanaka, N. Kitamoto, and M. K. Estes. 1996. Attachment and entry of recombinant Norwalk virus capsids to cultured human and animal cell lines. J. Virol. 70:6589-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wobus, C. E., L. B. Thackray, and H. W. T. Virgin. 2006. Murine norovirus: a model system to study norovirus biology and pathogenesis. J. Virol. 80:5104-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wyatt, R. G., H. B. Greenberg, D. W. Dalgard, W. P. Allen, D. L. Sly, T. S. Thornhill, R. M. Chanock, and A. Z. Kapikian. 1978. Experimental infection of chimpanzees with the Norwalk agent of epidemic viral gastroenteritis. J. Med. Virol. 2:89-96. [DOI] [PubMed] [Google Scholar]
- 46.Zheng, D. P., T. Ando, R. L. Fankhauser, R. S. Beard, R. I. Glass, and S. S. Monroe. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312-323. [DOI] [PubMed] [Google Scholar]