Abstract
Adeno-associated virus (AAV) is a human parvovirus that replicates only in cells coinfected with a helper virus, such as adenovirus or herpes simplex virus type 1 (HSV-1). We previously showed that nine HSV-1 factors are able to support AAV rep gene expression and genome replication. To elucidate the strategy of AAV replication in the presence of HSV-1, we undertook a proteomic analysis of cellular and HSV-1 factors associated with Rep proteins and thus potentially recruited within AAV replication compartments (AAV RCs). This study resulted in the identification of approximately 60 cellular proteins, among which factors involved in DNA and RNA metabolism represented the largest functional categories. Validation analyses indicated that the cellular DNA replication enzymes RPA, RFC, and PCNA were recruited within HSV-1-induced AAV RCs. Polymerase δ was not identified but subsequently was shown to colocalize with Rep within AAV RCs even in the presence of the HSV-1 polymerase complex. In addition, we found that AAV replication is associated with the recruitment of components of the Mre11/Rad50/Nbs1 complex, Ku70 and -86, and the mismatch repair proteins MSH2, -3, and -6. Finally, several HSV-1 factors were also found to be associated with Rep, including UL12. We demonstrated for the first time that this protein plays a role during AAV replication by enhancing the resolution of AAV replicative forms and AAV particle production. Altogether, these analyses provide the basis to understand how AAV adapts its replication strategy to the nuclear environment induced by the helper virus.
Adeno-associated virus (AAV) is a human parvovirus that is currently used as a gene transfer vector (14). AAV particles consist of a small icosahedral capsid protecting a single 4.7-kb single-stranded DNA (ssDNA) genome with two open reading frames, rep and cap, surrounded by inverted terminal repeats (ITRs). The ITRs are the only sequences required in cis for genome replication and packaging. The rep gene encodes four nonstructural Rep proteins: Rep78, -68, -52, and -40. The two larger isoforms, Rep78 and -68, have origin binding, helicase, and site-specific endonuclease activities and are involved in AAV gene expression and genome processing, including replication and site-specific integration (39). The two smaller Rep isoforms are not required for AAV DNA replication but are involved in the control of viral gene expression and packaging of viral DNA (30).
When wild-type (wt) AAV infects a cell in the absence of a helper virus, it enters latency. Latent AAV genomes persist in cells either as episomes or as integrated genomes, preferentially at a specific locus (named AAVS1) on human chromosome 19. In most instances, no detectable viral gene expression or genome replication occurs unless the cell is co- or superinfected by a helper virus, such as adenovirus, herpes simplex virus type 1 (HSV-1), or HSV-2. Under these conditions, AAV replication and assembly take place in large intranuclear domains called replication compartments (RCs) that frequently colocalize with replication domains formed by the helper virus itself (81). The viral genome replicates by leading-strand synthesis and generates new ssDNA molecules by a strand displacement mechanism that occurs after strand- and site-specific cleavage of viral DNA by Rep78/68 within the ITRs (39).
Studies conducted on the relationship between AAV and its helper viruses are important not only to identify helper activities that can be used to produce recombinant AAV vectors but also to understand how AAV adapts its replication strategy to the helper virus and to the nuclear environment in general. Adenovirus helper functions have historically been the first and most extensively studied functions. These studies have shown that adenovirus helps AAV by stimulating viral gene expression and by enhancing AAV genome replication, mostly indirectly (19). Indeed, early studies showed that the adenovirus polymerase (E2b) is dispensable for AAV replication (8) and that the viral DNA-binding protein (DBP), the product of the E2a gene, is able to modestly enhance the processivity of AAV genome replication in vitro (77). More recently, the adenovirus proteins E1b55k and E4orf6 were shown to stimulate AAV genome replication by degrading the cellular Mre11/Rad50/Nbs1 (MRN) complex that restricts AAV genome replication during adenovirus coinfection (32). The concept that AAV genome replication can rely mostly, if not uniquely, on direct help from cellular factors was further strengthened by the demonstration that purified proteins such as replication protein A (RPA), replication factor C (RFC), proliferating cell nuclear antigen (PCNA), minichromosome maintenance (MCM) proteins, and DNA polymerase δ (Pol δ) were sufficient to replicate the AAV genome in vitro in the presence of Rep (40-41, 43). The involvement of these cellular proteins during AAV genome replication was also confirmed by the proteomic analysis of factors associated with Rep proteins during adenovirus-induced AAV replication (42).
Interestingly, studies conducted on HSV-1 helper activities suggest that the strategy of AAV replication may vary depending on the helper virus. Indeed, previous studies showed that the HSV-1 helicase-primase (HP) complex (UL5/8/52) and DBP (ICP8) could replicate transfected AAV-2 plasmids (80) and that the helicase activity, but not primase activity, of the HP complex was required for this effect (62, 66). More recently, a comprehensive study of HSV-1 helper activities demonstrated that the HSV-1 immediate-early proteins ICP0, ICP4, and ICP22 could stimulate rep gene expression, probably by diminishing intrinsic antiviral effects (1, 18). In addition, the HSV-1 DNA polymerase encoded by UL30, along with its associated processivity factor (UL42), although not strictly required, was demonstrated to significantly increase AAV replication levels induced in the presence of the HP complex and ICP8. Interestingly, the HSV-1 HP complex, DBP, and polymerase were also shown to be sufficient to replicate AAV DNA in vitro in the presence of Rep proteins without any cellular protein (78). Altogether, these observations indicate that in the context of an HSV-1 coinfection, AAV relies extensively on viral activities provided by the helper that directly participate in AAV genome replication.
To further elucidate the strategy of AAV replication in the presence of HSV-1, we undertook a proteomic analysis to identify the cellular and HSV-1 factors associated with Rep proteins and, consequently, potentially recruited within AAV RCs. To analyze Rep-associated proteins in the presence and absence of HSV-1 DNA replication, this analysis was performed using wt HSV-1 and an HSV-1 mutant in which the DNA polymerase encoded by the UL30 gene is absent (HSVΔUL30). This study resulted in the identification of approximately 60 cellular proteins, among which the largest functional categories corresponded to factors involved in DNA and RNA metabolism. Immunofluorescence analyses confirmed that in the presence of HSV-1, a basal set of cellular DNA replication enzymes, including RPA, RFC, and PCNA, was recruited within AAV RCs, with the exception of the MCM helicases. The cellular DNA polymerases, in particular Pol δ, were not identified by this analysis but subsequently were shown to be recruited in AAV RCs even in the presence of the HSV-1 polymerase complex. In addition, our results indicate that AAV replication induced by HSV-1 is associated with the recruitment of DNA repair factors, including components of the MRN complex, the Ku proteins, PARP-1, and factors of the mismatch repair (MMR) pathway. Finally, several HSV-1 proteins, most notably the UL12 protein, were also identified within AAV RCs. Our analyses confirmed the association between UL12 and Rep and demonstrated for the first time that this viral exonuclease plays a critical role during AAV replication by enhancing the formation of discrete AAV replicative forms and the production of AAV particles.
Altogether, these results indicate that in the presence of HSV-1, AAV may replicate by using a basal set of cellular DNA replication enzymes but also relies extensively on HSV-1-derived proteins for its replication, including UL12, a newly discovered helper factor. These results suggest that AAV may be able to differentially adapt its replication strategy to the nuclear environment induced by the helper virus.
MATERIALS AND METHODS
Cell lines, virus strains, and infection of cells.
African green monkey kidney fibroblasts (Vero), human cervical epithelial cells (HeLa), and derived cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM; Sigma) supplemented with 10% fetal calf serum (FCS; HyClone) and 1% penicillin-streptomycin (5,000 U/ml; Invitrogen). The HeLaAAVtCR cellular clone used in this study has been described previously (1). Briefly, it contains a modified AAV-2 genome (AAVtCR) in which the cap gene has been removed and the sequence coding for AAV Rep proteins fused in frame at its N terminus with coding sequence for the mCherry fluorescent protein and the streptavidin and calmodulin binding peptide (SBP and CBP) tags derived from the pNTAP-A plasmid (Interplay mammalian tandem affinity purification [TAP] system; Stratagene). The HSV-1 strains used in the study were wt HSV-1 (17 syn+), HSVΔUL30 (HP66; derived from the KOS strain), provided by D. Coen (Harvard University, Boston, MA), and HSVΔUL12 (AN-1) (82). The wt, ΔUL30, and ΔUL12 HSV-1 stocks were produced and titrated by standard procedures on Vero PolB3 cells expressing the HSV-1 UL30 gene (provided by C. Hwang, SUNY Health Science Center, Syracuse, NY) or Vero 6.5 cells expressing the UL12 gene. HeLa and HeLaAAVtCR cells were infected just before confluence with HSV-1 at the indicated multiplicity of infection (MOI) in DMEM-2% FCS. Times postinfection (p.i.) were calculated from the time of addition of the virus.
Plasmids.
The HSV-1 plasmids used were described previously (1). Briefly, the pRF plasmid encodes the viral HP complex (UL5/8/52) and the ssDBP ICP8 (UL29). The pTF3pol plasmid codes for the HSV-1 proteins ICP0, ICP4, and ICP22 and the viral polymerase complex (UL30/UL42). The pSAKUL12 and pSAKUL12.5 plasmids contain the UL12 open reading frame (ORF) under the control of the cytomegalovirus promoter (36). The pSAKUL12 plasmid has a mutation of the internal UL12.5 start codon (35). The pUL12exo− mutant construct encodes a mutated UL12 protein (D340E) that has lost its exonuclease activity (21).
Optimized TAP procedure.
Nuclear extracts were prepared from approximately 3 × 108 HeLa and HeLaAAVtCR cells infected with either wt HSV-1 or HSVΔUL30 for 20 h at an MOI of 5 PFU/cell. Cells were resuspended in a cold hypotonic buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 2 mM MgCl2, and 5 mM dithioerythritol [DTE]) in the presence of a protease inhibitor mixture (Roche Applied Science) and then gently lysed by addition of 0.7% Nonidet P-40. The nuclei were pelleted by centrifugation at 4°C, resuspended in lysis buffer (TAP system; Stratagene, La Jolla, CA), and disrupted by ultrasonication on ice. Debris was removed by centrifugation, and supernatants containing the soluble nuclear proteins were harvested.
Purification of AAVtCRep fusion proteins and potential interactors was performed as suggested by the supplier (Stratagene, La Jolla, CA), with some modifications. All purification steps were conducted at 4°C in the presence of a protease inhibitor cocktail (Sigma) and phenylmethylsulfonyl fluoride (PMSF). Cleared nuclear protein extracts were incubated overnight under gentle rotation with streptavidin beads previously saturated with bovine serum albumin (BSA). Beads were washed extensively with streptavidin buffer. Bound complexes were eluted with streptavidin elution buffer, adjusted with 4 volumes of calmodulin buffer, and incubated overnight under gentle rotation with calmodulin beads previously saturated with BSA. Beads were washed with calmodulin buffer, and protein complexes bound to calmodulin beads were eluted with SDS-PAGE loading buffer. The presence of AAV Rep protein in the purified complexes was verified by Western blot analysis using the anti-Rep 303.9 antibody (87).
Sample preparation for mass spectrometry (in-gel trypsin digestion).
The proteins present in the complexes purified from nuclear extracts prepared from 80 × 106 cells were subsequently separated by a short 1-cm migration in a 12% SDS-PAGE gel. The gel was then stained with colloidal Coomassie blue, and each lane was manually cut into 10 1-mm-wide slices. Each gel slice was incubated with trypsin for in-gel protein digestion and prepared automatically (EVO150; Tecan). Samples were washed several times by incubation in 25 mM NH4HCO3 for 15 min and then in 50% (vol/vol) acetonitrile containing 25 mM NH4HCO3 for 15 min. Gel pieces were dehydrated with 100% acetonitrile and then incubated with 7% H2O2 for 15 min before being washed again with the destaining solutions described above. A total of 0.15 μg of modified trypsin (sequencing grade; Promega) in 25 mM NH4HCO3 was added to the dehydrated gel pieces for overnight incubation at 37°C. Peptides were then extracted from gel pieces in three 15-min sequential extraction steps in 30 μl of 50% acetonitrile, 30 μl of 5% formic acid, and finally, 30 μl of 100% acetonitrile. The pooled supernatants were then dried under vacuum.
Protein identification by nano-LC-MS/MS and data analysis.
The dried extracted peptides were resuspended in 4% acetonitrile and 0.5% trifluoroacetic acid and analyzed by online nano-liquid chromatography-tandem mass spectrometry (nano-LC-MS/MS) (Ultimate 3000 [Dionex] and LTQ-Orbitrap [Thermo Fischer Scientific] instruments). The nano-LC method consisted of a 40-min gradient ranging from 5% to 55% acetonitrile in 0.1% formic acid at a flow rate of 300 nl/min. Peptides were sampled on a 300-μm by 5-mm PepMap C18 precolumn and separated on a 75-μm by 150-mm C18 column (Gemini C18; Phenomenex). MS and MS/MS data were acquired using Xcalibur (Thermo Fischer Scientific) and were processed automatically using Mascot Daemon software (Matrix Science). Consecutive searches against the SwissProt/Trembl database were performed for each sample, using an in-house version of Mascot 2.0. Peptide modifications allowed during the search were acetylation (N-terminal), dioxidation (M), oxidation (M), and trioxidation (C). Proteins were identified by a score higher than the query threshold (P < 0.05) and were automatically validated with in-house software (IRMa; peptides with a rank superior to 1 and a score of <40 were discarded) (15). Raw MS/MS data were screened manually to remove misidentified proteins and redundancies and then analyzed using the VirHostNet interactome database (http://pbildb1.univ-lyon1.fr/virhostnet) in order to reconstitute likely protein-protein interaction networks. Some additional information was derived from the UniProtKB/SwissProt database and specialized literature.
Immunofluorescence analysis.
Analyses were performed as described previously, using Alexa Fluor 488-conjugated secondary antibodies (Molecular Probes) (1). Images were collected on an Axioplan2 LSM510 confocal microscope (Zeiss) and processed using LSM Image Browser software (Zeiss). The primary antibodies used (see Table 2) were DEK (610948; BD Biosciences), DNA-PKcs (SC 9051), hnRNP C1/C2 (10294; Abcam), Hsc70 (ab19136), ICP8 (39-S; ATCC), Ku70 (ab10878), MCM2 (sc-56321), MCM7 (ab2360), MRE11 (ab397), mtSSB (HPA002866; Sigma), MSH2 (ab52266), MSH3 (BD61139), MSH6 (sc1243), NBS1 (ab398), PARP1 (P7605; Sigma), PCNA (ab2426), PHB (ab28172), Pol δ (ab38338), RAD50 (ab89), RFC2 (ab3615), RPA2 (ab2175), RuvBL2 (ab36569), SMC1A (ab9262), UL12/12.5 (BWp12; a gift from Joel Bronstein and Peter Weber [6]), UL30 (mABC-4; a gift from C. Knopf), and UL42 (sc-53331) antibodies. Each experiment was reproduced several times, and the images shown are representative of the overall effects observed under each condition.
TABLE 2.
Summary of validation analyses
| Protein and functional category | Result of validation analysisa or reference |
||||||
|---|---|---|---|---|---|---|---|
| Identified by LC-MS/MS |
IF on AAV RCs induced by: |
WB | Co-IP | ||||
| wt | ΔUL30 | wt | ΔUL30 | HSV plasmids | |||
| Cellular proteins | |||||||
| Proteins involved in DNA metabolism | |||||||
| DEK | − | + | − | − | |||
| DNA-PKcs | − | − | − | − | − | ||
| Ku70 | + | + | + | + | + | + | +; 42 |
| MCM2 | − | − | − | − | − | 42 | |
| MCM7 | − | − | +/− | +/− | +/− | 42 | |
| MRE11 | − | − | + | + | + | ||
| MSH2 | − | + | + | + | + | + | + |
| MSH3 | + | + | + | + | + | ||
| MSH6 | + | − | + | + | + | ||
| NBS1 | − | − | + | + | + | ||
| PARP1 | − | + | + | + | +/− | + | + |
| PCNA | + | + | + | + | + | + | +; 42 |
| Pol δ | − | − | +/− | +/− | + | 42 | |
| RAD50 | + | + | + | + | + | + | |
| RFC2 | + | + | + | + | + | + | |
| RPA1/2 | + | + | + | + | + | + | 42 |
| RuvBL2 | + | − | − | − | + | ||
| SMC1A | + | + | + | + | +/− | ||
| Hsc70 | + | − | +/− | +/− | − | + | |
| Protein involved in RNA metabolism | |||||||
| hnRNP C1/C2 | + | + | +/− | +/− | + | ||
| Mitochondrial proteins | |||||||
| mtSSB | + | + | − | − | − | + | |
| PHB | − | + | +/− | +/− | − | + | |
| HSV-1 proteins | |||||||
| ICP4 | − | + | 1 | + | |||
| ICP8 | + | + | + | + | 26, 66 | + | |
| UL12/UL12.5 | + | + | + | + | + | + | + |
| UL30 | + | − | + | NR | 1 | ||
| UL42 | + | + | + | + | |||
Summary of validation analyses, performed by immunofluorescence (IF) assay of infected cells, Western blotting (WB) of TAP complexes, and reverse coimmunoprecipitation (co-IP). For IF analyses, “+” indicates colocalization with AAV RCs, “+/−” indicates unclear or faint colocalization with AAV RCs, and “−” indicates absence from AAV RCs. NR, not relevant.
Immuno-FISH analysis.
Fluorescence in situ hybridization (FISH) analyses were conducted as previously described (1), using a mixture of four nonoverlapping digoxigenin (DIG)-labeled probes of 400 to 500 bp covering the rep gene to detect AAV sequences. HSV-1 DNA was detected using a mixture of three biotin-labeled probes produced by nick translation (nick translation kit; Roche) of HSV-1 cosmids (a gift from F. Catez). Labeled HSV and AAV probes were detected using streptavidin-Alexa Fluor 647 (Molecular Probes) diluted 1/1,000 and sheep anti-DIG-fluorescein antibody (4 μg/μl; Roche). Rep proteins were detected using undiluted mouse anti-Rep 303.9 monoclonal antibody (87) and then Alexa Fluor 555-conjugated anti-mouse secondary antibody (Molecular Probes), used at a 1/1,000 dilution. Images were collected using a Spectral TCS SP5 AOBS DM6000 confocal microscope (Leica) and were processed with ImageJ software (NIH).
Western blot analysis.
Proteins from cell lysates and from purified complexes were resolved by 12% SDS-PAGE. Proteins in the gels were then transferred to nitrocellulose membranes. Membranes were incubated with a primary antibody, and proteins were revealed by chemiluminescence (Super Signal WestDura substrate; Pierce), using a peroxidase-conjugated anti-rabbit, anti-mouse, or anti-goat antibody at a 1/5,000 dilution (Sigma).
Coimmunoprecipitation.
Nuclear extracts in 50 mM Tris-HCl, pH 7.2, 150 mM NaCl, and protease inhibitors (Complete; Roche Molecular Biochemicals) were incubated overnight at 4°C with antibodies directed against several of the proteins identified by mass spectrometry in this study (6). Immune complexes were immobilized on protein A Sepharose beads (CL4B; Amersham Pharmacia Biotech) for 30 min at 4°C. The beads were collected and washed three times with the previous buffer. Bound proteins were eluted by addition of Laemmli buffer and 5 min of warming at 95°C. Proteins were then separated by SDS-PAGE and analyzed by Western blotting, using the antibodies used for coimmunoprecipitation or anti-Rep antibodies.
qPCR.
Primers used for quantitative PCRs (qPCRs) were Rep-F (5′-GCAAGACCGGATGTTCAAAT-3′), Rep-R (5′-CCTCAACCACGTGATCCTTT-3′), β-globin-F (5′-CCCTTGGACCCAGAGGTTCT-3′), and β-globin-R (5′-CGAGCACTTTCTTGCCATGA-3′). For all reaction mixtures, 10 μl of FastStart Universal SYBR green master mix (Rox; Roche) was used in a final volume of 20 μl (in a 96-well tray; Eurogentec), with a final concentration of 300 nM for each primer. Approximately 12.5 ng of DNA was added in a 5-μl volume. Reaction mixtures were always set up in duplicate. Each qPCR was performed under the following conditions: 10-min hot-start denaturation at 95°C and 40 amplification cycles (15 s at 95°C, 40 s at 60°C). The melting temperatures of the final double-stranded DNA (dsDNA) products were determined by gradual heating from 60°C to 95°C over 20 min. All qPCRs were performed with a StepOnePlus real-time PCR system (Applied Biosystems) and associated software. Absolute amounts of Rep and β-globin amplicons, in arbitrary units, were determined using serial dilutions of genomic DNA from uninfected HeLaAAVtCR cells as a standard. The data are expressed as Rep/β-globin ratios, fixed at 1 for uninfected HeLaAAVtCR cells.
RESULTS AND DISCUSSION
Purification of Rep-associated factors in HSV-1-induced AAV replication compartments.
In order to identify the cellular and viral factors recruited in AAV RCs, we developed a strategy based on the purification of protein complexes associated with the two larger AAV Rep proteins. Indeed, Rep78 and -68 are involved in AAV genome replication, through their DNA binding, helicase, and endonuclease activities (27), whereas the two smaller Rep isoforms are dispensable for AAV genome replication (11, 44). Previous studies have also shown that Rep68 and -78 can remain covalently attached to AAV genomes after cleavage (64). Therefore, we reasoned that by pulling down the two larger Rep proteins under nondenaturing conditions, we should also recover DNA-protein complexes containing factors that are directly or indirectly associated with Rep and/or AAV DNA. To efficiently purify Rep proteins and their nuclear interaction partners, we used the previously described HeLaAAVtCR cells (1), which contain approximately three integrated copies of a modified AAV-2 genome in which the rep ORF is fused at its 5′ terminus to sequences coding for two affinity purification tags (streptavidin and calmodulin binding peptides) and the fluorescent mCherry protein (Fig. 1A). The AAVtCR genome thus encodes two unmodified proteins, Rep52 and -40, and two fusion proteins, Rep78 and -68 (designated tCRep), that were previously demonstrated to be fully functional (1). Importantly, the cap gene was removed from this construct in order to focus the analysis on proteins involved in early replication events (i.e., viral gene expression and DNA replication) and to prevent sequestration of single-stranded AAV genomes within the viral capsids (71).
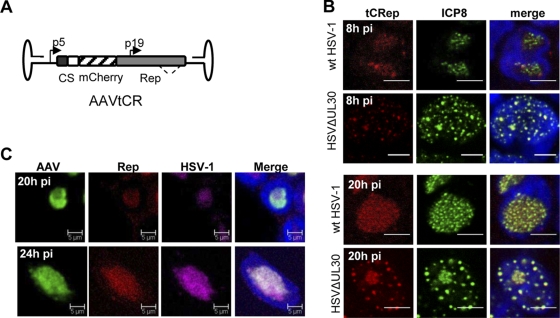
FIG. 1.
(A) Schematic view of the AAVtCR genome. p5 and p19 are the wt AAV-2 promoters, and C and S refer to the sequences coding for the calmodulin and streptavidin binding peptide tags. The dotted line indicates the position of alternative splicing in the rep gene. (B) Immunofluorescence analysis of HSV-1-infected HeLaAAVtCR cells to examine colocalization of tCRep and HSV-1 DBP ICP8 proteins. Infected cells were fixed at 8 and 20 h p.i., stained with an anti-ICP8 antibody, and analyzed by confocal microscopy. Bars, 5 μm. (C) Immuno-FISH analysis of HSV-1-infected HeLaAAVtCR cells. HeLaAAVtCR cells were infected for 20 or 24 h with wt HSV-1 (5 PFU/cell) and then analyzed using a biotin-labeled HSV-1 probe and a DIG-labeled AAV probe. After hybridization and washes, cells were additionally stained with an anti-Rep antibody and DAPI (4′,6-diamidino-2-phenylindole) and were analyzed by confocal microscopy. Bars, 5 μm.
Studies performed to visualize AAV Rep proteins upon infection with wt HSV-1 or HSVΔUL30 revealed the presence of fluorescent tCRep proteins as early as 8 h p.i. and their colocalization with HSV-1 ssDBP ICP8 protein, thus confirming previous observations (66) (Fig. 1B). The Rep pattern was different depending on the helper virus used. In the case of wt HSV-1, Rep and ICP8 accumulated in a large compartment that increased in size over time. With HSVΔUL30, Rep and ICP8 formed small foci early after infection and a larger but compact compartment at later times. These larger foci were previously demonstrated to contain AAV DNA in addition to Rep and ICP8 proteins (1). Because a previous study had suggested that AAV RCs are distinct from those formed by HSV-1 (20), we additionally analyzed AAV and HSV-1 DNA localization by immuno-FISH. At 20 h p.i., in cells where both HSV-1 and AAV DNAs were detected, two main patterns were observed: in some cells, the two viruses were found to replicate in partially overlapping domains (Fig. 1C, upper panels), and in other cells, the two replication domains coincided. The latter pattern was observed in nearly all cells at 24 h p.i. (Fig. 1C, lower panels). These observations suggest that as previously documented, HSV-1 and AAV replicate initially in adjacent domains that progressively fuse to form a unique replication compartment. Kinetic studies associated with a precise quantification of each phenotype will indicate whether these two phenotypes correspond to different stages during HSV-1 and AAV coinfection.
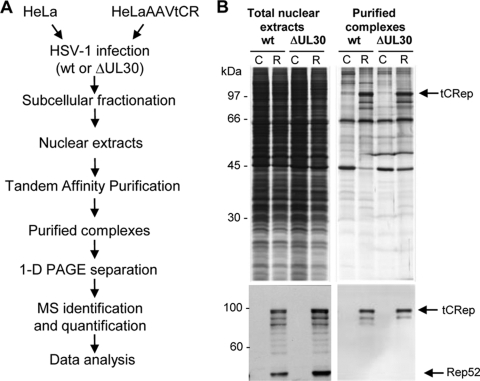
For TAP of Rep-associated complexes, HeLaAAVtCR cells were infected with either HSVΔUL30 or wt HSV-1. The HSVΔUL30 mutant was chosen to analyze Rep-associated proteins in the absence of HSV-1 DNA replication. The wt HSV-1 strain was used to perform the same analysis under conditions in which AAV replication is optimal (1). HeLa cells infected with the same HSV-1 strains were used as the corresponding negative controls. Our previous analysis indicated that with both HSV-1 strains, AAV DNA synthesis reached its peak between 20 and 24 h p.i. (data not shown). Therefore, infected cells were collected at 20 h p.i., and nuclear extracts were subjected to two consecutive purifications, first on streptavidin and then on calmodulin resin (Fig. 2A). Comparison of the protein profiles for a fraction of the purified material (Fig. 2B, upper panel) revealed that during this purification process, there was a progressive clearance of the extracts associated with an enrichment in tCRep78/68 proteins, as shown by silver staining and Western blotting, specifically in the HeLaAAVtCR-derived extracts (Fig. 2B).
FIG. 2.
(A) Outline of the experimental procedure used to purify nuclear complexes associated with AAV Rep proteins. See Materials and Methods for details of the TAP steps with streptavidin and calmodulin resins. (B) Sequential analysis of the purified extracts. Proteins present either in total nuclear extracts or in the Rep-containing complexes after elution from the second calmodulin resin were resolved by SDS-PAGE, followed by silver staining (upper panels) or Western blotting using an anti-Rep antibody (lower panels). C, control infected HeLa cells; R, HeLaAAVtCR cells expressing Rep upon HSV-1 infection; wt, wt HSV-1; ΔUL30, HSVΔUL30.
Identification of Rep-associated factors by mass spectrometry analysis and validation studies.
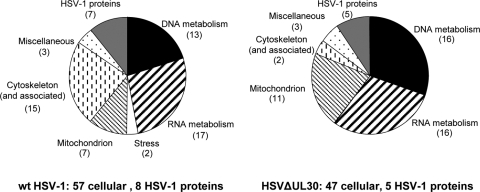
Proteins eluted from the calmodulin resin at the end of the large-scale TAP procedure were subjected to a semiquantitative LC-MS/MS analysis after separation in a preparative gel by a short migration. This resulted in the identification of 138 (112 cellular and 26 viral) and 140 (129 cellular and 11 viral) proteins in the Rep-containing complexes purified from HeLaAAVtCR cells infected with wt HSV-1 and HSVΔUL30, respectively. The lists were first double checked manually for redundant proteins, and then the proteins purified from HeLaAAVtCR cells were compared with those purified from the corresponding infected HeLa control cells. The identified proteins were classified according to their known functions and included in the final list (Table 1) if they met the following criteria: (i) the number of peptides found in the corresponding HeLa control sample was either null or at least four times smaller than that in the Rep-containing sample infected with the same HSV-1 strain, (ii) the protein was identified by at least two different specific peptides, and (iii) the protein was not known to be associated closely with the calmodulin interaction network. This resulted in a list of 57 and 47 cellular proteins, as well as 8 and 5 viral proteins, identified from complexes purified from HeLaAAVtCR cells infected with wt HSV-1 and HSVΔUL30, respectively. For both conditions, the largest functional categories corresponded to cellular factors involved in DNA metabolism, including DNA replication, repair, and chromatin modification, and RNA metabolism, with proteins involved in transcription, splicing/export, and translation (Fig. 3). Importantly, the final list of proteins also included several HSV-1 proteins, some of which were previously described as implicated in HSV-1-induced AAV replication. Interestingly, a significant number of proteins listed in Table 1 were previously found in a similar proteomic analysis performed during AAV and adenovirus coinfection, and some of them were shown to be involved directly in AAV replication (40-42).
TABLE 1.
Identification and classification of proteins in Rep-associated complexesa
| Protein and functional category | UniProt ID | Presence in HSV |
Function | Presence or homolog in Ad-AAV | |
|---|---|---|---|---|---|
| wt | ΔUL30 | ||||
| Cellular proteins | |||||
| Proteins involved in DNA metabolism | |||||
| DEK | P35659 | − | + | X, DR, A | − |
| H1E | A3R0T7 | + | − | X | H1C |
| H2B | Q99877 | − | + | X | − |
| H4 | P62805 | + | − | X | − |
| Ku70 | P12956 | + | + | DR, Tc | + |
| Ku86 | P13010 | + | + | DR, Tc | + |
| MSH2 | P43246 | − | + | DR | − |
| MSH3 | P20585 | + | + | DR | − |
| MSH6 | P52701 | + | − | DR | − |
| PARP1 | P09874 | − | + | DR | + |
| PCNA | P12004 | + | + | R, DR | + |
| RAD50 | Q92878 | + | + | DR | + |
| RFC2 | P35250 | + | + | R, DR | − |
| RFC3 | P40938 | + | + | R, DR | − |
| RFC4 | P35249 | − | + | R, DR | − |
| RPA1 | P27694 | + | + | R, DR | + |
| RPA2 | P15927 | − | + | R, DR | + |
| RPA3 | P35244 | − | + | R, DR | − |
| RuvBL2 | Q9Y230 | + | − | Tc, DR | RuvBL1 |
| SMC1A | Q14683 | + | + | X, DR | SMC2 |
| Proteins involved in RNA metabolism | |||||
| 40S RPS4X | P62701 | + | − | Tl, Rib | + |
| 40S RPS9 | P46781 | − | + | Tl, Rib | + |
| 40S RPS10 | P46783 | + | − | Tl, Rib | − |
| 40S RPS18 | P62269 | + | − | Tl, Rib | + |
| 60S P0 | P05388 | − | + | Tl, Rib | + |
| 60S RPL7 A | P62424 | − | + | Tl, Rib | + |
| 60S RPL9 | P32969 | − | + | Tl, Rib | − |
| 60S RPL10 | P27635 | − | + | Tl, Rib | − |
| 60S RPL11 | P62913 | + | − | Tl, Rib | + |
| 60S RPL13 | P26373 | + | − | Tl, Rib | + |
| 60S RPL23 A | P62750 | − | + | Tl, Rib | + |
| 60S RPL24 | P83731 | − | + | Tl, Rib | − |
| ABCE1 | P61221 | + | − | RNA | + |
| CPSF5 | O43809 | − | + | RNAP | − |
| DDX17 | Q92841 | + | − | RNA | − |
| EF1A1 | P68104 | + | − | Tl, NCE | EF1A2 |
| FL2 | Q13045 | + | − | Tc, CS | − |
| hnRNP A3 | P51991 | + | + | RNAP | − |
| hnRNP A/B | Q53F64 | + | + | RNAP | − |
| hnRNPC1/C2 | P07910 | + | + | RNAP | − |
| hnRNP D0 | Q14103 | + | − | RNAP | − |
| hnRNP G | P38159 | − | + | RNAP | − |
| hnRNP H | P31943 | + | − | RNAP | − |
| hnRNP M | P52272 | + | − | RNAP | − |
| JUN | P05412 | − | + | Tc | − |
| JUNB | P17275 | − | + | Tc | − |
| PURA | Q00577 | − | + | Tc, R, DR | − |
| RBM14 | Q96PK6 | + | − | Tc | − |
| RPABC1 | P19388 | − | + | Tc | − |
| SFPQ | P23246 | + | − | RNAP, DR | − |
| Mitochondrial proteins | |||||
| ACAD9 | Q9H845 | + | + | M | − |
| ADT2 | P05141 | − | + | Mt-ME | + |
| ADT3 | P12236 | + | − | Mt-ME, A | + |
| ATPA | P25705 | − | + | Mt-ME | + |
| ATPB | P06576 | − | + | Mt-ME | + |
| ATPG | P36542 | − | + | Mt-ME | + |
| ATPO | P48047 | + | − | Mt-ME | − |
| CMC1 | O75746 | + | − | Mt-ME | − |
| CMC2 | Q9UJS0 | − | + | Mt-ME | − |
| EFTU | P49411 | + | − | Mt-Tl | + |
| HSP79B | P38646 | + | − | PA, Ch | + |
| M2OM | Q02978 | − | + | Mt-ME | + |
| MPCP | Q00325 | − | + | Mt-ME | + |
| mtSSB | Q04837 | + | + | Mt-R | − |
| PHB | P35232 | − | + | PA, Ch | + |
| VDAC1 | P21796 | − | + | Mt-ME, A | + |
| Cytoskeletal proteins (and associated proteins) | |||||
| Keratin 13 | A8K2H9 | + | − | CS | − |
| LMNAC | P02545 | + | − | CS, X, DR | − |
| LMNB1 | P20700 | + | − | CS, X, DR | − |
| LMNB2 | Q03252 | + | − | CS, X, DR | − |
| LMO7 | Q8WWI1 | + | − | CS | − |
| MPRIP | Q6WCQ1 | + | − | CS | − |
| MYLK2 | Q9H1R3 | + | + | CS | − |
| Myosin 10 | P35580 | + | − | CS | − |
| MYL6 | P60660 | + | − | CS | + |
| MYL12A/B | P19105 | + | − | CS | − |
| SPTA2 | Q13813 | + | − | CS, Sc | + |
| SPTB2 | Q01082 | + | − | CS, Sc | + |
| STOM | P27105 | − | + | CS | STML2 |
| TMOD3 | Q9NYL9 | + | − | CS | − |
| β-Tubulin IVb | Q8IWP6 | + | − | CS | β-Tub2b, 5 |
| Vimentin | P08670 | + | − | CS | + |
| Miscellaneous proteins | |||||
| Annexin A5 | P08758 | + | − | B | − |
| ARL8B | Q9NVJ2 | − | + | V, M, CS | ARF1, 4 |
| Nup153 | P49790 | + | − | NCE | Nup85,358 |
| PHGDH | O43175 | + | − | M | + |
| VA0D1 | P61421 | − | + | V | − |
| VATB2 | P21281 | − | + | V | − |
| Stress proteins | |||||
| HSC70 | P11142 | + | − | Ch | HSP70 |
| HSP76 | P17066 | + | − | Ch | − |
| HSV-1 proteins | |||||
| gE | P04488 | + | − | Gp, Fc | |
| ICP4 | P08392 | − | + | Tc | |
| ICP8 | P04296 | + | + | R, DR | |
| TK | P03176 | + | + | M | |
| UL12/12.5 | P04294 | + | + | DR | |
| UL30 | P04293 | + | − | R | |
| UL42 | P10226 | + | + | R | |
| UL46 | P10230 | + | − | Tg, Tc | |
The table indicates the specific proteins identified in HeLaAAVtCR cells infected with wt HSV-1 or HSVΔUL30, their known functions, and whether they were previously found in adenovirus- and AAV-coinfected cells (42). A, apoptosis; B, blood; Ch, chaperone; CS, cytoskeleton; DR, DNA repair and recombination; Fc, Fc receptor; Gp, glycoprotein; M, metabolism; ME, membranes and transmembrane exchanges; Mt, mitochondrion; NCE, nucleocytoplasmic exchange; PA, control of cell proliferation and aging; R, DNA replication; Rib, ribosome; RNA, RNA metabolism, unspecified; RNAP, mRNA processing; Sc, secretion; Tc, transcription; Tg, tegument; Tl, translation; V, vesicles; X, chromatin and chromosomes. The list of identified proteins is also available on the VirHostNet website (http://pbildb1.univ-lyon1.fr/virhostnet/login.php).
FIG. 3.
Functional classification of proteins copurified with Rep from HeLaAAVtCR cells infected with wt HSV-1 or HSVΔUL30 and identified by mass spectrometry analysis. See Table 1 for definitions of the functional categories.
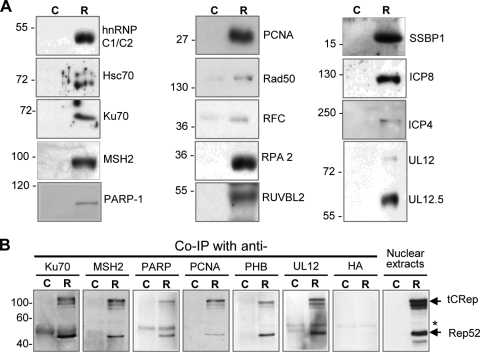
For this study, Western blot analyses performed on the purified Rep-containing complexes validated the presence of 15 selected proteins (Fig. 4A and Table 2). In addition, reverse coimmunoprecipitations were also conducted with antibodies recognizing several cellular and viral factors to verify their association with Rep (Fig. 4B and Table 2). These analyses confirmed the association of Rep with PCNA and Ku70 (42) and demonstrated, for the first time, its presence in purified complexes containing PARP-1, PHB, MSH2, and UL12. It remains to be determined whether these interactions are direct or mediated by other factors and/or DNA.
FIG. 4.
Western blot analyses of purified complexes. (A) Protein complexes were purified from HeLa (C) or HeLaAAVtCR (R) cells infected with wt HSV-1 and then analyzed by Western blotting, using antibodies against a panel of cellular and viral proteins. (B) Reverse coimmunoprecipitation of tCRep in purified complexes containing selected cellular and viral proteins. Control HeLa (C) cells and HeLaAAVtCR (R) cells were infected with wt HSV-1 for 20 h (MOI = 5 PFU/cell). Nuclear extracts from 2 × 106 infected cells were subjected to coimmunoprecipitation, using UL12, Ku70, MSH2, PARP-1, PCNA, and PHB polyclonal antibodies as indicated. The anti-HA antibody was used as a negative control. The immunoprecipitates were then immunoblotted with an anti-Rep antibody (303.9). The arrows indicate the positions of the tCRep and Rep52 proteins, and the asterisk shows a nonspecific band.
In silico analysis of cellular interaction pathways.
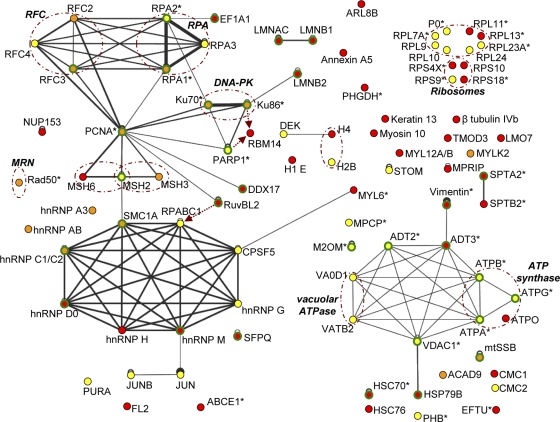
In order to reconstitute likely interaction networks between cellular proteins identified in AAV RCs, the final list of cellular proteins specific to complexes purified from HeLaAAVtCR extracts (Table 1) was submitted to the VirHostNet database (http://pbildb1.univ-lyon1.fr/virhostnet/login.php), which lists direct and experimentally validated interactions between human proteins (Fig. 5). Not surprisingly, the two main networks that emerged from this analysis were the DNA replication and repair network (upper left) and the RNA splicing network (lower left). Other minor networks identified included ribosomal proteins (upper right), mitochondrial factors (lower right), and cytoskeleton constituents.
FIG. 5.
In silico reconstitution of Rep-associated interaction networks. Likely protein-protein interactions were reconstituted using the cellular protein list from Table 1 and analysis tools provided by the VirHostNet interactome website (http://pbildb1.univ-lyon1.fr/virhostnet/login.php), complemented with additional information derived from the UniProtKB/SwissProt database and specialized literature. The color codes refer to proteins specifically identified from HeLaAAVtCR cells infected with wt HSV-1 (red), HSVΔUL30 (yellow), or both helper viruses (orange). Green circles, proteins for which a physical or functional interaction with a protein from another virus has already been reported; black lines, VirHostNet-derived interactions, with the link width correlating with the number of different reports describing the interaction found in the database (black circles correspond to self-interacting proteins); dashed brown arrows, UniProtKB/SwissProt-derived interactions; asterisks, proteins identified in Rep-containing complexes purified from adenovirus- and AAV-coinfected cells (42); dashed brown circles, proteins constituting a known functional complex.
Only minimal differences were found between the two HSV-1 helper strains. Notably, a larger number of cytoskeletal proteins were recovered in the presence of wt HSV-1. Their predominant identification from cells infected with wt HSV-1 might be the consequence of the more pronounced cytopathic effect induced by this helper virus than that with the replication-defective HSVΔUL30 strain.
Regarding the DNA replication machinery, components of RPA and RFC complexes were recovered, as well as PCNA, but no cellular DNA polymerase was recovered, even in the case of HSVΔUL30-induced AAV replication, where a cellular enzyme is necessarily required. RNA metabolism factors identified could be divided into two major categories, i.e., factors involved in splicing and ribosomal proteins; a few transcription factors were also identified. Most of the splicing factors identified are hnRNPs. Since most of these factors associate with mRNAs during the process of transcription and mark immature mRNAs (37), this finding was not unexpected, as viral RCs are active sites of transcription, and thus newly synthesized viral mRNAs and associated complexes are probably major components of these intranuclear domains (81). Rep proteins themselves are involved in AAV genome transcription and have been shown to affect the ratio of spliced versus unspliced AAV mRNAs (47). An increasing number of studies have suggested that various hnRNPs are involved in the replication of numerous viruses, including herpesviruses (7, 23, 24, 33). Interestingly, some of the hnRNPs recovered were also reported to be able to associate with DNA by recognizing structural motifs and to regulate several aspects of DNA metabolism, such as chromosome maintenance and DNA replication and repair (25). For instance, hnRNP A3 was recently reported to bind the single-stranded telomeric repeat, thus contributing to its protection from nuclease attacks (67). Interestingly, an hnRNP A/B-related protein was shown to bind the single-stranded genome of feline parvovirus and to modulate virus replication (73). Among the various hnRNPs found in this study, the association of hnRNP C1/C2 with Rep was validated by Western blotting (Fig. 4A). The other main category of RNA metabolism factors was composed of ribosomal proteins. Association of ribosomal constituents with AAV RCs could be mediated through the known interaction between Rep and the major nucleolar component B23/NPM (4). However, NPM was not recovered, and since only a minority of the ribosomal proteins was identified, it is likely that these proteins were derived from contamination of the purified extracts with these highly prevalent nuclear proteins. Accordingly, several ribosomal proteins were also identified in complexes purified from cells coinfected with AAV and adenovirus (42).
Another significant network was that of mitochondrial proteins. In particular, several mitochondrial factors involved in ATP synthesis were specifically retrieved from cells infected with HSVΔUL30, similar to what was previously reported in the case of adenovirus- and AAV-coinfected cells (42). This is surprising, since no specific localization of Rep to mitochondria has ever been reported. Since AAV, like other parvoviruses, has been shown to induce apoptosis (55, 72), it is tempting to speculate that the identification of these factors may be linked to a possible interaction of AAV Rep proteins with mitochondrial constituents.
Because we were interested mainly in the analysis of factors involved in the early steps of AAV replication, further analyses were focused on only a subset of cellular and viral factors that were found in this screen.
Factors involved in AAV DNA replication.
Adenovirus-induced AAV replication is performed mainly by cellular factors, and previous studies have shown that PCNA, RFC, Pol δ, RPA, and MCM constitute the minimal set of cellular proteins able to drive AAV DNA replication in vitro (40, 41, 44). Most of these factors were also found in a recent proteomic analysis of Rep-associated proteins in adenovirus- and AAV-coinfected cells (42) (Fig. 5 and Table 1). However, at the beginning of this study, it was unknown whether the same set of cellular proteins, with the exception of RPA, was also involved during HSV-1-induced AAV replication.
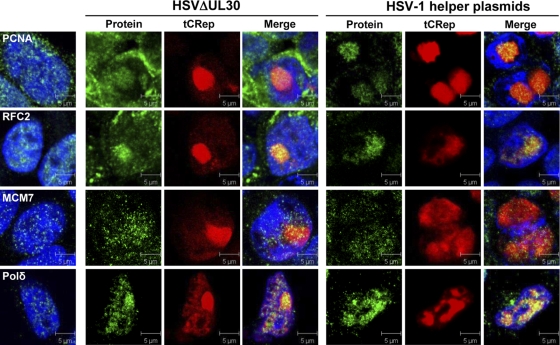
In our proteomic analysis using wt HSV and HSVΔUL30 as helpers, we identified members of the RPA and RFC complexes as well as PCNA as Rep-interacting partners (Table 1 and Fig. 4). In contrast, neither the MCM proteins nor Pol δ or any other cellular DNA polymerase was identified. To further analyze these data, immunofluorescence studies were conducted to determine whether these proteins were recruited in AAV RCs (Fig. 6 and Table 2). To discriminate between factors recruited by replicating HSV-1 and AAV DNAs, these analyses were performed in cells infected with HSVΔUL30. However, HSV-1 is known to carry a protein that binds to the Fc domain of human, rabbit, and goat antibodies (29, 83), thus resulting in significant nonspecific cytoplasmic staining in analyses performed at late stages of infection (14 to 20 h p.i.). Thus, additional analyses were also performed on cells transfected with a set of HSV-1 helper plasmids previously reported to be able to induce AAV replication as efficiently as that induced by wt HSV-1 (1). These analyses showed a colocalization of RFC, PCNA (Fig. 6), and RPA (not shown) with Rep. These results did not demonstrate a direct or indirect interaction with Rep but indicated that these cellular factors were recruited in HSV-1-induced AAV RCs. In contrast, our immunofluorescence study showed that MCM remained more uniformly distributed in the nucleus and did not clearly colocalize with Rep (Fig. 6 and Table 2). This is in agreement with the fact that MCM proteins were not identified by MS in this study. This result suggests that the MCM helicase complex does not participate in HSV-1-induced AAV replication, in contrast to what was recently shown in the presence of adenovirus (41, 42). Similar activities could be provided by other viral or cellular proteins. The previous study on adenovirus-induced AAV replication showed that the Ku proteins, although not essential, can partially substitute for MCM in in vitro replication assays (42). Therefore, it is possible that the Ku proteins directly or indirectly provide strand displacement activity during HSV-1-induced AAV DNA replication. Indeed, Ku86 and -70 were found in Rep-associated complexes (Table 1 and Fig. 4) and colocalized in AAV RCs (Fig. 7 and Table 2). Alternatively, this activity could also be provided by the HSV-1 HP complex (UL5/8/52), which was previously demonstrated to be essential for HSV-1 helper activity, in particular via its helicase subunit (62, 80). However, our analysis did not retrieve any of the members of the HP complex. It is possible either that the association of the HP complex with AAV genomes is not stable enough for it to be recovered during the purification procedure or that this complex is required only at early replication steps.
FIG. 6.
Colocalization of cellular DNA replication factors with Rep in HSV-1-induced AAV RCs. HeLaAAVtCR cells were either infected with HSVΔUL30 (MOI = 5 PFU/cell) and fixed at 9 or 14 h p.i. or transfected with 0.25 pmol/35-mm well of each HSV-1 helper plasmid (pTF3pol and pRF) and fixed at 48 h posttransfection. Alexa Fluor 488-conjugated donkey anti-mouse, anti-rabbit, and anti-goat secondary antibodies were used. DNA was stained with TO-PRO-3 iodide (Invitrogen). Images were taken using an Axioplan2 LSM510 confocal microscope (Zeiss). Green, protein of interest; red, tCRep; blue, TO-PRO-3 staining. The left panels correspond to results for noninfected and nontransfected control HeLaAAVtCR cells. Bars, 5 μm. The cytoplasmic background visible for some antibodies in HSV-infected cells is due to antibody binding to the HSV-encoded Fc receptor (29, 83).
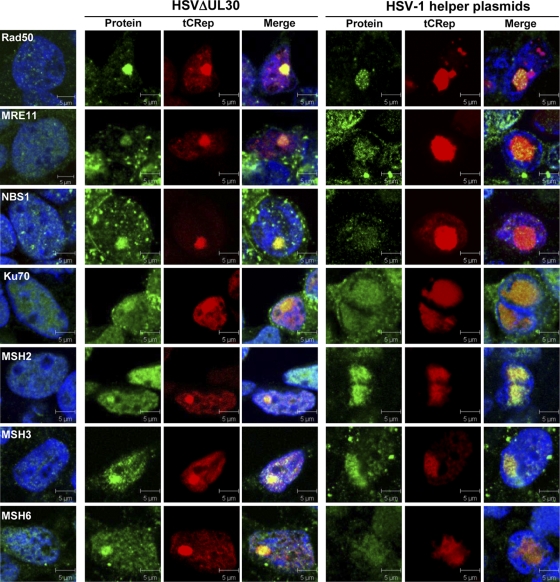
FIG. 7.
Colocalization of cellular DNA repair factors with Rep in AAV RCs. Cells were processed as described in the legend to Fig. 6 and then stained with antibodies recognizing the indicated cellular DNA repair factors. Green, protein of interest; red, tCRep; blue, TO-PRO-3 staining. The left panels indicate the staining observed in noninfected and nontransfected HeLaAAVtCR cells. Bars, 5 μm.
It was surprising that no cellular polymerase was identified by MS, even in the case of cells infected with HSVΔUL30, where a cellular enzyme is necessarily required (Table 1). Previous studies have indicated that Pol δ is the cellular enzyme involved in AAV genome replication in the presence of adenovirus and that this enzyme is also able to drive AAV replication in vitro (40, 41). In the case of HSV-1, the situation is more complex. Although the viral polymerase complex (UL30/42) is not strictly essential, it was shown to be able to replicate AAV DNA in vitro and to potently enhance AAV replication in live cells (1, 78, 80). In this regard, it is interesting that both UL30 and UL42 were identified by MS (Table 1) and colocalized within AAV RCs (Table 2). Therefore, we examined whether Pol δ could be visualized in AAV RCs. The recruitment of Pol δ within AAV RCs was observed in HeLaAAVtCR cells not only infected with HSVΔUL30 but also transfected with HSV-1 helper plasmids (Fig. 6) or infected with wt HSV-1 (data not shown). Altogether, these findings suggest that Pol δ is present within HSV-1-induced AAV RCs, even in the presence of the viral polymerase. This finding raises the questions of the relative extent to which AAV uses the cellular polymerase versus its viral analog when both are present and of the mode of AAV replication with these enzymes.
Finally, two additional cellular factors were of particular interest: prohibitin (PHB1) and mtSSB/SSBP1, the mitochondrial ssDBP. Prohibitin is a mitochondrial and nuclear protein which displays several functions, including inhibition of DNA synthesis (68, 74). Interestingly, a recent study indicates that this protein negatively controls the onset of cellular DNA replication by physically interacting with MCM proteins (53). In our study, PHB1 was identified in Rep-containing complexes purified from cells infected with HSVΔUL30 (Table 1) and was also found to be associated with Rep in reverse coimmunoprecipitation experiments (Fig. 4B). Immunofluorescence analysis indicated that in HeLaAAVtCR cells infected with either HSV-1 strain, the protein was present in the nucleus, but not exclusively within AAV RCs (Table 2). However, a similar pattern was not observed in HeLaAAVtCR cells transfected with HSV-1 helper plasmids, in which the protein was localized mostly in the cytoplasm. The second interesting factor that was identified in samples from cells infected with wt and ΔUL30 HSV-1 strains was mtSSB (Table 1 and Fig. 4A). Following infection with Epstein-Barr virus, this protein has been shown to be relocalized to the nucleus, where it stimulates viral replication (84). Surprisingly, however, the protein was not detected by immunofluorescence in the nuclei of either infected or transfected HeLaAAVtCR cells (Table 2). Both of these discrepancies could be linked to lower sensitivities of the PHB and mtSSB antibodies used for the immunofluorescence analysis. Additional studies will be needed to determine whether mtSSB and PHB can modulate AAV replication.
Factors involved in DNA repair.
Viruses that replicate in the nucleus, such as adenovirus and HSV-1, have been shown to significantly interfere with the DNA repair machinery and the DNA damage response (DDR) to inhibit the cellular defenses and to usurp some cellular functions for their own replication (10). In particular, several studies have highlighted that with respect to DNA repair, adenovirus and HSV-1 use different strategies to accomplish their replicative cycles. HSV-1 notably recruits and exploits the MRN complex to accomplish its replicative cycle (31, 85). This complex is involved in the recognition of dsDNA breaks and in downstream signaling to molecules involved in DNA repair through both homologous and nonhomologous recombination pathways (61). In addition, many other DNA repair proteins were found in ICP8-associated complexes, including the DNA-PK complex, XRCC4, Rad50, PARP1, the MMR proteins MSH2, -3, and -6, and the repair helicases WRN and BLM (69). In contrast, adenovirus degrades and/or inactivates MRN and ligase IV in order to prevent the concatemerization of the viral genomes and the inhibition of its replication (2, 3, 9, 48, 65, 79). Importantly, the ability of adenoviral proteins to degrade and/or delocalize the MRN complex contributes to the helper activities required for AAV replication (58).
Our proteomic analysis retrieved several factors involved in the DNA damage response, including the Rad50 protein, Ku70 and -86, PARP1, MSH2/3/6, RuvBL2, and SMC1A (Table 1 and Fig. 4). The identification of Rad50 within Rep-containing complexes is of particular interest given the previously documented positive role of the MRN complex during HSV-1 replication (31, 60, 85; N. Balasubramanian and S. K. Weller, unpublished data). Our immunofluorescence analysis confirmed the colocalization of Rad50 with Rep and showed that both Mre11 and Nbs1, the two other members of the MRN complex, were similarly recruited within AAV RCs induced upon infection with HSVΔUL30 (Fig. 7). Similar results were also observed when the cells were transfected with HSV-1 helper plasmids, further indicating that the recruitment of this complex is not dependent upon the presence of a replication-competent HSV-1 genome. It is not possible at the moment to conclude whether the MRN complex displays a positive or negative effect on AAV replication induced by HSV-1. Nevertheless, it is tempting to speculate that the same complex differentially regulates AAV genome replication depending on the nature of the helper virus used.
Other factors of interest identified in our study were the MMR factors MSH2, -3, and -6. These three proteins participate in the formation of two distinct complexes that remove mismatches arising during DNA synthesis (28). The MSH2/6 complex (MutSα) is the most abundant and is involved mainly in the recognition of base-base mismatches generated by errors of DNA polymerases. The MSH2/3 complex (MutSβ) recognizes longer insertion/deletion loops generated, notably, during replication of microsatellite regions (28). In addition, this pathway is also involved in the cellular DNA damage response and in the control of homologous recombination (HR) events. Indeed, in mammalian cells, MSH2, -3, and -6 prevent recombination between divergent sequences (16, 63). Our analyses confirmed the recruitment of MSH2, -3, and -6 in AAV RCs (Table 2 and Fig. 7) and the association of MSH2 with Rep (Fig. 4). These results are in agreement with previous studies suggesting that HSV-1 replication may be dependent upon HR events requiring both viral and cellular proteins (86). Indeed, MSH proteins have been identified in the replication compartments of various herpesviruses (13, 69, 75). Factors such as MRN and MSH are both involved in the control of HR and most likely present in AAV RCs because they are recruited by HSV-1-encoded factors for its own replication. However, this does not exclude the possibility that these factors may also be involved in the control of AAV replication induced by HSV-1.
Data from a recent study on adenovirus-induced AAV replication indicated the presence of both Ku70/80 and DNA-PKcs within Rep-associated complexes (42), and further analyses showed that DNA-PKcs mediates AAV-induced DDR in the presence of adenovirus (12, 57). DNA-PKcs was not identified in our proteomic analysis, despite the presence of Ku (Table 1). In addition, immunofluorescence analyses confirmed the absence of DNA-PKcs in HSV-1-induced AAV RCs (data not shown). This result was not surprising, since previous studies have shown that ICP0, an HSV-1 immediate-early factor, can induce the proteasomal degradation of DNA-PKcs in a cell line-dependent manner (45). This event is considered central to the strategy used by HSV-1 to inactivate the nonhomologous end-joining (NHEJ) DNA repair pathway initiated by the recognition of dsDNA breaks by the DNA-PK complex. However, recent studies have also raised the possibility of end-joining events in the absence of classical NHEJ factors, including joining of broken ends by the direct recruitment of XRCC4 and ligase IV by Ku70/80 in the absence of DNA-PKcs (34). Interestingly, knockdown of either ligase IV or XRCC4 has been shown to affect HSV-1 genome replication and viral yields (38). Therefore, it is possible that the Ku proteins found in AAV RCs (Fig. 7 and Table 2) may be implicated in the recruitment of downstream factors involved in NHEJ. Further studies will indicate whether these factors are indeed recruited within AAV RCs as well as the impact of potential NHEJ events on AAV genome replication. Finally, another important DNA repair signaling protein found in our analysis was PARP-1, which was found to be recruited in AAV RCs and associated with Rep (Table 2 and Fig. 4). This multifunctional factor, previously found associated with several viruses that replicate in the nucleus, has also been implicated in the regulation of dsDNA break repair pathway choice, in particular by promoting alternative NHEJ mechanisms (56, 61). Interestingly, Mre11, a component of MRN, was also recently found to be involved in alternative NHEJ (49, 88). The potential role of this alternative repair pathway during the AAV life cycle, and particularly during AAV DNA replication, remains to be defined.
Identification of HSV-1 helper proteins.
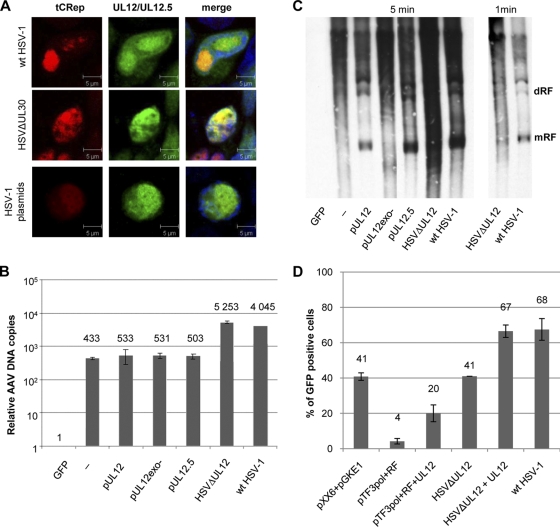
As expected, several known HSV-1 helper factors were identified in this study. In contrast, a previous analysis of adenovirus-induced AAV replication resulted in the identification of a single adenoviral helper protein (42). This finding additionally illustrates the more direct role of HSV-1 in AAV replication than that of adenovirus. Among the helper factors retrieved in our study were the ICP8, ICP4, UL30, and UL42 proteins. Other factors known to help AAV, such as ICP0 and the HP complex, were not identified, despite previous evidence of their biological effect (1, 18, 80). It is possible, as noted above, that these factors intervene only at early steps of the AAV replicative cycle, further explaining why they were not identified from complexes purified from cells at 20 h postinfection. Alternatively, helper factors such as ICP0 might stimulate AAV replication in an indirect way without actually physically associating with Rep complexes. Other HSV-1 factors not already known to exert any helper activity were also identified in the present study. In particular, the UL12 protein was identified for both helper viruses by the presence of a significant number of peptides. This early factor is a 5′-3′ exonuclease that has been shown to be involved in HSV-1 genome maturation. Indeed, previous studies have shown that this exonuclease forms a complex with the HSV-1 ICP8 protein that is able to mediate a robust strand exchange activity in vitro (51, 52). This recombinase activity plays an important role in the generation of mature progeny viral DNA that can be packaged efficiently into newly formed virions (21, 22, 46). The UL12 ORF also encodes a truncated protein, UL12.5, that similarly displays 5′-3′ exonuclease and strand exchange activities but is localized predominantly in the mitochondria, where it induces degradation of host mitochondrial DNA (50, 54). The MS analysis did not indicate which UL12 isoform was part of the purified complexes, since all identified peptides mapped in the region common to the two proteins. However, both UL12 and UL12.5 were detected in Rep-associated complexes by Western blotting (Fig. 4A). Reverse coimmunopurification experiments confirmed the interaction between UL12 and Rep (Fig. 4B). It is likely that this interaction is mediated by HSV-1 DBP, since this protein is able to interact directly with both UL12 and Rep (26, 66, 70). Also, immunofluorescence studies showed that UL12 colocalized with Rep in AAV RCs induced by infection with either wt HSV-1 or HSVΔUL30 (Fig. 8A). To determine whether UL12 had a helper activity, we analyzed its effect on AAV genome replication in HeLaAAVtCR cells either infected with a wt or UL12-defective (HSVΔUL12) HSV-1 strain or transfected with the set of HSV-1 helper plasmids together with a plasmid encoding UL12. Quantitative PCR analysis indicated that UL12 did not affect AAV genome replication in terms of the amount of viral DNA synthesized in infected or transfected cells (Fig. 8B). In contrast, Southern blot analysis indicated an altered pattern of AAV replication in the absence of UL12 (Fig. 8C). Indeed, the absence of this factor resulted in a reduction of the typical AAV replicative forms, the double-stranded monomer and dimer forms, as well as an increase in the smear-like background associated with high-molecular-weight forms that suggested the presence of a ladder of replication intermediates. Interestingly, a typical replicative pattern could be restored fully by complementation with a plasmid encoding either UL12 or UL12.5 but not with a plasmid encoding an exonuclease-negative UL12 mutant.
FIG. 8.
Involvement of HSV-1 UL12 protein in AAV replication. (A) Colocalization of UL12 with Rep within AAV RCs. HeLaAAVtCR cells were either infected with the indicated HSV-1 strain or transfected with the HSV-1 helper plasmids and pSAKUL12 plasmid and then stained with an anti-UL12 antibody and TO-PRO-3. Bars, 5 μm. (B) Analysis of the effect of UL12 on AAV DNA replication. Total DNA was extracted from HeLaAAVtCR cells either infected with wt HSV-1 or HSVΔUL12 or transfected with HSV-1 helper plasmids and a plasmid encoding either UL12, UL12.5, or an exonuclease-negative UL12 mutant (pUL12exo−) and then quantified by qPCR, using rep-specific primers. Data presented are means with error bars calculated for three independent experiments. (C) The same samples used in panel B were analyzed by Southern blotting, using a DIG-labeled rep probe. mRF, monomer replicative form; dRF, dimer replicative form. (D) Effect of UL12 on rAAV-GFP particle production. HeLa cells transfected with rAAV-GFP and rep-cap-expressing plasmids were either cotransfected with HSV-1 helper plasmids or infected with HSVΔUL12 in the absence or presence of a plasmid encoding UL12. As a control, the cells were cotransfected with adenoviral helper plasmids (pXX6 and pGKE1) or infected with wt HSV-1. Cell lysates were then used to infect HeLa cells in the presence of wt adenovirus, and GFP-positive cells were counted by flow cytometry at 24 h p.i. Mean values with error bars, calculated for at least three independent experiments, are presented.
These results are reminiscent of a report by Blumel et al. showing that the six core HSV-encoded replication factors can induce concatemer formation on simian virus 40 (SV40) genomes if SV40 large T antigen is also provided (5). Since SV40 replication normally results in the appearance of two circular daughter molecules, it is noteworthy that the presence of HSV replication proteins can alter the mode of replication to generate concatemeric DNA. Thus, when HSV is used as the helper, it is possible that the mechanism of AAV DNA replication is altered and that UL12 may be required to effectively produce progeny genomes suitable for packaging.
To further analyze the effect of UL12 on the AAV life cycle, we finally examined its effect on the production of infectious AAV particles. For this purpose, recombinant AAV vector particles coding for green fluorescent protein (rAAV-GFP) were produced in HeLa cells by using either the set of helper HSV-1 plasmids or the HSVΔUL12 virus complemented or not with the UL12-expressing plasmid. As a positive control, rAAV-GFP particles were also produced using either adenoviral helper plasmids or wt HSV-1. Cell lysates from transfected/infected cells were then used to infect HeLa cells, which were analyzed by flow cytometry to count the number of GFP-positive cells. The results shown in Fig. 8D indicate that HSV-1 helper plasmids induced the formation of infectious rAAV-GFP particles at a level that was approximately 10-fold lower than that found with adenoviral helper plasmids. Addition of UL12 reproducibly resulted in a 5-fold increase in the number of infectious rAAV-GFP particles produced. Interestingly, UL12 also led to an increase in the number of infectious rAAV-GFP particles when it was used to complement the HSVΔUL12 virus. These results clearly indicate that UL12 can increase the number of infectious AAV particles produced, further suggesting that its effect on AAV DNA replication enhances the production of DNA forms that are suitable for encapsidation. Altogether, these data indicate that UL12 can be considered an additional HSV-1 helper factor that, although not essential, can critically enhance the AAV productive cycle.
Several lines of evidence suggest that HSV-1 may replicate its genome in two stages, involving UL9-dependent initiation from a viral origin of replication and then replication via a recombination-driven and origin-independent mechanism (86). The UL12/ICP8 recombinase certainly plays an important role in this process. The participation of UL12 in HR was also suggested by recent data indicating that this viral factor is able to bind to some components of the MRN complex (Balasubramanian and Weller, unpublished data). In contrast, the current model of AAV genome replication does not involve any recombination event, despite previous indirect evidence for HR in vitro, in cultured cells, and in vivo, in tissues. Indeed, recombination events have been observed between mutant AAV genomes under conditions enabling replication, and multiple AAV variants have been detected in individual tissues from nonhuman primates (17, 59, 76). On the basis of these data, it is tempting to speculate that in the context of an HSV-1 infection, HR may control AAV genome replication, in particular by enhancing the resolution of high-molecular-weight concatemers into discrete replicative forms, thus providing more DNA substrates for packaging into assembled capsids.
Altogether, these results indicate that in the presence of HSV-1, AAV may replicate by using a basal set of cellular DNA replication enzymes but also relies on HSV-1-derived proteins for its replication, including UL12, a newly discovered helper factor that intervenes with AAV genome replication at a qualitative level. These findings provide the basis to further understand how AAV adapts its replication strategy to the nuclear environment induced by the helper virus.
Acknowledgments
We thank Bernard Lopez for critically reading the manuscript, Juergen Kleinschmidt for providing the anti-Rep antibodies, and Frédéric Catez for providing the HSV probes used for FISH analyses. We are also grateful to the PLATIM and qPCR teams of IFR128 for excellent technical assistance and to Vincent Lotteau, Benoît De Chassey, and Laurène Meyniel for helpful discussions and for help with data analysis on the VirHostNet website.
This work was supported by CNRS, INSERM, Université Claude Bernard Lyon-1, and Ecole Normale Supérieure de Lyon. It was funded by grants from the Association Française contre les Myopathies (AFM), to A.S. and A.G., and from the NIH (R01 AI069146), to S.K.W.
Footnotes
Published ahead of print on 23 June 2010.
REFERENCES
- 1.Alazard-Dany, N., A. Nicolas, A. Ploquin, R. Strasser, A. Greco, A. L. Epstein, C. Fraefel, and A. Salvetti. 2009. Definition of herpes simplex virus type 1 helper activities for adeno-associated virus early replication events. PLoS Pathog. 5:e1000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Araujo, F. D., T. H. Stracker, C. T. Carson, D. V. Lee, and M. D. Weitzman. 2005. Adenovirus type 5 E4orf3 protein targets the Mre11 complex to cytoplasmic aggresomes. J. Virol. 79:11382-11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, A., K. J. Rohleder, L. A. Hanakahi, and G. Ketner. 2007. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 81:7034-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bevington, J. M., P. G. Needham, K. C. Verrill, R. F. Collaco, V. Basrur, and J. P. Trempe. 2007. Adeno-associated virus interactions with B23/nucleophosmin: identification of sub-nucleolar virion regions. Virology 357:102-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumel, J., S. Graper, and B. Matz. 2000. Structure of simian virus 40 DNA replicated by herpes simplex virus type 1. Virology 276:445-454. [DOI] [PubMed] [Google Scholar]
- 6.Bronstein, J. C., and P. C. Weber. 1996. Purification and characterization of herpes simplex virus type 1 alkaline exonuclease expressed in Escherichia coli. J. Virol. 70:2008-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunner, J. E., K. J. Ertel, J. M. Rozovics, and B. L. Semler. 2010. Delayed kinetics of poliovirus RNA synthesis in a human cell line with reduced levels of hnRNP C proteins. Virology 400:240-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter, B. J. 1990. Adeno-associated virus helper functions, p. 255-282. In P. Tijssen (ed.), Handbook of parvoviruses, vol. 1. CRC Press, Boca Raton, FL. [Google Scholar]
- 9.Cathomen, T., and M. D. Weitzman. 2000. A functional complex of adenovirus proteins E1B-55kDa and E4orf6 is necessary to modulate the expression level of p53 but not its transcriptional activity. J. Virol. 74:11407-11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaurushiya, M. S., and M. D. Weitzman. 2009. Viral manipulation of DNA repair and cell cycle checkpoints. DNA Repair (Amsterdam) 8:1166-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chejanovsky, N., and B. J. Carter. 1989. Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral DNA replication. Virology 173:120-128. [DOI] [PubMed] [Google Scholar]
- 12.Collaco, R. F., J. M. Bevington, V. Bhrigu, V. Kalman-Maltese, and J. P. Trempe. 2009. Adeno-associated virus and adenovirus coinfection induces a cellular DNA damage and repair response via redundant phosphatidylinositol 3-like kinase pathways. Virology 392:24-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daikoku, T., A. Kudoh, Y. Sugaya, S. Iwahori, N. Shirata, H. Isomura, and T. Tsurumi. 2006. Postreplicative mismatch repair factors are recruited to Epstein-Barr virus replication compartments. J. Biol. Chem. 281:11422-11430. [DOI] [PubMed] [Google Scholar]
- 14.Daya, S., and K. I. Berns. 2008. Gene therapy using adeno-associated virus vectors. Clin. Microbiol. Rev. 21:583-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dupierris, V., C. Masselon, M. Court, S. Kieffer-Jaquinod, and C. Bruley. 2009. A toolbox for validation of mass spectrometry peptides identification and generation of database: IRMa. Bioinformatics 25:1980-1981. [DOI] [PubMed] [Google Scholar]
- 16.Elliott, B., and M. Jasin. 2001. Repair of double-strand breaks by homologous recombination in mismatch repair-defective mammalian cells. Mol. Cell. Biol. 21:2671-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, G., M. R. Alvira, S. Somanathan, Y. Lu, L. H. Vandenberghe, J. J. Rux, R. Calcedo, J. Sanmiguel, Z. Abbas, and J. M. Wilson. 2003. Adeno-associated viruses undergo substantial evolution in primates during natural infection. Proc. Natl. Acad. Sci. U. S. A. 100:6081-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geoffroy, M. C., A. L. Epstein, E. Toublanc, P. Moullier, and A. Salvetti. 2004. Herpes simplex virus type 1 ICP0 protein mediates activation of adeno-associated virus type 2 rep gene expression from a latent integrated form. J. Virol. 78:10977-10986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geoffroy, M. C., and A. Salvetti. 2005. Helper functions required for wild type and recombinant adeno-associated virus growth. Curr. Gene Ther. 5:265-271. [DOI] [PubMed] [Google Scholar]
- 20.Glauser, D. L., R. Strasser, A. S. Laimbacher, O. Saydam, N. Clement, R. M. Linden, M. Ackermann, and C. Fraefel. 2007. Live covisualization of competing adeno-associated virus and herpes simplex virus type 1 DNA replication: molecular mechanisms of interaction. J. Virol. 81:4732-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstein, J. N., and S. K. Weller. 1998. The exonuclease activity of HSV-1 UL12 is required for in vivo function. Virology 244:442-457. [DOI] [PubMed] [Google Scholar]
- 22.Goldstein, J. N., and S. K. Weller. 1998. In vitro processing of herpes simplex virus type 1 DNA replication intermediates by the viral alkaline nuclease, UL12. J. Virol. 72:8772-8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guang, S., A. M. Felthauser, and J. E. Mertz. 2005. Binding of hnRNP L to the pre-mRNA processing enhancer of the herpes simplex virus thymidine kinase gene enhances both polyadenylation and nucleocytoplasmic export of intronless mRNAs. Mol. Cell. Biol. 25:6303-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hadian, K., M. Vincendeau, N. Mausbacher, D. Nagel, S. M. Hauck, M. Ueffing, A. Loyter, T. Werner, H. Wolff, and R. Brack-Werner. 2009. Identification of a heterogeneous nuclear ribonucleoprotein-recognition region in the HIV Rev protein. J. Biol. Chem. 284:33384-33391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He, Y., and R. Smith. 2009. Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell. Mol. Life Sci. 66:1239-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heilbronn, R., M. Engstler, S. Weger, A. Krahn, C. Schetter, and M. Boshart. 2003. ssDNA-dependent colocalization of adeno-associated virus Rep and herpes simplex virus ICP8 in nuclear replication domains. Nucleic Acids Res. 31:6206-6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Im, D. S., and N. Muzyczka. 1990. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell 61:447-457. [DOI] [PubMed] [Google Scholar]
- 28.Jiricny, J. 2006. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell. Biol. 7:335-346. [DOI] [PubMed] [Google Scholar]
- 29.Johansson, P. J., E. B. Myhre, and J. Blomberg. 1985. Specificity of Fc receptors induced by herpes simplex virus type 1: comparison of immunoglobulin G from different animal species. J. Virol. 56:489-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King, J. A., R. Dubielzig, D. Grimm, and J. A. Kleinschmidt. 2001. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 20:3282-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lilley, C. E., C. T. Carson, A. R. Muotri, F. H. Gage, and M. D. Weitzman. 2005. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl. Acad. Sci. U. S. A. 102:5844-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lilley, C. E., R. A. Schwartz, and M. D. Weitzman. 2007. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 15:119-126. [DOI] [PubMed] [Google Scholar]
- 33.Malik, P., and J. B. Clements. 2004. Protein kinase CK2 phosphorylation regulates the interaction of Kaposi's sarcoma-associated herpesvirus regulatory protein ORF57 with its multifunctional partner hnRNP K. Nucleic Acids Res. 32:5553-5569. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Mari, P. O., B. I. Florea, S. P. Persengiev, N. S. Verkaik, H. T. Bruggenwirth, M. Modesti, G. Giglia-Mari, K. Bezstarosti, J. A. Demmers, T. M. Luider, A. B. Houtsmuller, and D. C. van Gent. 2006. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl. Acad. Sci. U. S. A. 103:18597-18602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez, R., J. N. Goldstein, and S. K. Weller. 2002. The product of the UL12.5 gene of herpes simplex virus type 1 is not essential for lytic viral growth and is not specifically associated with capsids. Virology 298:248-257. [DOI] [PubMed] [Google Scholar]
- 36.Martinez, R., L. Shao, J. C. Bronstein, P. C. Weber, and S. K. Weller. 1996. The product of a 1.9-kb mRNA which overlaps the HSV-1 alkaline nuclease gene (UL12) cannot relieve the growth defects of a null mutant. Virology 215:152-164. [DOI] [PubMed] [Google Scholar]
- 37.Martinez-Contreras, R., P. Cloutier, L. Shkreta, J. F. Fisette, T. Revil, and B. Chabot. 2007. hnRNP proteins and splicing control. Adv. Exp. Med. Biol. 623:123-147. [DOI] [PubMed] [Google Scholar]
- 38.Muylaert, I., and P. Elias. 2007. Knockdown of DNA ligase IV/XRCC4 by RNA interference inhibits herpes simplex virus type I DNA replication. J. Biol. Chem. 282:10865-10872. [DOI] [PubMed] [Google Scholar]
- 39.Muzyczka, N., and K. I. Berns. 2001. Parvoviridae: the viruses and their replication, p. 2327-2359. In D. M. Knipe et al. (ed.), Fields virology, vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 40.Nash, K., W. Chen, W. F. McDonald, X. Zhou, and N. Muzyczka. 2007. Purification of host cell enzymes involved in adeno-associated virus DNA replication. J. Virol. 81:5777-5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nash, K., W. Chen, and N. Muzyczka. 2008. Complete in vitro reconstitution of adeno-associated virus DNA replication requires the minichromosome maintenance complex proteins. J. Virol. 82:1458-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nash, K., W. Chen, M. Salganik, and N. Muzyczka. 2009. Identification of cellular proteins that interact with the adeno-associated virus rep protein. J. Virol. 83:454-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ni, T.-H., W. F. McDonald, I. Zolothukin, T. Melendy, S. Waga, B. Stillman, and N. Muzyczka. 1998. Cellular proteins required for adeno-associated virus DNA replication in the absence of adenovirus coinfection. J. Virol. 72:2777-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ni, T. H., X. Zhou, D. M. McCarty, I. Zolotukhin, and N. Muzyczka. 1994. In vitro replication of adeno-associated virus DNA. J. Virol. 68:1128-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parkinson, J., S. P. Lees-Miller, and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73:650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Porter, I. M., and N. D. Stow. 2004. Virus particles produced by the herpes simplex virus type 1 alkaline nuclease null mutant ambUL12 contain abnormal genomes. J. Gen. Virol. 85:583-591. [DOI] [PubMed] [Google Scholar]
- 47.Qiu, J., and D. J. Pintel. 2002. The adeno-associated virus type 2 Rep protein regulates RNA processing via interaction with the transcription template. Mol. Cell. Biol. 22:3639-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rass, E., A. Grabarz, I. Plo, J. Gautier, P. Bertrand, and B. S. Lopez. 2009. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 16:819-824. [DOI] [PubMed] [Google Scholar]
- 50.Reuven, N. B., S. Antoku, and S. K. Weller. 2004. The UL12.5 gene product of herpes simplex virus type 1 exhibits nuclease and strand exchange activities but does not localize to the nucleus. J. Virol. 78:4599-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reuven, N. B., A. E. Staire, R. S. Myers, and S. K. Weller. 2003. The herpes simplex virus type 1 alkaline nuclease and single-stranded DNA binding protein mediate strand exchange in vitro. J. Virol. 77:7425-7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reuven, N. B., S. Willcox, J. D. Griffith, and S. K. Weller. 2004. Catalysis of strand exchange by the HSV-1 UL12 and ICP8 proteins: potent ICP8 recombinase activity is revealed upon resection of dsDNA substrate by nuclease. J. Mol. Biol. 342:57-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rizwani, W., M. Alexandrow, and S. Chellappan. 2009. Prohibitin physically interacts with MCM proteins and inhibits mammalian DNA replication. Cell Cycle 8:1621-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saffran, H. A., J. M. Pare, J. A. Corcoran, S. K. Weller, and J. R. Smiley. 2007. Herpes simplex virus eliminates host mitochondrial DNA. EMBO Rep. 8:188-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmidt, M., S. Afione, and R. M. Kotin. 2000. Adeno-associated virus type 2 Rep78 induces apoptosis through caspase activation independently of p53. J. Virol. 74:9441-9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schreiber, V., F. Dantzer, J. C. Ame, and G. de Murcia. 2006. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell. Biol. 7:517-528. [DOI] [PubMed] [Google Scholar]
- 57.Schwartz, R. A., C. T. Carson, C. Schuberth, and M. D. Weitzman. 2009. Adeno-associated virus replication induces a DNA damage response coordinated by DNA-dependent protein kinase. J. Virol. 83:6269-6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwartz, R. A., J. A. Palacios, G. D. Cassell, S. Adam, M. Giacca, and M. D. Weitzman. 2007. The Mre11/Rad50/Nbs1 complex limits adeno-associated virus transduction and replication. J. Virol. 81:12936-12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Senapathy, P., and B. J. Carter. 1984. Molecular cloning of adeno-associated virus variant genomes and generation of infectious virus by recombination in mammalian cells. J. Biol. Chem. 259:4661-4666. [PubMed] [Google Scholar]
- 60.Shirata, N., A. Kudoh, T. Daikoku, Y. Tatsumi, M. Fujita, T. Kiyono, Y. Sugaya, H. Isomura, K. Ishizaki, and T. Tsurumi. 2005. Activation of ataxia telangiectasia-mutated DNA damage checkpoint signal transduction elicited by herpes simplex virus infection. J. Biol. Chem. 280:30336-30341. [DOI] [PubMed] [Google Scholar]
- 61.Shrivastav, M., L. P. De Haro, and J. A. Nickoloff. 2008. Regulation of DNA double-strand break repair pathway choice. Cell Res. 18:134-147. [DOI] [PubMed] [Google Scholar]
- 62.Slanina, H., S. Weger, N. D. Stow, A. Kuhrs, and R. Heilbronn. 2006. Role of the herpes simplex virus helicase-primase complex during adeno-associated virus DNA replication. J. Virol. 80:5241-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith, J. A., L. A. Bannister, V. Bhattacharjee, Y. Wang, B. C. Waldman, and A. S. Waldman. 2007. Accurate homologous recombination is a prominent double-strand break repair pathway in mammalian chromosomes and is modulated by mismatch repair protein Msh2. Mol. Cell. Biol. 27:7816-7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Snyder, R. O., R. J. Samulski, and N. Muzyczka. 1990. In vitro resolution of covalently joined AAV chromosome ends. Cell 60:105-113. [DOI] [PubMed] [Google Scholar]
- 65.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348-352. [DOI] [PubMed] [Google Scholar]
- 66.Stracker, T. H., G. D. Cassell, P. Ward, Y. M. Loo, B. van Breukelen, S. D. Carrington-Lawrence, R. K. Hamatake, P. C. van der Vliet, S. K. Weller, T. Melendy, and M. D. Weitzmann. 2004. The Rep protein of adeno-associated virus type 2 interacts with single-stranded DNA-binding proteins that enhance viral replication. J. Virol. 78:441-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tanaka, E., H. Fukuda, K. Nakashima, N. Tsuchiya, H. Seimiya, and H. Nakagama. 2007. hnRNP A3 binds to and protects mammalian telomeric repeats in vitro. Biochem. Biophys. Res. Commun. 358:608-614. [DOI] [PubMed] [Google Scholar]
- 68.Tatsuta, T., K. Model, and T. Langer. 2005. Formation of membrane-bound ring complexes by prohibitins in mitochondria. Mol. Biol. Cell 16:248-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taylor, T. J., and D. M. Knipe. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 78:5856-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thomas, M. S., M. Gao, D. M. Knipe, and K. L. Powell. 1992. Association between the herpes simplex virus major DNA-binding protein and alkaline nuclease. J. Virol. 66:1152-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Timpe, J., J. Bevington, J. Casper, J. D. Dignam, and J. P. Trempe. 2005. Mechanisms of adeno-associated virus genome encapsidation. Curr. Gene Ther. 5:273-284. [DOI] [PubMed] [Google Scholar]
- 72.Timpe, J. M., K. C. Verrill, B. N. Black, H. F. Ding, and J. P. Trempe. 2007. Adeno-associated virus induces apoptosis during coinfection with adenovirus. Virology 358:391-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang, D., and C. R. Parrish. 1999. A heterogeneous nuclear ribonucleoprotein A/B-related protein binds to single-stranded DNA near the 5′ end or within the genome of feline parvovirus and can modify virus replication. J. Virol. 73:7761-7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang, S., G. Fusaro, J. Padmanabhan, and S. P. Chellappan. 2002. Prohibitin co-localizes with Rb in the nucleus and recruits N-CoR and HDAC1 for transcriptional repression. Oncogene 21:8388-8396. [DOI] [PubMed] [Google Scholar]
- 75.Wang, Y., H. Li, Q. Tang, G. G. Maul, and Y. Yuan. 2008. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: involvement of host cellular factors. J. Virol. 82:2867-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ward, P. 2005. Replication of adeno-associated virus DNA, p. 189-211. In J. R. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 77.Ward, P., F. B. Dean, M. E. O'Donnell, and K. I. Berns. 1998. Role of the adenovirus DNA-binding protein in in vitro adeno-associated virus DNA replication. J. Virol. 72:420-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ward, P., M. Falkenberg, P. Elias, M. Weitzman, and R. M. Linden. 2001. Rep-dependent initiation of adeno-associated virus type 2 DNA replication by a herpes simplex virus type 1 replication complex in a reconstituted system. J. Virol. 75:10250-10258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weiden, M. D., and H. S. Ginsberg. 1994. Deletion of the E4 region of the genome produces adenovirus DNA concatemers. Proc. Natl. Acad. Sci. U. S. A. 91:153-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weindler, F. W., and R. Heilbronn. 1991. A subset of herpes simplex virus replication genes provides helper functions for productive adeno-associated virus replication. J. Virol. 65:2476-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weitzman, M. D., K. J. Fisher, and J. M. Wilson. 1996. Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J. Virol. 70:1845-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weller, S. K., M. R. Seghatoleslami, L. Shao, D. Rowse, and E. P. Carmichael. 1990. The herpes simplex virus type 1 alkaline nuclease is not essential for viral DNA synthesis: isolation and characterization of a lacZ insertion mutant. J. Gen. Virol. 71:2941-2952. [DOI] [PubMed] [Google Scholar]
- 83.Westmoreland, D., and J. F. Watkins. 1974. The IgG receptor induced by herpes simplex virus: studies using radioiodinated IgG. J. Gen. Virol. 24:167-178. [DOI] [PubMed] [Google Scholar]
- 84.Wiedmer, A., P. Wang, J. Zhou, A. J. Rennekamp, V. Tiranti, M. Zeviani, and P. M. Lieberman. 2008. Epstein-Barr virus immediate-early protein Zta co-opts mitochondrial single-stranded DNA binding protein to promote viral and inhibit mitochondrial DNA replication. J. Virol. 82:4647-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wilkinson, D. E., and S. K. Weller. 2004. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J. Virol. 78:4783-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wilkinson, D. E., and S. K. Weller. 2003. The role of DNA recombination in herpes simplex virus DNA replication. IUBMB Life 55:451-458. [DOI] [PubMed] [Google Scholar]
- 87.Wistuba, A., S. Weger, A. Kern, and J. A. Kleinschmidt. 1995. Intermediates of adeno-associated virus type 2 assembly: identification of soluble complexes containing Rep and Cap proteins. J. Virol. 69:5311-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xie, A., A. Kwok, and R. Scully. 2009. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 16:814-818. [DOI] [PMC free article] [PubMed] [Google Scholar]