Abstract
Physiological bone remodeling is a highly coordinated process responsible for bone resorption and formation and is necessary to repair damaged bone and to maintain mineral homeostasis. In addition to the traditional bone cells (osteoclasts, osteoblasts, and osteocytes) that are necessary for bone remodeling, several immune cells have also been implicated in bone disease. This minireview discusses physiological bone remodeling, outlining the traditional bone biology dogma in light of emerging osteoimmunology data. Specifically discussed in detail are the cellular and molecular mechanisms of bone remodeling, including events that orchestrate the five sequential phases of bone remodeling: activation, resorption, reversal, formation, and termination.
Keywords: Bone, Cytokine, Macrophage, Transforming Growth Factor beta (TGFbeta), Wnt Pathway, Bone Remodeling, Osteoblasts, Osteoclasts, Osteocytes, Osteoimmunology
Introduction
Bone is a dynamic tissue that undergoes continual adaption during vertebrate life to attain and preserve skeletal size, shape, and structural integrity and to regulate mineral homeostasis. Two processes, remodeling and modeling, underpin development and maintenance of the skeletal system. Bone modeling is responsible for growth and mechanically induced adaption of bone and requires that the processes of bone formation and bone removal (resorption), although globally coordinated, occur independently at distinct anatomical locations. Bone remodeling is responsible for removal and repair of damaged bone to maintain integrity of the adult skeleton and mineral homeostasis. This tightly coordinated event requires the synchronized activities of multiple cellular participants to ensure that bone resorption and formation occur sequentially at the same anatomical location to preserve bone mass. This work will review the cellular participants and molecular mechanisms that coordinate the five distinct phases of bone remodeling and includes an appraisal of immune cells and their role in regulating normal bone physiology.
Cells Involved in Bone Remodeling
Osteoclasts
Osteoclasts are terminally differentiated myeloid cells that are uniquely adapted to remove mineralized bone matrix. These cells have distinct morphological and phenotypic characteristics that are routinely used to identify them, including multinuclearity and expression of tartrate-resistant acid phosphatase and the calcitonin receptor (1). CSF-1 (colony-stimulating factor 1; also known as macrophage colony-stimulating factor) and RANKL (receptor activator of NF-κB ligand) are critical cytokines required for survival, expansion, and differentiation of osteoclast precursor cells in vitro (2). In vivo, the requirement of these cytokines for osteoclastic bone resorption has been demonstrated in mouse models that lacked functional CSF-1 and RANKL. The op/op mouse has a spontaneous thymidine insertion and subsequent frameshift that creates a stop codon in the Csf-1 gene and lacks functional CSF-1, resulting in osteopetrosis (dense bones) (3). Tnfs11 is the gene for RANKL, and its genetic ablation in mice also causes osteopetrosis (4). The osteopetrotic phenotype in these mouse models was caused by complete absence of osteoclasts (albeit a transient absence in op/op mice) (3, 4). Osteoprotegerin (OPG)2 is a soluble decoy receptor for RANKL and a physiological negative regulator of osteoclastogenesis; loss of functional OPG in mice results in animals with osteoporosis (brittle bones) due to excessive osteoclastogenesis (5). The current paradigm dictates that the RANKL/OPG expression ratio determines the degree of osteoclast differentiation and function (6).
A cascade of transcription factors is required to ultimately direct myeloid cells toward an osteoclast fate. Expression of the ETS transcription factor PU.1 early during myeloid cell differentiation is essential for development of osteoclasts and other mature myeloid cells. Mice lacking PU.1 fail to develop osteoclasts and macrophages, and in vitro osteoclast differentiation correlates with increasing PU.1 expression (7). The AP-1 transcription factor c-Fos is also essential for osteoclastogenesis. Mice deficient in c-Fos are osteopetrotic and lack osteoclasts but have increased macrophages (8). This implies that the requirement for c-Fos is secondary to PU.1, and although essential for osteoclastogenesis, c-Fos is not necessary for macrophage differentiation. Loss of c-Fos shuttles myeloid precursors away from an osteoclast fate and redirects them toward macrophage commitment. The transcription factors MITF (microphthalmia-associated transcription factor) and NFATc1 (nuclear factor of activated T-cells, cytoplasmic 1) are also required for osteoclast formation and expression of functionally relevant osteoclast genes, including tartrate-resistant acid phosphatase (9), cathepsin K, and the calcitonin receptor (10, 11). Although PU.1, MITF, and c-Fos work in concert with NFATc1 to orchestrate terminal osteoclast differentiation and function, NFATc1 is a critical switch because its presence is both necessary and sufficient for osteoclastogenesis to occur (12).
Despite the current knowledge of transcription factors involved in osteoclastogenesis, the definitive physiological in vivo osteoclast precursor in mice and humans remains elusive. Certainly, a defining characteristic of osteoclast precursors is expression of c-Fms and RANK (receptor activator of NF-κB) (13), which are the cognate receptors for CSF-1 and RANKL, respectively. However, in vitro, many murine myeloid cells at various stages of lineage development can differentiate into osteoclasts (14). Identification of the definitive population of in vivo osteoclast precursors in both rodents and humans is continuing. Currently, in the murine system, a quiescent endosteal myeloid population, referred to as cell cycle-arrested quiescent osteoclast precursors, which are c-Fms- and RANK-positive but F4/80-negative (a mature macrophage marker), is a likely candidate during physiological osteoclastogenesis (15).
Osteoblasts
Osteoblasts are specialized bone-forming cells that express parathyroid hormone (PTH) receptors and have several important roles in bone remodeling: expression of osteoclastogenic factors, production of bone matrix proteins, and bone mineralization (16). Osteoblastic cells comprise a diverse population of cells that include immature osteoblast lineage cells and differentiating and mature matrix-producing osteoblasts. In vitro, phenotypic osteoblast heterogeneity is associated with cell differentiation (17). The stage of osteoblast differentiation also influences the functional contribution of these cells to in vivo bone remodeling. Mice deficient in osteoblasts are also deficient in osteoclasts (18); however, conditional depletion of mature osteoblasts in vivo only ablates bone formation, whereas osteoclastic bone resorption persists (19). These data suggest that immature osteoblasts direct osteoclastogenesis, whereas mature osteoblasts perform the matrix production and mineralization functions.
Osteoblasts develop from pluripotent mesenchymal stem cells that have the potential to differentiate into adipocytes, myocytes, chondrocytes, and osteoblasts under the direction of a defined suite of regulatory transcription factors. Osteoblast differentiation is controlled by the master transcription factor RUNX2 (runt-related transcription factor 2; also known as CBFA1 (core-binding factor A1)) (20). RUNX2 null mice have a cartilaginous skeleton and completely lack mineralized tissue due to arrest of osteoblast maturation (18, 21).
Osteocytes
During bone formation, a subpopulation of osteoblasts undergoes terminal differentiation and becomes engulfed by unmineralized osteoid, at which time they are referred to as osteoid-osteocytes (22). Following mineralization of the bone matrix, these entombed cells are called osteocytes and form a network extending throughout mineralized bone. Osteocytes are cocooned in fluid-filled cavities (lacunae) within the mineralized bone and are highly abundant, accounting for 90–95% of all bone cells (23). Osteocytes have long dendrite-like processes that extend throughout canaliculi (tunnels) within the mineralized matrix. These dendrite-like processes interact with other osteocytes within mineralized bone and also interact with osteoblasts on the bone surface (24). Osteocytes respond to mechanical load, and this network is thought to be integral in the detection of mechanical strain and associated bone microdamage (microscopic cracks or fractures within the mineralized bone) that accumulates as a result of normal skeletal loading and fatigue (25). Data have been obtained that support the idea that osteocytes initiate and direct the subsequent remodeling process that repairs damaged bone.
Immune Cells Involved in Physiological Bone Remodeling
Despite the close anatomical localization of bone with bone marrow, the dynamic cross-talk between the skeletal and immune systems has been underappreciated and underinvestigated. Interest in immune cell regulation of bone dynamics initially focused on pathological diseases. For example, activated T-cells have been implicated in pathological diseases that result in bone destruction, including ovariectomy-induced bone loss (26). Mast cells have been connected with the bone marrow fibrosis that occurs in chronic hyperparathyroidism (27). However, it is becoming clear that immune cells also participate in bone homeostasis during normal physiology.
T-cells and B-cells
T- and B-lymphocytes are central components of the adaptive immune system that facilitate recognition and destruction of pathogens. Mice lacking either B- or T-cells have osteoporotic bones, suggesting that these immune cells participate in the maintenance of bone homeostasis during basal physiology (28). Mechanistically, mature B-cells produce >50% of total bone marrow-derived OPG, which would contribute significantly to restraining osteoclastogenesis during normal physiology. However, the role of T-cells in regulating bone remodeling during homeostasis is less clear. Based on data that T-cell-deficient CD40 knock-out and CD40L knock-out mice are osteoporotic (28), it has been proposed that T-cells work cooperatively with B-cells and enhance OPG production via CD40/CD40L co-stimulation. However, the relevance of this mechanism to homeostatic bone remodeling remains obscure given that T-cell activation is required for expression of CD40L and that there are very few activated T-cells in bone marrow under basal conditions.
Megakaryocytes
Derived from hematopoietic stem cells, megakaryocytes reside within bone marrow and produce thrombocytes (known as platelets) that are essential for normal blood clotting. Transgenic mice deficient in the transcription factors GATA-1 and NF-E2 (nuclear factor, erythroid-derived 2; involved in megakaryocyte differentiation) have increased megakaryocyte numbers and elevated bone volume (29). In vitro, megakaryocytes enhance osteoblast proliferation and differentiation, express RANKL and OPG, and secrete an unknown soluble anti-osteoclastic factor (30). Overall, these data suggest that megakaryocytes have the potential to direct both the resorption and formation arms of bone remodeling. However, expression of GATA-1 or NF-E2 is not restricted to megakaryocytes, and more definitive in vivo studies are required to confirm a role for these cells in physiological bone remodeling.
Osteomacs
Osteomacs are resident tissue macrophages that reside on or within three cells of endosteal and periosteal surfaces. Resident tissue macrophages compose ∼10–15% of most tissues and are important for tissue development, homeostasis, and repair (31). In mice, osteomacs are identified using the pan macrophage protein F4/80, which is not expressed by osteoclasts, and their anatomical location in close proximity to the bone surface. In human bone, osteomacs can be identified by expression of the myeloid marker CD68, their distinctive stellate morphology, and location in close proximity to bone surfaces (32). In vitro, osteomacs are required for full functional differentiation, including mineralization, of osteoblasts. In vivo, osteomacs form a canopy over mature matrix-producing osteoblasts at sites of bone modeling, an ideal anatomical location from which to regulate this process. Depletion of macrophages in vivo results in complete loss of endosteal osteomacs and their associated osteoblasts, suggesting that osteomacs are needed to maintain mature osteoblasts (32).
Bone Remodeling: The Process
Bone remodeling occurs over several weeks and is performed by clusters of bone-resorbing osteoclasts and bone-forming osteoblasts arranged within temporary anatomical structures known as “basic multicellular units” (BMUs). Traversing and encasing the BMU is a canopy of cells that creates a bone-remodeling compartment (33). The phenotype of the canopy cell is still under debate. Evidence in humans suggests that it is a bone-lining cell, whereas in the mouse, osteomacs traverse BMUs during physiological bone remodeling (34). Functionally, it has been proposed that the canopy structure, and subsequent bone-remodeling compartment, generates a unique microenvironment to facilitate “coupled” osteoclast resorption and osteoblast formation and ensures minimal net change in bone volume during physiological bone remodeling (35).
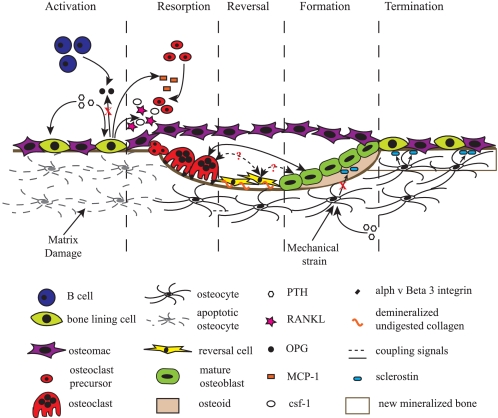
An active BMU consists of a leading front of bone-resorbing osteoclasts. Reversal cells, of unclear phenotype, follow the osteoclasts, covering the newly exposed bone surface, and prepare it for deposition of replacement bone. Osteoblasts occupy the tail portion of the BMU and secrete and deposit unmineralized bone matrix known as osteoid and direct its formation and mineralization into mature lamellar bone. This unique spatial and temporal arrangement of cells within the BMU is critical to bone remodeling, ensuring coordination of the distinct and sequential phases of this process: activation, resorption, reversal, formation, and termination, which are discussed below and illustrated schematically in Fig. 1.
FIGURE 1.
Schematic representation of a BMU and the associated bone-remodeling process. Prior to activation, the resting bone surface is covered with bone-lining cells, including preosteoblasts intercalated with osteomacs. B-cells are present in the bone marrow and secrete OPG, which suppresses osteoclastogenesis. Activation, the endocrine bone-remodeling signal PTH binds to the PTH receptor on preosteoblasts. Damage to the mineralized bone matrix results in localized osteocyte apoptosis, reducing the local TGF-β concentration and its inhibition of osteoclastogenesis. Resorption, in response to PTH signaling, MCP-1 is released from osteoblasts and recruits preosteoclasts to the bone surface. Additionally, osteoblast expression of OPG is decreased, and production of CSF-1 and RANKL is increased to promote proliferation of osteoclast precursors and differentiation of mature osteoclasts. Mature osteoclasts anchor to RGD-binding sites, creating a localized microenvironment (sealed zone) that facilitates degradation of the mineralized bone matrix. Reversal, reversal cells engulf and remove demineralized undigested collagen from the bone surface. Transition signals are generated that halt bone resorption and stimulate the bone formation process. Formation, formation signals and molecules arise from the degraded bone matrix, mature osteoclasts, and potentially reversal cells. PTH and mechanical activation of osteocytes reduce sclerostin expression, allowing for Wnt-directed bone formation to occur. Termination, sclerostin expression likely returns, and bone formation ceases. The newly deposited osteoid is mineralized, the bone surfaces return to a resting state with bone-lining cells intercalated with osteomacs, and the remodeling cycle concludes.
Activation Phase
The first stage of bone remodeling involves detection of an initiating remodeling signal. This signal can take several forms, e.g. direct mechanical strain on the bone that results in structural damage or hormone (e.g. estrogen or PTH) action on bone cells in response to more systemic changes in homeostasis.
Daily activity places ongoing mechanical strain on the skeleton, and it is thought that osteocytes sense changes in these physical forces and translate them into biological signals that initiate bone remodeling (23). Damage to the bone matrix (25) or limb immobilization (36) results in osteocyte apoptosis and increased osteoclastogenesis (36). Under basal conditions, osteocytes secrete transforming growth factor β (TGF-β), which inhibits osteoclastogenesis. Focal osteocyte apoptosis lowers local TGF-β levels, removing the inhibitory osteoclastogenesis signals and allowing osteoclast formation to proceed (37).
The calciotropic hormone PTH is an endocrine remodeling signal generated to maintain calcium homeostasis. PTH is secreted by the parathyroid glands in response to reduced serum calcium and acts peripherally on kidneys and bone and indirectly on the intestine to maintain serum calcium homeostasis. In the bone microenvironment, PTH activates a seven-transmembrane G-protein-coupled receptor, the PTH receptor, on the surface of osteoblastic cells (38). Binding of PTH to its receptor activates protein kinase A, protein kinase C, and calcium intracellular signaling pathways in these cells (39) and induces a wave of transcriptional responses that produce/modulate secretion of molecules that recruit osteoclast precursors, induce osteoclast differentiation and activation, and establish bone resorption.
Resorption Phase
Osteoblasts respond to signals generated by osteocytes or direct endocrine activation signals discussed above and recruit osteoclast precursors to the remodeling site. In response to PTH-induced bone remodeling, osteoblasts produce the chemokine MCP-1 (monocyte chemoattractant protein-1) in vivo, which is a chemoattractant for osteoclast precursors and enhances RANKL-induced osteoclastogenesis in vitro (40). In addition to recruitment of osteoclast precursors, osteoblast expression of the master osteoclastogenesis cytokines, CSF-1, RANKL, and OPG, is also modulated in response to PTH. OPG expression is reduced, and CSF-1 and RANKL production is increased to promote osteoclast formation and subsequent activity (41). CSF-1 and RANKL work in concert; CSF-1 promotes proliferation and survival of osteoclast precursors and directs spreading, motility, and cytoskeletal organization in mature cells (42). RANKL also promotes proliferation of osteoclast precursors and additionally coordinates the differentiation of osteoclast precursors to multinucleated osteoclasts, promotes resorption activity, and prolongs the life of the mature cells (43). Matrix metalloproteinases (MMPs), including MMP-13, are also secreted from osteoblasts in response to mechanical (44) and endocrine (45) remodeling signals. MMPs degrade the unmineralized osteoid that lines the bone surface and expose RGD adhesion sites within mineralized bone that are necessary to facilitate osteoclast attachment. Osteoclasts anchor to these RGD-binding sites via αvβ3 integrin molecules (46) and create an isolated microenvironment beneath the cell known as the “sealed zone.” Hydrogen ions are pumped into the sealed zone, and dissolution of mineralized matrix occurs in this acidic space, producing Howship's resorption lacunae (47). The remaining organic bone matrix is then degraded by a collection of collagenolytic enzymes with a low pH optimum (in particular, cathepsin K) (48).
Reversal Phase
Following osteoclast-mediated resorption, the Howship lacunae remain covered with undigested demineralized collagen matrix (49). A mononuclear cell of undetermined lineage removes these collagen remnants and prepares the bone surface for subsequent osteoblast-mediated bone formation. Initially, this “reversal” cell was proposed to be a monocytic phagocyte based on morphological assessment (50). However, more recently, it was reported that the reversal cell is from the osteoblast lineage, based on cell morphology, positive expression for alkaline phosphatase, and the absence of the monocyte macrophage marker MOMA-2 (monocyte + macrophage antibody-2) on these cells (49). In light of the recent characterization of F4/80+ osteomacs and their association with BMUs (34), a more detailed assessment of the reversal cell phenotype needs to be undertaken because macrophages can express alkaline phosphatase (51), and MOMA-2 and F4/80 detect different macrophage populations (52). From a functional perspective, the likely explanation is that both osteomacs and mesenchymal bone-lining cells work together to facilitate events during the reversal phase. Osteomacs are likely responsible for removal of matrix debris during the reversal phase. Indeed, macrophages can produce MMPs (53), the enzymes required for matrix degradation, and are professional phagocytic cells. Macrophages can also produce osteopontin (54), which is incorporated into mineralized tissue. However, the mesenchymal bone-lining cells are more ideally equipped to deposit the collagenous matrix that forms along osteopontin-rich cement lines within Howship lacunae (49). The final role of the reversal cells may be to receive or produce coupling signals that allow transition from bone resorption to bone formation within the BMU.
Formation Phase
The nature of the coupling signal that coordinates this transition and directs bone formation precisely to sites of bone resorption remains controversial. Initially, it was proposed that the coupling molecule(s) were stored in the bone matrix and liberated during bone resorption. Insulin-like growth factors I and II and TGF-β are all such factors, and regulation of active TGF-β appears to be a key signal for recruitment of mesenchymal stem cells to sites of bone resorption (55). However, in mice and humans that have functionally defective osteoclasts, unable to resorb bone, osteoblast bone formation is preserved even in the absence of released matrix-bound growth factors. These observations have led to the hypothesis that osteoclasts produce the coupling factor(s) (56). Several candidate coupling mechanisms have been proposed, including the soluble molecule sphingosine 1-phosphate and the cell-anchored EphB4·ephrin-B2 bidirectional signaling complex. Sphingosine 1-phosphate is secreted by osteoclasts, induces osteoblast precursor recruitment, and promotes mature osteoblast survival (57). EphB4 receptors are expressed on osteoblasts, whereas osteoclasts express the ligand ephrin-B2. Forward signaling through EphB4 into osteoblasts enhances osteogenic differentiation, and reverse signaling through ephrin-B2 into osteoclast precursors suppresses osteoclast differentiation by inhibiting the osteoclastogenic c-Fos/NFATc1 cascade (58). This EphB4·ephrin-B2 signaling complex provides a unique opportunity to activate bone formation and inhibit bone resorption simultaneously at this critical transition point of the remodeling process. However, the anatomical constraints within the BMU mean that direct cell contact between osteoclasts and osteoblasts is not always possible, and indeed, osteoblast recruitment and matrix deposition continue long after osteoclasts have vacated a resorption site. Therefore, several mechanisms, including both direct contact and soluble signals, may be required to achieve coupling.
Mechanical stimulation and the endocrine signal PTH can exert bone formation signals via osteocytes. Under resting conditions, osteocytes express sclerostin (59), a soluble molecule that binds to LRP5/6 (low density lipoprotein receptor-related protein-5/6) and directly prevents Wnt signaling (60), an inducer of bone formation. Mechanical strain on bone and PTH signaling, via PTH receptors on osteocytes (61), inhibit osteocyte expression of sclerostin (62), removing inhibition of Wnt signaling and allowing Wnt-directed bone formation to occur via its receptor and coreceptor, LRP5/6. This anabolic Wnt signaling pathway is critical in establishing basal bone mineral density; however, the specifics of how mechanical strain and PTH signaling exert opposing effects in the early and late stages of physiological bone remodeling remain to be determined.
Once mesenchymal stem cells or early osteoblast progenitors have returned to the resorption lacunae, they differentiate and secrete molecules that ultimately form replacement bone. Collagen type I is the primary organic component of bone. Non-collagenous proteins, including proteoglycans, glycosylated proteins such as tissue nonspecific alkaline phosphatase, small integrin-binding ligand (SIBLING) proteins, Gla-containing proteins (matrix Gla protein and osteocalcin), and lipids compose the remaining organic material (63). For bone to assume its final form, hydroxylapatite is incorporated into this newly deposited osteoid. The precise mechanism of mineralization remains to be fully elucidated; however, tissue nonspecific alkaline phosphatase (64), nucleotide pyrophosphatase phosphodiesterase, and ANK (progressive ankylosis) are involved in generating the optimal extracellular concentration of inorganic phosphate that allows mineralization to proceed (65).
Termination Phase
When an equal quantity of resorbed bone has been replaced, the remodeling cycle concludes. The termination signal(s) that inform the remodeling machinery to cease work are largely unknown, although a role for osteocytes is emerging. The loss of sclerostin expression, which occurred to initiate osteoblastic bone formation, likely returns toward the end of the remodeling cycle. Following mineralization, mature osteoblasts undergo apoptosis, revert back to a bone-lining phenotype or become embedded in the mineralized matrix, and differentiate into osteocytes. The resting bone surface environment is reestablished and maintained until the next wave of remodeling is initiated.
Conclusion
The current physiological bone-remodeling paradigm is incomplete, and deficiencies in understanding the mechanisms that couple bone resorption and formation are a barrier to successfully treating the numerous pathological bone diseases that result in bone loss. Many diseases of bone have an associated immune component. Complete confirmation and understanding of the role that immune cells, such as the osteomacs, have in bone remodeling are essential to facilitate the ongoing search for improved therapeutics for bone disease.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants DK47420 and DK48109 (to N. C. P.). This work was also supported by National Health and Medical Research Council Grants 455941 and 631484 (to L. J. R.). This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- OPG
- osteoprotegerin
- PTH
- parathyroid hormone
- BMU
- basic multicellular unit
- TGF-β
- transforming growth factor β
- MMP
- matrix metalloproteinase.
REFERENCES
- 1.Teitelbaum S. L., Ross F. P. (2003) Nat. Rev. Genet. 4, 638–649 [DOI] [PubMed] [Google Scholar]
- 2.Lacey D. L., Timms E., Tan H. L., Kelley M. J., Dunstan C. R., Burgess T., Elliott R., Colombero A., Elliott G., Scully S., Hsu H., Sullivan J., Hawkins N., Davy E., Capparelli C., Eli A., Qian Y. X., Kaufman S., Sarosi I., Shalhoub V., Senaldi G., Guo J., Delaney J., Boyle W. J. (1998) Cell 93, 165–176 [DOI] [PubMed] [Google Scholar]
- 3.Yoshida H., Hayashi S., Kunisada T., Ogawa M., Nishikawa S., Okamura H., Sudo T., Shultz L. D., Nishikawa S. (1990) Nature 345, 442–444 [DOI] [PubMed] [Google Scholar]
- 4.Kong Y. Y., Yoshida H., Sarosi I., Tan H. L., Timms E., Capparelli C., Morony S., Oliveira-dos-Santos A. J., Van G., Itie A., Khoo W., Wakeham A., Dunstan C. R., Lacey D. L., Mak T. W., Boyle W. J., Penninger J. M. (1999) Nature 397, 315–323 [DOI] [PubMed] [Google Scholar]
- 5.Simonet W. S., Lacey D. L., Dunstan C. R., Kelley M., Chang M. S., Lüthy R., Nguyen H. Q., Wooden S., Bennett L., Boone T., Shimamoto G., DeRose M., Elliott R., Colombero A., Tan H. L., Trail G., Sullivan J., Davy E., Bucay N., Renshaw-Gegg L., Hughes T. M., Hill D., Pattison W., Campbell P., Sander S., Van G., Tarpley J., Derby P., Lee R., Boyle W. J. (1997) Cell 89, 309–319 [DOI] [PubMed] [Google Scholar]
- 6.Hofbauer L. C., Khosla S., Dunstan C. R., Lacey D. L., Boyle W. J., Riggs B. L. (2000) J. Bone Miner. Res. 15, 2–12 [DOI] [PubMed] [Google Scholar]
- 7.Tondravi M. M., McKercher S. R., Anderson K., Erdmann J. M., Quiroz M., Maki R., Teitelbaum S. L. (1997) Nature 386, 81–84 [DOI] [PubMed] [Google Scholar]
- 8.Grigoriadis A. E., Wang Z. Q., Cecchini M. G., Hofstetter W., Felix R., Fleisch H. A., Wagner E. F. (1994) Science 266, 443–448 [DOI] [PubMed] [Google Scholar]
- 9.Luchin A., Purdom G., Murphy K., Clark M. Y., Angel N., Cassady A. I., Hume D. A., Ostrowski M. C. (2000) J. Bone Miner. Res. 15, 451–460 [DOI] [PubMed] [Google Scholar]
- 10.Hu R., Sharma S. M., Bronisz A., Srinivasan R., Sankar U., Ostrowski M. C. (2007) Mol. Cell. Biol. 27, 4018–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumoto M., Kogawa M., Wada S., Takayanagi H., Tsujimoto M., Katayama S., Hisatake K., Nogi Y. (2004) J. Biol. Chem. 279, 45969–45979 [DOI] [PubMed] [Google Scholar]
- 12.Takayanagi H., Kim S., Koga T., Nishina H., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., Wagner E. F., Mak T. W., Kodama T., Taniguchi T. (2002) Dev. Cell 3, 889–901 [DOI] [PubMed] [Google Scholar]
- 13.Arai F., Miyamoto T., Ohneda O., Inada T., Sudo T., Brasel K., Miyata T., Anderson D. M., Suda T. (1999) J. Exp. Med. 190, 1741–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacquin C., Gran D. E., Lee S. K., Lorenzo J. A., Aguila H. L. (2006) J. Bone Miner. Res. 21, 67–77 [DOI] [PubMed] [Google Scholar]
- 15.Mizoguchi T., Muto A., Udagawa N., Arai A., Yamashita T., Hosoya A., Ninomiya T., Nakamura H., Yamamoto Y., Kinugawa S., Nakamura M., Nakamichi Y., Kobayashi Y., Nagasawa S., Oda K., Tanaka H., Tagaya M., Penninger J. M., Ito M., Takahashi N. (2009) J. Cell Biol. 184, 541–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karsenty G. (2008) Annu. Rev. Genomics Hum. Genet. 9, 183–196 [DOI] [PubMed] [Google Scholar]
- 17.Gori F., Hofbauer L. C., Dunstan C. R., Spelsberg T. C., Khosla S., Riggs B. L. (2000) Endocrinology 141, 4768–4776 [DOI] [PubMed] [Google Scholar]
- 18.Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R. T., Gao Y. H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., Kishimoto T. (1997) Cell 89, 755–764 [DOI] [PubMed] [Google Scholar]
- 19.Corral D. A., Amling M., Priemel M., Loyer E., Fuchs S., Ducy P., Baron R., Karsenty G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13835–13840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franceschi R. T., Xiao G., Jiang D., Gopalakrishnan R., Yang S., Reith E. (2003) Connect. Tissue Res. 44, Suppl. 1, 109–116 [PMC free article] [PubMed] [Google Scholar]
- 21.Otto F., Thornell A. P., Crompton T., Denzel A., Gilmour K. C., Rosewell I. R., Stamp G. W., Beddington R. S., Mundlos S., Olsen B. R., Selby P. B., Owen M. J. (1997) Cell 89, 765–771 [DOI] [PubMed] [Google Scholar]
- 22.Palumbo C. (1986) Cell Tissue Res. 246, 125–131 [DOI] [PubMed] [Google Scholar]
- 23.Bonewald L. F. (2007) Ann. N.Y. Acad. Sci. 1116, 281–290 [DOI] [PubMed] [Google Scholar]
- 24.Kamioka H., Honjo T., Takano-Yamamoto T. (2001) Bone 28, 145–149 [DOI] [PubMed] [Google Scholar]
- 25.Verborgt O., Tatton N. A., Majeska R. J., Schaffler M. B. (2002) J. Bone Miner. Res. 17, 907–914 [DOI] [PubMed] [Google Scholar]
- 26.Weitzmann M. N., Pacifici R. (2007) Ann. N.Y. Acad. Sci. 1116, 360–375 [DOI] [PubMed] [Google Scholar]
- 27.Lowry M. B., Lotinun S., Leontovich A. A., Zhang M., Maran A., Shogren K. L., Palama B. K., Marley K., Iwaniec U. T., Turner R. T. (2008) Endocrinology 149, 5735–5746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y., Toraldo G., Li A., Yang X., Zhang H., Qian W. P., Weitzmann M. N. (2007) Blood 109, 3839–3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kacena M. A., Shivdasani R. A., Wilson K., Xi Y., Troiano N., Nazarian A., Gundberg C. M., Bouxsein M. L., Lorenzo J. A., Horowitz M. C. (2004) J. Bone Miner. Res. 19, 652–660 [DOI] [PubMed] [Google Scholar]
- 30.Lorenzo J., Horowitz M., Choi Y. (2008) Endocr. Rev. 29, 403–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hume D. A. (2008) Mucosal Immunol. 1, 432–441 [DOI] [PubMed] [Google Scholar]
- 32.Chang M. K., Raggatt L. J., Alexander K. A., Kuliwaba J. S., Fazzalari N. L., Schroder K., Maylin E. R., Ripoll V. M., Hume D. A., Pettit A. R. (2008) J. Immunol. 181, 1232–1244 [DOI] [PubMed] [Google Scholar]
- 33.Hauge E. M., Qvesel D., Eriksen E. F., Mosekilde L., Melsen F. (2001) J. Bone Miner. Res. 16, 1575–1582 [DOI] [PubMed] [Google Scholar]
- 34.Pettit A. R., Chang M. K., Hume D. A., Raggatt L. J. (2008) Bone 43, 976–982 [DOI] [PubMed] [Google Scholar]
- 35.Andersen T. L., Sondergaard T. E., Skorzynska K. E., Dagnaes-Hansen F., Plesner T. L., Hauge E. M., Plesner T., Delaisse J. M. (2009) Am. J. Pathol. 174, 239–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aguirre J. I., Plotkin L. I., Stewart S. A., Weinstein R. S., Parfitt A. M., Manolagas S. C., Bellido T. (2006) J. Bone Miner. Res. 21, 605–615 [DOI] [PubMed] [Google Scholar]
- 37.Heino T. J., Hentunen T. A., Väänänen H. K. (2002) J. Cell. Biochem. 85, 185–197 [DOI] [PubMed] [Google Scholar]
- 38.Jüppner H., Abou-Samra A. B., Freeman M., Kong X. F., Schipani E., Richards J., Kolakowski L. F., Jr., Hock J., Potts J. T., Jr., Kronenberg H. M. (1991) Science 254, 1024–1026 [DOI] [PubMed] [Google Scholar]
- 39.Swarthout J. T., D'Alonzo R. C., Selvamurugan N., Partridge N. C. (2002) Gene 282, 1–17 [DOI] [PubMed] [Google Scholar]
- 40.Li X., Qin L., Bergenstock M., Bevelock L. M., Novack D. V., Partridge N. C. (2007) J. Biol. Chem. 282, 33098–33106 [DOI] [PubMed] [Google Scholar]
- 41.Ma Y. L., Cain R. L., Halladay D. L., Yang X., Zeng Q., Miles R. R., Chandrasekhar S., Martin T. J., Onyia J. E. (2001) Endocrinology 142, 4047–4054 [DOI] [PubMed] [Google Scholar]
- 42.Insogna K. L., Sahni M., Grey A. B., Tanaka S., Horne W. C., Neff L., Mitnick M., Levy J. B., Baron R. (1997) J. Clin. Invest. 100, 2476–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burgess T. L., Qian Y., Kaufman S., Ring B. D., Van G., Capparelli C., Kelley M., Hsu H., Boyle W. J., Dunstan C. R., Hu S., Lacey D. L. (1999) J. Cell Biol. 145, 527–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang C. M., Chien C. S., Yao C. C., Hsiao L. D., Huang Y. C., Wu C. B. (2004) J. Biol. Chem. 279, 22158–22165 [DOI] [PubMed] [Google Scholar]
- 45.Partridge N. C., Jeffrey J. J., Ehlich L. S., Teitelbaum S. L., Fliszar C., Welgus H. G., Kahn A. J. (1987) Endocrinology 120, 1956–1962 [DOI] [PubMed] [Google Scholar]
- 46.McHugh K. P., Hodivala-Dilke K., Zheng M. H., Namba N., Lam J., Novack D., Feng X., Ross F. P., Hynes R. O., Teitelbaum S. L. (2000) J. Clin. Invest. 105, 433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teitelbaum S. L. (2000) Science 289, 1504–1508 [DOI] [PubMed] [Google Scholar]
- 48.Saftig P., Hunziker E., Wehmeyer O., Jones S., Boyde A., Rommerskirch W., Moritz J. D., Schu P., von Figura K. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13453–13458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Everts V., Delaissé J. M., Korper W., Jansen D. C., Tigchelaar-Gutter W., Saftig P., Beertsen W. (2002) J. Bone Miner. Res. 17, 77–90 [DOI] [PubMed] [Google Scholar]
- 50.Tran Van P., Vignery A., Baron R. (1982) Cell Tissue Res. 225, 283–292 [DOI] [PubMed] [Google Scholar]
- 51.Heinemann D. E., Siggelkow H., Ponce L. M., Viereck V., Wiese K. G., Peters J. H. (2000) Immunobiology 202, 68–81 [DOI] [PubMed] [Google Scholar]
- 52.Kraal G., Rep M., Janse M. (1987) Scand. J. Immunol. 26, 653–661 [DOI] [PubMed] [Google Scholar]
- 53.Newby A. C. (2008) Arterioscler. Thromb. Vasc. Biol. 28, 2108–2114 [DOI] [PubMed] [Google Scholar]
- 54.Takahashi F., Takahashi K., Shimizu K., Cui R., Tada N., Takahashi H., Soma S., Yoshioka M., Fukuchi Y. (2004) Lung 182, 173–185 [DOI] [PubMed] [Google Scholar]
- 55.Tang Y., Wu X., Lei W., Pang L., Wan C., Shi Z., Zhao L., Nagy T. R., Peng X., Hu J., Feng X., Van Hul W., Wan M., Cao X. (2009) Nat. Med. 15, 757–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin T. J., Sims N. A. (2005) Trends Mol. Med. 11, 76–81 [DOI] [PubMed] [Google Scholar]
- 57.Pederson L., Ruan M., Westendorf J. J., Khosla S., Oursler M. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20764–20769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao C., Irie N., Takada Y., Shimoda K., Miyamoto T., Nishiwaki T., Suda T., Matsuo K. (2006) Cell Metab. 4, 111–121 [DOI] [PubMed] [Google Scholar]
- 59.van Bezooijen R. L., Roelen B. A., Visser A., van der Wee-Pals L., de Wilt E., Karperien M., Hamersma H., Papapoulos S. E., ten Dijke P., Löwik C. W. (2004) J. Exp. Med. 199, 805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X., Zhang Y., Kang H., Liu W., Liu P., Zhang J., Harris S. E., Wu D. (2005) J. Biol. Chem. 280, 19883–19887 [DOI] [PubMed] [Google Scholar]
- 61.Fermor B., Skerry T. M. (1995) J. Bone Miner. Res. 10, 1935–1943 [DOI] [PubMed] [Google Scholar]
- 62.Robling A. G., Niziolek P. J., Baldridge L. A., Condon K. W., Allen M. R., Alam I., Mantila S. M., Gluhak-Heinrich J., Bellido T. M., Harris S. E., Turner C. H. (2008) J. Biol. Chem. 283, 5866–5875 [DOI] [PubMed] [Google Scholar]
- 63.Robey P., Boskey A. (2008) in Primer on the Metabolic Bone Diseases and Disorders of Mineral Metabolism (Rosen C. ed) 7th Ed., pp. 32–38, American Society of Bone and Mineral Research, Washington, D.C. [Google Scholar]
- 64.Murshed M., Harmey D., Millán J. L., McKee M. D., Karsenty G. (2005) Genes Dev. 19, 1093–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harmey D., Hessle L., Narisawa S., Johnson K. A., Terkeltaub R., Millán J. L. (2004) Am. J. Pathol. 164, 1199–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.