Abstract
Leukotrienes (LTs), derived from arachidonic acid (AA) released from the membrane by the action of phospholipase A2, are potent lipid mediators of the inflammatory response. In 1983, Dahlén et al. demonstrated that LTC4, LTD4, and LTE4 mediate antigen-induced constriction of bronchi in tissue obtained from subjects with asthma (Dahlén, S. E., Hansson, G., Hedqvist, P., Björck, T., Granström, E., and Dahlén, B. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 1712–1716). Over the last 25+ years, substantial progress has been made in understanding how LTs exert their effects, and a broader appreciation for the numerous biological processes they mediate has emerged. LT biosynthesis is initiated by the action of 5-lipoxygenase (5-LOX), which catalyzes the transformation of AA to LTA4 in a two-step reaction. Ca2+ targets 5-LOX to the nuclear membrane, where it co-localizes with the 5-LOX-activating protein FLAP and, when present, the downstream enzyme LTC4 synthase, both transmembrane proteins. Crystal structures of the AA-metabolizing LOXs, LTC4 synthase, and FLAP combined with biochemical data provide a framework for understanding how subcellular organizations optimize the biosynthesis of these labile hydrophobic signaling compounds, which must navigate pathways that include both membrane and soluble enzymes. The insights these structures afford and the questions they engender are discussed in this minireview.
Keywords: Eicosanoid, Leukotriene, Membrane Trafficking, Protein Structure, Protein Translocation, Protein-Protein Interactions, X-ray Crystallography
Introduction
The polyunsaturated fatty acid arachidonic acid (AA)2 serves as a precursor to prostaglandins, leukotrienes (LTs), and other eicosanoids derived from the 20-carbon substrate. The proteins that constitute the LT biosynthetic pathway are located in several cellular compartments and include extra- and intracellular as well as membrane and soluble proteins. Products of the intracellular pathway are exported by specific pumps for further elaboration in extra- or transcellular biosynthesis or for access to the extracellular receptors they target. The complex compartmentalized biosynthetic pathway for LTs offers numerous opportunities for control of pathway flux through dynamic control of the locations of key proteins. We focus in this minireview on the early events in the LT biosynthetic pathway, specifically a major branch point in LT biosynthesis that determines whether cysteinyl-LTs that result from the pathway that requires conjugation with glutathione are produced or whether the non-cysteinyl-LT LTB4 is synthesized.
The Reaction Pathway
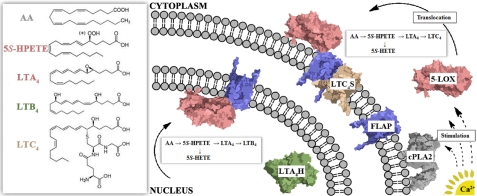
The biosynthesis of LTs is initiated by the action of 5-lipoxygenase (5-LOX), which promotes the transformation of AA to LTA4. The action of 5-LOX is triggered by calcium-dependent membrane binding, which targets it to the nuclear membrane. There it acquires its substrate, liberated from membrane phospholipids by the action of Ca2+-stimulated cytosolic phospholipase A2, from the transmembrane protein FLAP (five-lipoxygenase-activating protein). Like all LOXs, 5-LOX catalyzes the regio- and stereospecific peroxidation of a polyunsaturated fatty acid (1, 2). The enzyme transforms the substrate AA to the 5S-isomer of hydroperoxyeicosatetraenoic acid (5S-HPETE). However, in contrast to other members of this superfamily, 5-LOX can further metabolize the hydroperoxy product to an allylic epoxide, specifically LTA4. Studies have shown that both in vitro (3) and in vivo (4), the efficiency of LTA4 production is improved with membrane binding, i.e. 5-LOX successfully carries the two-step reaction to completion, and the ratio of LTA4 to intermediate (5S-HPETE) produced is increased. (An alternative fate for 5S-HPETE is reduction to the corresponding hydroxyeicosatetraenoic acid.) Subsequently, LTA4 is transformed to either LTB4 by the action of LT hydrolase or LTC4 by the action of LTC4 synthase. The former is a soluble enzyme that traffics between the nucleus and cytoplasm, and the latter is a membrane-embedded enzyme that is localized to the outer, but not inner, nuclear membrane (Fig. 1).
FIGURE 1.
Initial events in LT biosynthesis. Ca2+ promotes membrane binding of both cytosolic phospholipase A2 (cPLA2; gray) and 5-LOX (salmon). The AA (see box) generated by phospholipase A2 is the substrate for 5-LOX. The 5-LOX product (LTA4) is converted to either LTC4 or LTB4 depending on the location of 5-LOX. *, OH for the corresponding hydroxyeicosatetraenoic acid (5S-HETE), produced by reduction of 5S-HPETE. LTA4H, LTA4 hydrolase; LTC4S, LTC4 synthase.
Protein Structures
5-LOX
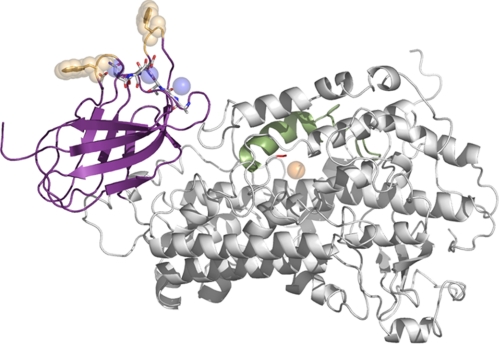
Although there is no structure available for 5-LOX, structures for several plant (5–8) and animal (9–11) enzymes (Fig. 2) have been determined. The LOX framework defined by these structures contains two distinct domains. The first of these is the smaller (∼120 amino acids) PLAT (polycystin-1/lipoxygenase/alpha-toxin) domain originally termed “C2-like” because of its resemblance to the Ca2+-dependent C2 domains described in phospholipases and other Ca2+-dependent membrane-binding proteins such as synaptotagmin (12–15). The structures of two PLAT domains that share Ca2+-binding amino acids with 5-LOX (gangrene α-toxin (16, 17) and 8R-LOX from Plexaura homomalla (10)) revealed three Ca2+ ions positioned by the conserved amino acids. These structures and a body of biochemical data suggest that 5-LOX similarly harbors three Ca2+-binding sites in the N-terminal PLAT domain. Mutation of the putative Ca2+-binding amino acids abrogates membrane binding by 5-LOX (18–21).
FIGURE 2.
8R-LOX. The schematic shows 8R-LOX from P. homomalla (Protein Data Bank code 2FNQ). The membrane-targeting PLAT domain is in purple, and the catalytic domain is in white. Blue spheres mark the Ca2+ ions positioned by amino acids conserved in 5-LOX, which are included in stick rendering. White, carbon; red, oxygen; blue, nitrogen. Trp residues in the PLAT domain found in the putative membrane insertion loops common to 8R- and 5-LOXs are show as gold spheres. The catalytic iron is shown as an orange sphere, and the position of the terminal amino acid is in red. The arched helix, which shelters the active site, is in green.
The remainder of the protein forms the catalytic domain (∼550 amino acids in animal enzymes and 750 in plant enzymes); it is primarily α-helical and houses the catalytic iron. The iron is positioned by both side and main chain contacts: conserved histidines fill the coordination sphere as well as the main chain carboxylate of the C terminus. Another structurally distinct conserved feature in this domain, previously described in detail by Minor et al. (6) for soybean LOX L-1, is an arched helix that shields access to the catalytic iron. The curvature of the helix is a consequence of a reverse-turn insertion that contains an invariant glycine. Just prior to the reverse turn is an invariant Leu that has been proposed to control access of O2 to the substrate (22, 23) and to position the substrate pentadiene for attack (11). The PLAT and catalytic domains appear to constitute independently folded domains, and there are reports of LOX truncations (lacking the N-terminal domain) that are catalytically active (24, 25). However, the structure of a truncated form of 12S-LOX (Protein Data Bank code 3D3L) suggests that the PLAT domain may critically influence the three-dimensional structure of the catalytic domain. The 12S-LOX fragment lacks the PLAT domain and the first helix (α1) and loop of the catalytic domain, and in this structure, α2 is not positioned as it is in any intact LOX structure. Furthermore, α2 crosses over the site where the arched helix is found; as a consequence, the arched helix is also not properly positioned. (In fact, the authors were unable to model it in the electron density map.) Because α1 makes no contacts with α2 in the full-length LOX structures, the structure of the truncated LOX may indicate that α2 is constrained by the N-terminal domain. In the absence of the physical restraint imposed by the PLAT domain, neither α2 nor the arched helix is orientated correctly in the 12S-LOX fragment.
As the substrate for animal LOXs is AA, the individual LOX isoforms must discriminate among three chemically equivalent methylene carbons to generate a single regio- and stereospecific product. Structures are available for rabbit reticulocyte 15-LOX (9, 26) and P. homomalla 8R-LOX (10, 11), enzymes that have ∼40% sequence identity to human 5-LOX. As one can infer from the names, the former produces 15-HPETE (the S-isomer) and the latter 8R-HPETE. These structures have led to models for product specificity in the 15S- and 8R-enzymes, but the product of 5-LOX represents a significant challenge to extrapolation of these models. Experimental results are consistent with a mechanism that requires abstraction of hydrogen at the central carbon of a pentadiene, followed by the antarafacial peroxidation of the free radical intermediate, i.e. hydrogen abstraction and oxygenation must occur on opposite sides of the pentadiene (for review, see Ref. 28). The site of peroxidation is either C+2 or C−2 relative to the position of hydrogen abstraction. For the models inferred from the rabbit and coral structures that explain the products of these enzymes, the stereochemistry of the product is consistent with the carboxylate of the AA tethered by surface amino acids (carboxylate out). However, the stereochemistry of the 5-LOX product and the fact that peroxidation occurs on the opposite side of the substrate to hydrogen abstraction necessitate an inverse orientation of substrate (carboxylate in). Furthermore, the consecutive reactions catalyzed by 5-LOX require hydrogen abstraction from two of the three pentadienes in the substrate (attack at C7 and C10). The 8R- and 15-LOXs perform abstraction at C10 and C13, respectively. Thus, despite compelling experimental data testing a model for 5-LOX product specificity (e.g. Ref. 29), in the absence of a structure of 5-LOX or other LOX with “inverse entry,” it is difficult to predict with certainty the structural basis for 5-LOX specificity.
FLAP and LTC4 Synthase
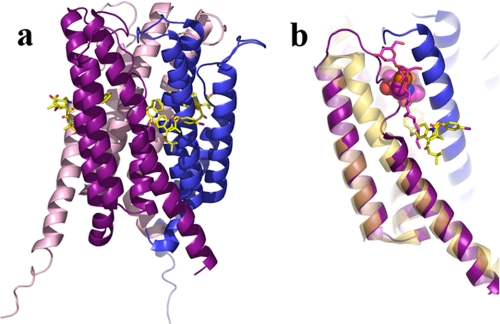
Targeted to the membrane by the binding of Ca2+, 5-LOX is localized to the nuclear membrane, where both its “helper” protein FLAP (Fig. 3) and LTC4 synthase, an enzyme that metabolizes the 5-LOX product, can be found. The two proteins, which span the bilayer, are members of the MAPEG (membrane-associated proteins in eicosanoid and glutathione metabolism) superfamily (30, 31). Crystal structures have been obtained for both (32–34), with that of LTC4 synthase at significantly higher resolution (2.15 Å (33) versus 4.0 Å (34) resolution). The proteins are trimers of four-helix bundles with active sites located at monomer interfaces. FLAP appears to represent a distinct subclass in the family, as unlike the other MAPEG family members, it does not have a catalytic activity or a binding site for GSH. All other MAPEG proteins either conjugate GSH with their substrate or metabolize GSH in the catalytic cycle. Furthermore, a structure-based sequence lineup of MAPEG proteins supports the premise that FLAP represents a unique subclass (35), and FLAP homologs from Danio rerio to Homo sapiens show specific hallmarks: distinct loop sequences that connect helices α1 and α2 and helices α3 and α4. These are the loops at the surface of the membrane, i.e. the regions of the protein most likely to interact with 5-LOX should a specific protein-protein interaction occur. A role in a specific interaction with a protein partner is consistent with the fact that the sequences of the surface loops of FLAP are highly conserved. In Fig. 3, the structure of a monomer of LTC4 synthase with the GSH co-substrate is superimposed on that of FLAP. In this orientation, it is clear that the N-terminal portion of FLAP helix α4 is “unwound” compared with the GSH-dependent MAPEG structures. The resulting extended loop region blocks access to the GSH-binding site region of the glutathione-dependent MAPEG proteins. Moreover, FLAP also lacks the conserved side chains that interact with GSH.
FIGURE 3.
FLAP and LTC4 synthase. a, schematic of a FLAP trimer (Protein Data Bank code 2Q7M). Monomers are in purple, blue, and pink. The inhibitor MK-591 is depicted in stick rendering. Yellow, carbon; red, oxygen; blue, nitrogen. b, superposition of a LTC4 synthase monomer (beige; code 2UUH) onto FLAP. GSH is in sphere rendering. Magenta, carbon; orange, sulfur. The stick rendering corresponds to a detergent molecule and is presumed to indicate the binding site for LTA4.
The structure of LTC4 synthase, which catalyzes the conjugation of GSH to LTA4, includes GSH, and the FLAP structure includes the competitive inhibitor of AA binding MK-591. Both ligands are located in a groove formed by helices α1 and α4 from neighboring protomers. Furthermore, both proteins have all three sites in the trimers occupied. The GSH subsite of the synthase is in a deeper portion of the groove, and LTA4 is thought to occupy the region of the site that contains a co-crystallized detergent molecule. This arrangement makes the LTA4 subsite more superficial with respect to the protein than the GSH subsite. However, the LT subsite extends along the surface of protein and penetrates deep into the membrane bilayer. The arrangement of the LTA4 and GSH subsites, with the GSH buried deeper in the protein, led to the suggestion that GSH binds first and induces a conformation that is amenable to LTA4 binding (33). However, previous kinetic data for this and other MAPEG proteins are consistent with a random binding order (36). In contrast, FLAP has a single binding cavity in the groove between monomers. In the x-ray structure of FLAP, the competitive inhibitor of AA is found in the region of the protein buried in the bilayer. Thus, the locations of the AA- and LTA4-binding sites in FLAP and LTC4 synthase deep in the bilayer are consistent with a model in which the proteins obtain their ligand/substrate directly from the bilayer by lateral diffusion.
LTA4 Hydrolase
The product of the 5-LOX reaction is also the substrate for LTA4 hydrolase. This soluble enzyme converts LTA4 into LTB4 and thus provides an alternative fate for the product of the 5-LOX reaction. The crystal structure of LTA4 hydrolase confirmed the prediction from sequence alignments that it is a member of the M1 amino peptidase superfamily (37, 38). These are zinc-containing amino peptidases typified by thermolysin. However, LTA4 hydrolase is roughly double the size of thermolysin, as it contains two additional domains: an all-β-domain at the N terminus and an α-helical domain at the C terminus (Fig. 4). These two domains extend from the thermolysin-like core to close over what would be the exposed peptide-binding site in an M1 peptidase. The triangular arrangement of the three domains allows access to the catalytic zinc via a central channel. In addition to the hydrolase activity, the enzyme also has peptidase activity, but a biological role for the peptidase activity remains to be identified. It is interesting to note that the LTA4 hydrolase-“modified” M1 peptidase structure is not unique to an enzyme of the LT biosynthetic pathway. A similar structure has been described for a cold-active amino peptidase from Colwellia psychrerythraea (39).
FIGURE 4.
LTA4 hydrolase. a, thermolysin-like (M1 peptidase) core of LTA4 hydrolase. Zn2+ is a gray sphere. b, LTA4 hydrolase (Protein Data Bank code 1HS6). Purple, N-terminal domain; green, C-terminal domain. The arrow marks the approximate position of the active-site access channel.
Compartmentalization
What Determines the Fate of the 5-LOX Product?
Clustering of sequential catalytic activities is thought to provide a mechanism to enhance biosynthetic efficiency, and reversible clustering of six enzymes in the de novo biosynthetic pathway for purines has been shown to occur in the cytoplasm in response to changes in purine levels (40). The proteins of the LT biosynthetic pathway are found in the nucleus, cytoplasm, inner and outer nuclear membranes, and extracellular space and, as a result, are not free to reversibly cluster. In addition, some steps in the pathway that further modify the LTs are transcellular (e.g. Ref. 41). Consequently, regulation of pathway flux can be executed by the subcellular targeting of key enzymes. For example, as described below, targeting of 5-LOX to the outer nuclear membrane is thought to promote the formation of the cysteinyl-LTs, whereas 5-LOX targeting to the inner nuclear membrane results in the production of LTB4 (37, 38).
The LTC4 synthase and LTA4 hydrolase activities represent a bifurcation of the LT biosynthetic pathway: the former directs the 5-LOX product into the production of cysteinyl-LTs such as LTC4 and LTD4 and the latter to LTB4. These LTs are distinct in the biological responses they trigger; thus, an intriguing aspect of the regulation of LT biosynthesis is how the relative rates of cysteinyl-LT and LTB4 production are controlled. Compartmentalization of enzyme activities has been proposed to play a significant role in the regulation of LT production. Prior to activation, 5-LOX can reside in the cytoplasm or the nucleus (due to the presence of multiple nuclear location sequences (42, 43)); furthermore, its cellular location is dynamic and can vary with cell type. One mechanism of control of 5-LOX location is phosphorylation. For instance, phosphorylation of Ser-271 interferes with the export of 5-LOX from the nucleus (44). Thus, cell stimulation results in the Ca2+-dependent targeting of 5-LOX to the inner and/or outer nuclear membrane(s), depending upon the compartment(s) in which the unstimulated enzyme resides.
Recall that 5-LOX is presented its substrate by its helper protein FLAP, located in the nuclear membrane. Although FLAP is not required for 5-LOX activity or its translocation to the membrane, in vivo, the cellular capacity for LT synthesis correlates with FLAP expression (45–47). Furthermore, inhibitors of FLAP are effective inhibitors of LT production (48). MK-591, the drug with which FLAP was crystallized, inhibits LT biosynthesis through binding to FLAP. Recall also that LTC4 synthase is a structurally similar membrane protein. However, in contrast to FLAP, which is distributed on both the inner and outer nuclear membranes, LTC4 synthase is restricted to the outer nuclear membrane (49). Studies by Mandal et al. (50) revealed two populations of FLAP in the nuclear membrane: FLAP that is unassociated and FLAP that is associated with LTC4 synthase. Because FLAP is more abundant than LTC4 synthase, there is no LTC4 synthase that is not in complex with FLAP. The authors proposed the existence of two pools of FLAP, one to channel LTA4 to the synthase and the other to allow access to the LT by LTA4 hydrolase. In subsequent work, the authors demonstrated activation-dependent assembly of a complex of 5-LOX and FLAP (51). In their model, the outer nuclear membrane complex is composed of 5-LOX, FLAP, and LTC4 synthase, whereas on the inner membrane, which is devoid of the synthase, the complex consists of 5-LOX and FLAP. Interaction of FLAP with both LTC4 synthase and 5-LOX has been suggested by others (52).
Implications for Substrate Transfer
The proposed AA-binding site in FLAP is deep within the hydrophobic region of the bilayer. This placement appears to preclude a direct interaction between the donor- and acceptor-binding sites and suggests that accessibility to FLAP-bound AA by 5-LOX is limited, even when the enzyme is membrane-bound. The only part of 5-LOX demonstrated to penetrate the bilayer is the putative membrane insertion loop that contains Trp-75 in the PLAT domain. This amino acid has been proposed to insert ∼9 Å deep into the bilayer (53). Thus, 5-LOX is tethered to the membrane with calcium activation, but its substrate is bound deep within the hydrophobic bilayer to the helper protein FLAP. How the AA travels from its position in the bilayer to the catalytic domain of LOX, which does not localize to the membrane in the absence a functional PLAT domain, is unknown. Note from Fig. 2 that the overall structure of LOX is an elongated ellipsoid. The membrane-tethering PLAT domain encompasses the first third of the ellipsoid, and the catalytic domain encompasses the remaining two-thirds. The entrance to the active site is roughly in the middle of the catalytic domain, ∼30 Å removed from the PLAT-catalytic domain interface. Thus, direct passage of substrate from the bilayer to the active site of the soluble enzyme (as occurs in some monotypic membrane-associated enzymes (54)) can occur only if the catalytic domain abuts the membrane. A green fluorescent protein-labeled catalytic domain-only construct does not translocate to the nuclear membrane under conditions that mobilize the similarly tagged PLAT domain (18). However, this observation does not rule out an association of the catalytic domain with the membrane once it is tethered by the PLAT domain. Another aspect of substrate transfer to consider is the orientation of AA in FLAP. The structures of both FLAP and LTC4 synthase suggest that AA and LTA4 bind with the hydrophobic tails innermost in the membrane and the carboxylate groups at the membrane surface. This may require that the initial interaction of 5-LOX with FLAP-bound AA is via the substrate carboxylate. Such a transfer mechanism might utilize a positively charged amino acid at the entrance to the active site.
LTA4 hydrolase, the alternative enzyme that metabolizes LTA4, is a soluble enzyme found in the nucleus and cytoplasm (55). Its distribution between these compartments is dynamic (56). Luo et al. (57) have shown that the cellular capacity for the synthesis of LTB4 correlates with the capacity for import of 5-LOX into the nucleus. Thus, the exclusion of LTC4 synthase from the inner nuclear membrane and the presence of the hydrolase in the nucleus result in a pool of LTA4 that does not have access to the cysteinyl-LT biosynthetic branch of the bifurcation but is available to the hydrolase. If LTC4 were not excluded from the inner nuclear membrane in cells expressing both LTC4 synthase and LTA4 hydrolase, the transmembrane synthase would have preferential access to substrate due to its proximity to FLAP and consequently 5-LOX. The clustering of FLAP, 5-LOX, and LTC4 synthase is likely to maximize the partitioning of the hydrophobic LTA4 in the membrane phase. The half-life of LTA4 is ∼10 s in aqueous milieu but is significantly extended in the presence of phospholipids (58). As a consequence, even the chemical nature of the phospholipid bilayer favors the LTC4 synthase reaction over that of the hydrolase, as the substrate need not enter an aqueous environment, where it is susceptible to non-enzymatic hydrolysis.
The bifurcation of the LT biosynthetic pathway after the production of LTA4 contributes to the production of LTs of diverse structures and functions. However, data suggest that downstream events in one pathway branch can have an effect on the flux through the alternative branch. LTC4 is exported from the cell by the membrane pump known as MRP1 (multidrug resistance protein-1); in mice lacking the multidrug export pump, LTB4 synthesis is increased. The interpretation of this result is that LTC4 accumulation inhibits LTC4 synthase activity, and thus, more LTA4 is available for metabolism by the hydrolase (59).
Concluding Remarks
The initial step in LT synthesis is catalyzed by the Ca2+-dependent membrane-binding enzyme 5-LOX. 5-LOX is translocated to the nuclear membrane, and whether it is located on the inner or outer nuclear membrane appears to determine the fate of its product, LTA4. Thus, at an early step in LT biosynthesis, compartmentalization is an important determinant in pathway flux within the branches of this intricate biosynthetic pathway for lipid mediators. Furthermore, compartmentalization can be regulated by post-translational modifications such as phosphorylation. Downstream enzymes and proteins in the pathway reside in multiple cellular compartments; thus, it is likely that a regulatory mechanism that requires mobility of a key enzyme is repeated farther down the line. An issue that was only briefly alluded to here but is likely to impact LT production is the lability of pathway intermediates. For example, LTA4 is subject to non-enzymatic hydrolysis, but the half-life in the cell can be extended by either its association with phospholipid membrane (58) or fatty acid-binding proteins (27, 60). Thus, on top of spatial restraints imposed by the multiple compartments occupied by LT biosynthetic enzymes are temporal constraints that further impact pathway flux. The abundance of structural and functional data on early events in LT biosynthesis helps simplify our perception of the complex pathway for LT biosynthesis.
Supplementary Material
This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- AA
- arachidonic acid
- LT
- leukotriene
- 5-LOX
- 5-lipoxygenase
- 5S-HPETE
- 5S-hydroperoxyeicosatetraenoic acid.
REFERENCES
- 1.Brash A. R. (1999) J. Biol. Chem. 274, 23679–23682 [DOI] [PubMed] [Google Scholar]
- 2.Kuhn H., Thiele B. J. (1999) FEBS Lett. 449, 7–11 [DOI] [PubMed] [Google Scholar]
- 3.Noguchi M., Miyano M., Matsumoto T., Noma M. (1994) Biochim. Biophys. Acta 1215, 300–306 [DOI] [PubMed] [Google Scholar]
- 4.Hill E., Maclouf J., Murphy R. C., Henson P. M. (1992) J. Biol. Chem. 267, 22048–22053 [PubMed] [Google Scholar]
- 5.Boyington J. C., Gaffney B. J., Amzel L. M. (1993) Science 260, 1482–1486 [DOI] [PubMed] [Google Scholar]
- 6.Minor W., Steczko J., Stec B., Otwinowski Z., Bolin J. T., Walter R., Axelrod B. (1996) Biochemistry 35, 10687–10701 [DOI] [PubMed] [Google Scholar]
- 7.Skrzypczak-Jankun E., Bross R. A., Carroll R. T., Dunham W. R., Funk M. O., Jr. (2001) J. Am. Chem. Soc. 123, 10814–10820 [DOI] [PubMed] [Google Scholar]
- 8.Youn B., Sellhorn G. E., Mirchel R. J., Gaffney B. J., Grimes H. D., Kang C. (2006) Proteins 65, 1008–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillmor S. A., Villaseñor A., Fletterick R., Sigal E., Browner M. F. (1997) Nat. Struct. Biol. 4, 1003–1009; Erratum (1998) Nat. Struct. Biol.5, 242 [DOI] [PubMed] [Google Scholar]
- 10.Oldham M. L., Brash A. R., Newcomer M. E. (2005) J. Biol. Chem. 280, 39545–39552 [DOI] [PubMed] [Google Scholar]
- 11.Neau D. B., Gilbert N. C., Bartlett S. G., Boeglin W., Brash A. R., Newcomer M. E. (2009) Biochemistry 48, 7906–7915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davletov B., Perisic O., Williams R. L. (1998) J. Biol. Chem. 273, 19093–19096 [DOI] [PubMed] [Google Scholar]
- 13.Dessen A., Tang J., Schmidt H., Stahl M., Clark J. D., Seehra J., Somers W. S. (1999) Cell 97, 349–360 [DOI] [PubMed] [Google Scholar]
- 14.Perisic O., Paterson H. F., Mosedale G., Lara-González S., Williams R. L. (1999) J. Biol. Chem. 274, 14979–14987 [DOI] [PubMed] [Google Scholar]
- 15.Chapman E. R., Davis A. F. (1998) J. Biol. Chem. 273, 13995–14001 [DOI] [PubMed] [Google Scholar]
- 16.Naylor C. E., Eaton J. T., Howells A., Justin N., Moss D. S., Titball R. W., Basak A. K. (1998) Nat. Struct. Biol. 5, 738–746 [DOI] [PubMed] [Google Scholar]
- 17.Naylor C. E., Jepson M., Crane D. T., Titball R. W., Miller J., Basak A. K., Bolgiano B. (1999) J. Mol. Biol. 294, 757–770 [DOI] [PubMed] [Google Scholar]
- 18.Chen X. S., Funk C. D. (2001) J. Biol. Chem. 276, 811–818 [DOI] [PubMed] [Google Scholar]
- 19.Chen X. S., Zhang Y. Y., Funk C. D. (1998) J. Biol. Chem. 273, 31237–31244 [DOI] [PubMed] [Google Scholar]
- 20.Hammarberg T., Provost P., Persson B., Rådmark O. (2000) J. Biol. Chem. 275, 38787–38793 [DOI] [PubMed] [Google Scholar]
- 21.Kulkarni S., Das S., Funk C. D., Murray D., Cho W. (2002) J. Biol. Chem. 277, 13167–13174 [DOI] [PubMed] [Google Scholar]
- 22.Knapp M. J., Klinman J. P. (2003) Biochemistry 42, 11466–11475 [DOI] [PubMed] [Google Scholar]
- 23.Knapp M. J., Seebeck F. P., Klinman J. P. (2001) J. Am. Chem. Soc. 123, 2931–2932 [DOI] [PubMed] [Google Scholar]
- 24.Maccarrone M., Salucci M. L., van Zadelhoff G., Malatesta F., Veldink G., Vliegenthart J. F., Finazzi-Agrò A. (2001) Biochemistry 40, 6819–6827 [DOI] [PubMed] [Google Scholar]
- 25.Walther M., Anton M., Wiedmann M., Fletterick R., Kuhn H. (2002) J. Biol. Chem. 277, 27360–27366 [DOI] [PubMed] [Google Scholar]
- 26.Choi J., Chon J. K., Kim S., Shin W. (2008) Proteins 70, 1023–1032 [DOI] [PubMed] [Google Scholar]
- 27.Zimmer J. S., Dyckes D. F., Bernlohr D. A., Murphy R. C. (2004) J. Lipid Res. 45, 2138–2144 [DOI] [PubMed] [Google Scholar]
- 28.Schneider C., Pratt D. A., Porter N. A., Brash A. R. (2007) Chem. Biol. 14, 473–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwarz K., Walther M., Anton M., Gerth C., Feussner I., Kuhn H. (2001) J. Biol. Chem. 276, 773–779 [DOI] [PubMed] [Google Scholar]
- 30.Jakobsson P. J., Morgenstern R., Mancini J., Ford-Hutchinson A., Persson B. (2000) Am. J. Respir. Crit. Care Med. 161, S20–S24 [DOI] [PubMed] [Google Scholar]
- 31.Jakobsson P. J., Morgenstern R., Mancini J., Ford-Hutchinson A., Persson B. (1999) Protein Sci. 8, 689–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ago H., Kanaoka Y., Irikura D., Lam B. K., Shimamura T., Austen K. F., Miyano M. (2007) Nature 448, 609–612 [DOI] [PubMed] [Google Scholar]
- 33.Martinez Molina D., Wetterholm A., Kohl A., McCarthy A. A., Niegowski D., Ohlson E., Hammarberg T., Eshaghi S., Haeggström J. Z., Nordlund P. (2007) Nature 448, 613–616 [DOI] [PubMed] [Google Scholar]
- 34.Ferguson A. D., McKeever B. M., Xu S., Wisniewski D., Miller D. K., Yamin T. T., Spencer R. H., Chu L., Ujjainwalla F., Cunningham B. R., Evans J. F., Becker J. W. (2007) Science 317, 510–512 [DOI] [PubMed] [Google Scholar]
- 35.Martinez Molina D., Eshaghi S., Nordlund P. (2008) Curr. Opin. Struct. Biol. 18, 442–449 [DOI] [PubMed] [Google Scholar]
- 36.Gupta N., Gresser M. J., Ford-Hutchinson A. W. (1998) Biochim. Biophys. Acta 1391, 157–168 [DOI] [PubMed] [Google Scholar]
- 37.Thunnissen M. M., Andersson B., Samuelsson B., Wong C. H., Haeggström J. Z. (2002) FASEB J. 16, 1648–1650 [DOI] [PubMed] [Google Scholar]
- 38.Thunnissen M. M., Nordlund P., Haeggström J. Z. (2001) Nat. Struct. Biol. 8, 131–135 [DOI] [PubMed] [Google Scholar]
- 39.Bauvois C., Jacquamet L., Huston A. L., Borel F., Feller G., Ferrer J. L. (2008) J. Biol. Chem. 283, 23315–23325 [DOI] [PubMed] [Google Scholar]
- 40.An S., Kumar R., Sheets E. D., Benkovic S. J. (2008) Science 320, 103–106 [DOI] [PubMed] [Google Scholar]
- 41.Zarini S., Gijón M. A., Ransome A. E., Murphy R. C., Sala A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8296–8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones S. M., Luo M., Healy A. M., Peters-Golden M., Brock T. G. (2002) J. Biol. Chem. 277, 38550–38556 [DOI] [PubMed] [Google Scholar]
- 43.Jones S. M., Luo M., Peters-Golden M., Brock T. G. (2003) J. Biol. Chem. 278, 10257–10263 [DOI] [PubMed] [Google Scholar]
- 44.Flamand N., Luo M., Peters-Golden M., Brock T. G. (2009) J. Biol. Chem. 284, 306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bennett C. F., Chiang M. Y., Monia B. P., Crooke S. T. (1993) Biochem. J. 289, 33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bennett A. F., Buckley P. D., Blackwell L. F. (1982) Biochemistry 21, 4407–4413 [DOI] [PubMed] [Google Scholar]
- 47.Coffey M. J., Wilcoxen S. E., Peters-Golden M. (1994) Am. J. Respir. Cell Mol. Biol. 11, 153–158 [DOI] [PubMed] [Google Scholar]
- 48.Evans J. F., Ferguson A. D., Mosley R. T., Hutchinson J. H. (2008) Trends Pharmacol. Sci. 29, 72–78 [DOI] [PubMed] [Google Scholar]
- 49.Christmas P., Weber B. M., McKee M., Brown D., Soberman R. J. (2002) J. Biol. Chem. 277, 28902–28908 [DOI] [PubMed] [Google Scholar]
- 50.Mandal A. K., Skoch J., Bacskai B. J., Hyman B. T., Christmas P., Miller D., Yamin T. T., Xu S., Wisniewski D., Evans J. F., Soberman R. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6587–6592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mandal A. K., Jones P. B., Bair A. M., Christmas P., Miller D., Yamin T. T., Wisniewski D., Menke J., Evans J. F., Hyman B. T., Bacskai B., Chen M., Lee D. M., Nikolic B., Soberman R. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20434–20439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strid T., Svartz J., Franck N., Hallin E., Ingelsson B., Söderström M., Hammarström S. (2009) Biochem. Biophys. Res. Commun. 381, 518–522 [DOI] [PubMed] [Google Scholar]
- 53.Pande A. H., Qin S., Tatulian S. A. (2005) Biophys. J. 88, 4084–4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Forneris F., Mattevi A. (2008) Science 321, 213–216 [DOI] [PubMed] [Google Scholar]
- 55.Brock T. G., Maydanski E., McNish R. W., Peters-Golden M. (2001) J. Biol. Chem. 276, 35071–35077 [DOI] [PubMed] [Google Scholar]
- 56.Brock T. G., Lee Y. J., Maydanski E., Marburger T. L., Luo M., Paine R., 3rd, Peters-Golden M. (2005) Am. J. Physiol. Lung Cell. Mol. Physiol. 289, L224–L232 [DOI] [PubMed] [Google Scholar]
- 57.Luo M., Jones S. M., Peters-Golden M., Brock T. G. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12165–12170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fiore S., Serhan C. N. (1989) Biochem. Biophys. Res. Commun. 159, 477–481 [DOI] [PubMed] [Google Scholar]
- 59.Schultz M. J., Wijnholds J., Peppelenbosch M. P., Vervoordeldonk M. J., Speelman P., van Deventer S. J., Borst , van der Poll T. (2001) J. Immunol. 166, 4059–4064 [DOI] [PubMed] [Google Scholar]
- 60.Dickinson Zimmer J. S., Voelker D. R., Bernlohr D. A., Murphy R. C. (2004) J. Biol. Chem. 279, 7420–7426 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.