Abstract
The CDX2 transcription factor is known to play a crucial role in inhibiting proliferation, promoting differentiation and the expression of intestinal specific genes in intestinal cells. The overall effect of CDX2 in intestinal cells has previously been investigated in conditional knock-out mice, revealing a critical role of CDX2 in the formation of the normal intestinal identity. The identification of direct targets of transcription factors is a key problem in the study of gene regulatory networks. The ChIP-seq technique combines chromatin immunoprecipitation (ChIP) with next generation sequencing resulting in a high throughput experimental method of identifying direct targets of specific transcription factors. The method was applied to CDX2, leading to the identification of the direct binding of CDX2 to several known and novel target genes in the intestinal cell. Examination of the transcript levels of selected genes verified the regulatory role of CDX2 binding. The results place CDX2 as a key node in a transcription factor network controlling the proliferation and differentiation of intestinal cells.
Keywords: Cell Differentiation, Chromatin Immunoprecipitation (ChIP), DNA Sequencing, Gene Expression, Gene Regulation, Gene Transcription, Intestine, Transcription, Transcription Factors, Transcription Target Genes
Introduction
The intestinal epithelium lining the gastrointestinal tract is composed of a self-renewing monolayer of cells. Cell division is confined to the crypts of Lieberkühn, a pocket-like invagination into the gut mucosa, where a stem cell population continuously provides renewal of the gut epithelium. The daughter cells undergo rapid amplification and subsequently undergo differentiation into one of the three different cell types: enterocytes, goblet, and entero-endocrine cells (1). The cells differentiate and cease proliferation as they migrate from the crypt to the lumenal surface. In the small intestine a fourth cell type, the Paneth cell, is produced. This cell type migrates to the base of the crypts. The human intestinal Caco-2 cell line undergoes spontaneous enterocyte-like differentiation when the cells reach confluence and is, therefore, widely used as a model to study intestinal cell maturation.
The proliferation and differentiation of the intestinal epithelium occurs in a sequential and spatially organized manner and is highly regulated at the transcriptional level. Several different signaling pathways are required and act in a combinatorial manner to maintain homeostasis in the normal intestinal epithelium. Some of the pathways identified to date include the Wnt pathway (2), the Notch pathway (3), the Hedgehog system (4), members of the transforming growth factor-β family including the bone morphogenetic proteins (5), and the phosphatidylinositol 3-kinase pathway (6). The intestine specific gene expression requires the tightly regulated activity of transcription factors, including the well characterized key factors HNF1α/β,2 HNF4α, GATA factors, ETS, CDX1, and CDX2 (7–13). These transcription factors often cooperate to control the expression of the intestinal specific genes; until now, in particular HNF1α, GATA4–6, and CDX2 have been found to interact in order to regulate the activity of several promoters in the intestine (7, 14–20).
The caudal related homeobox gene CDX2 is essential for intestinal differentiation. The expression of this transcription factor in adults is restricted to the small intestine and colon (21, 22). The intestine-specific expression is partly controlled by cooperation between HNF4α, GATA6, β-catenin, and TCF7L2 (23). The CDX2 protein is present in all cells along the crypt-villous axis, although there is a post-translational control of CDX2 transcriptional activity, and there are several different pathways that regulate the activity of the protein by phosphorylation of different sites in CDX2 (21, 24). The mitogen-activated protein kinase (MAPK) family member p38 has been shown to directly interact with CDX3 (the hamster homologue of CDX2) and by phosphorylation activating the CDX2 protein during differentiation (25). Another part of the MAPK pathway, extracellular signal-regulated kinase (ERK), has been shown to phosphorylate the CDX2 protein at Ser-60, leading to a reduced CDX2 transcriptional activity in crypt and lower villous cells (26). Another phosphorylation-dependent regulation of CDX2 is that of Ser-281 by cyclin-dependent kinase 2. This coordinates its polyubiquitination and degradation by the proteasome (27).
The important role of CDX2 in differentiation is seen when forced expression of CDX2 in an undifferentiated intestinal cell line (IEC-6) leads to arrest of proliferation and initializes differentiation (28). The essential role of CDX2 in mammalian embryonic development is evidenced by the inactivation of both Cdx2 alleles (Cdx2−/−), which leads to early mortality in mouse embryos (29). The heterozygotic Cdx2+/− mice show an increased susceptibility for tumors, underlining the important role of Cdx2 in proliferation and differentiation. A recent study used conditional Cdx2 knock-out mice to confirm the essential role of Cdx2 in intestinal development and furthermore showed that Cdx2 is a key regulator of several transcription factors involved in intestinal development (30).
The regulation of gene expression by transcription factors is a vital mechanism for controlling cell proliferation and differentiation. Thus, the identification of transcription factor binding sites is essential in understanding the regulatory circuits that control cellular processes such as cell division and differentiation. In this study we have used the in vivo method of isolation of transcription factor target DNA through chromatin immunoprecipitation and have combined this with next generation sequencing (ChIP-seq) to map the binding sites of CDX2 in Caco-2 cells. We have identified many novel CDX2 in vivo binding sites that may be involved in gene regulation during differentiation. We have also confirmed many of the potential CDX2 binding sites using quantitative PCR (ChIP-qPCR), thereby validating the quality of our dataset. These results provide novel insight into the transcriptional network containing CDX2, HNF4α, and HNF1α, which control the differentiation of intestinal epithelia.
EXPERIMENTAL PROCEDURES
Plasmid Construction
All primers used in this study were purchased from MWG Biotech. The primers in supplemental Data S1 were used to amplify the following promoter and enhancer sequences from human genomic DNA (Roche Applied Science): CDX2 promoter (GenBankTM number NM_001265) position −1212 to +36; CDX2 enhancer position +8104 to +8418; HNF4α promoter (GenBankTM number NM_175914) position −452 to +75; HNF4α enhancer position +86875 to +87241; hephaestin (HEPH) promoter (GenBankTM number NM_138737) position −710 to +23; MEP1A promoter (GenBankTM number NM_005588) position −835 to +99. HBP1 promoter (GenBankTM number NM_012257) position −509 to +16. For each promoter the PCR product was cloned in pCR®2.1 (Invitrogen). The cloned PCR fragment was cloned into pGL4.10 (Promega) using the XhoI and HindIII for the promoters or into pGL4.10 containing the corresponding promoter using the BamHI site for the enhancers. All the constructed plasmids were sequenced and analyzed on an ABI 3100 sequencer (Applied Biosystems).
Cell Cultures and Transfections
The cells were grown, and reporter gene transfections were carried out as described in Boyd et al. (12). The cells were stably transfected with SureSilencingTM shRNA plasmid for Human CDX2 (SABiosciences Corp.) according to the manufacturer's recommendations. The stable clones were isolated as monoclonal clones, and the amount of functional CDX2 protein was determined by adding extraction buffer (31) and using the whole cell extracts in electrophoretic mobility shift assay (EMSA).
Quantitative Reverse Transcription-PCR
cDNA from stable Caco2 cell clones transfected with either shRNA plasmid against CDX2 (CDX2 knock down Caco2) or a random shRNA plasmid (negative control) were extracted from differentiated cells (10 days after confluence) as previously described (34). Gene-specific PCR primer pairs against cDNAs from potential CDX2 target genes were found in PrimerBank (35) (listed in supplemental Data S1). The expression levels were measured with the LighCycler 480 SYBR Green I Master kit (Roche Applied Science) on a Lightcycler 480II (Roche Applied Science).
ChIP Assay and ChIP-seq
Caco-2 cells were cultured as described above until confluence, 4 days after confluence, and 10 days after confluence. The ChIP procedure was performed as previously described (12) with an antibody against CDX2 (Biogenex) and HA (negative control). ChIP-seq was performed, with immunoprecipitated DNA from cells 10 days after confluence according to the manufacturer's instructions (Illumina Inc.).
Bioinformatics
The image files generated by the Genome Analyzer were processed to extract DNA sequence data and mapped to the human genome using the Illumina Analysis Pipeline (bed files with chromosomal positions of the sequences are available on request).
The detection of CDX2 peaks was performed using CisGenome (32) with a window of 300 bp, a minimum of 10 tags, and a false discovery rate level <0.001. Using these parameters 604 peaks were identified as potential CDX2 binding sites. Overrepresented functional categories (GO categories) from the Gene Ontology were determined using the program PinkThing. The PinkThing program was also used to assign the 604 CDX2 ChIP-seq peaks to the nearest genes. All sequence analyses were conducted based on the Homo sapiens NCBI 34 genome assembly accessed from Ensemble.
ChIP qPCR Analysis
Verification of enrichment due to immunoprecipitation was done as previously described (12). The primers listed in supplemental Data S1 were used to amplify human genomic sequences at CDX2-target loci. Quantification of the ChIP DNA was done using the method described by Frank et al. (33).
EMSA
Preparation of nuclear extracts and EMSAs were performed as previously described (36). 2 μg of antibodies against CDX-2 (Biogenex) were used in the supershift analyses. Oligonucleotides used were the SIF1-CDX2 site from the human sucrase-isomaltase promoter: 5′-TGGGTGCAATAAAACTTTATGAGTA-3′ and 5′-TTACTCATAAAGTTTTATTGCACCC-3′ (22, 37). Determination of CDX2 protein levels was done by allowing the Caco-2 cells to differentiate (day 10), harvesting the cells and extracting the proteins with buffer as described in Schöler et al. (31). Protein quantification was done with the Bradford assay (Bio-Rad), and equal amounts of protein were used in an EMSA.
RESULTS
CDX2 ChIP-seq
The CDX2 and HA ChIP DNA was sequenced using the Illumina DNA sequencing technology. The ChIP-seq results were aligned to the human genome (see supplemental Data S2) and analyzed with the CisGenome software (32) to identify the CDX2-enriched DNA fragments along with the nearest gene. The analysis results in a list with 604 potential CDX2 target genes (supplemental Data S3). Using the PinkThing program, we assigned the 604 identified CDX2 ChIP-seq peaks to the nearest genes, and the target sites were grouped depending on their position relative to the nearest gene. This analysis reveals that more than half of the CDX2 ChIP-seq peaks are located in enhancer positions either in distant enhancers (147), 3′ end (95), or introns (176) (supplemental Data S4A). Furthermore, the analysis by PinkThing revealed that the list of potential CDX2 target genes has an overrepresentation of GO terms within the categories of pattern specification and transcription regulator activity (supplemental Data S4B). A manual inspection of the list of 604 CDX2 ChIP-seq peaks shows that many known CDX2 target genes are present, although some known CDX2 target genes are not present on the list (e.g. SI and LI-Cadherin (CDH17)). The list, thus, represents a stringent version of the predicted CDX2 target genes from the ChIP-seq results. We, therefore, decided to explore some of the targets from the CisGenome list along with some chosen targets based on the known functions of CDX2 and a manual inspection of the CDX2 ChIP-seq tags.
CDX2 ChIP-qPCR during Different Stages of Caco-2 Differentiation
The ChIP-seq results indicated that several known and novel genes are regulated by CDX2 in Caco-2 cells. To investigate the role of CDX2 during differentiation in the regulation of putative target genes, we selected a series of genes for further analysis. We fixed and harvested Caco-2 cells at different stages during differentiation and used these cells in a CDX2 immunoprecipitation followed by a quantitative PCR with gene specific primers.
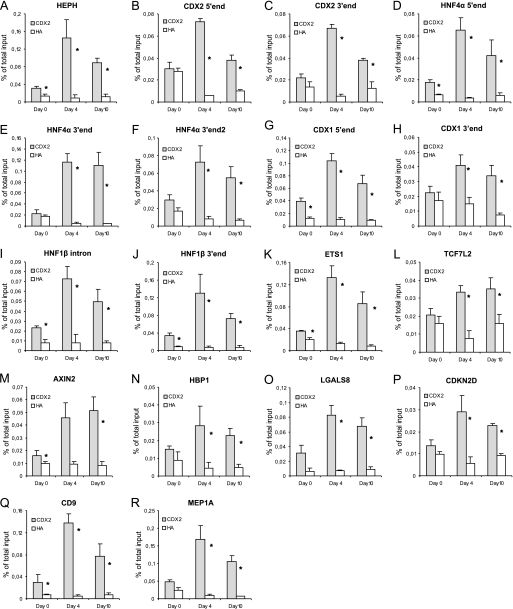
The general tendency is that the CDX2 ChIP procedure on undifferentiated cells at confluence (day 0) results in the range from 0 up to 4.8-fold enrichment of potential CDX2 target genes in the CDX2 immunoprecipitation as compared with the HA control (Fig. 1). When compared with the two later stages of differentiation, this is quite a low recovery, which can be explained by CDX2 mainly being involved in differentiation and that it is activated by phosphorylation so that at day 0, the majority of the CDX2 protein in the cells are not bound to DNA. The recovery of CDX2-bound DNA fragments in Fig. 1 can be divided into two categories, where one contained the DNA with little CDX2 bound in undifferentiated cells (enrichment <2 in Fig. 1, B, C, E, F, H, K, L, M, N, and P), whereas the remaining DNA had an enrichment above 2 in the undifferentiated cells. It could be speculated that the genomic regions that do show some enrichment regulate gene products that are necessary for the early events of differentiation and that their regulatory elements contain strong CDX2 binding sites that are able to recruit CDX2.
FIGURE 1.
CDX2 binding is dependent on the level of differentiation. A–R, regulatory regions for 13 different genes (HEPH, CDX2, HNF4α, CDX1, HNF1β, ETS1, TCF7L2, Axin2, HBP1, LGALS8, CDKN2D, CD9, and MEP1A) were selected for investigation based on the CisGenome list of potential CDX2 target genes (supplemental Data S3) and manual inspection of ChIP-seq tags (supplemental data S2). Shown are real-time quantitative PCR with CDX2- and HA (negative control)-ChIP DNA from Caco-2 cells at different stages during differentiation. Day 0 = undifferentiated, Day 4 = initial differentiation, Day 10 = fully differentiated. Data are expressed as mean values ± S.E. (n = 3). CDX2 versus HA ChIP DNA; *, p < 0.05. Day 0 versus Day 4, p < 0.05 in (A–R). Day 0 versus Day 10, p < 0.05 for (A–R, except L and N). Results are representative of at least three independent experiments.
Between day 0 and day 4 there was a strong increase in the binding of CDX2 protein to the target sequences, which is in good agreement with our finding that CDX2 mRNA and protein levels increase during this period (Fig. 2A and data not shown). All the examined DNA regions had a statistically significant increase in the amount of recovered DNA by the CDX2 ChIP between day 0 and day 4 (Fig. 1). Notice in particular that the positive control HEPH had a 7-fold increase in recovered DNA, HNF4α promoter and enhancer had a 6.6- and 18.6-fold increase respectively, and CDX2 promoter and enhancer had a 12.2- and 7.7-fold increase, respectively. This dramatic increase in CDX2 binding during differentiation indicates an important role for CDX2 regulation of these genes during differentiation.
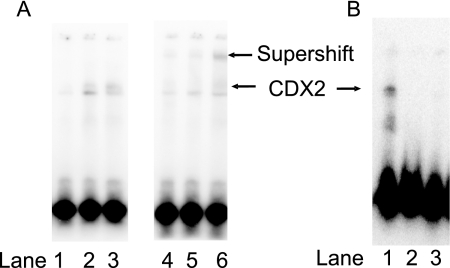
FIGURE 2.
Analysis of CDX2 expression in Caco-2 cells. A, the binding activity of CDX2 protein was determined by EMSA during differentiation (days 0, 4, and 10); lanes 1, 2, and 3, respectively. Lanes 4, 5, and 6 corresponds to days 0, 4, and 10, where the CDX2-DNA complex was supershifted with specific CDX2 antibody. B, the binding activity of CDX2 protein in the normal (lane 1) and two CDX2 knockdown Caco-2 strains (lane 2 and 3) was determined by EMSA. The CDX2 knockdown strain 2 (lane 3) was used in this study.
Between day 4 and day 10 the Caco-2 cells completed differentiation to resemble mature enterocytes. During this period the level of CDX2 protein only increased slightly (Fig. 2A). The level of CDX2 binding did not increase on any of the DNA fragments containing putative CDX2 binding sites; in fact, several of the DNA segments were recovered at a lower level at day 10 than day 4 (Fig. 1).
CDX2 ChIP and qPCR on Control and CDX2 Knockdown Caco-2 Cells
To further investigate the role of CDX2 in the regulation of the selected genes, we used CDX2 shRNA plasmid to generate stable Caco-2 clones with reduced CDX2 protein expression. Clones of CDX2 shRNA-transfected cells were screened for the level of CDX2 protein using whole cell extracts in EMSA with a known CDX2 binding sequence from the sucrase-isomaltase promoter (22). The amount of CDX2 protein was greatly reduced in the CDX2 knockdown Caco-2 cells when compared with the control (Fig. 2B).
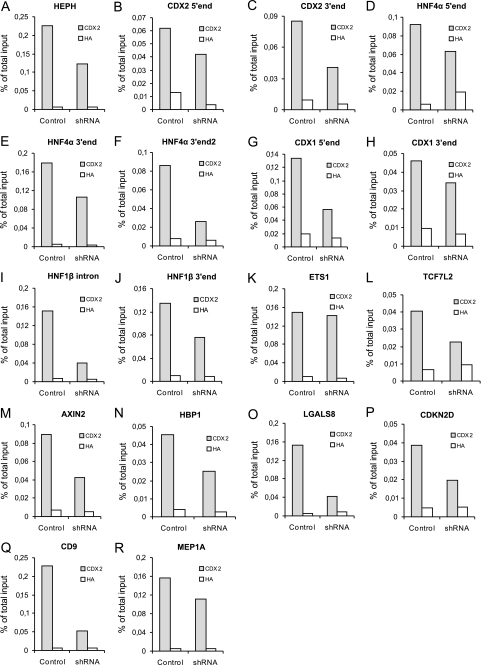
In general, our results show that the knockdown of CDX2 results in a reduction in the CDX2 ChIP signal of the CDX2 target sequences. Interestingly, the signals from the target sequences are in all cases greater in the CDX2 immunoprecipitation compared with the HA immunoprecipitation, which shows that even though CDX2 knockdown Caco-2 cells have a severely reduced level of CDX2 protein, there is still enough to allow some CDX2 binding to target sequences.
The effect of the CDX2 knockdown on the recovery rate can roughly be divided into three categories. The first category contained the CDX2 target sequences, where the CDX2 immunoprecipitation recovered more than 60% target DNA in the CDX2 knockdown strain as compared with the control. This was the case for the DNA sequences from CDX2 5′ end, HNF4α 5′ end, CDX1 3′ end, ETS1, and MEP1A (Fig. 3, B, D, H, K, and R, respectively). These DNA sequences show strong CDX2 binding despite the reduction in CDX2 protein in the CDX2 knockdown strain.
FIGURE 3.
CDX2 knockdown affects expression of selected genes. A–R, real-time quantitative PCR with CDX2- and HA (negative control)-ChIP DNA from differentiated Caco-2 cells with either normal CDX2 (control) or reduced CDX2 (shRNA) expression. The same regions as in Fig. 2 were analyzed. Results are representative of at least three independent experiments.
The second category contains the CDX2 target sequences with an intermediate CDX2 binding strength, which have a recovery rate of 40–60% target DNA in the CDX2 knockdown strain as compared with the control strain. The DNA sequences in this category included HEPH, CDX2 3′ end, HNF4α 3′ end, CDX1 5′ end, HNF1β 3′ end, TCF7L2, AXIN2, HBP1, and CDKN2D (Fig. 3, A, C, E, G, J, L, M, N, and P, respectively).
The last category of DNA sequences had a very low recovery rate (<40% target DNA when compared with the control) in the CDX2 knockdown strain as a result of the CDX2 immunoprecipitation. This category contained HNF4α 3′ end2, HNF1β intron, LGALS8, and CD9 (Fig. 3, F, I, O, and Q, respectively).
In summary, the reduction of CDX2 protein in the CDX2 shRNA knockdown strain resulted in a decreased CDX2 immunoprecipitation recovery of all of our putative CDX2 target sequences investigated here. There seemed to be significant differences in the ability to recruit CDX2 when the level of CDX2 was lowered in the cells, thus indicating different binding affinities of CDX2 to the regulatory DNA elements.
Analysis of the Effect of CDX2 on Promoter Activity of Selected Target Genes
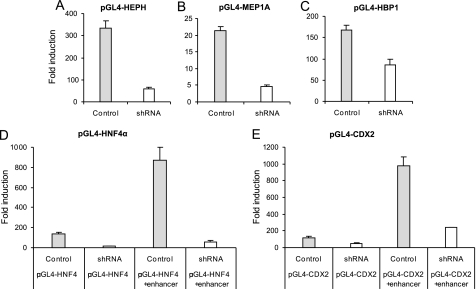
To analyze the effect of the CDX2 binding in regulation of gene expression, we chose DNA sequences predicted by the CDX2 ChIP-seq to contain CDX2 binding sites and created promoter/enhancer reporter gene expression plasmids containing these DNA fragments in control of the expression of the luciferase gene. These plasmids were transfected into Caco-2 cells with normal (control) and reduced (shRNA) CDX2 protein levels. The regulatory DNA elements chosen were restricted to being proximal promoters except in the cases of HNF4α and CDX2, where the 3′-located enhancers were included in the plasmids containing the corresponding promoter.
The insertion of a regulatory DNA sequence with CDX2 binding sites into the pGL4.10 plasmid resulted in an increased expression of the luciferase gene in all seven cases examined here (Fig. 4, A–E). The fold induction of luciferase activity varied between 21- and 982-fold above background. These measurements are an indication of how strong the promoter/enhancer is at regulating gene expression and, thus, an indication of how much of the corresponding gene product is produced. Notice in particular the high activity of the HEPH promoter (333× above background) and the dramatic effect of including the enhancer of the HNF4α and CDX2 gene (Fig. 4, A, D, and E, respectively).
FIGURE 4.
CDX2 influences the transcription of promoter/enhancer constructs. The promoters of HEPH, MEP1A, HBP1, HNF4α, and CDX2 were used to create promoter reporter gene constructs. For HNF4α and CDX2, the corresponding enhancer element was cloned into the promoter reporter gene constructs. A–E, Caco-2 cells with normal CDX2 (Control) or reduced CDX2 (shRNA) were transfected with reporter plasmid and the expression vector CMV-βGal. After 48 h luciferase were quantified and normalized to the β-galactosidase activity. -Fold induction was calculated against the basic reading from the pGL4.10. Data are expressed as mean values ± S.E. (n = 4). The difference between control and shRNA for each expression construct is significant, p < 0.01. The results are representative of at least two independent experiments.
The effect on luciferase expression of the DNA sequences was measured in the CDX2 knockdown Caco-2 strain. A specific effect of reducing the level of CDX2 protein can be observed, and the results are divided into two categories. The first contains the DNA sequences that show a roughly 2-fold reduction in expression activity when the level of CDX2 protein was reduced. This was the case for the HBP1 and CDX2 promoter (Fig. 4, C and E). The second category contains HEPH, MEP1A, HNF4α promoter, HNF4α promoter and enhancer, and CDX2 promoter and enhancer (Fig. 4, A, B, D, and E), which all showed a larger than 2-fold reduction in activity when measured in the CDX2 knockdown strain.
In summary, a 2-fold reduction in luciferase activity is an indication of a medium dependence on CDX2 for the transcriptional activity of the DNA sequence. A greater reduction in luciferase expression was observed with the remaining DNA sequences; therefore, these are more strongly dependent on CDX2 for their transcriptional activity.
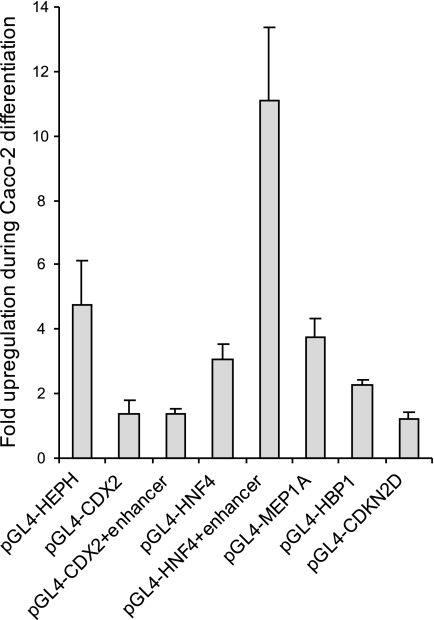
We wished to further examine the role of CDX2 on the promoter activity of the potential target genes. We previously developed an assay by which we can measure promoter activity during differentiation of Caco-2 cells using transient transfection (38). We, therefore, assayed selected potential CDX2 target promoters using the reporter gene plasmids. The -fold change in expressed luciferase activity was measured between undifferentiated cells and differentiated Caco-2 cells. In this period the level of active CDX2 protein increased (Fig. 2A) (26). The expression of luciferase from all of the promoters increased during the differentiation of the Caco-2 cells in the range from 1.3- to 11-fold (Fig. 5). These results indicate that the increase in active CDX2 protein during differentiation results in an increased transcriptional activity from the investigated promoters that have CDX2 binding sites.
FIGURE 5.
The activity of the promoter/enhancer constructs increases during differentiation. The promoter/enhancer reporter gene constructs for HEPH, MEP1A, HBP1, CDKN2D, HNF4α, and CDX2 were transfected into Caco-2 cells with the expression vector CMV-βGal. After 2 and 10 days luciferase were quantified and normalized to the β-galactosidase activity. -Fold induction was calculated against the reading from day 2. Data are expressed as mean values +S.E. (n = 4).
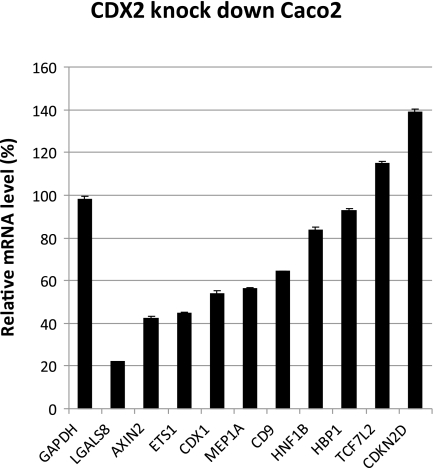
To analyze the effect of CDX2 on the endogenous gene expression, we examined the transcript levels of selected genes in the two Caco-2 strains with either normal or reduced levels of CDX2 protein (Fig. 6). The transcript levels of the glyceraldehyde-3-phosphate dehydrogenase gene was included to assure that the reduced level of CDX2 protein did not change the overall transcript level in the cells. The reduction of CDX2 protein by shRNA led to a decreased transcript level of all but two of the potential CDX2 target genes, thus, further validating the proposed positive regulation of these genes by CDX2 (Fig. 6).
FIGURE 6.
Expression of selected genes in the CDX2 knockdown Caco-2 cells. The levels of mRNA were measured by real-time PCR and normalized to the expression in Caco-2 cells transfected with a negative control shRNA plasmid. n = 3.
DISCUSSION
The analysis with the CisGenome program of the CDX2 ChIP-seq tags resulted in a list of 604 potential CDX2 target genes. This list includes several known CDX2 target genes including the HOX genes (30, 39), which are known to function downstream of CDX2 in the molecular pathway that establishes the anteroposterior axis during development. The list also excluded some of the known CDX2 targets; this is a consequence of the procedure performed on the data by the CisGenome program. This analysis only predicts a potential target gene if the appropriate amount of tags is present within a certain stretch of DNA. We chose these parameters based on a manual inspection of some of the known CDX2 targets, and the end result is a compromise to obtain a high quality list with few false positives. The analysis of the position of the 604 ChIP-seq peaks relative to the nearest genes revealed that more than half (418) of the peaks are located in the enhancer regions. This could be interpreted as the strongest binding of CDX2 being present in the enhancer regions. Using sensitive qPCR analyses of ChIP signals located in promoters not included on the list from the CisGenome program allowed us to demonstrate that indeed significant CDX2 binding to these promoters is indeed present (Fig. 1). The CisGenome list (supplemental Data S3) is, therefore, a stringent list of the highest CDX2 ChIP signals in our analysis, and some biological relevant signals are not included on this list. Another interesting finding is the fact that the overrepresented GO-terms on our list of CDX2 target genes belong to the category of development, thus, underlining the important role of CDX2 in this process.
The HEPH promoter has been shown to be directly regulated by CDX2 (40) and was, therefore, used as a positive control in our experiments. Hephaestin was initially identified as the defective gene in sex-linked anemia (sla) mice, which has a block in intestinal iron transport. The protein product is a transmembrane-bound ferroxidase that is necessary for the intestinal transport of iron by the enterocytes (41). During the first days of differentiation of Caco-2 cells, there is an increase in the amount of HEPH promoter DNA isolated with the CDX2 immunoprecipitation (Fig. 1A). We then examined the effect of CDX2 knockdown, which resulted in an almost 50% reduction in recovery of HEPH promoter DNA in our CDX2 ChIP as measured by qPCR (Fig. 3A). In the transfection experiments the hephaestin promoter in the CDX2 knockdown strain has a luciferase activity that is 5.7-fold reduced when compared with that measured in the control strain (Fig. 4A). In conclusion, these results confirm that hephaestin has CDX2 bound to the promoter DNA and that CDX2 influences the expression from the promoter. Furthermore, the results show that our experiments are suitable for examining the binding and effect of CDX2 on a known CDX2 target promoter.
CDX2 Autoregulation
Previously, CDX2 has been found to activate the expression from its own promoter (42). To investigate the autoregulation of CDX2 expression, we examined the CDX2 ChIP-seq signals and, as expected, found strong signals from the CDX2 promoter. In addition to the CDX2 signals from the promoter, we found an even stronger set of CDX2 signals from a 3′ end region that has not previously been reported bound by CDX2. During differentiation of Caco-2 cells, we observed similar patterns for both promoter and enhancer elements, where CDX2 binding is non-significant in undifferentiated cells and increases during the first days of differentiation (Fig. 1, B and C). Furthermore, for both regulatory elements we found that the CDX2 knockdown strain exhibits a reduction in recovered DNA in the CDX2 ChIP when compared with the control (Fig. 3, B and C). The ability of the regulatory elements to control expression of luciferase was shown to be quite high above background and dependent on the level of CDX2 protein in the Caco-2 cells (Fig. 4E). We, thus, conclude that CDX2 is found to activate its own expression; however, the 3′ end enhancer generally contributes more significantly to the transcriptional autoregulation, which is a novel finding.
CDX2 Is a Central Node in the Intestinal Transcription Factor Network
HNF4α is a transcription factor that is essential for the normal colon development and expression of several intestinal genes (43). Previous work by Lussier et al. (44) has shown that CDX2 is essential in initiating differentiation and that this is coordinated with the activation of HNF4α and HNF1α expression, which in turn requires CDX2 action. Furthermore, we have previously shown that HFN4α regulates the expression of CDX2; thus, a loop that is necessary for the initiation and completion of differentiation of intestinal cells appears to exist (12). The promoter of HNF4α has previously been shown to contain a functional CDX2 binding site (30). In this work we analyzed the promoter along with two novel CDX2 binding sites in a 3′ end enhancer element. There was a strong increase in the CDX2 binding to the HNF4α regulatory elements during differentiation of Caco-2 cells (Fig. 1, D–F). The shRNA knockdown of CDX2 resulted in a significant reduction in CDX2 ChIP recovery of all three CDX2 binding sites (Fig. 3, D–F). In the transfection experiments we have shown that both regulatory DNA elements have a significant effect on the expression. Furthermore, we demonstrated that both the promoter and the enhancer elements are strongly dependent on CDX2 for their activity (Fig. 4D). In conclusion, we have shown CDX2 binding to both promoter and 3′ end enhancer elements of HNF4α and that the CDX2 binding has a positive effect on the transcriptional activation potential of both regulatory elements. Especially interesting is the fact that the effect on reporter gene expression from the enhancer element is several-fold greater than from the promoter and that CDX2 binding to the enhancer has a greater stimulatory effect on expression than CDX2 binding to the promoter.
The CDX1 protein is highly similar to CDX2 and binds to many of the same DNA elements as CDX2 (for review, see Ref. 45). The role of CDX1 has been controversial with regard to tumor development, but recent results indicate that CDX1 is dispensable for normal gut development and with only a limited effect on intestinal cancer (46). The expression of CDX1 has previously been shown to be affected by CDX2, and a putative CDX2 binding site was identified in the 5′ end (30). According to the ChIP-seq results, a second CDX2 binding site is potentially located in an enhancer element in the 3′ end of the CDX1 gene. During the early Caco-2 differentiation, both CDX1 regulatory elements showed an increase in CDX2 binding, although the CDX2 binding to the enhancer was somewhat lower than that on the promoter (Fig. 1, G and H). The reduction of CDX2 protein in the shRNA strain resulted in reduction in recovery of both CDX1 promoter and enhancer DNA as measured with qPCR on the CDX2 ChIP DNA (Fig. 3, G and H). The endogenous transcript level of CDX1 was reduced when the level of CDX2 protein was decreased by shRNA (Fig. 6), thus, providing evidence that CDX2 is a positive regulator of CDX1 expression.
To further analyze the role of CDX2 in the intestinal transcription factor network, the interaction of CDX2 with two enhancer elements of the HNF1β gene was investigated. HNF1β is necessary for normal embryonic development and the activation of the transcription factors HNF4α, HNF1α, and HNF3γ (47). The two potential CDX2 binding elements in the HNF1β gene showed both a significant CDX2 binding at day 0 (Fig. 1, I and J) and an increase in CDX2 immunoprecipitated DNA between day 0 and day 4 during Caco-2 differentiation. The qPCR showed a reduction in recovery of both the intronic and 3′ end element by CDX2 ChIP in our CDX2 knockdown cells (Fig. 3, I and J). The transcription of the HNF1β gene is reduced in the CDX2 knockdown strain (Fig. 6), thus, indicating that CDX2 is a positive regulator of this gene. In conclusion, we have identified two regulatory DNA elements near the HNF1β gene that are bound by CDX2 in increasing amounts during the beginning of differentiation and that CDX2 up-regulates the expression of this important transcription factor.
The v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) (ETS1) protein is a member of the ETS-domain family of transcription factors. This large family of ETS proteins plays diverse roles in cellular differentiation, proliferation, development, transformation, and apoptosis (48). ETS1 has been proven to be a proto-oncoprotein even though there are reports that ETS1 have both pro- and anti-apoptotic effects (49, 50). When comparing the CDX2 immunoprecipitation from day 0 and day 4, there was an increase in the CDX2 binding to the ETS1 DNA (Fig. 1K). The reduced CDX2 protein level did not result in a reduction in the amount of CDX2 immunoprecipitated ETS1 intron DNA (Fig. 3K). These results clearly show that CDX2 is bound to the intron of the ETS1 gene. Further validating the role of CDX2 in regulating the ETS1 gene is the fact that the level of ETS1 gene expression was reduced when the level of CDX2 protein was decreased (Fig. 6). Interestingly, ETS transcription factors have been speculated to be involved in the regulation of CDX2 expression (9), so a regulatory loop between ETS1 and CDX2 could exist to regulate the differentiation process.
To summarize, we have shown that the CDX2 protein binds to regulatory DNA elements of several essential transcription factors involved in the normal differentiation of Caco-2 cells, and we have shown the effect of CDX2 binding to several of these regulatory elements. The majority of the examined transcription factors bound by CDX2 are involved in promoting differentiation, whereas CDX1 and ETS1, on the other hand, are primarily known as activators of proliferation. The function of CDX2 regulation of proliferation-promoting genes could be an indication of the role of CDX2 during embryogenesis. The quantity of identified CDX2 regulated transcription factors place CDX2 as a central component in the transcription factor network controlling the differentiation of intestinal cells.
The Role of CDX2 in Wnt Signaling and Cell Cycle Regulation
Another important transcription factor bound by CDX2 is the TCF7L2, which is a nuclear transcription factor that interacts with β-catenin, which in turn mediates the canonical WNT signaling pathway. This signaling pathway has been shown to be critical for normal embryogenesis, cell proliferation, and motility as well as maintenance of the stem cells in the crypt of the small intestine (51, 52). In undifferentiated Caco-2 cells, the TCF7L2 promoter was not bound by CDX2, whereas there is was a slight increase in CDX2 binding during differentiation (Fig. 1L). The TCF7L2 promoter exhibited a reduction in CDX2 binding (Fig. 3L) when the level of CDX2 protein was decreased. The endogenous level of TCF7L2 transcription rose when the protein level of CDX2 was reduced (Fig. 6), thus, indicating that CDX2 does not activate the TCF7L2 gene expression.
Two other proteins with functions in the Wnt signaling pathway were identified in this study as potential CDX2 targets. The first protein is axis inhibitor 2 (AXIN2), which is involved in the formation of a multiprotein complex containing adenomatosis polyposis coli (APC), glycogen synthase kinase-3β, β-catenin, and AXIN2; this leads to the degradation of β-catenin in the Wnt signaling pathway (53, 54). During differentiation of Caco-2 cells, the CDX2 immunoprecipitation of AXIN2 DNA was quite low at day 0 but increased during the later stages (Fig. 1M). The reduction of CDX2 in the shRNA strain also led to a reduction of recovered AXIN2 (Fig. 3M). The transcription of Axin2 was 40% reduced when the level of CDX2 protein was lowered by shRNA (Fig. 6). It is quite reasonable that the expression of AXIN2 is up-regulated by CDX2 during differentiation as the AXIN2 protein is involved in degradation of β-catenin and thereby inhibits proliferation.
The third protein involved in Wnt signaling is the HBP1 protein, which has homology to the high mobility class of transcription factors and has been shown to function as a transcriptional repressor that regulates cell cycle arrest during differentiation (55). In addition, HBP1 has been shown to suppress the Wnt signaling pathway by inhibiting the lymphoid enhancer factor/T-cell factor (LEF/TCF) transcription factors (56). The CDX2 ChIP showed no enrichment of HBP1 in undifferentiated Caco-2 cells followed by a slight increase in signal during differentiation (Fig. 1N). The reduced level of CDX2 protein in the knockdown strain resulted in a reduction in HBP1 promoter-bound CDX2 (Fig. 3N) and a reduction in expressed luciferase from the promoter (Fig. 4C). In addition the reduced level of CDX2 protein also resulted in a slight decrease in endogenous HBP1 expression (Fig. 6) This result corroborates the role of CDX2 acting as an inhibitor of proliferation.
Lectin, galactoside-binding, soluble, 8 (LGALS8) is a member of the galectin family that is involved in inhibition of cell growth and transformation while promoting apoptosis (57, 58). The LGALS8 can inhibit cell adhesion and cell cycle progression and induce growth arrest or apoptosis (59, 60). During the early phase of differentiation, the CDX2 signal from the LGALS8 DNA increased (Fig. 1O). The shRNA reduction of CDX2 resulted in a significant reduction of CDX2 immunoprecipitated LGALS8, indicating that this DNA has a weak binding site for CDX2 (Fig. 3O). The reduction of CDX2 protein resulted in a significant reduction in LGALS8 endogenous transcription (Fig. 6). The regulation of LGALS8 by CDX2 could be a clue in explaining how CDX2 is involved in stopping proliferation and how the shedding of differentiated cells into the lumen is regulated.
Another protein involved in cell cycle regulation is the cyclin-dependent kinase inhibitor 2D (CDKN2D/p19INK4d), which is a member of the INK4 family of CDK inhibitors (61). The CDK inhibitor members of the INK4 family specifically bind to CDK4/6, the kinases of the D-type cyclins implicated in phosphorylation of the retinoblastoma tumor suppressor protein (pRB) and thereby in the progression through the G1 phase of the cell cycle (for review, see Ref. 62). The CDKN2D promoter showed a significant binding of CDX2 early in Caco-2 differentiation, and this binding was dependent on the amount of CDX2 present (Figs. 1P and 3P). The endogenous transcription of CDKN2D was increased when the level of CDX2 protein was decreased (Fig. 6), thus indicating that CDX2 functions as a repressor of this gene. This could indicate a role of CDX2 in inhibition of cell cycle progression.
In summary, we have shown CDX2 binding and regulation of proteins involved in proliferation and cell cycle regulation. It is interesting that CDX2 appears to repress the expression of TCF7L2 when at the same time CDX2 seems to affect the expression of two inhibitors of the Wnt signaling pathway and in general is known to inhibit proliferation. When seen in the light of TCF7L2, which has been shown to activate CDX2 expression (23), then this new data could indicate the presence of a regulatory loop. Recent studies by Guo et al. (63) show that Cdx2 is able to bind β-catenin and, thus, silence the β-catenin/TCF target gene expression. In addition to the results described so far, the CDX2 ChIP-seq results showed a significant likelihood for CDX2 to be bound to APC, glycogen synthase kinase-3β, and possible β-catenin regulatory DNA elements (supplemental data S3). Thus, CDX2 appears to bind to several regulatory elements for genes involved in the Wnt signaling pathway, so it would be very interesting to examine the downstream targets in the Wnt signaling pathway in order to examine the effect of the CDX2 binding to regulatory elements in control of components in the Wnt signaling pathway.
CDX2 Regulation of Membrane-associated Proteins
Membrane-associated proteins can have diverse functions in the multicellular organism. In this study we have looked at the possible role of CDX2 in regulating two different membrane-associated proteins believed to be involved in migration among others. The results indicate a possible role for CDX2 in regulating the migration of the enterocytes along the crypt-villus axis during differentiation. The first protein is the CD9 antigen (CD9), which is a cell-surface molecule and a member of the transmembrane-4 family. It can interact with members of the integrin family and other membrane proteins, thus, participating in cell proliferation, migration, and adhesion (64, 65). The regulatory DNA element exhibited a dramatic increase of bound CDX2 during the early stage of differentiation (Fig. 1Q). The reduced CDX2 protein level in the shRNA strain led to a reduction of CD9 intron DNA fragment immunoprecipitated by the CDX2 ChIP (Fig. 3Q). The reduced level of CDX2 protein also resulted in a reduction in CD9 expression (Fig. 6). Thus, CDX2 binds to regulatory elements of the CD9 gene and positively regulates the expression.
The second CDX2-regulated membrane-associated protein is a member of the meprins. This group of proteins are metalloendopeptidases capable of hydrolyzing a variety of peptide and protein substrates (66). MEP1A has via its protease activity on extracellular matrix components been speculated to contribute to tumor progression by facilitating migration (67). The expression of Mep1a has previously been shown to be affected in Cdx2 conditional knock-out mice (30). The CDX2 immunoprecipitation shows an increase in recovered MEP1A promoter DNA between day 0 and day 4 (Fig. 1R). Furthermore, the amount of recovered MEP1A promoter DNA is reduced in the shRNA strain as compared with the negative control (Fig. 3R). The luciferase activity expressed from the MEP1A promoter was reduced in the shRNA strain compared with the control strain. Further confirming the role of CDX2 as a positive regulator of MEP1A, expression is seen when the level of CDX2 protein was decreased, and the measured endogenous level of MEP1A transcription was reduced (Fig. 6).
Comparison to Functional Analysis of Cdx2 Knock-out
Our study has identified several known and novel direct targets of CDX2; another Cdx2 study examined the effect on gene expression in conditional knock-out mice (30).
A comparison of the identified target genes shows that both studies find that CDX2 activates the expression of HEPH, CDX2, HNF4α, CDX1, and MEP1A. In general, the knock-out mice show a larger effect of Cdx2 knock-out on gene expression than what we observed in the transfection experiments (Fig. 4). We speculate that this is caused by the fact that Gao et al. (30) performed a conditional knock-out, whereas we have conducted a knockdown where the cells still retained some active CDX2 protein and we examined the direct effect of CDX2 binding.
There are also distinct genes found in either study that are regulated by CDX2, which we assume to be caused by the different nature of the two set of experiments. We have looked at the direct binding and effect of CDX2 on regulatory DNA elements, and although we are aware of the fact that not all binding events lead to effects on expression, we have provided evidence that selected genes are affected by the level of CDX2 protein (Fig. 6). This makes us confident that many of the CDX2 ChIP-seq-predicted target genes will prove to be bona fide CDX2 targets. In the conditional Cdx2 knock-out mice Gao et al. (30) looked at genes with an altered expression; these are not necessarily direct Cdx2 targets but could be affected by downstream effects of lacking Cdx2 in the cell, especially because Cdx2 is shown to regulate several other transcription factors, and in fact Gao et al. (30) conclude that the Cdx2−/Cdx2− intestinal cell type more resembles keratinocytes.
Conclusion
We found that the binding of CDX2 is regulated during differentiation of Caco-2 cells, with the maximum level reached at the early stage of differentiation at day 4 after confluence. Our results show that low amounts of active CDX2 present in undifferentiated cells seem to be bound to few targets; when differentiation initiates, then more CDX2 protein is able to bind to its DNA targets. This result is also observed in our CDX2 knockdown strain where the regulatory DNA elements containing strong CDX2 binding sites are able to retain CDX2 binding, whereas the weaker CDX2 binding sites lose the CDX2 binding. An explanation of these observations could be that the genes bound by CDX2 are mainly necessary for the early events of differentiation but not for the terminal differentiation, and it is also possible that CDX2 binding is needed to initiate expression of these genes but perhaps not to maintain expression during differentiation.
The information about CDX2 binding to DNA elements, either directly or in a transcription factor complex, is the first step in examining the regulatory role of CDX2 as the transcriptional regulation is dependent on CDX2 being located at the DNA. The effect of the bound CDX2 is obviously not examined by the ChIP method because the binding of CDX2 is not the same as measuring the effect on transcription. To examine the effect of CDX2 binding on expression, it is necessary to either measure the transcript of the regulated gene or place the regulatory DNA element in control of the expression of a reporter gene. Initially we chose the latter method to investigate the effect of CDX2 binding on some of the regulatory DNA elements. The 2-fold reduction in luciferase activity observed in most cases is an indication of a medium dependence on CDX2. When a more dramatic effect of the CDX2 knockdown is observed on some regulatory DNA elements, then these elements are more strongly dependent on CDX2 for transcriptional activity. To further study the effect of CDX2 on the expression of potential target genes, we examined the endogenous transcript levels of selected target genes in cells with either normal or reduced CDX2 protein levels. These analyses confirmed that CDX2 mainly functions as a positive regulator of transcription. Another interesting finding is that the effect of CDX2 on the two regulatory elements of HNF4α appears to be most pronounced on the enhancer rather than the promoter, whereas the CDX2 autoregulation on the contrary seems to be equally influenced by the CDX2 activity on both the promoter and enhancer element.
Supplementary Material
Acknowledgments
We thank Lotte Laustsen and Pernille Smith for excellent technical assistance. This project was an activity under the Cell Biology Cluster at the Panum Institute, University of Copenhagen.
This work was supported by the Novo Nordisk Foundation, the Lundbeck Foundation, and the Danish Research Council.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Data S1–S4.
- HNF
- hepatocyte nuclear factor
- ChIP-seq
- ChIP with next generation sequencing
- qPCR
- quantitative PCR
- HEPH
- hephaestin
- AXIN2
- axis inhibitor 2
- LGALS8
- Lectin, galactoside-binding, soluble, 8
- CDK
- cyclin-dependent kinase.
REFERENCES
- 1.Marshman E., Booth C., Potten C. S. (2002) Bioessays 24, 91–98 [DOI] [PubMed] [Google Scholar]
- 2.Pinto D., Gregorieff A., Begthel H., Clevers H. (2003) Genes Dev. 17, 1709–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fre S., Huyghe M., Mourikis P., Robine S., Louvard D., Artavanis-Tsakonas S. (2005) Nature 435, 964–968 [DOI] [PubMed] [Google Scholar]
- 4.Berman D. M., Karhadkar S. S., Maitra A., Montes De Oca R., Gerstenblith M. R., Briggs K., Parker A. R., Shimada Y., Eshleman J. R., Watkins D. N., Beachy P. A. (2003) Nature 425, 846–851 [DOI] [PubMed] [Google Scholar]
- 5.Hardwick J. C., Van Den Brink G. R., Bleuming S. A., Ballester I., Van Den Brande J. M., Keller J. J., Offerhaus G. J., Van Deventer S. J., Peppelenbosch M. P. (2004) Gastroenterology 126, 111–121 [DOI] [PubMed] [Google Scholar]
- 6.Laprise P., Chailler P., Houde M., Beaulieu J. F., Boucher M. J., Rivard N. (2002) J. Biol. Chem. 277, 8226–8234 [DOI] [PubMed] [Google Scholar]
- 7.Boudreau F., Rings E. H., van Wering H. M., Kim R. K., Swain G. P., Krasinski S. D., Moffett J., Grand R. J., Suh E. R., Traber P. G. (2002) J. Biol. Chem. 277, 31909–31917 [DOI] [PubMed] [Google Scholar]
- 8.Gautier-Stein A., Domon-Dell C., Calon A., Bady I., Freund J. N., Mithieux G., Rajas F. (2003) Nucleic Acids Res. 31, 5238–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jedlicka P., Sui X., Sussel L., Gutierrez-Hartmann A. (2009) Am. J. Pathol. 174, 1280–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krasinski S. D., Van Wering H. M., Tannemaat M. R., Grand R. J. (2001) Am. J. Physiol. Gastrointest. Liver Physiol. 281, G69–G84 [DOI] [PubMed] [Google Scholar]
- 11.Sauvaget D., Chauffeton V., Citadelle D., Chatelet F. P., Cywiner-Golenzer C., Chambaz J., Pinçon-Raymond M., Cardot P., Le Beyec J., Ribeiro A. (2002) J. Biol. Chem. 277, 34540–34548 [DOI] [PubMed] [Google Scholar]
- 12.Boyd M., Bressendorff S., Møller J., Olsen J., Troelsen J. T. (2009) BMC Gastroenterol. 9, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stegmann A., Hansen M., Wang Y., Larsen J. B., Lund L. R., Ritié L., Nicholson J. K., Quistorff B., Simon-Assmann P., Troelsen J. T., Olsen J. (2006) Physiol. Genomics 27, 141–155 [DOI] [PubMed] [Google Scholar]
- 14.Balakrishnan A., Stearns A. T., Rhoads D. B., Ashley S. W., Tavakkolizadeh A. (2008) Surgery 144, 168–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Escaffit F., Boudreau F., Beaulieu J. F. (2005) J. Cell. Physiol. 203, 15–26 [DOI] [PubMed] [Google Scholar]
- 16.Fang R., Olds L. C., Sibley E. (2006) Gene Expr. Patterns 6, 426–432 [DOI] [PubMed] [Google Scholar]
- 17.Jonckheere N., Vincent A., Perrais M., Ducourouble M. P., Male A. K., Aubert J. P., Pigny P., Carraway K. L., Freund J. N., Renes I. B., Van Seuningen I. (2007) J. Biol. Chem. 282, 22638–22650 [DOI] [PubMed] [Google Scholar]
- 18.Mitchelmore C., Troelsen J. T., Spodsberg N., Sjöström H., Norén O. (2000) Biochem. J. 346, 529–535 [PMC free article] [PubMed] [Google Scholar]
- 19.Valente A. J., Zhou Q., Lu Z., He W., Qiang M., Ma W., Li G., Wang L., Banfi B., Steger K., Krause K. H., Clark R. A., Li S. (2008) Free Radic. Biol. Med. 44, 430–443 [DOI] [PubMed] [Google Scholar]
- 20.Wang L., Klopot A., Freund J. N., Dowling L. N., Krasinski S. D., Fleet J. C. (2004) Am. J. Physiol. Gastrointest. Liver Physiol. 287, G943–G953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.James R., Kazenwadel J. (1991) J. Biol. Chem. 266, 3246–3251 [PubMed] [Google Scholar]
- 22.Suh E., Chen L., Taylor J., Traber P. G. (1994) Mol. Cell. Biol. 14, 7340–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benahmed F., Gross I., Gaunt S. J., Beck F., Jehan F., Domon-Dell C., Martin E., Kedinger M., Freund J. N., Duluc I. (2008) Gastroenterology 135, 1238–1247 [DOI] [PubMed] [Google Scholar]
- 24.Silberg D. G., Swain G. P., Suh E. R., Traber P. G. (2000) Gastroenterology 119, 961–971 [DOI] [PubMed] [Google Scholar]
- 25.Houde M., Laprise P., Jean D., Blais M., Asselin C., Rivard N. (2001) J. Biol. Chem. 276, 21885–21894 [DOI] [PubMed] [Google Scholar]
- 26.Rings E. H., Boudreau F., Taylor J. K., Moffett J., Suh E. R., Traber P. G. (2001) Gastroenterology 121, 1437–1450 [DOI] [PubMed] [Google Scholar]
- 27.Boulanger J., Vézina A., Mongrain S., Boudreau F., Perreault N., Auclair B. A., Lainé J., Asselin C., Rivard N. (2005) J. Biol. Chem. 280, 18095–18107 [DOI] [PubMed] [Google Scholar]
- 28.Suh E., Traber P. G. (1996) Mol. Cell. Biol. 16, 619–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chawengsaksophak K., James R., Hammond V. E., Köntgen F., Beck F. (1997) Nature 386, 84–87 [DOI] [PubMed] [Google Scholar]
- 30.Gao N., White P., Kaestner K. H. (2009) Dev. Cell 16, 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schöler H. R., Hatzopoulos A. K., Balling R., Suzuki N., Gruss P. (1989) EMBO J. 8, 2543–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji H., Jiang H., Ma W., Johnson D. S., Myers R. M., Wong W. H. (2008) Nat. Biotechnol. 26, 1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frank S. R., Schroeder M., Fernandez P., Taubert S., Amati B. (2001) Genes Dev. 15, 2069–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schjoldager K. T., Maltesen H. R., Balmer S., Lund L. R., Claesson M. H., Sjöström H., Troelsen J. T., Olsen J. (2008) Am. J. Physiol. Gastrointest. Liver Physiol. 294, G1335–G1343 [DOI] [PubMed] [Google Scholar]
- 35.Spandidos A., Wang X., Wang H., Seed B. (2010) Nucleic Acids Res. 38, D792–D799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Troelsen J. T., Mitchelmore C., Olsen J. (2003) Gene 305, 101–111 [DOI] [PubMed] [Google Scholar]
- 37.Traber P. G., Wu G. D., Wang W. (1992) Mol. Cell. Biol. 12, 3614–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Troelsen J. T., Olsen J., Møller J., Sjöström H. (2003) Gastroenterology 125, 1686–1694 [DOI] [PubMed] [Google Scholar]
- 39.Shimizu T., Bae Y. K., Hibi M. (2006) Development 133, 4709–4719 [DOI] [PubMed] [Google Scholar]
- 40.Hinoi T., Gesina G., Akyol A., Kuick R., Hanash S., Giordano T. J., Gruber S. B., Fearon E. R. (2005) Gastroenterology 128, 946–961 [DOI] [PubMed] [Google Scholar]
- 41.Vulpe C. D., Kuo Y. M., Murphy T. L., Cowley L., Askwith C., Libina N., Gitschier J., Anderson G. J. (1999) Nat. Genet. 21, 195–199 [DOI] [PubMed] [Google Scholar]
- 42.Xu F., Li H., Jin T. (1999) J. Biol. Chem. 274, 34310–34316 [DOI] [PubMed] [Google Scholar]
- 43.Garrison W. D., Battle M. A., Yang C., Kaestner K. H., Sladek F. M., Duncan S. A. (2006) Gastroenterology 130, 1207–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lussier C. R., Babeu J. P., Auclair B. A., Perreault N., Boudreau F. (2008) Am. J. Physiol. Gastrointest. Liver Physiol. 294, G418–G428 [DOI] [PubMed] [Google Scholar]
- 45.Guo R. J., Suh E. R., Lynch J. P. (2004) Cancer Biol. Ther. 3, 593–601 [DOI] [PubMed] [Google Scholar]
- 46.Bonhomme C., Calon A., Martin E., Robine S., Neuville A., Kedinger M., Domon-Dell C., Duluc I., Freund J. N. (2008) Oncogene 27, 4497–4502 [DOI] [PubMed] [Google Scholar]
- 47.Barbacci E., Reber M., Ott M. O., Breillat C., Huetz F., Cereghini S. (1999) Development 126, 4795–4805 [DOI] [PubMed] [Google Scholar]
- 48.Li R., Pei H., Watson D. K. (2000) Oncogene 19, 6514–6523 [DOI] [PubMed] [Google Scholar]
- 49.Baillat D., Laitem C., Leprivier G., Margerin C., Aumercier M. (2009) Biochem. Biophys. Res. Commun. 378, 213–217 [DOI] [PubMed] [Google Scholar]
- 50.Seth A., Watson D. K. (2005) Eur. J. Cancer 41, 2462–2478 [DOI] [PubMed] [Google Scholar]
- 51.Korinek V., Barker N., Moerer P., van Donselaar E., Huls G., Peters P. J., Clevers H. (1998) Nat. Genet. 19, 379–383 [DOI] [PubMed] [Google Scholar]
- 52.Polakis P. (2000) Genes Dev. 14, 1837–1851 [PubMed] [Google Scholar]
- 53.Behrens J., Jerchow B. A., Würtele M., Grimm J., Asbrand C., Wirtz R., Kühl M., Wedlich D., Birchmeier W. (1998) Science 280, 596–599 [DOI] [PubMed] [Google Scholar]
- 54.Mai M., Qian C., Yokomizo A., Smith D. I., Liu W. G. (1999) Genomics 55, 341–344 [DOI] [PubMed] [Google Scholar]
- 55.Tevosian S. G., Shih H. H., Mendelson K. G., Sheppard K. A., Paulson K. E., Yee A. S. (1997) Genes Dev. 11, 383–396 [DOI] [PubMed] [Google Scholar]
- 56.Sampson E. M., Haque Z. K., Ku M. C., Tevosian S. G., Albanese C., Pestell R. G., Paulson K. E., Yee A. S. (2001) EMBO J. 20, 4500–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barondes S. H., Cooper D. N., Gitt M. A., Leffler H. (1994) J. Biol. Chem. 269, 20807–20810 [PubMed] [Google Scholar]
- 58.Perillo N. L., Pace K. E., Seilhamer J. J., Baum L. G. (1995) Nature 378, 736–739 [DOI] [PubMed] [Google Scholar]
- 59.Hadari Y. R., Arbel-Goren R., Levy Y., Amsterdam A., Alon R., Zakut R., Zick Y. (2000) J. Cell Sci. 113, 2385–2397 [DOI] [PubMed] [Google Scholar]
- 60.Arbel-Goren R., Levy Y., Ronen D., Zick Y. (2005) J. Biol. Chem. 280, 19105–19114 [DOI] [PubMed] [Google Scholar]
- 61.Okuda T., Hirai H., Valentine V. A., Shurtleff S. A., Kidd V. J., Lahti J. M., Sherr C. J., Downing J. R. (1995) Genomics 29, 623–630 [DOI] [PubMed] [Google Scholar]
- 62.Sherr C. J., Roberts J. M. (1999) Genes Dev. 13, 1501–1512 [DOI] [PubMed] [Google Scholar]
- 63.Guo R. J., Funakoshi S., Lee H. H., Kong J., Lynch J. P. (2010) Carcinogenesis 31, 159–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tachibana I., Hemler M. E. (1999) J. Cell Biol. 146, 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okochi H., Mine T., Nashiro K., Suzuki J., Fujita T., Furue M. (1999) J. Clin. Gastroenterol. 29, 63–67 [DOI] [PubMed] [Google Scholar]
- 66.Bertenshaw G. P., Turk B. E., Hubbard S. J., Matters G. L., Bylander J. E., Crisman J. M., Cantley L. C., Bond J. S. (2001) J. Biol. Chem. 276, 13248–13255 [DOI] [PubMed] [Google Scholar]
- 67.Lottaz D., Maurer C. A., Hahn D., Büchler M. W., Sterchi E. E. (1999) Cancer Res. 59, 1127–1133 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.