Abstract
The process by which the human immunodeficiency virus type 1 (HIV-1) conical core dissociates is called uncoating, but not much is known about this process. Here, we show that the uncoating process requires the interaction of the capsid (CA) protein with the peptidyl-prolyl isomerase Pin1 that specifically recognizes the phosphorylated serine/threonine residue followed by proline. We found that the HIV-1 core is composed of some isoforms of the CA protein with different isoelectric points, and one isoform is preferentially phosphorylated in the Ser16-Pro17 motif. The mutant virus S16A/P17A shows a severely attenuated HIV-1 replication and an impaired reverse transcription. The S16A/P17A change increased the amount of particulate CA cores in the cytosol of target cells and correlated with the restriction of HIV-1 infection. Glutathione S-transferase pulldown assays demonstrated a direct interaction between Pin1 and the HIV-1 core via the Ser16-Pro17 motif. Suppression of Pin1 expression by RNA interference in a target cell results in an attenuated HIV-1 replication and increases the amount of particulate CA cores in the cytosol of target cells. Furthermore, heat-inactivated, inhibitor-treated, or W34A/K63A Pin1 causes an attenuated in vitro uncoating of the HIV-1 core. The Pin1-dependent uncoating is inhibited by antisera raised against a CA peptide phosphorylated at Ser16 or treatment of the HIV-1 core with alkaline phosphatase. These findings provide insights into this obscure uncoating process in the HIV-1 life cycle and a new cellular target for HIV-1 drug development.
Keywords: HIV, Prolyl Isomerase, Protein Phosphorylation, Protein Stability, Protein-Protein Interactions, Capsid Core, Capsid Protein, Core Disassembly, Uncoating
Introduction
The HIV-14 CA protein must assemble properly and should be sufficiently stable to protect the genomic RNA but must disassemble to allow genomic RNA release after fusion. Because both the assembly and disassembly processes cannot act on the CA proteins at the same time, an important question concerns the mechanisms responsible for the switch from the assembly to disassembly mode. However, not much has been known about the disassembly process so far because this process has been assumed to occur immediately after fusion (1, 2).
Recent studies suggested that cell factors may assist the disassembly process, commonly referred to as uncoating (3, 4). Cellular activation has long been considered a requirement for HIV-1 infection of CD4+ T cells, because HIV-1 fails to infect resting CD4+ T cells from peripheral blood. Infection is aborted either during reverse transcription (5) or before nuclear import of the viral preintegration complex (6). It was shown that HIV-1 can enter resting CD4+ T cells efficiently, but only short incomplete reverse transcripts are formed. Auewarakul et al. (3) demonstrated that the uncoating of HIV-1 is efficiently induced by lysate from activated CD4+ lymphocytes, whereas resting CD4+ lymphocyte lysate cannot uncoat the HIV-1 core. Furthermore, Warrilow et al. (4) reported that endogenous reverse transcription is inefficient in comparison with reverse transcription in cell infection, as exemplified by the poor efficiency of the production of intravirion late reverse transcription products. This observation also suggests a requirement for a contribution from the host cell environment for optimal HIV-1 reverse transcription to proceed by an ordered disassembly of the HIV-1 CA core. These findings suggest that the uncoating activity that releases the CA protein from the HIV-1 core is due to cellular factors.
We previously reported that the HIV-1 core is composed of some isoforms of the CA protein with different isoelectric points (7). The difference in isoelectric points among CA protein isoforms might be due to multiple post-translational modifications, such as phosphorylation. Indeed, several reports have provided evidence that the CA protein is predominantly phosphorylated at serine residues rather than threonine/tyrosine residues (8, 9). Phosphorylation often plays an important role in modulating protein activity and protein-protein interactions and promoting the disassembly of cellular macromolecular assembly such as the nuclear pore complex. Protein kinases catalyze post-translational phosphorylation, and many kinases are themselves regulated by phosphorylation. The phosphorylation-dependent modulation of protein-protein interactions is the pivotal molecular switch for the regulation of signaling processes within eukaryotic cells. Furthermore, current studies point toward mitotic phosphorylation of nucleoporins as the trigger of the nuclear pore complex disassembly process (10–12). These findings led to a hypothesis that phosphorylation induces the dissociation of CA-CA protein interactions and promotes the disassembly of the HIV-1 core.
In this study, we show that the serine residue in the Ser16-Pro17 motif of a CA protein isoform is phosphorylated and that peptidyl-prolyl isomerase Pin1 specifically recognizes the Ser16-Pro17 motif of the HIV-1 core. Furthermore, we demonstrated that Pin1 prolyl isomerase activity is required for the disassembly of the HIV-1 core. These results reveal a novel regulation step of HIV-1 infection.
EXPERIMENTAL PROCEDURES
Virus Preparation
HIV-1LAV-1 and HIV-1JRFL were prepared as previously described (7, 13). The supernatant from the culture medium of CEM/LAV-1 or CEM-CCR5/JRFL was filtered through a 0.22-μm-pore size disposable filter and then centrifuged at 43,000 × g for 3 h at 4 °C. The obtained pellet was resuspended in PBS(−) (0.02% KH2PO4, 0.29% Na2HPO4·12H2O, 0.8% NaCl, 0.02% KCl) and then centrifuged at 100,000 × g for 1 h at 4 °C. The resulting pellet was resuspended in 10 mm Tris-HCl buffer (pH 8.0). Virions were treated with or without 1 mg/ml subtilisin (ICN Biomedicals Inc., Costa Mesa, CA) in 10 mm Tris-HCl (pH 8.0) and 1 mm EDTA-2Na. The virions were then purified using PBS(−) by column chromatography with a 10-ml column of Sepharose CL-4BTM at room temperature in a biosafety hood. Fractions containing the virus were pooled and centrifuged at 100,000 × g for 1 h at 4 °C. The resulting pellet derived from HIV-1LAV-1 or HIV-1JRFL was boiled for 1 min and lysed in 200 μl of lysis buffer (9.5 m urea, 2% (w/v) Nonidet P-40, 2% ampholine (pH 6–8), and 5% 2-mercaptoethanol). For the MAGIC-5 assay and quantitative analysis of HIV-1 reverse transcription, pNL4-3-based mutants and replication-defective viruses were prepared as follows. The HIV-1 proviral DNA constructs pNL4-3 (14), containing full-length HIV-1, and pNL4-3Δenv, in which there is a truncation in env, were the control viruses utilized in this study. The 1136-bp fragment containing the BssHII and SpeI sites of pNL4-3 or pNL4-3Δenv was subcloned into pT7Blue (Novagen, Inc., Madison, WI). Point mutations were introduced into the Ser16-Pro17 motif in the HIVNL4–3 or HIVNL4–3Δenv CA protein by site-directed mutagenesis. The fragments were sequenced to confirm mutations. The mutant BssHII-SpeI fragment was recloned into the full-length HIV expression plasmid pNL4-3BssHII-SpeI or pNL4-3ΔenvBssHII-SpeI, which does not have the backbone BssHII-SpeI fragment. All of the mutations were verified by sequencing. The mutant viruses (HIVNL4–3(S16A/P17A), HIVNL4–3ΔenvWT, and HIVNL4–3Δenv(S16A/P17A)) were produced by the transient transfection of HEK293 cells using pNL4-3(S16A/P17A) or pNL4-3Δenv and pNL4-3Δenv(S16A/P17A) with the pNL4-3-based env expression vector. Four days after transfection, the virus-containing supernatant was collected and clarified by filtration through 0.45-μm-pore size filters. Virus production and release were monitored by measuring virion-associated RT activity as previously described (7).
Peptide Mass Fingerprinting
An aliquot of the viral lysate was subjected to two-dimensional polyacrylamide gel electrophoresis in accordance with the method of O'Farrell (15). The sample was loaded using a 15-cm immobilized pH gradient tube gel (pH 6–8) (Daiichi Pure Chemicals Co., Ltd.). The proteins were focused for 1 h at 400 V followed by a step gradient (1000 V for 18 h; 2000 V for 1 h). After the second-dimensional run (10–20% PAG large gel; Daiichi Pure Chemicals Co., Ltd.), the gel was stained with Coomassie Brilliant Blue (CBB) R-250 or ProQ diamond (Invitrogen). CBB-stained gel pieces were excised from the gel and immersed in 100 μl of acetonitrile, dried under vacuum centrifugation, rehydrated in 25 μl of trypsin solution (3.9 ng/μl trypsin in 170 mm ammonium bicarbonate, 40% H218O, 30% acetonitrile) or 25 μl of Glu-C solution (10 ng/μl Glu-C in 55 mm NaH2PO4 and 40% H218O), and then incubated for 60 min. The unabsorbed enzyme solution was removed, and the gel pieces were incubated for 12 h at 37 °C. The digested peptides were purified using ZipTip (Nihon Millipore Ltd., Tokyo, Japan) and then analyzed by MALDI-TOF mass spectrometry as previously described (7). Phosphorylated peptides were analyzed as follows. Alkaline phosphatase (Wako Pure Chemical Industries, Ltd.) liberated the phosphate group from phosphorylated peptides. Alkaline phosphatase reactions were carried out directly on the MALDI target. The sample was mixed with 12 ng/ml ALP from Escherichia coli (Wako Pure Chemical Industries) that had been dissolved in 10 mm ammonium bicarbonate. The reactions were carried out for 5 min at room temperature followed by the addition of 0.5 μl of α-cyano-4-cinnamic acid or 2,5-dihydroxybenzoic acid saturated in 40% acetonitrile, 0.1% trifluoroacetic acid.
Preparation of HIV-1 p24-derived Peptide-Keyhole Limpet Hemocyanin (KLH) Conjugates and Antibody Screening
A p24-derived linear decapeptide (pSP, H2N-HQAIpSPRTLN-COOH) was synthesized using an automatic peptide synthesizer. The molecular mass was determined by MALDI-TOF mass spectrometry (Burker Franzen Analytik). The amino group of His1 was coupled to KLH (pSP-KLH) through bis(sulfosuccinimidyl)suberate (Thermo Fisher Scientific Inc.). Ten BALB/c mice were immunized intraperitoneally with 200 μg of pSP-KLH in Freund's adjuvant at 1-week intervals and administered an intravenous boost of 40 μg of pSP-KLH 3 days prior to splenectomy. The blood samples were collected prior to euthanasia and centrifuged at 1500 × g for 5 min, and the anti-pS16/P17 serum was harvested. Then the mice were euthanized, and their spleens were removed for in vitro hybridoma cell production. The hybridomas were generated by a standard method, by which splenocytes were fused with P3U1 cells and selected in hypoxanthine-, aminopterin-, and thymidine-supplemented media. In the screening, supernatants were tested for reactivity to ELISA plates coated with pSP. The hybridoma that produced high titers of anti-pS16/P17 mAb (3/D-6) was then cloned.
Preparation of HIV-1 CA Cores and Detection of Phosphorylated CA Protein in the HIV Core
The WT HIV CA cores were purified in accordance with the method of Auewarakul et al. (3) with some modifications. Briefly, 3 ml of purified cell-free viruses was overlayered on a discontinuous sucrose density gradient. The gradient was composed of 1 ml of 50% sucrose at the bottom and 1 ml of 10% sucrose containing 0.1% Igepal CA-630 on the top. The overlayered gradient was then centrifuged at 100,000 × g in a Swing Rotor P65ST (HitachiKoki Co., Ltd.) for 2 h at 4 °C. The pellets containing viral cores were then resuspended in the uncoating assay buffer (50 mm NaH2PO4, pH 7.2, 150 mm NaCl, and 200 μm sodium orthovanadate) and pooled. The CA cores were quantitated by measuring HIV-1 p24 antigen with RETRO-TEK HIV-1 p24 antigen ELISA (ZeptoMetrix Corp.). To examine the Ser16 phosphorylation of CA core, the pellets containing viral cores were then lysed with 62.5 mm Tris-HCl, 2% SDS, 10% glycerol (pH 6.8), and 10% 2-mercaptoethanol. The lysate from HIV cores was subjected to SDS-PAGE, and separated products were transferred to a polyvinylidene difluoride membrane for Western blotting. The membrane was stained with normal mouse serum and anti-pS16/P17 serum raised against pSP-KLH with or without pSP.
Viral Infectivity Assay
Jurkat and CEM cells (1 × 105) were exposed to the HEK293 cell-derived WT virus or S16A/P17A variant (100 ng of p24 antigen) for 2 h at 37 °C, washed with PBS(−), and cultured for 96 h. Virus infectivity was monitored by measurement of the p24 antigen from cell culture supernatants using RETRO-TEK HIV-1 p24 antigen ELISA (ZeptoMetrix Corp.).
Viral Entry Assay and Quantitative Analysis of HIV-1 Reverse Transcription during Acute Infection
The entry level of WT or mutant viruses was determined as follow. Viral entry assay was performed using the protocol of Kawano et al. (16) with modifications. MAGIC-5 cells (1 × 106 cells) were exposed to the virus (213.9 ng of HIV-1 RT) at 37 °C for 2 h. The cells were washed with PBS(−) and then incubated for 5 min at 37 °C in PBS(−) containing 0.25% trypsin. After trypsinization, the cells were washed twice with PBS(−) and pelleted. Cell-associated CA antigen content was determined by Western immunoblot analysis using an anti-p24 mAb (Immuno Diagnostics, Inc.). Furthermore, de novo synthesized HIV-1 cDNA was analyzed with or without Pin1 siRNA treatment in accordance with the method of Ikeda et al. (17).
Electron Microscopy of Virus
A viral preparation was fixed with 0.1 m sodium phosphate buffer (pH 7.4) containing 2% paraformaldehyde and 2.5% glutaraldehyde at 4 °C for 2 h prior to postfixation with 1% osmium tetroxide for 30 min at 4 °C. After dehydration in ethanol, the specimens were embedded in epoxy resin.
MAGIC-5 Assay and Cytopathic Effect (CPE) Assay
Viral infectivity was determined using MAGIC-5 cells as previously described in Ref. 18. MAGIC-5 cells (1 × 104 cells) were incubated in suspensions of WT, mutant, or single-round replication-defective viruses with 20 μg/ml DEAE dextran for 2 h and then cultured in a medium (200 μl) for 48 h. The cells were fixed, and HIV-1-infected cells identified by their blue staining were counted by conventional methods. To further examine whether a decrease in endogenous Pin1 expression level decreases viral infectivity, both MAGIC-5 assay and CPE assay were performed. 60 nm small interfering RNA against Pin1 (Pin1 C1 sense siRNA, GCCAUUUGAAGACGCCUCGdTdT; Pin1 C1 antisense siRNA, GAGGCGUCUUCAAAUGGCdTdT; Pin1 C2 sense siRNA, CUGGCCUCACAGUUCAGCGdTdT; Pin1 C2 antisense siRNA, CGCUGAACUGUGAGGCCAGdTdT; Pin1 C3 sense siRNA, GCCGAGUGUACUACUUCAAdTdT; or Pin1 C3 antisense siRNA, UUGAAGUAGUACACUCGGCdTdT) or control siRNA (sense siRNA against GL2 Luciferase, CGUACGCGGAAUACUUCGAdTdT; or antisense siRNA, UCGAAGUAUUCCGCGUACGdTdT) was transfected into the MAGIC-5 cells (1 × 104 or 2 × 105 cells) using Lipofectamine 2000. The cells were infected with HIV-1LAV-1 (12 ng of p24 antigen). HIV-1LAV-1-infected cells identified by their blue staining were counted by conventional methods. In CPE assay, the cells were infected with a single-round replication-defective HIV-1 (HIVNL4–3ΔenvWT, 11 ng of HIV RT).
Fate-of-Capsid Assay
Fate-of-capsid assay was performed in accordance with the method of Sodroski and co-workers (19) with some modifications. Briefly, recombinant HIV-1 virus-like particles (WT HIV-1(VSV-G) and S16A/P17A HIV-1(VSV-G)) were produced by cotransfection of 293T cells with pHCMV-G and pNL4-3Δenv or pNL4-3Δenv carrying the S16A/P17A mutation (pNL4-3Δenv(S16A/P17A)) as described in Ref. 20. To examine whether the infectivity defect of the S16A/P17A variant is linked to a dysfunction of uncoating, MAGIC-5 (4.5 × 106) cells were seeded in 60-cm2 flasks and the next day incubated with either 5 ml of WT HIV-1(VSV-G) or 5 ml of S16A/P17A HIV-1(VSV-G) incubated at 37 °C for 4 h after 30 min of incubation at 4 °C. On the other hand, to examine whether Pin1 depletion is linked to a dysfunction of uncoating, MAGIC-5 cells (4.0 × 106) were transfected with Pin1 C1 and C2 siRNA (600 nm) or control siRNA (600 nm) using NeonTM transfection system (Invitrogen) and incubated for 44 h. The cells were further incubated with 5 ml of WT HIV-1(VSV-G) at 37 °C for 4 h after 30 min of incubation at 4 °C. These cells were washed three times with ice-cold PBS and detached by incubating with 2 ml of proteinase K (7 mg/ml in Dulbecco's modified Eagle's medium) for 5 min at 4 °C. The cells were washed once in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and twice in PBS. The pellets were resuspended in 2 ml of hypotonic lysis buffer (10 mm Tris-HCl, pH 8.0, 10 mm KCl, and 1 mm EDTA) and placed on ice for 15 min. The cells were lysed using a 7-ml Dounce homogenizer and 15 strokes with pestle B. Cell debris was removed by centrifugation for 3 min at 2,000 × g. After centrifugation, 1 ml of lysate was layered onto a 3.5-ml of 50% sucrose cushion (made in PBS) and centrifuged at 125,000 × g for 2 h at 4 °C in a Swing Rotor P65ST (HitachiKoki Co., Ltd). After centrifugation, 150 μl from the topmost part of the supernatant was collected and made 1× in SDS sample buffer. The pellets were resuspended in 10 μl of 1× SDS sample buffer. The samples were subjected to SDS-PAGE and Western blotting for CA proteins.
Determination of Pin1-CA Interaction
The interaction between Pin1 and the CA core was determined by GST-Pin1 pulldown assay. Recombinant Pin1 proteins were produced as N-terminal GST-tagged proteins as previously described (21). The CA core was treated with ALP (Wako Pure Chemical Industries) or heat-denatured ALP for 2 h (final concentration, 0.024 μg/μl ALP) in a buffer containing 50 mm NaH2PO4 (pH 7.2), 150 mm NaCl, 200 μm sodium orthovanadate, 10% glycerol, and 1% Triton X-100 and then incubated with 30 μl of beads containing GST or GST-Pin1 proteins for 2 h at 4 °C. Immunoblotting analysis was performed using an anti-p24 mAb (Immuno Diagnostics, Inc.). Furthermore. a fraction of the ALP-treated CA core was subjected to Western blot analysis using an anti-pS16/P17 mAb raised against a synthetic phosphoserine-containing peptide, pSP, or an anti-p24 mAb.
Suppression of Pin1 Expression by siRNA
MAGIC-5 cells were transfected with Pin1 C3 siRNA (30–120 nm) or control siRNA (30–120 nm). The cells were infected with the WT virus and harvested 2 h after infection. To confirm the level of Pin1 expression, immunoblot analysis was carried out using the chemiluminescence protocol. The mouse monoclonal and goat polyclonal antibodies used were as follows: anti-Pin1 (R & D Systems, Inc.), anti-actin (OncogeneTM research product), and anti-lactate dehydrogenase (Chemicon) antibodies.
Uncoating Assay
The WT HIV CA core was purified as described above. The purified CA cores (2 ng) were resuspended in the uncoating assay buffer (50 mm NaH2PO4, pH 7.2, 150 mm NaCl, 200 μm sodium orthovanadate) and incubated with 0.114 μg/μl GST-Pin1, 0.114 μg/μl heat-denatured GST-Pin1, 0.114 μg/μl GST-Pin1 treated with juglone (molar ratio of GST-Pin1:juglone = 1:10; juglone was preincubated with GST-Pin1 at 37 °C for 1 h and then removed by gel filtration) or 0.114 μg/μl GST-Pin1(W34A/K63A) at 37 °C for 1 h. To examine whether the extent of CA disassembly was enhanced by Pin1, the pellets from the uncoating reactions were subjected to Western blot analysis using the anti-p24 mAb. Furthermore, a fraction of purified CA cores was pretreated with anti-pS16/P17 sera or normal sera. Another fraction of purified CA cores was treated with ALP (final concentration, 0.345 μg/μl) or heat-denatured ALP (final concentration, 0.345 μg/μl). The resulting CA cores were incubated with 0.114 μg/μl GST-Pin1 at 37 °C for 1 h. Each core suspension was then diluted in 5 ml of uncoating assay buffer and centrifuged at 100,000 × g for 2 h at 4 °C. The extent of CA release is quantified by p24 ELISA (ZeptoMetrix Corporation; the detection limit for the p24 antigen by ELISA was 7.8 pg/ml as provided by the manufacturer) because the undissociated cores were pelleted, and the dissociated CA protein was in the supernatant after ultracentrifugation. The relative percentage of disassembly efficiency (RPDE) is given by RPDE (%) = 100 × (CAS/CAT)/(CASC/CATC), where CAS is the concentration of the CA protein in the supernatant, CAT is the concentration of total CA protein in the supernatant plus pellet in the disassembly experiments of the CA core using boiled, juglone-treated GST-Pin1, or GST-Pin1(W34A/K63A); CASC is the concentration of the CA protein in the supernatant; and CATC is the concentration of total CA protein in the supernatant plus pellet in the control experiment using GST-Pin1. Likewise, the RPDE of CA cores pretreated with anti-pS16/P17 sera or ALP is given by RPDE (%) = 100 × (CAS/CAT)/(CASC/CATC), where CAS is the concentration of CA protein in the supernatant; CAT is the concentration of total CA protein in the supernatant plus pellet in the disassembly experiments of CA core pretreated with anti-pS16/P17 sera or ALP using the GST-Pin1; CASC is the concentration of CA protein in the supernatant; and CATC is the concentration of total CA protein in the supernatant plus pellet in the control experiments of CA core pretreated with normal mouse sera or heat-denatured ALP using the GST-Pin1.
RESULTS
A CA Protein Isoform Is Selectively Phosphorylated at Serine Residue 16 in the Ser16-Pro17 Motif and Incorporated into the HIV Core
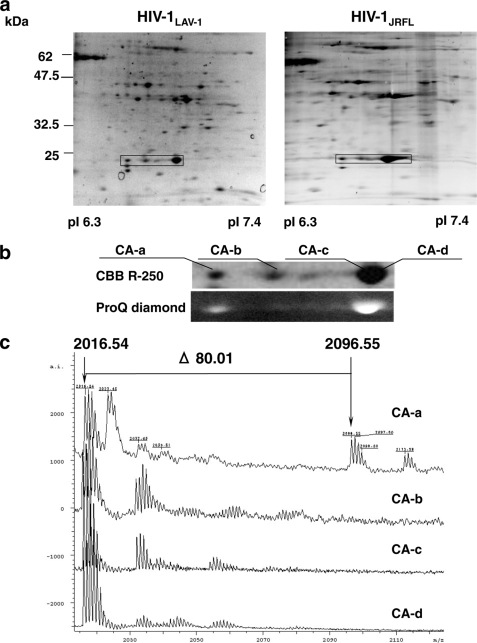
Some isoforms of the HIV-1 CA protein associated with CXCR4-tropic HIV-1LAV-1 or CCR5-tropic HIV-1JRFL were identified by proteomics using two-dimensional gel electrophoresis and MALDI-TOF mass spectrometry (Fig. 1a and supplemental Table S1). We found that four isoforms of the HIV-1LAV-1 CA protein (CA-a (pI 6.70), CA-b (pI 6.80), CA-c (pI 6.91), and CA-d (pI 6.95)) are inside the virion (Fig. 1b). The differences between the pI values of the four isoforms of the CA protein may be due to post-translational modifications, such as phosphorylation and formylation (8, 9, 22, 23). Indeed, the CA proteins are composed of phosphorylated and unphosphorylated (Fig. 1b, lower panel) and formylated isoforms (23). Peptide mass fingerprinting data were used to identify the probable co/post-translational modifications. Then the specific phosphorylation of the N-terminal tryptic peptide (1PIVQNIQGQMVHQAISPR18) of CA-a was estimated because the spectrum corresponding to the N-terminal tryptic peptide exhibited a peak at m/z 2016.54 and the mass spectrum corresponding to the phosphorylated form exhibited a peak at m/z 2096.55 (Fig. 1c). The phosphorylation was confirmed by the liberation of a phosphate group from the N-terminal peptide of CA-a. The peak of [M+H]+ at m/z 2016.50 corresponded to the peptide (1PIVQNIQGQMVHQAISPR18) whose phosphate group was cleaved off by an alkaline phosphatase (ALP) (Fig. 1d). These results show that the serine residue in the Ser16-Pro17 motif of CA-a is selectively phosphorylated. To further determine whether CA-a is actually present in the CA core, the WT CA core was purified in accordance with the method of Auewarakul et al. (3) and was subjected to Western blot analysis using anti-pS16/P17 serum with or without pSP. As shown in Fig. 1e, CA-a is actually incorporated into the CA core.
FIGURE 1.
A serine residue in the Ser16-Pro17 motif of a CA isoform is selectively phosphorylated. a, CBB R-250-stained two-dimensional gel images of HIV-1LAV-1 and HIV-1JRFL. The rectangles indicate CA isoforms in virions. b, CBB R-250 and ProQ diamond staining of CA isoforms of HIV-1LAV-1. c, spectra of N-terminal tryptic peptides derived from CA isoforms of HIV-1LAV-1. The peak of [M+H]+ at m/z 2096.55 corresponds to the phosphorylated form of the N-terminal tryptic peptide Pro1-Arg18 of CA-a. d, liberation of phosphate group from N-terminal phosphorylated Pro1-Arg18 derived from CA-a using alkaline phosphatase. The lower and upper panels show samples treated and not treated with alkaline phosphatase, respectively. The phosphorylated Pro1-Arg18 disappeared after the phosphatase treatment. e, incorporation of phosphorylated CA-a into HIV core. The purified HIV core is subjected to Western immunoblot analysis using normal sera or anti-pS16/P17 serum with or without pSP, H2N-HQAIpSPRTLN-COOH.
Mutation at the Ser16-Pro17 Motif Affects Uncoating Process
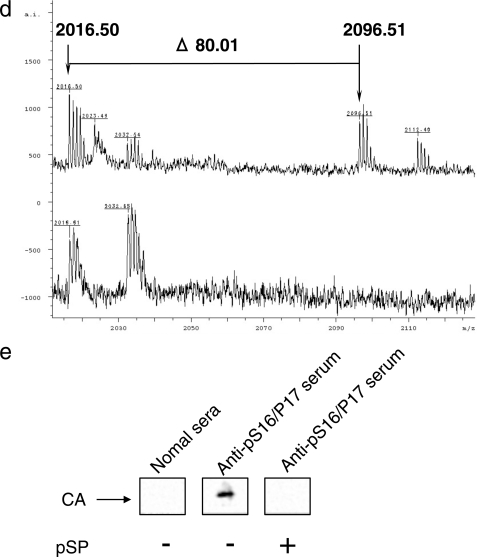
To study the role of the phosphorylated Ser16 that precedes Pro, we replaced Ser16 with glutamic acid (S16E) or alanine (S16A) to mimic phosphorylated or unphosphorylated Ser, respectively. We then tested the infectivity of the mutants by MAGIC-5 assay (21). The results showed that the S16E variant had the same infectivity as the wild type (Fig. 2a, WT). In contrast, the S16A variant had a decreased infectivity (Fig. 2a, p < 0.01). Assuming that glutamic acid mimics the negative charge of phosphorylated Ser16 in only a fraction of the CA protein in a viral core, the results obtained for S16E or S16A suggest that phosphorylation at Ser16 plays a critical role in maintaining viral infectivity. To examine whether the phenotype of the S16A variant was specific to the selected amino acid changes, we produced another variant with a phosphorylatable amino acid residue, changing Ser to Thr (S16T). The S16T variant showed a remarkably decreased infectivity compared with the WT virus (Fig. 2a, p < 0.01), suggesting that the Ser16 residue in the highly conserved Ser16-Pro17 motif (supplemental Fig. S1) plays an important role in maintaining the viral infectivity. To further investigate why Ser16 in a fraction of the CA protein in the virion needs to be phosphorylated, we generated another variant (S16A/P17A) because single-point mutations of Ser16 in the Ser16-Pro17 motif may provide an alternate binding site for conventional peptidyl-prolyl cis-trans isomerases, such as cyclophilins, or FK-506-binding proteins, which catalyze the cis-trans isomerization of Xaa-Pro peptide bonds (where Xaa is the preceding amino acid) in oligopeptides or proteins. The S16A/P17A variant also had a markedly decreased infectivity in MAGIC-5 cells (Fig. 2a, p < 0.01) and could not replicate efficiently in T cell lines, Jurkat cells, and CEM cells (Fig. 2b), although the core yield of the S16A/P17A variant from HEK293 cells was very similar to that of WT HIV-1, and the phenotypes of the CA core were normal (Fig. 2c). Furthermore, to determine whether the lack of infectivity of S16A/P17A was due to a defect in the reverse transcription process, we carried out a viral entry assay and quantitative real time PCR analysis using primers designed to specifically amplify early or late products of reverse transcription. The viral entry assay indicated that the entry levels of S16A/P17A and WT were the same because the amounts of S16A/P17A CA protein in subcellular fractions were very similar to that of WT CA protein (Fig. 2d). However, compared with WT, S16A/P17A produced the same level of the early (R/U5) form of the viral cDNA product (Fig. 2e) but showed a significant decrease in the level of the late (R/gag) form of the viral cDNA product (Fig. 2f, p < 0.01). To further examine whether the infectivity defect of the S16A/P17A variant is linked to a dysfunction of uncoating, cell-based fate-of-capsid uncoating assay developed by Sodroski's group (19) was performed. This assay allows us to measure the amounts of cores and soluble free forms of the CA protein present in the cytosol of infected cells. Moreover, the appearance of CA cores in the cytosol inversely correlates with the proper conversion of CA cores to soluble CA in target cells. The amounts of pelletable CA cores in the cells infected with the S16A/P17A mutant virus increased against those detected in the cells infected with WT virus (Fig. 2g, lower panel, Pellet). Furthermore, the levels of the CA protein in the supernatants from MAGIC-5 cells incubated with the S16A/P17A mutant virus were lower than those detected in the cells infected with WT virus (Fig. 2g, upper panel, Sup.). These results suggest that the infectivity defect of the S16A/P17A variant is linked to a dysfunction of the uncoating process.
FIGURE 2.
The S16A/P17A CA mutant shows impaired uncoating process. a, effect of various mutations in Ser16-Pro17 motif on viral infectivity. Viral infectivity was determined using MAGIC-5 cells as previously described in Ref. 18. MAGIC-5 cells (1 × 104 cells) were exposed to an equal amount of WT or mutant viruses (corresponding to 3 ng RT). Statistical analysis using the analysis of variance test was performed. The error bars denote the standard deviation. b, replication kinetics of S16A/P17A variant in Jurkat and CEM cells. c, effect of alanine mutation of Ser16-Pro17 motif on viral core phenotype. The arrows indicate electron-dense cores typical of intact HIV. d, effect of alanine mutation of Ser16-Pro17 motif on viral entry efficiency. Viral entry assay was performed as described under “Experimental Procedures.” e and f, quantitative analysis of HIV-1 reverse transcription during acute infection using early stage primers (e, R/U5) and late stage primers (f, R/gag). Briefly, MAGIC-5 cells (1 × 106 cells) were exposed to WT or S16A/P17A virus (2.5 ng of HIV-1 RT) and harvested at 1 day postinfection. The cells were washed with PBS(−), and nucleic acids were extracted. Quantitative analysis was performed using the same primers in accordance with the method of Ikeda et al. (17). The significant differences (unpaired t test) are indicated as follows: *, p < 0.01; n.s., not significant. The error bars denote the standard deviation. g, effect of Ser16-Pro17 motif on fate of CA core in infected cells. MAGIC-5 cells were incubated with WT HIV-1(VSV-G) or S16A/P17A HIV-1(VSV-G) at 4 °C for 30 min and then at 37 °C for 4 h. The cell lysates were analyzed as described under “Experimental Procedures.” The supernatants and pellets were Western blotted for CA proteins.
Pin1 within the Target Cell Affects an Early Phase in HIV-1 Replication
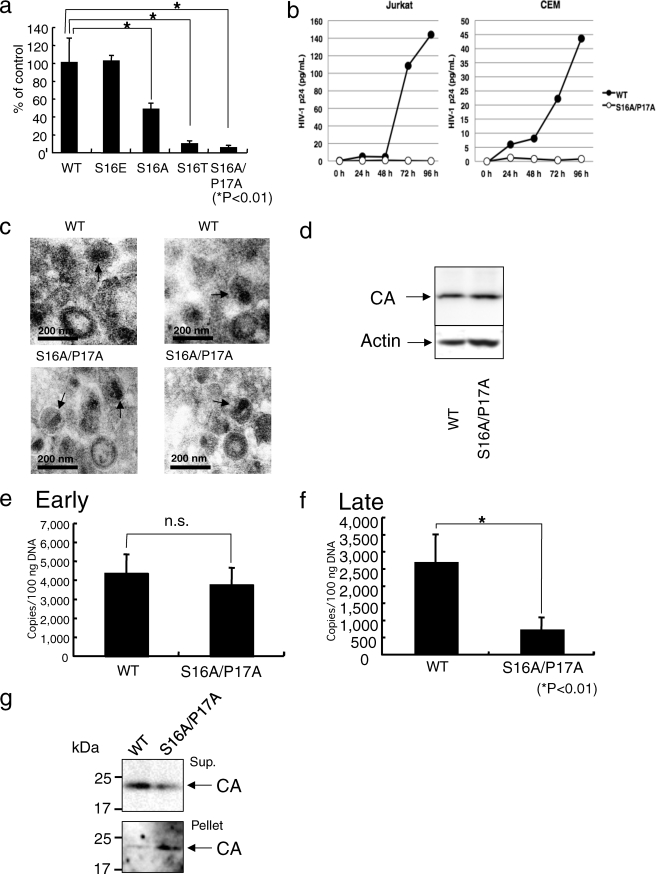
Pin1 is a peptidyl-prolyl isomerase that specifically recognizes phosphorylated serine/threonine followed by proline via its WW domain and then catalyzes a conformational change of the bound substrate in a phosphorylation-dependent manner (24, 25). Pin1 has been shown to regulate the stability and localization of its substrates during transcriptional activation, cell cycle progression, and cell death, and a deregulated expression or loss of function of Pin1 leads to the progression of serious human diseases such as cancer and Alzheimer disease (24, 26–28). We therefore considered that Pin1 might bind to and regulate the function of the CA core. We first investigated whether Pin1 directly binds to the CA core by GST-Pin1 pulldown assay. Glutathione beads containing GST or GST-Pin1 were incubated with the CA core purified in accordance with the method described under “Experimental Procedures.” To demonstrate that cores were indeed successfully purified, we performed a comparative immunoblot analysis of core preparations. As expected, the membrane-associated matrix (MA) protein (supplemental Fig. S2a) and the HIV surface glycoprotein gp120 (supplemental Fig. S2b) were substantially depleted in the core preparation. These results indicated that CA core is sufficiently purified. Therefore, in Pin1 pulldown assay, the binding proteins were then analyzed by immunoblotting with an anti-CA antibody. Fig. 3a shows that GST-Pin1, but not GST, was bound to the CA core. To investigate whether the binding of the CA core is directly due to the phosphorylation of Ser16 in the CA core, WT core particles were treated with ALP or the heat-denatured ALP. The treatment removed phosphate groups from phosphorylated Ser16 (Fig. 3b) and eliminated the Pin1 binding activity of the CA core (Fig. 3c). There are three highly conserved Ser-Pro motifs (Ser16-Pro17, Ser33-Pro34, and Ser146-Pro147) and a conserved Thr48-Pro49 motif in the CA protein (supplemental Fig. S1). To examine whether Ser33, Thr48, or Ser146 in CA core is phosphorylated, the mixture of CA protein isoforms were subjected to proteome analysis (supplemental Fig. S3). The data demonstrated that Ser33, Thr48, and Ser146 in CA core were not phosphorylated. These results suggested that Pin1 binds preferentially to the phosphorylated Ser16-Pro17 motif.
FIGURE 3.
Interactions of Pin1 with the CA core and Pin1 within the target cells regulate HIV-1 infectivity. a, the CA core was subjected to GST-Pin1 pulldown assay and immunoblotted with anti-p24 mAb (upper panel). CBB staining of GST and GST-Pin1 (lower panel). b, treatment of CA core with ALP or heat-denatured ALP. The ALP- and heat-denatured ALP-treated CA cores were subjected to Western immunoblot analysis using anti-pS16/P17 mAb or anti-p24 mAb. The treatment removed phosphate groups from phosphorylated Ser16. c, effect of ALP on interaction between Pin and CA core. The ALP-treated CA core was subjected to GST-Pin1 pulldown assay and immunoblotted with anti-p24 mAb (upper panel). CBB staining of GST-Pin1 is shown (lower panel). d and e, the suppression of Pin1 expression by 60 nm Pin1 C3 siRNA (d, upper panel, the expressions of actin and lactate dehydrogenase (LDH) were not affected by Pin1-specific siRNA)) in MAGIC-5 cells (1 × 104 cells) results in an attenuated HIV-1LAV-1 replication (e). The significant differences (unpaired t test) are indicated as follows: *, p < 0.01. The error bars denote the standard deviation. f–i, the effects of Pin1-specific siRNAs were evaluated on the basis of CPE observations. The cells (2 × 105 cells) were pretreated with 60 nm control siRNA (f), Pin1 C1 (g), Pin1 C1+C2 (h), and Pin1 C3 (i) and then infected with single-round replication-defective HIV-1 (HIVNL4–3ΔenvWT). Pin1-specific siRNAs were capable of inhibiting CPE induced by HIV infection. j, effect of Pin1 siRNAs on reverse transcription. The knockdown of Pin1 showed that there was a significant decrease in the level of the late (R/gag) form of the viral cDNA product. Statistical analysis using the analysis of variance test was performed. The error bars denote the standard deviation. N.D., not detected. The detection limit of late products of HIV-1 reverse transcription was 400 copies/reaction in the assay. k, Pin1 C1+C2-induced suppression of Pin1 expression. MAGIC-5 cells were transfected with Pin1 C1+C2 siRNA (600 nm) using the NeonTM transfection system (upper panel, the expression of actin was not affected by Pin1 C1+C2). l, effect of Pin1 depletion on fate of CA core in infected cells. Pin1 knockdown cells treated with Pin1 C1+C2 siRNA were incubated with WT HIV-1(VSV-G) at 4 °C for 30 min and then at 37 °C for 4 h. The cell lysates were analyzed as described under “Experimental Procedures.” The supernatants and pellets were analyzed by Western blotting for the CA proteins.
Because Pin1 has CA core binding activity, we next determined whether a decrease in endogenous Pin1 expression level decreases HIV-1 infectivity. We transfected MAGIC-5 cells with siRNA against Pin1 or control siRNA. As shown in the top panel of Fig. 3d, Pin1 siRNA (Pin1 C3) specifically decreased the Pin1 expression level in MAGIC-5 cells without altering actin or lactate dehydrogenase expression level. The cells were infected with HIV-1LAV-1, and β-galactosidase activity was measured as an indicator of infection. In Fig. 3e, 60 nm Pin1 siRNA (Pin1 C3)-treated MAGIC-5 cells were infected with HIV-1LAV-1 (12 ng of p24 antigen) under the condition in which no HIV-1LAV-1-induced CPE was observed in 60 nm control siRNA-treated MAGIC-5 cells. Interestingly, a decrease in endogenous Pin1 expression level decreased HIV-1LAV-1 infectivity (Fig. 3e, p < 0.01). Furthermore, the effects of Pin1-targeted siRNA treatment on HIV-1 replication were evaluated on the basis of cytopathic effect (y) observations (Fig. 3, f–i) and quantitative real time PCR analysis using primers designed to specifically amplify late products of reverse transcription. 60 nm Pin1 C1-, C1+C2-, or C3-treated MAGIC-5 cells was infected with a single-round replication-defective HIV-1 under the condition in which a single-round replication-defective HIV-1-induced CPE was observed in 60 nm control siRNA-treated MAGIC-5 cells (Fig. 3, f–i). After treatment with siRNAs, MAGIC-5 cells were infected with single-round replication-defective HIVNL4–3ΔenvWT (11 ng of HIV RT). CPE in infected cells treated with the control siRNA was observed at 46 h postinfection (Fig. 3f). On the other hand, CPE was hardly observed in cells treated with 60 nm Pin1 C1, C1+C2, or C3 (Fig. 3, g–i, respectively). This indicates that Pin1-targeted siRNAs are capable of inhibiting CPE induced by HIV infection. Furthermore, Fig. 3j shows that there was a significant decrease in the level of the late (R/gag) form of the viral cDNA product.
To further examine whether Pin1 depletion is linked to a dysfunction of uncoating, cell-based fate-of-capsid uncoating assay was performed. The amounts of pelletable CA cores in the Pin1 knockdown cells infected with WT HIV-1(VSV-G) increased against those detected in the control siRNA-treated cells infected with WT HIV-1(VSV-G) (Fig. 3, k and l, lower panels, Pellet). Furthermore, the levels of the CA protein in the supernatants from the Pin1 knockdown cells infected with WT HIV-1(VSV-G) were lower than those detected in the control siRNA-treated cells infected with the WT HIV-1(VSV-G) (Fig. 3l, upper panel, Sup.). These results indicate that Pin1 depletion is linked to a dysfunction of uncoating.
Pin1 Induces Disassembly of the CA Core
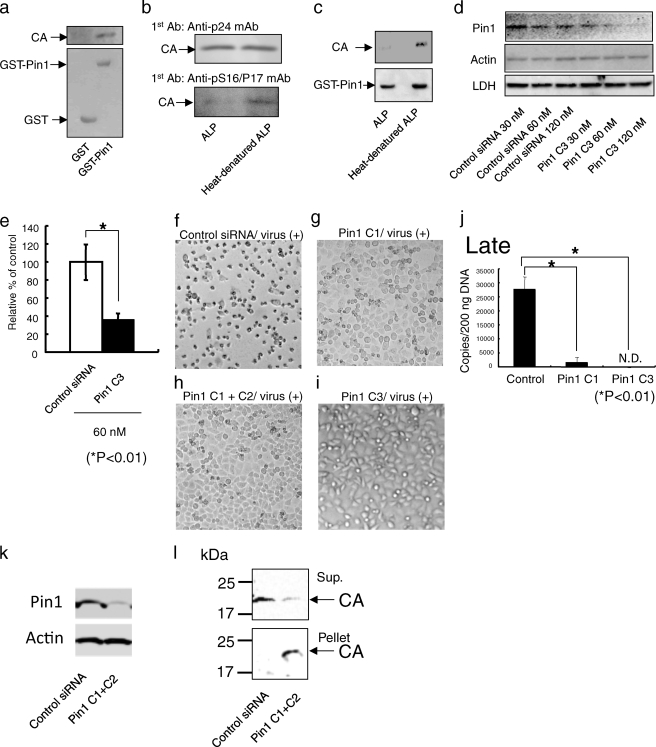
Pin1 might regulate the uncoating of the CA core. To test this possibility, we attempted to study this process in vitro by detecting the disassembly of the CA core in the presence of Pin1. Purified CA cores were diluted with an uncoating assay buffer and incubated with GST-Pin1, heat-denatured, inhibitor-treated, or W34A/K63A GST-Pin1. Following incubation, the CA cores were subjected to ultracentrifugation to separate the free CA protein from the intact CA core. CA core dissociation was found to be GST-Pin1-dependent (Fig. 4a, lane 2), and the effect was attenuated when Pin1 was denatured (Fig. 4b, p < 0.01) or treated with the Pin1 inhibitor juglone (Fig. 4c, p < 0.01) and catalytically inactived the W34A/K63A Pin1 mutant (Fig. 4d, p < 0.01). Furthermore, the uncoating assay demonstrated that the Pin1-dependent disassembly was competed with the anti-pS16/P17 serum (Fig. 4e, p < 0.01) and blocked by the pretreatment of the core with ALP (Fig. 4e, p < 0.01). These results support the idea that Pin1 facilitates the HIV-1 uncoating by interacting with the CA core.
FIGURE 4.
In vitro uncoating of CA core by GST-Pin1. a, in vitro uncoating assay was performed as described under “Experimental Procedures.” The extent of uncoating was visualized by Western blot analysis using anti-p24 mAb after the undisassembled cores have been pelleted in an ultracentrifuge. CA cores in the presence of GST-Pin1 disassembled in vitro (lane 2). The quantities of the CA protein in the pellets in the presence of uncoating assay buffer were similar to that of the input CA protein (lanes 1 and 3). b, after incubating HIV-1 cores with GST-Pin1, heat-denatured GST-Pin1, or uncoating assay buffer for 1 h, the relative percentage of disassembly efficiency is given by the equation shown under “Experimental Procedures.” Pin1 induces disassembly of the CA cores. c, in vitro uncoating activities after incubating HIV-1 cores with juglone-treated Pin1 for 1 h. The treatment of GST-Pin1 with juglone results in an attenuated in vitro uncoating. d, effect of GST-Pin1 (W34A/K63A) on uncoating. In vitro uncoating activities after incubating GST-Pin1 (W34A/K63A) with CA cores for 1 h. e, in vitro competitive uncoating assay. The pretreatment of CA cores with anti-pS16/p17 serum results in an attenuated in vitro uncoating. f, in vitro uncoating activities after incubating HIV CA cores with ALP for 2 h. Pin1-dependent disassembly was blocked by the pretreatment of the cores with ALP. b–f, statistical analysis using the analysis of variance test was performed. The error bars denote the standard deviation. The significant differences are indicated as follows: *, p < 0.01.
DISCUSSION
Much is known about the structure of the HIV-1 CA protein (29–31), but the mechanism and requirement of HIV-1 uncoating in postentry events have remained poorly defined. However, recent studies showed that the HIV-1 uncoating activity, which releases the CA protein from HIV-1 cores and is required for the activation of reverse transcriptase, is due to cellular factors (3, 4). The studies suggest that the ordered uncoating process of HIV-1 is not spontaneous and is regulated by cellular factors. Therefore, we employed a series of biochemical approaches to clarify the mechanism of HIV-1 CA core uncoating.
In this study, we showed that Pin1 assists the uncoating of the HIV core. Proteome analysis suggests that the Ser residue in the Ser16-Pro17 motif of CA-a is preferentially phosphorylated (Fig. 1, c and d). Interestingly, three Ser-Pro motifs (Ser16-Pro17, Ser33-Pro34, and Ser146-Pro147) are highly conserved across HIV-1 clades (supplemental Fig. S1), and only the Ser16-Pro17 motif is conserved among HIV-1, HIV-2, and simian immunodeficiency virus (SIV) (supplemental Fig. S4). As shown in Figs. 2g and 3l, cell-based fate-of-capsid uncoating assay demonstrated that the infectivity defect of the S16A/P17A variant is linked to a dysfunction of the uncoating process, and Pin1 depletion in the target cells is linked to a dysfunction of uncoating. Furthermore, GST-Pin1 pulldown assay and in vitro uncoating assay indicated that Pin1 facilitates HIV-1 uncoating by interacting with the CA core through the Ser16-Pro17 motif (Figs. 3, b and c, and 4, e and f). These results suggest that only a fraction of the CA protein phosphorylated at Ser16-Pro17 motif serves as a molecular switch to signal CA core uncoating because both the assembly and disassembly pathways cannot act on the CA core at the same time.
To investigate how the motif of CA-a is phosphorylated, Western immunoblot analysis with monoclonal anti-pS16/P17 mAb was carried out, which demonstrated that Ser16 was phosphorylated inside HIV-1LAV-1 virions but not inside CEM/LAV-1 cells, although the specific kinase that phosphorylates Ser16 is currently unknown (supplemental Fig. S5). As shown in Fig. 3e, a decrease in endogenous Pin1 expression level decreases HIV-1 infectivity. These findings suggest that the CA core phosphorylated inside the viral particle after release from the infected cells associates directly with Pin1 after entry into host cells for the uncoating process. A better understanding of these findings is crucial not only for elucidating the uncoating process at the early stage of the HIV-1 life cycle but also for identifying new anti-HIV drugs.
Supplementary Material
Acknowledgments
We thank A. Muneoka (Shin Nippon Biomedical Laboratories, Ltd.) for the technical support during electron microscopy. We thank Dr. R. Swanstrom (Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill) for support in preparing pNL4-3Δenv(S16A/P17A) with the pNL4-3-based env expression vector and helpful discussions.
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and a Health Science Research Grant from the Ministry of Health, Labour, and Welfare of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S5.
- HIV-1
- human immunodeficiency virus type 1
- CA
- capsid
- GST
- glutathione S-transferase
- CPE
- cytopathic effect
- ALP
- alkaline phosphatase
- RT
- reverse transcriptase
- PBS
- phosphate-buffered saline
- CBB
- Coomassie Brilliant Blue
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- KLH
- keyhole limpet hemocyanin
- ELISA
- enzyme-linked immunosorbent assay
- WT
- wild type
- mAb
- monoclonal antibody
- siRNA
- small interfering RNA
- RPDE
- relative percentage of disassembly efficiency
- MA
- matrix
- SIV
- simian immunodeficiency virus.
REFERENCES
- 1.Goff S. P. (2001) J. Gene Med. 3, 517–528 [DOI] [PubMed] [Google Scholar]
- 2.Dvorin J. D., Malim M. H. (2003) Curr. Top. Microbiol. Immunol. 281, 179–208 [DOI] [PubMed] [Google Scholar]
- 3.Auewarakul P., Wacharapornin P., Srichatrapimuk S., Chutipongtanate S., Puthavathana P. (2005) Virology 337, 93–101 [DOI] [PubMed] [Google Scholar]
- 4.Warrilow D., Meredith L., Davis A., Burrell C., Li P., Harrich D. (2008) J. Virol. 82, 1425–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zack J. A., Arrigo S. J., Weitsman S. R., Go A. S., Haislip A., Chen I. S. (1990) Cell 61, 213–222 [DOI] [PubMed] [Google Scholar]
- 6.Stevenson M., Stanwick T. L., Dempsey M. P., Lamonica C. A. (1990) EMBO J. 9, 1551–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misumi S., Fuchigami T., Takamune N., Takahashi I., Takama M., Shoji S. (2002) J. Virol. 76, 10000–10008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Veronese F. D., Copeland T. D., Oroszlan S., Gallo R. C., Sarngadharan M. G. (1988) J. Virol. 62, 795–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mervis R. J., Ahmad N., Lillehoj E. P., Raum M. G., Salazar F. H., Chan H. W., Venkatesan S. (1988) J. Virol. 62, 3993–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macaulay C., Meier E., Forbes D. J. (1995) J. Biol. Chem. 270, 254–262 [DOI] [PubMed] [Google Scholar]
- 11.Favreau C., Worman H. J., Wozniak R. W., Frappier T., Courvalin J. C. (1996) Biochemistry 35, 8035–8044 [DOI] [PubMed] [Google Scholar]
- 12.Glavy J. S., Krutchinsky A. N., Cristea I. M., Berke I. C., Boehmer T., Blobel G., Chait B. T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3811–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ott D. E., Coren L. V., Kane B. P., Busch L. K., Johnson D. G., Sowder R. C., 2nd, Chertova E. N., Arthur L. O., Henderson L. E. (1996) J. Virol. 70, 7734–7743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adachi A., Gendelman H. E., Koenig S., Folks T., Willey R., Rabson A., Martin M. A. (1986) J. Virol. 59, 284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Farrell P. H. (1975) J. Biol. Chem. 250, 4007–4021 [PMC free article] [PubMed] [Google Scholar]
- 16.Kawano Y., Tanaka Y., Misawa N., Tanaka R., Kira J. I., Kimura T., Fukushi M., Sano K., Goto T., Nakai M., Kobayashi T., Yamamoto N., Koyanagi Y. (1997) J. Virol. 71, 8456–8466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikeda T., Nishitsuji H., Zhou X., Nara N., Ohashi T., Kannagi M., Masuda T. (2004) J. Virol. 78, 11563–11573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hachiya A., Aizawa-Matsuoka S., Tanaka M., Takahashi Y., Ida S., Gatanaga H., Hirabayashi Y., Kojima A., Tatsumi M., Oka S. (2001) Antimicrob. Agents Chemother. 45, 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stremlau M., Perron M., Lee M., Li Y., Song B., Javanbakht H., Diaz-Griffero F., Anderson D. J., Sundquist W. I., Sodroski J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5514–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yee J. K., Miyanohara A., LaPorte P., Bouic K., Burns J. C., Friedmann T. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 9564–9568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen M., Stukenberg P. T., Kirschner M. W., Lu K. P. (1998) Genes Dev. 12, 706–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laurent A. G., Krust B., Rey M. A., Montagnier L., Hovanessian A. G. (1989) J. Virol. 63, 4074–4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuchigami T., Misumi S., Takamune N., Takahashi I., Takama M., Shoji S. (2002) Biochem. Biophys. Res. Commun. 293, 1107–1113 [DOI] [PubMed] [Google Scholar]
- 24.Wulf G., Finn G., Suizu F., Lu K. P. (2005) Nat. Cell Biol. 7, 435–441 [DOI] [PubMed] [Google Scholar]
- 25.Lu K. P., Hanes S. D., Hunter T. (1996) Nature 380, 544–547 [DOI] [PubMed] [Google Scholar]
- 26.Ryo A., Nakamura M., Wulf G., Liou Y. C., Lu K. P. (2001) Nat. Cell Biol. 3, 793–801 [DOI] [PubMed] [Google Scholar]
- 27.Zacchi P., Gostissa M., Uchida T., Salvagno C., Avolio F., Volinia S., Ronai Z., Blandino G., Schneider C., Del Sal G. (2002) Nature 419, 853–857 [DOI] [PubMed] [Google Scholar]
- 28.Liou Y. C., Sun A., Ryo A., Zhou X. Z., Yu Z. X., Huang H. K., Uchida T., Bronson R., Bing G., Li X., Hunter T., Lu K. P. (2003) Nature 424, 556–561 [DOI] [PubMed] [Google Scholar]
- 29.Gamble T. R., Vajdos F. F., Yoo S., Worthylake D. K., Houseweart M., Sundquist W. I., Hill C. P. (1996) Cell 87, 1285–1294 [DOI] [PubMed] [Google Scholar]
- 30.Gamble T. R., Yoo S., Vajdos F. F., von Schwedler U. K., Worthylake D. K., Wang H., McCutcheon J. P., Sundquist W. I., Hill C. P. (1997) Science 278, 849–853 [DOI] [PubMed] [Google Scholar]
- 31.Gitti R. K., Lee B. M., Walker J., Summers M. F., Yoo S., Sundquist W. I. (1996) Science 273, 231–235 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.