Abstract
Frameshift mutations are particularly deleterious to protein function and play a prominent role in carcinogenesis. Most commonly these mutations involve the insertion or omission of a single nucleotide by a DNA polymerase that slips on a damaged or undamaged template. The mismatch DNA repair pathway can repair these nascent polymerase errors. However, overexpression of enzymes of the base excision repair (BER) pathway is known to increase the frequency of frameshift mutations suggesting competition between these pathways. We have examined the fate of DNA containing single nucleotide bulges in human cell extracts and discovered that several deaminated or alkylated nucleotides are efficiently removed by BER. Because single nucleotide bulges are more highly exposed we anticipate that they would be highly susceptible to spontaneous DNA damage. As a model for this, we have shown that chloroacetaldehyde reacts more than 18-fold faster with an A-bulge than with a stable A·T base pair to create alkylated DNA adducts that can be removed by alkyladenine DNA glycosylase. Reconstitution of the BER pathway using purified components establishes that bulged DNA is efficiently processed. Single nucleotide deletion is predicted to repair +1 frameshift events, but to make −1 frameshift events permanent. Therefore, these findings suggest an additional factor contributing to the bias toward deletion mutations.
Keywords: DNA Damage, DNA Enzymes, DNA Repair, Enzyme Mechanisms, Nucleic Acid Enzymology, Alkyladenine DNA Glycosylase, Base Excision Repair, Frameshift, Glycosylase, Uracil DNA Glycosylase
Introduction
The loss or gain of one or more base pairs is one of the most common types of genetic instability (1). Single nucleotide deletions or insertions occur more frequently than larger deletions or insertions and they cause frameshift mutations if they occur within the open reading frame of a gene. Genomic instability is proposed to be a hallmark of carcinogenesis and the loss or alteration of DNA replication or repair pathways is an early step in the progression of cancer (2, 3). Normally the mismatch DNA repair (MMR)3 pathway suppresses frameshift mutations by performing replication-coupled DNA repair, but many cancer cells inactivate this pathway through mutations or changes in promoter methylation that reduce expression of one or more proteins in this pathway (4–6). However, not all cases of increased frameshift frequency can be attributed to defects in MMR (7, 8). Here we consider the possibility that other DNA repair pathways might also play a role in frameshift mutagenesis.
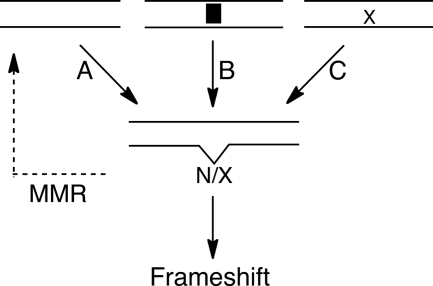
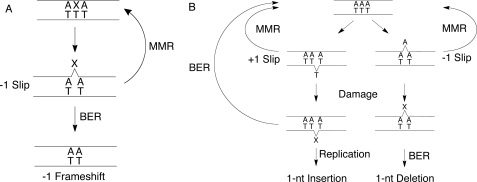
Many of the factors that influence the frequency of frameshift mutations are known (9, 10). Streisinger and co-workers (11) proposed that slipping of a primer/template pair within a polymerase active site could lead to a misaligned intermediate that would subsequently be extended to generate a bulged intermediate. Subsequent models for frameshift mutations (1, 6), including the effects of damaged templates and DNA intercalators (12, 13), have incorporated this essential feature (Fig. 1). It is generally assumed that another round of DNA replication is required to make this frameshift event permanent, with one copy that retains the original sequence and another copy that is mutant. Although both −1 and +1 frameshifts occur, human cells show a strong bias (3–6-fold) toward −1 frameshift mutations (14–16). This bias has been attributed to the intrinsic propensity for some polymerases to make more frequent −1 than +1 errors. For example, yeast polymerase δ generates a greater number of −1 slips (17). Here we have considered the possibility that alternative DNA repair pathways recognize frameshift intermediates and preferentially repair +1 events and/or make −1 events permanent.
FIGURE 1.
Polymerase slipping creates bulged intermediates that give rise to frameshift mutations. Arrows indicate DNA replication events. Slipping of polymerases generate a bulged nucleotide (N) that is less likely to be corrected by proofreading if it occurs in a homopolymeric sequence (A) (11). The probability of a polymerase slipping is increased by DNA intercalators (B) (12), or by a damaged template (C) (43, 45). Slipping on a damaged nucleotide (X) generates a damaged bulge. MMR can correct these errors, whereas an additional round of replication makes the frameshift permanent.
The base excision repair (BER) pathway is one DNA repair pathway that might compete against MMR, because the overexpression of BER enzymes result in increased frequency of point mutations and frameshift mutations, especially −1 frameshifts (18–20). The BER pathway repairs the majority of damaged bases that arise from spontaneous hydrolytic, oxidative, or alkylative reactions (for reviews, see Refs. 21–23). This pathway is initiated by glycosylases that locate sites of damage and excise the lesion base. A few of these enzymes also nick the DNA backbone via a lyase mechanism, but most release an apurinic/apyrimidinic (AP) product. The AP site is subsequently hydrolyzed by AP endonuclease 1 (APE1) to create a 3′-hydroxyl and a 5′-deoxyribose phosphate (dRP) group. Commonly the missing nucleotide is incorporated by DNA polymerase β, which also removes the 5′-dRP group via a lyase reaction. Finally, the single strand nick is ligated to complete the BER pathway. This pathway removes the damaged nucleotide and inserts a new nucleotide based upon base pairing with the opposing nucleotide. However, if the BER pathway were initiated on a single nucleotide bulge, then it would be expected to delete the bulged nucleotide.
In this study we tested the hypothesis that the human BER pathway can delete bulged nucleotides, by performing DNA repair assays in cell extracts and reconstituting the BER pathway with purified components. The results demonstrate that the BER pathway is active toward alkylated or deaminated nucleotide bulges, and confirm that the deletion of the damaged nucleotide closely follows the well characterized activities of the BER enzymes. Damaged single nucleotide bulges could arise from the slipping of a polymerase on a damaged template, or could result from spontaneous damage of an undamaged bulge. These observations suggest that BER may compete against MMR and that this competition could contribute to the bias toward single nucleotide deletions.
EXPERIMENTAL PROCEDURES
Recombinant Proteins
Human BER enzymes were expressed in Escherichia coli and purified by affinity and ion exchange chromatography. The affinity tags were removed and the purity was judged to be >95% by SDS-PAGE analysis with Coomassie staining. The purification of the catalytic domain of AAG (Δ80), lacking the first 79 amino acids (24), the catalytic domain of DNA ligase I (Δ232), lacking the first 232 amino acids (25), and the full-length APE1 have been previously described (26). Full-length DNA polymerase β was cloned into a modified pET28 vector containing amino-terminal His6 tag, followed by a TEV cleavage site. After TEV cleavage, the amino terminus is extended by two extra amino acids (GH). It was purified by a similar protocol as previously described (27). Protein concentrations were determined by the estimated extinction coefficients at 280 nm.
Oligonucleotide Substrates
DNA oligonucleotides were synthesized by commercial sources using standard phosphoramidite chemistry, and were purified and annealed as described (28). The sequence of 5′-fluorescein (FAM)-labeled oligonucleotide was 5′-FAM-CGATAGCATCCTXCCTTCTCTCCAT, in which X was either a normal nucleotide or a damaged nucleotide. The labeled oligonucleotide was annealed with a 24-mer complement to create a single nucleotide bulge, or with a 25-mer complement to create a central base pair (X·Y; supplemental Fig. S1).
Glycosylase Assay in Whole Cell Extracts
Glycosylase reactions were monitored at 37 °C in a buffer containing 50 mm NaHEPES (pH 7.5), 100 mm NaCl, 1 mm EDTA, 1 mm dithiothreitol, 0.1 mg/ml of bovine serum albumin, and 10% (v/v) glycerol. Reactions (20 μl) contained 10 nm FAM-labeled substrate and 3-μl aliquots were removed at the desired time points and quenched with 2 volumes of 0.3 m NaOH to obtain a final concentration of 0.2 m. AP sites formed from the glycosylase reaction were quantitatively converted to DNA breaks by heating at 70 °C for 15 min, followed by the addition of 3.3 volumes of formamide/EDTA loading buffer containing 0.05% (w/v) of bromphenol blue and xylene cyanol FF dyes. Samples were separated via denaturing PAGE and FAM fluorescence was quantified by imaging with a Typhoon trio (GE Healthcare) using 488 nm excitation and a 520BP40 emission filter. Fluorescence intensity of individual bands was determined with ImageQuant TL (GE Healthcare). The fraction of glycosylase product was determined by dividing the fluorescence intensity of the 12-mer product band by the sum of the 12-mer product and 25-mer substrate bands. The fraction was converted into concentration by multiplying by the total amount of DNA substrate present. Reactions were carried out in triplicate and the initial rates were measured by a linear fit. To confirm the identity of the 12-mer reaction product, we generated the authentic 12-mer from a single turnover reaction with 2 μm AAG and 10 nm I·T DNA. The reaction was quenched with NaOH after 15 min and treated as described above. Whole cell extracts (WCE) were obtained from Active Motif and were stored at −80 °C in lysis buffer (20 mm NaHEPES, pH 7.5, 350 mm NaCl, 20% glycerol, 1% Igepal-CA630, 1 mm MgCl2, 0.5 mm EDTA, 0.1 mm EGTA) until immediately before use.
Inhibition of U-bulge glycosylase activity was achieved by addition of 0.02 units of uracil glycosylase inhibitor (New England Biolabs) to 0.4 mg/ml of HeLa WCE as described above. Inhibition of the glycosylase activity toward I-bulge and ϵA-bulge bulge DNA (10 nm) was investigated by adding 100 nm unlabeled 25-mer competitor DNA that was either damaged (ϵA·T) or undamaged (A·T). To estimate the amount of AAG present in extracts, the standard glycosylase assay was performed with either 0.2 or 0.4 mg/ml of WCE, 10 nm I·T-labeled substrate, and 100 nm A·T unlabeled DNA. The glycosylase activity of T47D and HeLa toward the I·T substrate was linearly dependent on the amount of extract added (supplemental Fig. S6).
Alkylation of DNA by Chloroacetaldehyde
A coupled assay was employed to determine the susceptibility of a bulged base to alkylative damage from chloroacetaldehyde. 1 μm A·T or A-bulge oligonucleotide was incubated at 37 °C in 50 mm Na2HPO4 (pH 7.5), and 100 mm NaCl. 100 mm Chloroacetaldehyde (Sigma) was added as indicated. Reactions (4 μl) were incubated for 4 h at 37 °C, followed by the addition of 50 μl of TE buffer (10 mm Tris, pH 8.0, 1 mm EDTA) and 50 μl of hydrated diethyl ether. The organic layer was removed and a second ether extraction step was performed. DNA was precipitated by the addition of 150 μl of cold ethanol, 0.12 m sodium acetate. Samples were incubated for 30 min at −80 °C and centrifuged for 15 min. After removal of the supernatant, samples were washed with 0.5 ml of 95% ethanol and centrifuged for 10 min. The supernatant was removed and the samples were air-dried. The ϵA lesions that were formed by alkylation of the DNA were subsequently removed by AAG, which was found to have the same level of activity toward an ϵA·T mismatch as toward an ϵA-bulge (data not shown). Samples were resuspended in 20 μl of AAG reaction buffer, 50 mm NaMES (pH 6.1), 100 mm NaCl, 1 mm EDTA, 1 mm dithiothreitol, and 0.1 mg/ml of bovine serum albumin, and incubated with 2 μm AAG at 37 °C for 30 min. Samples were quenched in NaOH and analyzed by the standard glycosylase assay.
BER Assays in WCE
Assays for the processing of labeled mismatch and bulge DNA were performed at 37 °C in 50 mm NaHEPES (pH 7.5), 100 mm NaCl, 5 mm MgCl2, 4 mm ATP, and 25 μm of each dNTP. Typically 10 nm FAM-labeled mismatch or bulge substrate and 100 nm unlabeled (A·T) DNA were incubated with 0.4 mg/ml of extract (Active Motif) in a reaction volume of 20 μl. Samples were taken by quenching 3-μl aliquots into an equal volume of formamide with 20 mm EDTA, heated to 70 °C for 5 min, and separated by denaturing PAGE. AP sites remained intact during this procedure. The amount of repair that had occurred was determined by quenching reactions with EDTA (10 mm final). Recombinant AAG (2 μm) was added and reactions were incubated at 37 °C for 30 min. Finally, these reactions were quenched in 0.2 m NaOH and processed as described above to cleave abasic sites.
In Vitro BER Reconstitution
Recombinant human proteins were used to reconstitute BER activity on either an I·T mismatch or an I-bulge. Reactions were incubated at 37 °C under the standard conditions of 50 mm NaHEPES (pH 7.5), 100 mm NaCl, 5 mm MgCl2, 4 mm ATP, and 2.5 μm of each dNTP. Reactions contained 300 nm DNA substrate with 30 nm AAG, APE1, polymerase β, and ligase I, as indicated in a total volume of 20 μl. After 60 min the BER reaction was complete, at which point 3-μl aliquots were quenched in formamide/EDTA and analyzed as described above. We used this same protocol to follow the time course for multiple turnover BER activity, except that reactions contained 700 nm DNA and the putative physiological concentration (29) of APE1 (120 nm), polymerase β (20 nm), and ligase I (40 nm).
RESULTS
Glycosylase-catalyzed Excision of Bulged Nucleotides in HeLa WCE
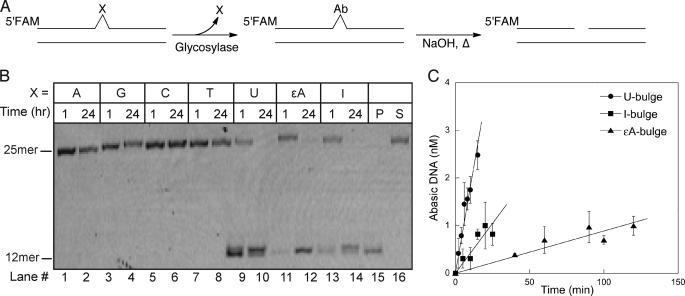
Oligonucleotide duplexes were incubated in human WCE to test whether DNA glycosylases can excise unpaired nucleotides (Fig. 2A). These oligonucleotides were 25 base pairs in length, with the central nucleotide unpaired (i.e. presented as a single nucleotide bulge). In addition to the four natural nucleotides, we also tested some common deaminated or alkylated nucleotides that are known to be present in chromosomal DNA. Deamination of C gives rise to uracil (U), deamination of A yields inosine (I), and alkylation of A can yield 1,N6-etheno-A (ϵA). Glycosylases do not require Mg2+ ions for activity, therefore we included EDTA in the glycosylase assay buffer. In the absence of Mg2+, most endonuclease and exonuclease activities were blocked and oligonucleotides remained intact for greater than 24 h (Fig. 2B).
FIGURE 2.
Human glycosylases excise alkylated and deaminated bases, but not undamaged bases, from one-nucleotide bulges. A, DNA glycosylase activity was monitored in HeLa WCE using fluorescently labeled oligonucleotides that contained single nucleotide bulges. AP products were cleaved under alkaline conditions and samples were analyzed by denaturing PAGE. B, a representative gel is shown. Undamaged (lanes 1–8) and damaged (lanes 9–14) DNA (10 nm) was incubated in 0.4 mg/ml of WCE for the indicated time. An I·T substrate was incubated in the presence or absence of recombinant AAG to provide size standards for the 25-mer intact substrate (S) and the 12-mer glycosylase-generated product (P). C, initial rates for excision of damaged nucleotides present in single nucleotide bulges were determined as described above. The mean ± S.D. is shown from 3 independent experiments.
There was no detectable glycosylase activity toward undamaged nucleotides in single nucleotide bulges in HeLa WCE (Fig. 2B, lanes 1–8). This could be due to extremely low rates of excision of natural nucleotides by human DNA glycosylases or to protection of these sites by other proteins present in the extract. In contrast, damaged nucleotides were rapidly excised (Fig. 2B, lanes 9–14). Significant excision was detected at 1 h, with complete processing of the bulge occurring within 24 h. If reactions were quenched in formamide, instead of hydroxide, the 12-mer product was not observed. This confirmed that all three damaged nucleotides were excised by monofunctional DNA glycosylases (supplemental Fig. S2). Initial rates of base excision in the HeLa extracts show that the U-bulge was most rapidly removed (Fig. 2C). The I-bulge was removed at a 4-fold slower rate, and the ϵA-bulge was removed at a 20-fold slower rate than the U-bulge. We next sought to identify the enzymes responsible for this activity on single nucleotide bulges.
Identification of DNA Glycosylases Acting at Bulged Nucleotides
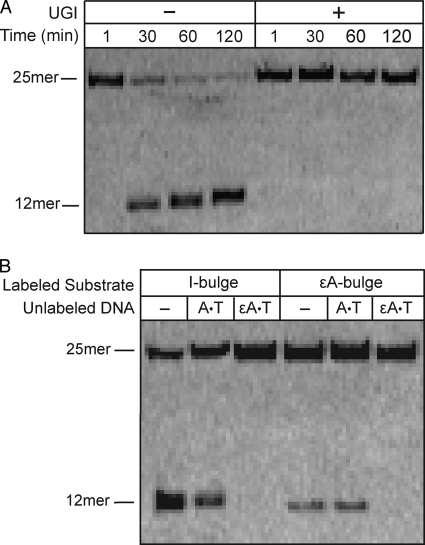
There are four human glycosylases that are known to excise U from DNA (30). The most active of these is uracil DNA glycosylase (UDG) (31). UDG is inhibited with high affinity and specificity by the bacteriophage-encoded UDG inhibitory protein (UGI), therefore we tested if UGI would block the excision of a U-bulge in HeLa WCE (32). Complete inhibition was observed, demonstrating that UDG was the glycosylase responsible for acting on a U-bulge (Fig. 3A).
FIGURE 3.
Deaminated bulged bases are excised by UDG and AAG. Glycosylase assays in HeLa cell extracts were performed as described in the legend to Fig. 2. A, U-bulge DNA was incubated in the presence or absence of UGI (0.02 units). UGI completely blocks U-bulge glycosylase activity, demonstrating that UDG is the enzyme responsible. B, fluorescently labeled I-bulge or ϵA-bulge DNA was incubated with no competitor DNA (−), undamaged DNA (A·T), or damaged DNA (ϵA·T) for 24 h. AAG is the only human glycosylase known to excise ϵA lesions, therefore the strong competition by ϵA-containing DNA suggests that AAG is responsible for both activities.
We expected that the I and ϵA-specific activities would both be attributed to alkyladenine DNA glycosylase (AAG), because this is the only human enzyme known to excise alkylated and deaminated purines from DNA (33–36). ϵA is formed by endogenous reactive species resulting from lipid peroxidation (37). Vinyl chloride exposure causes increased levels of ϵA via the P450 catalyzed production of chloroethylene oxide and chloroacetaldehyde (37, 38). AAG is the only protein found to bind ϵA·T DNA in cell extracts from human tissues (39, 40) and it binds with very high affinity (41, 42). Therefore, we tested if unlabeled ϵA·T DNA inhibits the excision of I and ϵA from single nucleotide bulges. As expected, the addition of 10-fold excess of ϵA·T decreased the excision of ϵA from the bulge to undetectable levels, whereas undamaged competitor DNA (A·T) had no effect (Fig. 3B). Similarly, the ϵA·T competitor completely blocked activity toward an I-containing single nucleotide bulge. This suggests that AAG is responsible for the excision of alkylated and deaminated purines present in a single nucleotide bulge.
Bulges containing damaged nucleotides can be formed when a DNA polymerase encounters a bulky lesion, such as ϵA, on the template strand. The Y-family polymerases are particularly prone to template slipping. For example, frequent −1 slips are observed during replication of ϵA-DNA by human polymerases μ, κ, and η (43, 44) and during replication of ϵG-DNA by Dpo4 (45, 46). However, other types of damaged nucleotides are not known to induce polymerase slipping. For these types of damage, which includes U and I, we considered the possibility that they might form subsequent to DNA replication.
Bulged Nucleotides Are More Susceptible to Damage
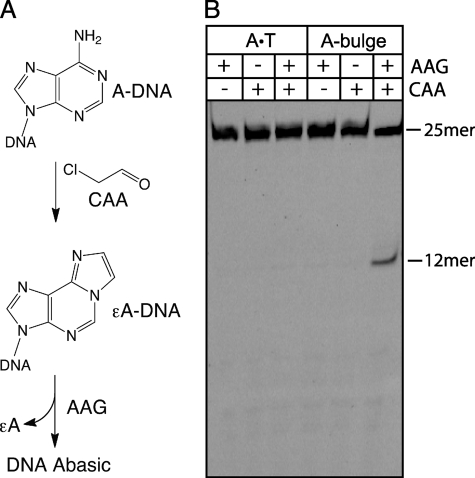
Single nucleotide bulges formed by polymerase slipping events are more exposed than Watson-Crick paired nucleotides (47, 48), suggesting that bulged nucleotides would also be damaged by oxidation and alkylation. We tested whether a bulged A is more susceptible to alkylative damage than an A·T base pair. As described above, alkylation of DNA by chloroacetaldehyde results in the formation of a variety of alkylation adducts, including ϵA (Fig. 4A). Thus, alkylation of an A-bulge would create a good substrate for AAG.
FIGURE 4.
Single nucleotide bulges are highly susceptible to alkylation damage. A, chloroacetaldehyde (CAA) reacts with A nucleotides in DNA to produce the alkylation adduct ϵA. Sites of ϵA formation were detected by the glycosylase activity of AAG. B, stably paired (A·T) or a single nucleotide A-bulge was incubated with chloroacetaldehyde (100 mm) for 4 h. After AAG-catalyzed excision of ϵA lesions and alkaline hydrolysis of AP sites, samples were analyzed by denaturing PAGE. Only the A-bulge showed detectable ϵA formation under these conditions. Quantification of duplicate experiments revealed that 18 ± 2% of the A-bulge is alkylated to form an ϵA bulge, whereas the A·T pair was not detectably alkylated (≤1%; supplemental Fig. S4). This indicates that the A-bulge is at least 18-fold more reactive than and A·T pair.
We exposed a fully paired duplex (A·T) and a single A-bulge to a high concentration of chloroacetaldehyde (100 mm) for 4 h. Excess alkylating agent was removed and the DNA was incubated with recombinant AAG in the standard glycosylase assay. Both alkylating agent and AAG were required to get the specific 12-mer product resulting from the glycosylase reaction at this bulged site (Fig. 4). Additional experiments showed that this alkylation reaction is dependent upon the concentration of chloroacetaldehyde and on the amount of time that DNA was exposed (supplemental Fig. S4). We observed that the bulged A has ≥18-fold increased reactivity relative to a paired A (Fig. 4 and supplemental Fig. S4).
Deletion of an I-bulge in Cell Extracts
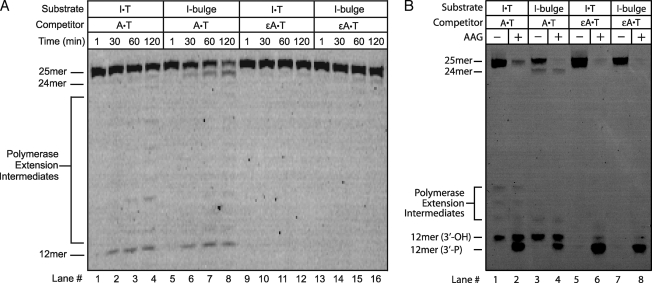
Above we demonstrated that DNA glycosylases catalyze the excision of lesions from a single nucleotide bulge to create a bulged AP site. If this intermediate can be processed by the major human APE1, then the BER pathway would be expected to cause a single nucleotide deletion. Therefore, we added the necessary cofactors to support the BER pathway, including dNTPs, ATP, and Mg2+, to HeLa WCE to test if the predicted one nucleotide deletion could be detected with either an I-bulge or a U-bulge substrate. Under these conditions, we observed a variety of BER intermediates that were consistent with the processing of these deaminated single nucleotide bulges by BER (supplemental Fig. S7). However, we observed a substantial level of 3′–5′ exonuclease activity in HeLa WCE that degraded the DNA oligonucleotides even when the BER pathway was inhibited. It should be noted that this exonuclease activity is an artifact of using short oligonucleotide substrates and is not expected to play a role when bulged nucleotides are present in chromosomal DNA. This exonucleolytic activity could be decreased by adding unlabeled competitor DNA (undamaged A·T 25-mer), presumably by providing additional DNA ends that would be substrates for the hydrolytic enzyme(s). We infer that there was insufficient glycosylase activity for the BER pathway to fully outcompete the degradation pathway(s) in HeLa WCE when short oligonucleotides are used as substrates. Therefore, we investigated several additional human cell lines in which the balance of BER and exonucleolytic degradation might differ.
Many cancer cell lines, especially breast cancers, have increased AAG expression (49, 50). We chose one of these cell lines (T47D), a ductal breast tumor, because AAG is reported to be expressed at a 3-fold higher level relative to HeLa cells (50). We first measured the level of glycosylase activity toward an I·T mismatch in HeLa and T47D WCE and confirmed the 3-fold higher level of AAG activity in T47D (supplemental Fig. S5). We next examined the repair of I·T and I-bulge DNA in T47D extracts under the BER conditions. The I·T mismatch was processed to build up a nicked DNA intermediate (Fig. 5A). The presence of the repaired 25-mer product was confirmed by treating it with excess recombinant AAG and alkaline cleavage, which fully cleaved any remaining lesion-containing DNA (Fig. 5B, lane 2). In the case of the I-bulge, a new 24-mer product was formed (Fig. 5A, lanes 5–8) that was similarly resistant to cleavage by recombinant AAG (Fig. 5B, lane 4). This suggests that WCE are capable of catalyzing a single nucleotide deletion with an I-bulge on a similar time scale to repair of an I·T mismatch. When ϵA·T competitor was added there was no repair of I·T or I-bulge (Fig. 5B, lanes 5–8). We also noticed small amounts of polymerase extension products under these conditions. These products were greatly reduced by aphidicolin, an inhibitor of polymerase α, δ, and ϵ, and did not occur if dNTPs were omitted from the reaction (supplemental Figs. S10 and S11).
FIGURE 5.
Base excision repair assays in T47D WCE demonstrate single nucleotide deletion. A, conditions were similar to those described in the legend to Fig. 2, but 5 mm MgCl2, 1 mm ATP, and 0.1 mm of each dNTP were added to support BER activity. The expected single nucleotide deletion product (24-mer) was detected within 30 min for the I-bulge substrate (lanes 5–8). The I·T mismatch was processed on the same time scale (lanes 1–4), as evidenced by the build-up of BER intermediates (12-mer). No BER intermediates or deletion products were observed when ϵA·T-containing unlabeled competitor DNA was included in the incubation (lanes 9–16). B, determination of extent of repair for reactions shown in A. After 120 min samples were split and either incubated with recombinant AAG (+AAG) to process the remaining I lesions, or immediately quenched in formamide/EDTA (−AAG). Alkaline hydrolysis of the abasic site occurs via β,δ-elimination to generate a 3′-phosphate (3′-P), making it possible to quantify the amount of I lesion present after incubation in WCE. In contrast, the APE1-catalyzed reaction in the extract yields a 3′-hydroxyl (3′-OH). The persistent 25-mer in lane 2 and 24-mer in lane 4 confirms that the I lesion was removed.
We also tested two additional cell lines that overexpress AAG (49, 50), a second breast ductal carcinoma (MCF-7 (51)) and a colon adenocarcinoma (HT-29 (52)). Incubation of the I-bulge substrate under BER conditions revealed a high level of AAG activity in both cell lines (supplemental Figs. S8 and S9). In addition to the 12-mer nicked BER intermediate, a 24-mer single nucleotide deletion product could be detected that was above the background of 3′–5′ exonucleolytic degradation. These observations indicate that extracts from cancer cells that have different origins share the ability to delete a single nucleotide I-bulge.
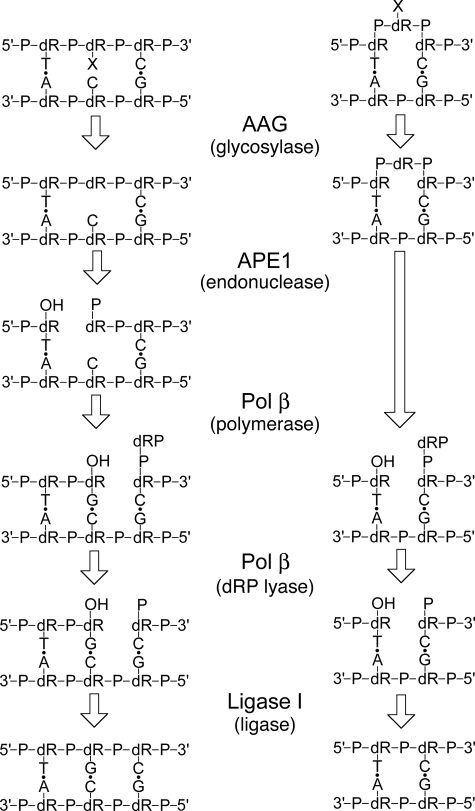
This proposed single nucleotide deletion of a bulged lesion is depicted in Fig. 6 and compared with the normal BER pathway. Only the short patch pathway is shown, but long patch BER with proliferating cell nuclear antigen-mediated strand displacement, flap cleavage by FEN1, and ligation would also be expected to form the single nucleotide deletion (21, 22). The key steps are the initiation of the pathway by a DNA repair glycosylase, as we have shown for AAG and UDG, and the subsequent hydrolysis of the AP-bulge by APE1, as shown below. This is because the dRP lyase and DNA ligase reactions are identical for the short patch BER and bulge deletion pathways (Fig. 6). To confirm the proposed pathway and to evaluate the relative rate of single nucleotide deletions we reconstituted the reaction in vitro with recombinant proteins.
FIGURE 6.
Proposed single nucleotide deletion pathway catalyzed by BER enzymes. The pathway for AAG-initiated short-patch BER is shown on the left and the proposed pathway for single nucleotide deletion is shown on the right. X denotes a damaged nucleotide. Note that only the glycosylase and endonuclease reactions differ for the two pathways. After APE1 cleavage the 5′-dRP intermediate is chemically identical to the intermediate generated after single nucleotide incorporation in the short-patch BER pathway.
Reconstitution of Single Nucleotide Deletion by Enzymes of the Short Patch BER Pathway
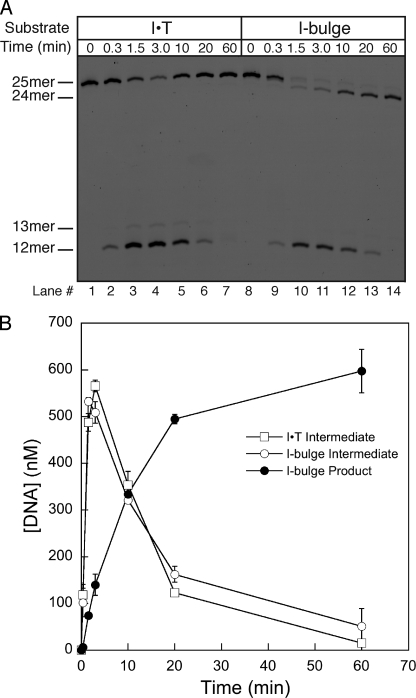
We examined multiple turnover repair reactions of either an I·T mismatch or an I-bulge under the standard BER assay conditions with recombinant AAG, APE1, polymerase β, and DNA ligase I. A representative time course is shown in which reactions were quenched in formamide/EDTA and analyzed by denaturing PAGE (Fig. 7). An early time point confirmed that this quench is sufficient under these conditions, because only intact substrate was observed (lanes 1 and 6). The nicked 12-mer DNA intermediate rapidly built up in reactions containing either an I·T mismatch or an I-bulge. In both cases, this intermediate was depleted by 60 min, indicating complete repair. The quantitative conversion of an I-bulge to a single nucleotide deletion product (24-mer, lane 10) demonstrates that all enzymes are active and capable of catalyzing this deletion reaction. Quantification of the nicked DNA intermediate that were formed from a mismatch and from a bulge shows similar reaction profiles (Fig. 7B). This indicates that both substrates are repaired with similar rates.
FIGURE 7.
Time dependence of repair demonstrates that the single nucleotide deletion pathway occurs at the same rate as the BER pathway. A, multiple turnover reconstituted BER reactions contained 700 nm I·T mismatch or bulge DNA, 350 nm AAG, 120 nm APE1, 20 nm polymerase β, and 50 nm DNA ligase I and the necessary nucleotide and Mg2+ cofactors. Reactions were quenched at the indicated time with formamide/EDTA loading buffer and analyzed by denaturing PAGE. B, the quantification of the formation of the 24-mer deletion product and the formation and breakdown of the 12-mer nicked DNA intermediate is from 3 independent experiments and the mean ± S.D. is shown. Identical rates of repair of the I·T mismatch and I-bulge were also observed at physiological concentration of repair enzymes (supplemental Fig. S12).
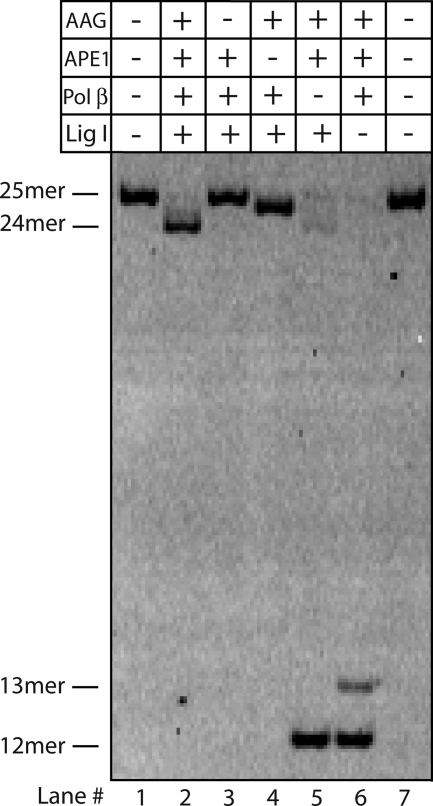
To confirm that these four proteins are necessary and sufficient for the single nucleotide deletion, we omitted each protein in turn (Fig. 8). If all proteins were present, the single nucleotide deletion was complete within 1 h (lane 2). When AAG was omitted, no detectable reaction occurred (lane 3). This shows that APE1 has little or no activity toward an I-bulge. Omission of APE1 led to the formation of the AP-DNA intermediate, which had slightly increased mobility under these conditions (lane 4). When polymerase β was omitted, a 12-mer single strand nick was formed. This intermediate is expected to retain the 5′-dRP group, and therefore is not a substrate for DNA ligase (lane 5). Finally, the omission of DNA ligase I also prevented ligation (lane 6). Both the 12-mer nick and a 13-mer product were observed. The 13-mer product is due to the incorporation of an additional nucleotide by polymerase β in the absence of ligase and this species is not observed when all proteins are present (lane 2) or in the absence of dNTP substrates (data not shown). These experiments demonstrate that the human BER enzymes efficiently catalyze the deletion of a damaged nucleotide bulge, and that this reaction occurs on the same time scale as the well characterized short patch BER pathway for the repair of damaged nucleotides present as a mismatch (Fig. 7).
FIGURE 8.
In vitro reconstitution of the single nucleotide deletion pathway with recombinant human BER proteins. Recombinant AAG, APE1, polymerase (pol) β, and DNA ligase I were necessary and sufficient for the single nucleotide deletion in vitro (lane 2). Omission of any single protein gave the expected product (Fig. 6). Note that the AP intermediate migrates between the 25-mer substrate and the 24-mer product (lane 4) and the omission of DNA ligase (lane 6) leads to some strand displacement by DNA polymerase β.
DISCUSSION
Reports that imbalanced levels of BER enzymes increase the frequency of frameshift mutations (20, 53) led us to test if the BER pathway acts on single nucleotide bulges. Such bulges can occur by a variety of mechanisms (Fig. 1). Using cell extracts and full reconstitution with recombinant proteins, we find that deaminated and alkylated nucleotides present in single nucleotide bulges are substrates for BER, resulting in rapid deletion of the damaged nucleotide. These observations suggest that competition between repair pathways can affect the frequency of frameshift mutations.
Glycosylases Excise Damaged Nucleotides Present in Single Nucleotide Bulges
We identified UDG and AAG as two glycosylases that efficiently recognize base lesions presented in a single nucleotide bulge. UDG has robust activity toward U in both single- and double-stranded DNA (31). In contrast, the activity of AAG toward I in a single-stranded substrate is ∼103-fold slower than toward I in a double-stranded substrate (28). Single turnover kinetics establish that AAG has a 3-fold preference for an I-bulge over an I·T mismatch (28). These observations demonstrate that the UDG and AAG active sites are extremely flexible and able to recognize substrate nucleotides in very different structural contexts.
This ability to accept bulged lesions may be common. Human NEIL1, a glycosylase that recognizes oxidized and fragmented bases, has recently been shown to excise lesions present in single nucleotide bulges (54). Further work is needed to evaluate the range of potential BER substrates.
There was no detectable glycosylase activity toward normal nucleotide bulges in HeLa WCE under multiple turnover conditions. However, AAG exhibits low levels of activity toward the normal nucleotides A and G, and this activity is greater when they are present in a mismatch (36, 55, 56). Under single turnover conditions we found that AAG has similar activity toward an undamaged purine bulge as toward the purine mismatch (supplemental Fig. S3). The lack of detectable glycosylase activity toward A- and G-bulges in cell extracts is consistent with the extremely low concentration of AAG. Under our assay conditions we predict that less than 0.1% of the bulge would be processed by AAG, which would be below our limit of detection. Although AAG activity on undamaged purine bulges could give rise to a very low incidence of deletions, a more likely scenario is that spontaneous damage to a bulged nucleotide would generate a good substrate for BER.
Bulged Nucleotides Are More Susceptible to Damage
The reactivity of single nucleotide bulges has not been extensively characterized. However, the nucleobases of single-stranded DNA are much more reactive than those in duplex DNA toward various types of damage. For example, deamination of C to U occurs ∼100-fold faster in single-stranded DNA than in duplex DNA (57, 58). It has also been observed that C mismatches have elevated spontaneous deamination rates relative to C·G pairs, and in some sequences the rates of deamination of the C mismatch approaches that of single-stranded DNA (59). Therefore, we predict that a C-bulge is likely to be similarly susceptible to deamination. Here we have tested the reactivity of a bulged A toward a DNA alkylating agent. We found that a single nucleotide bulge reacts at least 18-fold faster than a stably paired nucleotide with chloroacetaldehyde (Fig. 4). The greater reactivity of single nucleotide bulges toward endogenous and exogenous forms of base damage underscores the importance of rapidly correcting −1 and +1 polymerase slips.
Human BER Excises Damaged Nucleotides Present in Single Nucleotide Bulges
We tested whether the BER pathway can process glycosylase-initiated bulged AP sites in human cell extracts to effectively delete the damaged nucleotide. In extracts from two human breast cancer cell lines (T47D and MCF-7) and a colon cancer cell line (HT-29), we observed the expected single nucleotide deletion product (Fig. 5 and supplemental Figs. S8 and S9). Both AAG and UDG initiate repair of deaminated nucleotides by a mixture of short and long patch BER pathways (29, 60). It should be noted that both pathways are expected to result in the single nucleotide deletion. To further test the proposed model, we reconstituted the short patch BER pathway using recombinant proteins (Figs. 7 and 8). This confirms that APE1 efficiently processes the bulged AP site, which has important implications for the mutation spectrum of AP sites that are discussed below. The subsequent dRP lyase activity of polymerase β removes the 5′-dRP group and the nick is sealed by DNA ligase (Fig. 6).
The robust activity of APE1 toward a bulged AP site suggests intriguing similarities in substrate recognition between APE1 and the DNA glycosylases UDG and AAG. DNA glycosylases access their damaged nucleotide substrates via nucleotide flipping, in which the substrate nucleotide is flipped 180° out of the duplex into the enzyme active site (23, 61). Similarly, AP site recognition by APE1 involves flipping of the sugar into the active site pocket where endonucleolytic cleavage 5′ of the AP site can take place (62). APE1 is able to process AP sites in single-stranded DNA (63) and it can also catalyze endonucleolytic cleavage at bulky lesions, such as α-anomeric nucleotides (64). The finding that APE1 acts on a bulged AP site is further evidence for a great flexibility in substrate recognition.
In addition to the activity of APE1 toward a bulged AP site, at least one human bifunctional DNA glycosylase is able to act on this substrate. NEIL1, a bifuctional DNA glycosylase/AP lyase, has been shown to perform both glycosylase and lyase activities on a single nucleotide bulge (54). In this case, the action of polynucleotide kinase 3′-phosphatase is required to generate a nick that can be ligated (54, 65). Consistent with this finding, we observed that the bulged AP product from the AAG-catalyzed reaction was slowly hydrolyzed in the absence of Mg2+, presumably by an AP lyase present in HeLa cell extract (supplemental Fig. S2).
The ability of the BER pathway to delete a bulged AP site helps to explain the high frequency of frameshift mutations caused by AP sites (66). For example, overexpression of polymerase β is associated with an increased frequency of −1 frameshift events (18). Polymerase β is especially prone to bypass the AP site and read the adjacent nucleotide (18, 67, 68). We have provided evidence that the resulting AP bulges would be quickly processed to give a −1 frameshift.
Competition for Repair of Bulged Nucleotide Intermediates
A model for BER-mediated frameshift mutations is summarized in Fig. 9. Replication of a damaged template can cause the polymerase to bypass the lesion to produce a damaged, bulged nucleotide product (Fig. 9A). MMR can provide another opportunity for replication of the damaged template nucleotide by removing the newly synthesized strand. In contrast, BER activity would resolve the bulge to a −1 deletion. Alternatively, a polymerase could slip on an undamaged template to give rise to an undamaged bulge (Fig. 9B). MMR can repair both −1 and +1 slips to prevent these errors from becoming frameshift mutations. We found that AAG is able to excise normal purines present in bulges (supplemental Fig. S3). Although this level of activity is extremely low, stochastic action of AAG could nevertheless contribute a bias toward deletion particularly in the absence of functioning MMR. If the bulge remains undetected then it is highly susceptible to DNA damage. We have shown that for at least some types of oxidative and alkylative damage, BER is likely to compete with MMR (Fig. 9B). This competition will favor BER if there is a high burden of DNA damage or if the levels of repair activities are imbalanced (i.e. depletion of MMR enzymes or overexpression of BER enzymes). In the case of a +1 slip, this cycle of damage and deletion via BER would restore the original DNA sequence. Whereas, a −1 slip would be converted into a −1 frameshift mutation.
FIGURE 9.
Model for competition between MMR and BER in the processing of nascent frameshift mutations. A, a polymerase slips on a damaged template to create a bulged lesion nucleotide. The MMR pathway allows another attempt at replication of the damaged template, whereas the BER pathway would catalyze the removal of the damaged nucleotide resulting in a −1 frameshift mutation. B, a polymerase slips on an undamaged homopolymeric run, resulting in a −1 or +1 event. Subsequent damage allows it to be recognized by BER, which will repair a +1 event but make a −1 event permanent. MMR provides a high fidelity pathway for repairing either −1 or +1 events, but imbalances in MMR or BER or a high level of DNA damage are expected to increase the frequency of frameshift mutations. The low level of activity of AAG on undamaged purines could allow some deletions even in the absence of DNA damage (not shown).
Our results also shed light on observations that overexpression of BER enzymes can lead to mutator phenotypes (19). A model, collectively referred to as imbalanced BER, postulates that increased production of BER intermediates leads to persistent DNA repair intermediates that can ultimately result in mutations (20, 53, 69, 70). This model successfully accounts for many observations, but does not easily explain the finding that heterologous expression of human AAG in yeast leads to a 30-fold increase in the frequency of −1 frameshifts in a poly(A) tract and a 6-fold bias toward deletions over insertions (20). Our finding that AAG excises a damaged or undamaged A-bulge provides a mechanism whereby AAG could outcompete the yeast MMR pathway to recognize polymerase slipping errors.
This model also has implications for cancer, because the frequency of mutations is one of the driving forces for carcinogenesis (2, 3). Several studies have found a positive correlation between overexpression of AAG and an increase in mutation rates in human cells (20, 50, 71). AAG expression is commonly up-regulated in breast cancers and previous studies have found that these same cell lines exhibit elevated mutation rates, including increased frequency of −1 frameshift mutations (49, 50, 71). There is a well established link between defects in MMR and microsatellite instability, which includes increased frequency of frameshift mutations, and is commonly associated with hereditary colon cancers (72, 73). The overexpression of enzymes of the BER pathway, such as AAG, could increase the frequency of frameshift mutations by shifting the balance from MMR to BER. Our studies highlight additional factors that can contribute to the bias of −1 frameshift over +1 frameshift mutations in human cells, and offers an explanation for the frameshift mutations that accompany imbalances in the BER pathway.
Supplementary Material
Acknowledgments
We thank Tom Wilson for cDNA encoding polymerase β, and members of the O'Brien lab for critical comments on the manuscript.
Note Added in Proof
Another study has recently been published investigating the mechanism of how AAG causes frameshift mutations (74). The authors use heterologous expression of AAG in yeast cells to demonstrate that AAG glycosylase activity is important for the generation of frameshift mutations. Remarkably, active site mutants of AAG were identified that further increase the frequency of both −1 and +1 frameshift mutations, and it is proposed that AAG outcompetes other DNA repair pathways for binding to bulged DNA intermediates.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA122254 from the United States Public Health Service.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S12.
- MMR
- mismatch repair
- AAG
- alkyladenine DNA glycosylase also known as MPG, methylpurine DNA glycosylase
- AP
- apurinic/apyrimidinic
- APE1
- AP endonuclease 1
- BER
- base excision repair
- ϵA
- 1,N6-ethenoadenine
- dRP
- deoxyribose phosphate
- FAM
- 6-carboxyfluorescein (aminohexyl linker)
- I
- inosine
- U
- uracil
- UDG
- uracil DNA glycosylase
- UGI
- uracil glycosylase inhibitor
- WCE
- whole cell extract
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Kunkel T. A. (1990) Biochemistry 29, 8003–8011 [DOI] [PubMed] [Google Scholar]
- 2.Loeb L. A. (2001) Cancer Res. 61, 3230–3239 [PubMed] [Google Scholar]
- 3.Loeb L. A., Bielas J. H., Beckman R. A. (2008) Cancer Res. 68, 3551–3557 [DOI] [PubMed] [Google Scholar]
- 4.Iyer R. R., Pluciennik A., Burdett V., Modrich P. L. (2006) Chem Rev. 106, 302–323 [DOI] [PubMed] [Google Scholar]
- 5.Kunkel T. A., Erie D. A. (2005) Annu. Rev. Biochem. 74, 681–710 [DOI] [PubMed] [Google Scholar]
- 6.Greene C. N., Jinks-Robertson S. (2001) Genetics 159, 65–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson A. L., Loeb L. A. (1998) Genetics 148, 1483–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poynter J. N., Siegmund K. D., Weisenberger D. J., Long T. I., Thibodeau S. N., Lindor N., Young J., Jenkins M. A., Hopper J. L., Baron J. A., Buchanan D., Casey G., Levine A. J., Le Marchand L., Gallinger S., Bapat B., Potter J. D., Newcomb P. A., Haile R. W., Laird P. W. (2008) Cancer Epidemiol. Biomarkers Prev. 17, 3208–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunkel T. A. (1992) J. Biol. Chem. 267, 18251–18254 [PubMed] [Google Scholar]
- 10.Lovett S. T. (2004) Mol. Microbiol. 52, 1243–1253 [DOI] [PubMed] [Google Scholar]
- 11.Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. (1966) Cold Spring Harbor Symp. Quant. Biol. 31, 77–84 [DOI] [PubMed] [Google Scholar]
- 12.Shearman C. W., Forgette M. M., Loeb L. A. (1983) J. Biol. Chem. 258, 4485–4491 [PubMed] [Google Scholar]
- 13.Shearman C. W., Loeb L. A. (1983) J. Biol. Chem. 258, 4477–4484 [PubMed] [Google Scholar]
- 14.Colgin L. M., Hackmann A. F., Emond M. J., Monnat R. J., Jr. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1437–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z., Gerstein M. (2003) Nucleic Acids Res. 31, 5338–5348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor M. S., Ponting C. P., Copley R. R. (2004) Genome Res. 14, 555–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fortune J. M., Pavlov Y. I., Welch C. M., Johansson E., Burgers P. M., Kunkel T. A. (2005) J. Biol. Chem. 280, 29980–29987 [DOI] [PubMed] [Google Scholar]
- 18.Chan K., Houlbrook S., Zhang Q. M., Harrison M., Hickson I. D., Dianov G. L. (2007) Mutagenesis 22, 183–188 [DOI] [PubMed] [Google Scholar]
- 19.Frosina G. (2000) Eur. J. Biochem. 267, 2135–2149 [DOI] [PubMed] [Google Scholar]
- 20.Hofseth L. J., Khan M. A., Ambrose M., Nikolayeva O., Xu-Welliver M., Kartalou M., Hussain S. P., Roth R. B., Zhou X., Mechanic L. E., Zurer I., Rotter V., Samson L. D., Harris C. C. (2003) J. Clin. Invest. 112, 1887–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fortini P., Dogliotti E. (2007) DNA Repair 6, 398–409 [DOI] [PubMed] [Google Scholar]
- 22.David S. S., Williams S. D. (1998) Chem. Rev. 98, 1221–1262 [DOI] [PubMed] [Google Scholar]
- 23.Stivers J. T., Jiang Y. L. (2003) Chem. Rev. 103, 2729–2759 [DOI] [PubMed] [Google Scholar]
- 24.O'Brien P. J., Ellenberger T. (2003) Biochemistry 42, 12418–12429 [DOI] [PubMed] [Google Scholar]
- 25.Pascal J. M., O'Brien P. J., Tomkinson A. E., Ellenberger T. (2004) Nature 432, 473–478 [DOI] [PubMed] [Google Scholar]
- 26.Baldwin M. R., O'Brien P. J. (2009) Biochemistry 48, 6022–6033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Werneburg B. G., Ahn J., Zhong X., Hondal R. J., Kraynov V. S., Tsai M. D. (1996) Biochemistry 35, 7041–7050 [DOI] [PubMed] [Google Scholar]
- 28.Lyons D. M., O'Brien P. J. (2009) J. Am. Chem. Soc. 131, 17742–17743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sokhansanj B. A., Rodrigue G. R., Fitch J. P., Wilson D. M., 3rd (2002) Nucleic Acids Res. 30, 1817–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Visnes T., Doseth B., Pettersen H. S., Hagen L., Sousa M. M., Akbari M., Otterlei M., Kavli B., Slupphaug G., Krokan H. E. (2009) Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 563–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kavli B., Sundheim O., Akbari M., Otterlei M., Nilsen H., Skorpen F., Aas P. A., Hagen L., Krokan H. E., Slupphaug G. (2002) J. Biol. Chem. 277, 39926–39936 [DOI] [PubMed] [Google Scholar]
- 32.Wang Z., Mosbaugh D. W. (1988) J. Bacteriol. 170, 1082–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Engelward B. P., Weeda G., Wyatt M. D., Broekhof J. L., de Wit J., Donker I., Allan J. M., Gold B., Hoeijmakers J. H., Samson L. D. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 13087–13092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hang B., Singer B., Margison G. P., Elder R. H. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12869–12874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hitchcock T. M., Dong L., Connor E. E., Meira L. B., Samson L. D., Wyatt M. D., Cao W. (2004) J. Biol. Chem. 279, 38177–38183 [DOI] [PubMed] [Google Scholar]
- 36.O'Brien P. J., Ellenberger T. (2004) J. Biol. Chem. 279, 9750–9757 [DOI] [PubMed] [Google Scholar]
- 37.Bolt H. M. (2005) Crit. Rev. Toxicol. 35, 307–323 [DOI] [PubMed] [Google Scholar]
- 38.Guengerich F. P., Kim D. H., Iwasaki M. (1991) Chem. Res. Toxicol. 4, 168–179 [DOI] [PubMed] [Google Scholar]
- 39.Rydberg B., Dosanjh M. K., Singer B. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 6839–6842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singer B., Antoccia A., Basu A. K., Dosanjh M. K., Fraenkel-Conrat H., Gallagher P. E., Kuśmierek J. T., Qiu Z. H., Rydberg B. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 9386–9390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaerer O. D., Verdine G. L. (1995) J. Am. Chem. Soc. 117, 10781–10782 [Google Scholar]
- 42.Wolfe A. E., O'Brien P. J. (2009) Biochemistry 48, 11357–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y., Wu X., Guo D., Rechkoblit O., Taylor J. S., Geacintov N. E., Wang Z. (2002) J. Biol. Chem. 277, 44582–44587 [DOI] [PubMed] [Google Scholar]
- 44.Levine R. L., Yang I. Y., Hossain M., Pandya G. A., Grollman A. P., Moriya M. (2000) Cancer Res. 60, 4098–4104 [PubMed] [Google Scholar]
- 45.Zang H., Goodenough A. K., Choi J. Y., Irimia A., Loukachevitch L. V., Kozekov I. D., Angel K. C., Rizzo C. J., Egli M., Guengerich F. P. (2005) J. Biol. Chem. 280, 29750–29764 [DOI] [PubMed] [Google Scholar]
- 46.Zhang H., Beckman J. W., Guengerich F. P. (2009) J. Biol. Chem. 284, 35144–35153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patel D. J., Kozlowski S. A., Marky L. A., Rice J. A., Broka C., Itakura K., Breslauer K. J. (1982) Biochemistry 21, 445–451 [DOI] [PubMed] [Google Scholar]
- 48.Joshua-Tor L., Rabinovich D., Hope H., Frolow F., Appella E., Sussman J. L. (1988) Nature 334, 82–84 [DOI] [PubMed] [Google Scholar]
- 49.Vickers M. A., Vyas P., Harris P. C., Simmons D. L., Higgs D. R. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 3437–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cerda S. R., Turk P. W., Thor A. D., Weitzman S. A. (1998) FEBS Lett. 431, 12–18 [DOI] [PubMed] [Google Scholar]
- 51.Soule H. D., Vazguez J., Long A., Albert S., Brennan M. (1973) J. Natl. Cancer Inst. 51, 1409–1416 [DOI] [PubMed] [Google Scholar]
- 52.Keydar I., Chen L., Karby S., Weiss F. R., Delarea J., Radu M., Chaitcik S., Brenner H. J. (1979) Eur. J. Cancer 15, 659–670 [DOI] [PubMed] [Google Scholar]
- 53.Glassner B. J., Rasmussen L. J., Najarian M. T., Posnick L. M., Samson L. D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9997–10002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao X., Krishnamurthy N., Burrows C. J., David S. S. (2010) Biochemistry 49, 1658–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berdal K. G., Johansen R. F., Seeberg E. (1998) EMBO J. 17, 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Connor E. E., Wyatt M. D. (2002) Chem. Biol. 9, 1033–1041 [DOI] [PubMed] [Google Scholar]
- 57.Frederico L. A., Kunkel T. A., Shaw B. R. (1990) Biochemistry 29, 2532–2537 [DOI] [PubMed] [Google Scholar]
- 58.Lindahl T., Nyberg B. (1974) Biochemistry 13, 3405–3410 [DOI] [PubMed] [Google Scholar]
- 59.Frederico L. A., Kunkel T. A., Shaw B. R. (1993) Biochemistry 32, 6523–6530 [DOI] [PubMed] [Google Scholar]
- 60.Fortini P., Parlanti E., Sidorkina O. M., Laval J., Dogliotti E. (1999) J. Biol. Chem. 274, 15230–15236 [DOI] [PubMed] [Google Scholar]
- 61.Roberts R. J., Cheng X. (1998) Annu. Rev. Biochem. 67, 181–198 [DOI] [PubMed] [Google Scholar]
- 62.Mol C. D., Izumi T., Mitra S., Tainer J. A. (2000) Nature 403, 451–456 [DOI] [PubMed] [Google Scholar]
- 63.Marenstein D. R., Wilson D. M., 3rd, Teebor G. W. (2004) DNA Repair 3, 527–533 [DOI] [PubMed] [Google Scholar]
- 64.Gros L., Ishchenko A. A., Ide H., Elder R. H., Saparbaev M. K. (2004) Nucleic Acids Res. 32, 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karimi-Busheri F., Lee J., Tomkinson A. E., Weinfeld M. (1998) Nucleic Acids Res. 26, 4395–4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Q. M., Dianov G. L. (2005) DNA Repair 4, 263–270 [DOI] [PubMed] [Google Scholar]
- 67.Blanca G., Villani G., Shevelev I., Ramadan K., Spadari S., Hübscher U., Maga G. (2004) Biochemistry 43, 11605–11615 [DOI] [PubMed] [Google Scholar]
- 68.Efrati E., Tocco G., Eritja R., Wilson S. H., Goodman M. F. (1997) J. Biol. Chem. 272, 2559–2569 [DOI] [PubMed] [Google Scholar]
- 69.Coquerelle T., Dosch J., Kaina B. (1995) Mutat. Res. 336, 9–17 [DOI] [PubMed] [Google Scholar]
- 70.Posnick L. M., Samson L. D. (1999) J. Bacteriol. 181, 6763–6771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Okochi E., Watanabe N., Sugimura T., Ushijima T. (2002) Mutat. Res. 506–507, 101–111 [DOI] [PubMed] [Google Scholar]
- 72.Karran P. (1996) Semin. Cancer Biol. 7, 15–24 [DOI] [PubMed] [Google Scholar]
- 73.Shah S. N., Hile S. E., Eckert K. A. (2010) Cancer Res. 70, 431–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klapacz J., Lingaraju G. M., Guo H. H., Shah D., Moar-Shoshani A., Loeb L. A., Samson L. D. (2010) Mol. Cell 37, 843–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.