Abstract
Kinesin motor proteins use an ATP hydrolysis cycle to perform various functions in eukaryotic cells. Many questions remain about how the kinesin mechanochemical ATPase cycle is fine-tuned for specific work outputs. In this study, we use isothermal titration calorimetry and stopped-flow fluorometry to determine and analyze the thermodynamics of the human kinesin-5 (Eg5/KSP) ATPase cycle. In the absence of microtubules, the binding interactions of kinesin-5 with both ADP product and ATP substrate involve significant enthalpic gains coupled to smaller entropic penalties. However, when the wild-type enzyme is titrated with a non-hydrolyzable ATP analog or the enzyme is mutated such that it is able to bind but not hydrolyze ATP, substrate binding is 10-fold weaker than ADP binding because of a greater entropic penalty due to the structural rearrangements of switch 1, switch 2, and loop L5 on ATP binding. We propose that these rearrangements are reversed upon ATP hydrolysis and phosphate release. In addition, experiments on a truncated kinesin-5 construct reveal that upon nucleotide binding, both the N-terminal cover strand and the neck linker interact to modulate kinesin-5 nucleotide affinity. Moreover, interactions with microtubules significantly weaken the affinity of kinesin-5 for ADP without altering the affinity of the enzyme for ATP in the absence of ATP hydrolysis. Together, these results define the energy landscape of a kinesin ATPase cycle in the absence and presence of microtubules and shed light on the role of molecular motor mechanochemistry in cellular microtubule dynamics.
Keywords: ATPases, Calorimetry, Enzyme Kinetics, Enzyme Mechanisms, Kinesin, Microtubules, Molecular Motors, Site-directed Mutagenesis, Thermodynamics, Isothermal Titration Calorimetry
Introduction
Cytoskeletal motor proteins from the kinesin superfamily are enzymes that use ATP hydrolysis to perform various functions in eukaryotic cells including vesicle transport, regulation of microtubule (MT)2 dynamics, mitotic spindle assembly, and chromosome segregation. Kinesins share an ∼350-amino acid catalytic core domain that contains a conserved nucleotide binding site, an MT-binding region, a 14–20-amino acid segment called the neck linker, and a 9–12-residue region called the cover strand (reviewed in Refs. 1 and 2, and also see the Kinesin Home Page). The catalytic core coordinates movements of structural elements located at the active site (conserved phosphate-binding loop (P-loop, GQTXXGK(T/S)), switch 1 (Sw1, NXXSSR), and switch 2 (Sw2, DXXGXE)) with the MT-binding interface and the neck linker region (3). Numerous studies have investigated the ATPase mechanism of different kinesin monomers in the presence of MTs, including kinesin-1/conventional kinesin (4–6), kinesin-3/KIF1A (7, 8), kinesin-5/Eg5/KSP (9, 10), kinesin-10/NOD (11), and kinesin-14/Ncd/Kar3 (12–14). Current theory on the kinesin mechanism of force generation suggests that the nucleotide bound at the active site triggers a conformational change that is subsequently transmitted to adjacent regions of the core that interact with the MT. This structural communication causes changes in MT binding affinity and stabilization of the neck linker in alternate conformations (1). It is also known that the minor structural differences among different kinesins result in variations in the kinetic rate and equilibrium constants that govern their respective ATPase cycles. Nevertheless, it is unclear how different classes of kinesins are adapted to generate the specific work outputs necessary for their task(s) inside cells, and the complete structural mechanism remains under investigation.
One kinesin of particular interest is the human mitotic kinesin-5 (Eg5/KSP), which is involved in the organization and assembly of the mitotic spindle during cell division. In vivo, kinesin-5 forms a homotetrameric quaternary structure with a dimer positioned at each end of the coiled-coil stalk (15, 16). This structural arrangement allows the enzyme to slide along adjacent MTs during mitosis, thus contributing to the plus end-directed force that is necessary for forming and maintaining the mitotic spindle (17, 18). Previous studies have kinetically characterized the ATPase mechanism of dimeric (19) and monomeric kinesin-5 in the absence (20) and presence of MTs (9, 10, 21). The results of these kinetic studies have been correlated to changes in the neck linker orientation of kinesin-5 for force generation and processive motility along the MT (10, 19, 22). In this study, we have quantified the thermodynamics of individual steps in the kinesin-5 ATPase mechanism, adding to the limited data available on the energetics of ATPases (23, 24).
ITC is the most straightforward experimental method available to determine the thermodynamics of protein-ligand interactions. The ITC instrument simultaneously measures the heat change (ΔHITC), equilibrium binding constant (Ka), and binding stoichiometry (n) for protein-ligand complex formation. The standard Gibbs free energy change (ΔG°) and change in entropy (ΔS°) are then calculated from the observed constants. Experiments can be performed over a range of temperatures to determine the change in heat capacity (ΔCp) for the reaction, which reveals information about changes in protein solvation on binding (25). Much of the work in protein thermodynamics has been dedicated to correlating observed and calculated thermodynamic parameters with meaningful structural changes. For example, the magnitude and sign of the binding enthalpy (ΔH°) have been shown to reflect the loss of protein-solvent hydrogen bonds, the formation of protein-ligand hydrogen bonds, and the formation of salt bridges (25). On the other hand, changes in reaction entropy (ΔS°) are largely due to hydration effects and changes in degrees of freedom for the ligand and protein on binding (25). However, without the corresponding structural models, deconvolution of these individual contributions can be difficult.
Here, we present a detailed calorimetric analysis of the mechanochemical ATPase cycle of a motor protein. In conjunction with available x-ray crystallographic models of kinesin-5 in different nucleotide states (26, 27), the results have uncovered features of the ATPase cycle that provide insight into mechanochemical energy transduction in kinesins and could also be applied to myosins and G proteins.
EXPERIMENTAL PROCEDURES
Experimental Conditions
Experiments were performed in ITC buffer (20 mm HEPES, pH 7.2, with KOH, 5 mm magnesium acetate, 95 mm potassium acetate, 150 mm sucrose).
Cloning, Expression, and Protein Purification
The DNA construct coding for the motor domain of human kinesin-5 (amino acids 1–368) was synthesized by PCR using full-length cDNA as the template (generously provided by Dr. Anne Blangy) and ligated into pET16b for bacterial expression (referred to as E368(wt)). Site-directed mutagenesis was performed to generate E358(wt), E368(R234K), and E368(S233C/R234K) (supplemental Table S2), and these mutations were confirmed by DNA sequencing. Each construct was co-transformed into B834(DE3) cells with pRIL (Stratagene), and the kinesin-5 protein was expressed and purified in the nucleotide-free state as described previously (9, 20, 21). Briefly, MgATP was excluded from all column chromatography buffers, and the enriched kinesin-5 fractions from the nickel-nitrilotriacetic acid agarose column (Qiagen) were pooled, incubated with 10 mm EDTA, and buffer-exchanged using desalting columns (GE Healthcare). For all experiments containing MTs, an aliquot of purified bovine brain tubulin was thawed and cycled, and the MTs were stabilized with 20 μm Taxol (paclitaxel, Sigma-Aldrich).
Isothermal Titration Calorimetry
Titrations were performed with a MicroCal VP-ITC (GE/MicroCal Inc.) at temperatures ranging from 4 to 15 °C. For most experiments, kinesin-5 (150–590 μm) was loaded into the sample cell, and adenosine nucleotide (ADP, ATP, AMPPNP, or ATPγS at 1–4 mm) was loaded into the injection syringe (∼300 μl). Unless otherwise specified, the first injection (2 μl; 3.4-s duration; 600-s spacing) was followed by 29 injections (10 μl; 20-s duration; 600-s spacing between injections) with continuous stirring at 260 rpm. The sum of the heat evolved from the reaction, corrected for the small heats of dilution for each injection, was plotted against the molar ratio of reactants. The data were fit to a 1:1 binding model (25) using a non-linear least-squares algorithm incorporated in the Origin software provided with the instrument. The Gibbs free energy (ΔG°) and entropy of binding (ΔS°) were calculated using the thermodynamic relationships
where R is the gas constant, and T is the temperature in Kelvin.
Stopped-flow Fluorometry
The kinetics of nucleotide binding to kinesin-5, Pi release, and ADP release were measured using an SF-2004 KinTek stopped-flow instrument as described previously (9, 11, 20, 21). Competitive inhibition experiments using various adenine nucleotides and nucleotide analogs (AXP) were performed by rapidly mixing kinesin-5 or MT-kinesin-5 with a fixed concentration of mantATP and increasing unlabeled AXP concentrations in a stopped-flow instrument and monitoring the change in mant nucleotide fluorescence over time (excitation, 356 nm; emission, >400 nm). The amplitude of each transient was normalized and plotted against the competitive nucleotide concentration [AXP], and the data were fit to a hyperbolic equation for competitive inhibition
where Amplitude represents the normalized amplitude of each transient divided by the maximum amplitude with no added competitive inhibitor, and Ki,AXP is the AXP inhibition constant.
RESULTS
ADP Binding
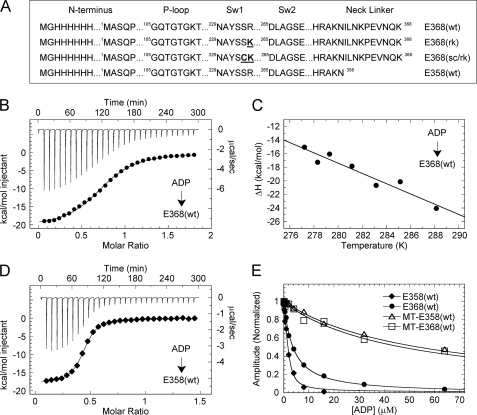
This study was initiated by modifying the expression construct for the monomeric motor domain of kinesin-5 (hereafter referred to as E368(wt)) to generate an N-terminally His-tagged enzyme, which was purified in the nucleotide-free state, as described in previous studies (20, 21). Because one goal of this study was to investigate the role of the C-terminal neck linker, the N-terminally His-tagged enzyme was used to prevent the tag from potentially confounding the results. The purified enzyme displayed a similar overall ATPase mechanism to that of a previously used monomeric kinesin-5 construct (E367(wt) with a C-terminal His-tag (20, 21)) (supplemental Fig. S1, A–C). Specifically, the kinetics of nucleotide binding for E368(wt) were similar to that of E367(wt) (supplemental Fig. S1C) (20). To determine the thermodynamics of individual steps in the ATPase mechanism (discussed below), wild-type kinesin-5 and several mutant enzymes (Fig. 1A) were utilized.
FIGURE 1.
Thermodynamics of ADP binding. A, kinesin-5 constructs used in this study with substitutions highlighted in bold. B, ITC titration of 1.0 mm ADP into 0.13 mm E368(wt) at 10 °C (top) and the peak-integrated heats of reaction versus molar ratio (bottom). The smooth line corresponds to a single-site binding model yielding ΔH = −20.7 kcal mol−1 and Kd = 7.5 μm. C, ΔH versus temperature for ADP binding to E368(wt), yielding ΔCp = −0.755 kcal mol−1 K−1. D, ITC titration of 0.6 mm ADP into 0.094 mm E358(wt) at 10 °C: ΔH = −17.8 kcal mol−1 and Kd = 0.79 μm. E, ADP-dependent competitive inhibition of mantATP binding (as indicated). Final concentrations are: 1 μm kinesin-5, 0 or 1.5 μm MTs, 0 or 6 μm Taxol, 1.5 μm mantATP, 0–128 μm ADP. Ki,ATP = 0.45, 3.44, 53.6, and 48.0 μm, respectively.
The thermodynamic parameters of ADP binding to nucleotide-free E368(wt) were measured by ITC. Fig. 1B shows representative data for ADP titrated into E368(wt) at 10 °C in HEPES buffer. The overall binding reaction was exothermic with a significant gain of free energy (ΔG = −6.6 kcal mol−1). This free energy change was characterized by a large enthalpic gain (ΔH = −20.6 kcal mol−1) coupled to a smaller entropic penalty (−TΔS = 14.0 kcal mol−1; Table 1). This experiment was repeated in PIPES buffer to rule out any heat contribution from proton transfer to the buffer upon ADP binding to E368(wt) (supplemental Fig. S2). These results corroborate with a recent study by Sheth et al. (28) who also investigated the thermodynamics of ADP binding to a similar monomeric kinesin-5. The observed ADP affinity (Kd = 7.5 μm) was similar to that observed in previous studies with kinesin-5 using mantADP binding kinetics (Kd,mantADP = 6.5 μm (20)). Together, these values indicate that kinesin binds ADP with relatively high affinity. However, it should be noted that the binding stoichiometries are less than unity despite the use of fresh, stable, nucleotide-free protein. A previous study characterizing kinesin-5 suggested that a “non-productive” subpopulation of enzyme existed in the absence of microtubules such that nucleotide binding and hydrolysis could not occur (20). We propose that our stoichiometry deviation from unity provides additional support for this non-productive kinesin-5 subpopulation.
TABLE 1.
Thermodynamic parameters for representative ITC experiments at 10 °C
± is the S.E. from the fit of the data.
| Cell | Syringe | ΔG | ΔH | −TΔS | Kd | Stoichiometrya |
|---|---|---|---|---|---|---|
| kcal mol−1 | kcal mol−1 | kcal mol−1 | μm | |||
| E368(wt) | ADP | −6.6 ± 0.03 | −20.7 ± 0.2 | 14.0 ± 0.2 | 7.5 ± 0.4 | 0.8 ± 0.005 |
| E368(wt) (PIPES buffer) | ADP | −6.4 ± 0.03 | −20.0 ± 0.1 | 13.6 ± 0.2 | 13.1 ± 0.5 | 0.6 ± 0.002 |
| E358(wt) | ADP | −7.9 ± 0.02 | −17.8 ± 0.1 | 9.9 ± 0.1 | 0.79 ± 0.03 | 0.4 ± 0.001 |
| E368(wt) | AMPPNP | −5.4 ± 0.01 | −8.5 ± 0.1 | 3.1 ± 0.1 | 64.5 ± 1.4 | 0.6 ± 0.003 |
| E368(wt) | ATPγS | −6.9 ± 0.04 | −24.0 ± 0.2 | 17.1 ± 0.3 | 4.8 ± 0.3 | 0.8 ± 0.006 |
| E368(wt) | ATP | −7.4 ± 0.01 | −27.7 ± 0.1 | 20.3 ± 0.1 | 2.1 ± 0.1 | 0.5 ± 0.001 |
| E368(rk) | ADP | −6.9 ± 0.01 | −20.1 ± 0.1 | 13.2 ± 0.1 | 4.6 ± 0.1 | 0.7 ± 0.002 |
| E368(rk) | ATPγS | −5.2 ± 0.01 | −21.6 ± 0.9 | 16.4 ± 0.1 | 97.1 ± 0.9 | 0.4 ± 0.001 |
| E368(rk) | ATP | −7.0 ± 0.02 | −39.0 ± 0.2 | 32.0 ± 0.2 | 3.2 ± 0.1 | 0.4 ± 0.002 |
| E368(sc/rk) | ADP | −6.1 ± 0.02 | −20.1 ± 0.1 | 14.0 ± 0.1 | 19.0 ± 0.1 | 0.8 ± 0.004 |
| E368(sc/rk) | ATP | −5.7 ± 0.02 | −24.7 ± 0.2 | 19.0 ± 0.2 | 43.5 ± 1.6 | 0.6 ± 0.004 |
a Deviation of stoichiometry from unity suggests non-productive enzyme.
To determine the change in the heat capacity (ΔCp) for the ADP binding interaction, the titration of ADP into E368(wt) was performed over a range of temperatures (Fig. 1C; supplemental Table S1). The linear regression analysis of ΔHobs plotted versus temperature yielded a sizable negative heat capacity (ΔCp = −0.755 kcal mol−1 K−1). For ligand-protein interactions, ΔCp < 0 has been correlated with burial of the hydrophobic surface area of the protein and release of solvent from the protein surface on complex formation (29, 30). NACCESS (version 2.1.1 (31)) was used to calculate the solvent-accessible surface area of E368(wt) (1II6 (26)) with MgADP absent and bound at the active site. The heat capacity change upon ADP binding was consistent with the burial of ∼420 Å2 of surface area on complex formation.
ITC was also used to explore the role of the neck linker (amino acids 359–368 for kinesin-5) in modulating ADP binding thermodynamics. Fig. 1D shows ITC data for ADP titrated into the truncated construct E358(wt) at 10 °C. An exothermic binding reaction was observed, with ΔG = −7.9 kcal mol−1. When compared with E368(wt), ΔH at −17.8 kcal mol−1 and −TΔS at 9.9 kcal mol−1 were notably reduced (14 and 29% reduction, respectively). Both effects are significant, but the larger reduction in −TΔS contributes to a 10-fold increase in ADP affinity for E358(wt) (Kd = 0.79 μm; Table 1). These results suggest that upon transition from the nucleotide-free state to the ADP-bound state, a disordered-to-ordered conformational change occurs that is dependent on the presence of the neck linker. In the absence of the neck linker, this transition does not occur, thus giving rise to a smaller entropic penalty for E358(wt) when compared with E368(wt).
To confirm the results from ITC experiments and also characterize ADP affinity in the presence of MTs, the ADP-dependent competitive inhibition of mantATP binding was monitored using stopped-flow fluorometry (Fig. 1E). For E368(wt) and E358(wt) in the absence of MTs, the observed ADP inhibition constants (Ki,ADP = 3.3 and 0.45 μm, respectively) were similar to the ADP binding constants determined by ITC. The ADP inhibition constant for E368(wt) is similar to the ADP binding affinity determined by ITC (Kd = 7.5 μm) in this study, and the 2-fold difference in affinity is within the S.D. for multiple stopped-flow and ITC measurements (Kd,ADP = 5.4 ± 2.7 μm, S.D.). In the presence of MTs, ADP affinity was reduced more than 10-fold for E368(wt) with Ki,ADP = 48 μm and 120-fold for E358(wt) with Ki,ADP = 54 μm. Control experiments were performed with E358(wt) to confirm that the MT-activated steady-state ATPase was not affected by removing the neck linker (supplemental Fig. S3). This reduced affinity for the MT-kinesin-5 complex was comparable with the apparent ADP dissociation constant previously obtained for MT-E367(wt) using [α-32P]ADP equilibrium binding experiments (9).
ADP Release
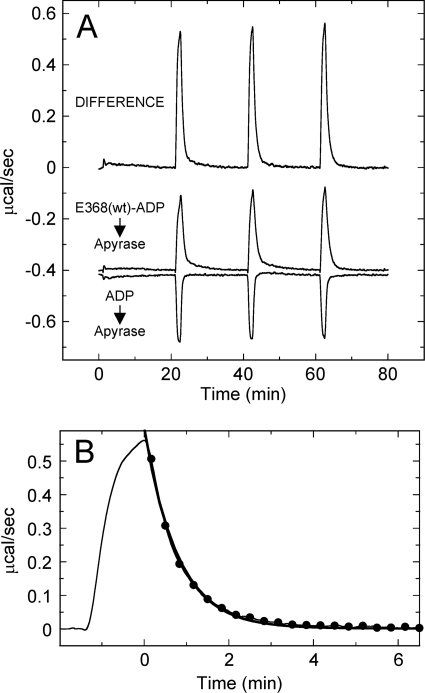
An innovative ITC experiment was used to determine the thermodynamics and kinetics of ADP product release from kinesin. Multiple 40-μl samples of the E368(wt)-ADP complex (1:1) were titrated into a solution of apyrase (2 units ml−1), and the heat of the reaction was monitored over time (Fig. 2A). This experiment assumes that once ADP is released from kinesin, it is rapidly cleaved into AMP + Pi by the apyrase. To control for the heat associated with the apyrase cleavage reaction and the heat of AMP + Pi binding to E368(wt), ADP was titrated into apyrase and AMP + Pi was titrated into nucleotide-free E368(wt), respectively. The overall ADP release reaction was endothermic, but the apyrase cleavage and AMP + Pi binding reactions were exothermic. The heats from these control reactions were subtracted from the E368(wt)-ADP reaction to yield the enthalpy change associated with ADP release, 18.7 ± 1.0 kcal mol−1 at 10 °C. These results were virtually opposite to the enthalpy change associated with ADP binding (ΔH = −20.6 kcal mol−1), which indicates that ADP release is an entropically driven step in the ATPase cycle and also likely symmetric to ADP binding.
FIGURE 2.
Thermodynamics and kinetics of ADP release by ITC. A, ITC titration of 0.1 mm E368(wt)-ADP into 2 units ml−1 apyrase. A control reaction is shown for 0.1 mm ADP titrated into apyrase. The difference isotherm corresponds to the ADP-only isotherm subtracted from the E368(wt)-ADP titration. The average integrated heat (ΔH) for ADP release from E368(wt) was 18.7 ± 1.0 kcal mol−1. B, zoom of the third injection and normalization of time to fit the exponential decay of the heat of reaction (–●–). The fit provides kobs = 1.28 min−1 (0.021 s−1).
This experimental approach also provides a means to directly monitor the kinetics of ADP release from kinesin by ITC. Fig. 2B highlights one of the injections in which the decrease in the heat of reaction over time was fitted to an exponential decay to yield the observed rate of 1.28 min−1 (0.02 s−1), which is identical to the previously observed rate of [α-32P]ADP release from E367(wt) (9). This methodology provides a quick and easy way of directly and simultaneously monitoring the kinetics and thermodynamics of the rate-limiting ADP release step for most kinesins.
ATP Binding
After the products of ATP hydrolysis are released from kinesin, ATP substrate must bind and be hydrolyzed to complete the mechanochemical cycle. Previous studies suggest that ATP binding to kinesin-5 occurs via a two-step mechanism in which a relatively slow isomerization step (∼1 s−1) that enhances ATP binding is followed by the rapid reactions (>10 s−1) of ATP hydrolysis and Pi release (20). This mechanism poses a challenge: how to measure the thermodynamics of ATP binding without contributions from ATP hydrolysis or Pi release. To solve this problem, three different sets of ITC titrations were performed. 1) AMPPNP, a non-hydrolyzable ATP analog, was titrated into E368(wt); 2) ATPγS, a slowly hydrolyzable ATP analog, was titrated into a slowly hydrolyzing single switch 1 mutant (R234K) (E368(rk)); and 3) ATP was titrated into a non-hydrolyzing double switch 1 mutant (S233C and R234K) (E368(sc/rk)) (Fig. 1A). To ensure that these nucleotide analogs and the substitutions at positions Ser-233 and Arg-234 only affected hydrolysis, stopped-flow and ITC control experiments were performed, and the results were compared with those of E368(wt) (supplemental Fig. S1, C–E; Table 1).
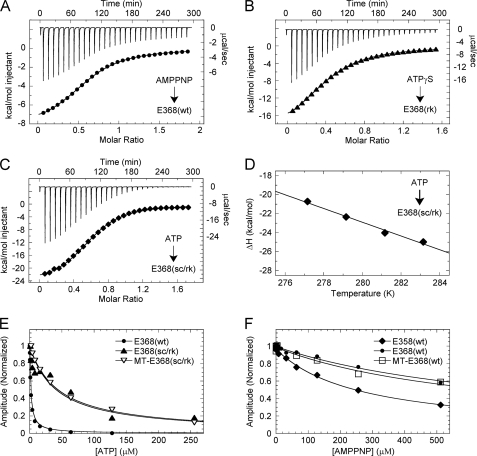
Fig. 3, A–C, show representative ITC data from the three different experiments performed to measure ATP binding thermodynamics as described above (Table 1). When compared with ADP binding, ATP binding was found to be 6-fold weaker (Kd,ATP = 43.5 μm). In addition, ATP binding is characterized by a more negative enthalpy of reaction (ΔΔH = 4.5 kcal mol−1) and a greater increase in entropy (Δ(−TΔS) = 5.6 kcal mol−1 K−1), corresponding to a smaller overall Gibbs free energy (ΔΔG = 1.1 kcal mol−1). These differences in the thermodynamic parameters are evidence that some structural elements have different conformations in the ATP-bound and ADP-bound states, leading to an increase in hydrogen bond/salt-bridge formation and an overall more ordered protein-ligand complex for the ATP-bound state.
FIGURE 3.
Thermodynamics of ATP binding. A, ITC titration of 4.0 mm AMPPNP into 0.49 mm E368(wt) at 10 °C (top) and the peak-integrated heats of reaction versus molar ratio (bottom). The smooth line corresponds to a single-site binding model yielding ΔH = −8.5 kcal mol−1 and Kd = 64.5 μm. B, ITC titration of 4.0 mm ATPγS into 0.59 mm E368(rk) at 10 °C: ΔH = −21.6 kcal mol−1 and Kd = 97 μm. C, ITC titration of 4.5 mm ATP into 0.59 mm E368(sc/rk) at 10 °C: ΔH = −24.7 kcal mol−1 and Kd = 43.5 μm. D, ΔH versus temperature for ATP binding to E368(sc/rk), yielding ΔCp = −0.72 kcal mol−1 K−1. E, ATP-dependent competitive inhibition of mantATP (as indicated). Final concentrations are: 3 μm kinesin-5, 0 or 3.5 μm MTs, 0 or 12 μm Taxol, 4 μm mantATP, 0–256 μm ATP. The fits of the data provide Ki,ATP at 2.2, 41.1, and 43.9 μm, respectively. F, AMPPNP-dependent competitive inhibition of mantATP binding (as indicated). Final concentrations: 1 μm kinesin-5, 0 or 1.5 μm MTs, 0 or 6 μm Taxol, 1.5 μm mantATP, 0–512 μm AMPPNP. Ki,AMPPNP = 264 μm, 782 μm, and 676 μm, respectively.
The change in heat capacity (ΔCp) for the interaction of ATP with E368(sc/rk) was determined by performing ITC experiments over a range of temperatures (Fig. 3D; supplemental Table S1). The linear fit of the data yielded a large negative heat capacity (ΔCp = −0.723 kcal mol−1 K−1) similar to the ADP binding heat capacity (Fig. 1C). We estimated the solvent-accessible surface area of E368(wt) with MgAMPPNP bound at the active site (3HQD (27)), and our change in heat capacity was consistent with ∼537 Å2 of surface area being buried upon complex formation.
AXP-dependent competitive inhibition experiments using stopped-flow fluorometry were performed to confirm the ATP binding measurements obtained by ITC and also to characterize ATP and AMPPNP binding in the presence of MTs. For E368(wt) in the absence of MTs, tight ATP-dependent inhibition (Ki,ADP = 2.2 μm) was observed, likely due to kinetic partitioning of the kinesin-5-ATP intermediate forward in the cycle as a result of rapid ATP hydrolysis and Pi release (Fig. 3E). However, the non-hydrolyzing switch 1 double-mutant E368(sc/rk) shows weak ATP affinity at 41 μm, which was comparable with the Kd,ATP observed in the corresponding ITC titration (Fig. 3C). Interestingly, in the presence of MTs, ATP affinity was largely unaffected (Ki,ADP = 44 μm) and comparable with the ADP affinity for the MT-E368(wt) complex (Ki,ADP = 48 μm; Fig. 1D). At 10 °C, the affinity of AMPPNP for E368(wt) (Kd = 64.5 μm) was also similar to the affinity of ATP for E368(sc/rk), suggesting that both titrations reflect an ATP-bound, non-hydrolyzing state. Fig. 3F shows that the observed AMPPNP affinity to E368(wt) was similar in the absence and presence of MTs (Ki,AMPPNP = 782 versus 676 μm at 25 °C, respectively). However, when the motor was truncated to eliminate the neck linker, a 3-fold increase in AMPPNP affinity (Ki,AMPPNP = 264 μm) in the absence of MTs was observed. Again, these results are consistent with a disordered-to-ordered conformational change in the neck linker upon AMPPNP binding to kinesin-5, although this conformational change is likely different from the one associated with ADP binding.
ATP Hydrolysis and Pi Release
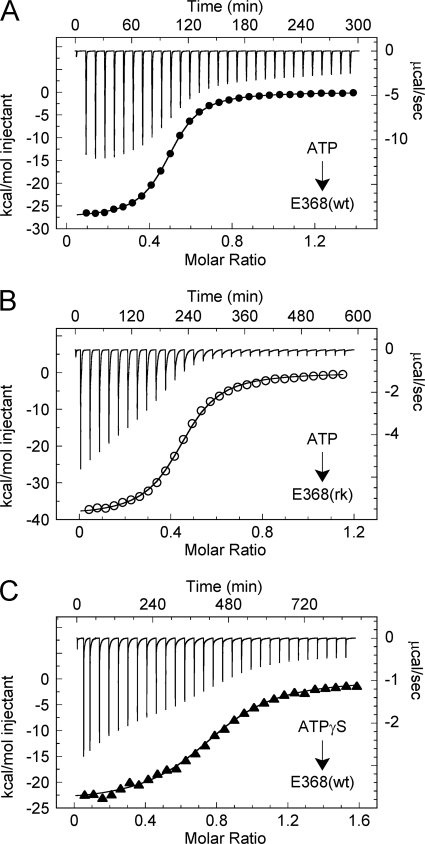
After ATP binding to kinesin-5, ATP hydrolysis and Pi product release occur to complete the cycle. Three independent ITC experiments were performed to monitor the thermodynamics of the combined steps of ATP binding, ATP hydrolysis, and Pi product release: 1) ATP titrated into E368(wt), 2) ATP titrated into E368(rk), and 3) ATPγS titrated into E368(wt) (Fig. 4, A–C). There are several prominent features in these thermodynamic profiles. First, we observed tight ATP affinity for each titration (Kd = 2–5 μm; ΔG = −6.9–7.3 kcal mol−1), which was similar to the ATP-dependent competitive inhibition constant at 2.2 μm (Fig. 3E). Second, the observed enthalpy (ΔH) for the titrations of ATP and ATPγS into E368(wt) were similar (ΔH = −27.7 and −24.0 kcal mol−1, respectively) and also comparable with the ΔH of ATP binding to E368(sc/rk) at this temperature (ΔH = −24.7 kcal mol−1; Fig. 3C; Table 1). The similarities between these data suggest that the non-covalent interactions upon substrate binding are comparable despite the slowed rate of ATPγS hydrolysis by E368(wt) or the lack of ATP hydrolysis by E368(sc/rk). Unexpectedly, the ΔH for ATP titrated into E368(rk) was very high (ΔH = −39.0 kcal mol−1) with a compensatory entropic penalty (−TΔS = 32.0 kcal mol−1) to give rise to the overall similar ATP affinity when compared with E368(wt). We hypothesize that the arginine-to-lysine substitution results in an alternate conformational change in Sw1 that traps the enzyme in an enthalpically favorable, entropically unfavorable state, which gives rise to the dramatic reduction in the rate of ATP hydrolysis. Third, the heats of reaction for the final 5–6 injections show differences between these titrations prior to baseline normalization; ATP into E368(wt) was approximately −10 kcal mol−1, whereas ATP into E368(rk) and ATPγS into E368(wt) were approximately −4 kcal mol−1. Together, these results suggest that ATP hydrolysis and Pi product release generate additional net heat between 4 and 10 kcal mol−1. Based on results from the ADP release experiments (Fig. 2), it appears that Pi release would be an endothermic reaction. Thus, this net exothermic heat from ATP turnover likely arises from the chemical hydrolysis step in the cycle.
FIGURE 4.
Thermodynamics of ATP hydrolysis. A, ITC titration of 1.1 mm ATP into 0.18 mm E368(wt) at 10 °C with spacing between injections at 600 s (top) and the peak-integrated heats of reaction versus molar ratio (bottom). The fit to a single-site binding model yields ΔH = −27.7 kcal mol−1 and Kd = 2.1 μm. B, ITC titration of 1.0 mm ATP into 0.20 mm E368(rk) at 10 °C with spacing at 900 s: ΔH = −39.0 kcal mol−1 and Kd = 3.2 μm. C, ITC titration of 0.7 mm ATPγS into 0.10 mm E368(wt) at 10 °C with spacing at 1200 s: ΔH = −24.0 kcal mol−1 and Kd = 4.8 μm.
DISCUSSION
The thermodynamic parameters that govern the ATPase mechanism of kinesin-5, a processive, motile kinesin, have been determined in the absence and presence of MTs. In the absence of MTs, the overall energy for ADP or ATP binding is characterized by a large favorable enthalpic contribution coupled to a smaller entropic penalty. Additionally, the individual roles of the neck linker and the MT in modulating kinesin nucleotide affinity were determined. This analysis revealed several new facets of the energy landscape governing kinesin mechanochemistry that provide a better understanding of how these molecular machines direct the energy of ATP hydrolysis for force production (Fig. 5).
FIGURE 5.
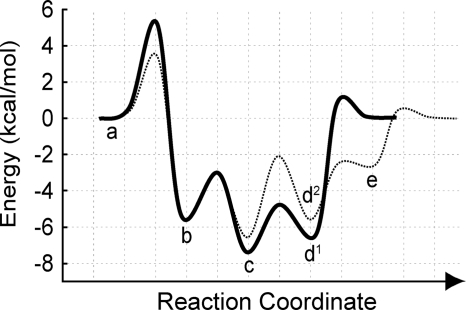
Model of the kinesin-5 energy landscape. The energy landscape for kinesin-5 in the absence (solid line) and presence (dashed line) of MTs is shown. The thermodynamically and kinetically defined intermediate states represent: (MT)kinesin + ATP (a), (MT)kinesin-ATP (b), (MT)kinesin-ADP-Pi (c), kinesin-ADP + Pi (d1), MT + kinesin-ADP + Pi (d2), and MT-kinesin-ADP (e). The magnitude of energy for each transition state was estimated from the Arrhenius equation using previously determined kinetic rate constants for the kinesin-5 ATPase mechanism (9, 20, 21).
Kinesin-5 nucleotide affinity was found to differ drastically between ADP product and ATP substrate (4–8 versus 40–100 μm, respectively) when the enzyme was mutated such that it can bind but not hydrolyze the substrate. This difference in affinity was dominated by a much larger entropic penalty (TΔS) for ATP binding (19 kcal mol−1) when compared with ADP binding (14 kcal mol−1), which can be explained by several conserved conformational changes that occur when kinesin binds ATP. A recent crystallographic model of a kinesin-AMPPNP complex (27) shows conformational changes in switch 1 and switch 2 that would likely reduce the structural dynamics of loops L9 and L11, respectively. In addition, loop L5 shows different conformations between the ADP (26) and ATP states (27), with the overall B-factor for this loop being reduced in the ATP state. Finally, the switch 2 cluster (α4-L12-α5) undergoes a rigid body movement upon ATP binding that is correlated with docking of the neck linker (27). Together, these ATP-dependent structural changes in the catalytic domain would contribute to an unfavorable reduction in entropy (disorder) that leads to weaker ATP affinity when the enzyme is unable to hydrolyze ATP.
Nevertheless, wild-type kinesin undergoes rapid ATP hydrolysis and Pi release (20), rendering the kinesin-ATP intermediate kinetically and thermodynamically invisible (Fig. 5). As described previously (20), the rate of ATP hydrolysis is much faster than the rate of formation of the kinesin-ATP complex, and the apparent reversal rate of ATP hydrolysis is much slower than the rate of the isomerization reversal due to kinetic partitioning of the kinesin-ADP-Pi complex forward with rapid Pi release. These facts imply that much tighter ATP binding affinity would be observed for the wild-type enzyme that can hydrolyze ATP. This hypothesis was confirmed using ITC by titrating ATP into E368(wt) (Fig. 4A), with ATP affinity shown to be 2–5 μm. For ATPγS titrated into E368(wt) (Fig. 4B), the enthalpy change (ΔH) was comparable with the ATP binding thermodynamics observed using the non-hydrolyzing E368(sc/rk) mutant (Table 1), yet the entropic penalty was dramatically reduced (Δ(TΔS) = −2 kcal mol−1). These data indicate that the increase in ATP affinity due to ATP hydrolysis and Pi release results from the reversal of the conformational changes that occur in the catalytic domain to form a tight kinesin-ATP complex, which would result in a favorable increase in entropy. Therefore, this fine-tuning of the overall thermodynamic equilibrium constants would produce diversity in the ATPase mechanism across the kinesin superfamily as well as other NTPase families such as myosins and G proteins.
Previous studies have looked at the importance of the kinesin neck linker in the mechanochemical ATPase cycle (27). Here, the role of the kinesin-5 neck linker in modulating nucleotide affinity was investigated by engineering a truncated construct lacking the amino acids for the neck linker (E358(wt); Fig. 1A). ITC and stopped-flow experiments yielded data indicating that the removal of the neck linker results in a 10-fold enhancement in ADP affinity (Fig. 1D) and a 3-fold enhancement in AMPPNP affinity (Fig. 3F). Data from ITC experiments show changes in both enthalpy (ΔH) and entropy (TΔS) for ADP binding to E358(wt) and E368(wt). However, the magnitude of the change in entropy was greater than the magnitude of the change in enthalpy, giving rise to a lower ΔG, and hence, a higher E358(wt) ADP affinity. This result suggests that in the absence of MTs, the kinesin-5 neck linker adopts a disordered conformation in the nucleotide-free state similar to kinesin-1 (6). However, concomitant with nucleotide binding, the neck linker undergoes a conformational change resulting in docking to the motor domain. Crystallographic models (26, 27) and spectroscopic analyses (10, 32) suggest that these docked neck linker conformations are different between the ADP- and ATP-bound states. The difference in neck linker docking onto the core domain may be reflected in the dissimilarity of nucleotide affinity enhancement observed in experiments with E358(wt) (10-fold for ADP affinity versus 3-fold for AMPPNP affinity).
A recent study by Sheth et al. (28) utilized a different monomeric construct of kinesin-5 that lacked the initial 14 amino acids at the N terminus, which was replaced by a hexahistidine tag. Recent studies by Khalil et al. (33) and Hwang et al. (34) demonstrate that the N-terminal region of kinesin (called the cover strand) interacts with the neck linker to form the so-called cover-neck bundle, which is essential for proper force production. The nucleotide binding affinities that Sheth et al. observed (28) were 10-fold tighter than the measured affinities for E368(wt) in this study, yet they were similar to the affinities measured for our truncated construct E358(wt). This discrepancy implies that both the N-terminal region and the neck linker of kinesin-5 play a role in modulating nucleotide affinity. Thus, it appears that in the absence of the N-terminal cover strand, the neck linker is unable to dock onto the motor domain upon nucleotide binding and remains disordered. Conversely, if the neck linker is missing, as in the E358(wt) construct, the N-terminal cover strand remains flexible and disordered. This flexible and disordered state is seen in the crystal structure of NOD, a kinesin that naturally lacks the neck linker (11). It is only when both the N-terminal cover strand and the neck linker are present that the disordered-to-ordered transition can occur, which thereby weakens nucleotide affinity.
The role of MT in regulating nucleotide binding to kinesin-5 was also investigated. Stopped-flow experiments revealed that the ATP affinity for E368(sc/rk), the construct incapable of hydrolysis, is similar both on and off the MT (Fig. 3, E and F). Previous studies show that the kinetics of the ATP-dependent isomerization in the kinesin-5 catalytic core were over 10-fold faster in the presence of MTs (9, 10, 20, 21). However, the data presented here suggest that the overall affinity for ATP does not change when kinesin is bound to the MT, indicating that the rate of the reversal of the isomerization reaction must also be faster. Therefore, it is likely that the MT lowers the energy of the transition state for tight ATP binding but does not affect the difference in energy between the free and bound states (Fig. 5). To date, the structural explanation for this isomerization in the kinesin catalytic core upon nucleotide binding remains elusive. Results given here provide a foundation for future studies to reveal the nature of this structural transition in the kinesin ATPase cycle.
In contrast to ATP binding to the MT-kinesin-5 complex, the ADP affinity was drastically weakened in the presence of MTs (4 μm for kinesin-5 versus 40 μm for MT-kinesin-5; Fig. 3E). Interestingly, the apparent affinity for ATP (without ATP hydrolysis) and ADP was equivalent when kinesin-5 was bound to the MT. Therefore, the overall energy of the ATP- or ADP-bound state of the complex was equivalent if the motor was unable to hydrolyze the ATP substrate. Using wild-type kinesin-5 that was capable of ATP hydrolysis, the apparent affinity for ATP increased to 7 μm in the presence of MTs based on the steady-state Michaelis constant (Km,ATP) (21), thus lowering the overall energy of the MT-kinesin-ADP-Pi intermediate (Fig. 5). It was observed for kinesin-5 that the transition from the MT-kinesin-ADP-Pi state to the MT-kinesin-ADP intermediate through Pi release was rate-limiting for the ATPase mechanism (21). Therefore, we propose that a significant change in energy occurs upon ATP hydrolysis, which results in a stable MT-kinesin intermediate state. This difference in energy may explain the difference in MT affinity for kinesin in this nucleotide state, which promotes active detachment of the motor from the filament. Upon kinesin-ADP-Pi dissociation from the MT, the kinesin would reverse the interactions of the switches with the nucleotide to promote Pi release. When the kinesin-ADP complex rebinds the MT, it undergoes a rapid conformational change to release the bound ADP product, thus completing the cycle (Fig. 5). Together, this energy landscape provides a model of the thermodynamic and kinetic profiles of a motile kinesin ATPase cycle and allows for future comparison with different kinesin, myosin, and G protein cycles to better understand the mechanism of how these enzymes function in mechanochemical energy transduction.
Supplementary Material
Acknowledgments
We thank Colette Quinn, Jolene Schuster, and Dean Wilcox for training, guidance, and usage of the VP-ITC. We also thank Dean Wilcox (Dartmouth College) and Dale Mierke (Dartmouth College) for insightful comments on this manuscript and our colleagues in the Kull laboratory for intellectual discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant F32AR054653 through the NIAMS (to J. C. C.). This work was also supported by the Dartmouth College Zabriskie Fellowship (to Y. C. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Figs. S1–S3, and Tables S1 and S2.
- MT
- microtubule
- AMPPNP
- adenosine 5′-(β,γ-imido)triphosphate
- ATPγS
- adenosine 5′-[γ-thio]triphosphate
- AMPPCP
- adenosine 5′-(β,γ-methyl)triphosphate
- AXP
- any adenine nucleotide including ADP, ATP, AMPPNP, ATPγS, or AMPPCP
- E368(wt)
- human kinesin-5 (Eg5/KSP) motor domain containing N-terminal 368 residues plus an N-terminal His10 tag
- E358(wt)
- Eg5 motor domain minus the neck linker
- mant
- 2′-(3 ′)-O-(N-methylanthraniloyl)
- ITC
- isothermal titration calorimetry
- PIPES
- 1,4-piperazinediethanesulfonic acid.
REFERENCES
- 1.Kull F. J., Endow S. A. (2002) J. Cell Sci. 115, 15–23 [DOI] [PubMed] [Google Scholar]
- 2.Vale R. D. (2003) Cell 112, 467–480 [DOI] [PubMed] [Google Scholar]
- 3.Marx A., Müller J., Mandelkow E. (2005) Adv. Protein Chem. 71, 299–344 [DOI] [PubMed] [Google Scholar]
- 4.Moyer M. L., Gilbert S. P., Johnson K. A. (1996) Biochemistry 35, 6321–6329 [DOI] [PubMed] [Google Scholar]
- 5.Ma Y. Z., Taylor E. W. (1997) J. Biol. Chem. 272, 717–723 [DOI] [PubMed] [Google Scholar]
- 6.Rosenfeld S. S., Jefferson G. M., King P. H. (2001) J. Biol. Chem. 276, 40167–40174 [DOI] [PubMed] [Google Scholar]
- 7.Okada Y., Hirokawa N. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 640–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nitta R., Kikkawa M., Okada Y., Hirokawa N. (2004) Science 305, 678–683 [DOI] [PubMed] [Google Scholar]
- 9.Cochran J. C., Sontag C. A., Maliga Z., Kapoor T. M., Correia J. J., Gilbert S. P. (2004) J. Biol. Chem. 279, 38861–38870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenfeld S. S., Xing J., Jefferson G. M., King P. H. (2005) J. Biol. Chem. 280, 35684–35695 [DOI] [PubMed] [Google Scholar]
- 11.Cochran J. C., Sindelar C. V., Mulko N. K., Collins K. A., Kong S. E., Hawley R. S., Kull F. J. (2009) Cell 136, 110–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pechatnikova E., Taylor E. W. (1997) J. Biol. Chem. 272, 30735–30740 [DOI] [PubMed] [Google Scholar]
- 13.Mackey A. T., Gilbert S. P. (2000) Biochemistry 39, 1346–1355 [DOI] [PubMed] [Google Scholar]
- 14.Mackey A. T., Gilbert S. P. (2003) J. Biol. Chem. 278, 3527–3535 [DOI] [PubMed] [Google Scholar]
- 15.Cole D. G., Saxton W. M., Sheehan K. B., Scholey J. M. (1994) J. Biol. Chem. 269, 22913–22916 [PMC free article] [PubMed] [Google Scholar]
- 16.Kashina A. S., Baskin R. J., Cole D. G., Wedaman K. P., Saxton W. M., Scholey J. M. (1996) Nature 379, 270–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawin K. E., LeGuellec K., Philippe M., Mitchison T. J. (1992) Nature 359, 540–543 [DOI] [PubMed] [Google Scholar]
- 18.Kapitein L. C., Peterman E. J., Kwok B. H., Kim J. H., Kapoor T. M., Schmidt C. F. (2005) Nature 435, 114–118 [DOI] [PubMed] [Google Scholar]
- 19.Krzysiak T. C., Gilbert S. P. (2006) J. Biol. Chem. 281, 39444–39454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cochran J. C., Gilbert S. P. (2005) Biochemistry 44, 16633–16648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cochran J. C., Krzysiak T. C., Gilbert S. P. (2006) Biochemistry 45, 12334–12344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valentine M. T., Fordyce P. M., Krzysiak T. C., Gilbert S. P., Block S. M. (2006) Nat. Cell Biol. 8, 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siligardi G., Hu B., Panaretou B., Piper P. W., Pearl L. H., Prodromou C. (2004) J. Biol. Chem. 279, 51989–51998 [DOI] [PubMed] [Google Scholar]
- 24.Pretz M. G., Albers S. V., Schuurman-Wolters G., Tampé R., Driessen A. J., van der Does C. (2006) Biochemistry 45, 15056–15067 [DOI] [PubMed] [Google Scholar]
- 25.Perozzo R., Folkers G., Scapozza L. (2004) J. Recept. Signal Transduct. Res. 24, 1–52 [DOI] [PubMed] [Google Scholar]
- 26.Turner J., Anderson R., Guo J., Beraud C., Fletterick R., Sakowicz R. (2001) J. Biol. Chem. 276, 25496–25502 [DOI] [PubMed] [Google Scholar]
- 27.Parke C. L., Wojcik E. J., Kim S., Worthylake D. K. (2010) J. Biol. Chem. 285, 5859–5867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheth P. R., Basso A., Duca J. S., Lesburg C. A., Ogas P., Gray K., Nale L., Mannarino A. F., Prongay A. J., Le H. V. (2009) Biochemistry 48, 11045–11055 [DOI] [PubMed] [Google Scholar]
- 29.Murphy K. P., Freire E. (1992) Adv. Protein Chem. 43, 313–361 [DOI] [PubMed] [Google Scholar]
- 30.Spolar R. S., Livingstone J. R., Record M. T., Jr. (1992) Biochemistry 31, 3947–3955 [DOI] [PubMed] [Google Scholar]
- 31.Lee B., Richards F. M. (1971) J. Mol. Biol. 55, 379–400 [DOI] [PubMed] [Google Scholar]
- 32.Maliga Z., Xing J., Cheung H., Juszczak L. J., Friedman J. M., Rosenfeld S. S. (2006) J. Biol. Chem. 281, 7977–7982 [DOI] [PubMed] [Google Scholar]
- 33.Khalil A. S., Appleyard D. C., Labno A. K., Georges A., Karplus M., Belcher A. M., Hwang W., Lang M. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19247–19252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hwang W., Lang M. J., Karplus M. (2008) Structure 16, 62–71 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.