Abstract
The antiosteoporotic treatment strontium ranelate (SrRan) was shown to increase bone mass and strength by dissociating bone resorption and bone formation. To identify the molecular mechanisms of action of SrRan on osteoblasts, we investigated its effects on calcineurin-NFAT (nuclear factor of activated T cells) signaling, an important calcium sensitive pathway controlling bone formation. Using murine MC3T3-E1 and primary murine osteoblasts, we demonstrate that SrRan induces NFATc1 nuclear translocation, as shown by immunocytochemical and Western blot analyses. Molecular analysis showed that SrRan increased NFATc1 transactivation in osteoblasts, an effect that was fully abrogated by the calcineurin inhibitors cyclosporin A or FK506, confirming that SrRan activates NFATc1 signaling in osteoblasts. This has functional implications because calcineurin inhibitors blunted the enhanced osteoblast replication and expression of the osteoblast phenotypic markers Runx2, alkaline phosphatase, and type I collagen induced by SrRan. We further found that SrRan increased the expression of Wnt3a and Wnt5a as well as β-catenin transcriptional activity in osteoblasts, and these effects were abolished by calcineurin inhibitors. The Wnt inhibitors sFRP1 and DKK1 abolished SrRan-induced osteoblast gene expression. Furthermore, blunting the Wnt5a receptor Ryk or RhoA that acts downstream of Ryk abrogated cell proliferation and osteoblast gene expression induced by SrRan. These results indicate that activation of NFATc1 and downstream canonical and non-canonical Wnt signaling pathways mediate SrRan-induced osteoblastic cell replication and differentiation, which provides novel insights into the mechanisms of action of this antiosteoporotic agent in osteoblastogenesis.
Keywords: Beta-Catenin, Bone, Calcineurin, Calcium, Wnt Pathway, Osteoblast, Strontium
Introduction
The maintenance of bone mass is dependent on bone remodeling activity, which involves the balanced effects of bone-resorbing osteoclasts and bone-forming osteoblasts. Excessive bone resorption relative to bone formation results in decreased bone mass, altered bone quality, and increased incidence of fractures in osteoporosis (1). Available treatments for osteoporosis include antiresorptive or anabolic treatments, which induce variable effects on bone mass and fracture incidence (2). An antiosteoporotic treatment, strontium ranelate (SrRan),3 was shown to promote bone formation and to reduce bone resorption in vitro (3). In vivo, SrRan was found to induce a positive bone balance in experimental osteopenic models (3–5). This treatment was also shown to decrease the risk of vertebral and non-vertebral fractures in patients with postmenopausal osteoporosis (6–8). In several in vitro models, SrRan was shown to promote osteoblast replication, differentiation, and survival (9–13). Nevertheless, the molecular mechanisms of action of SrRan on bone-forming cells are still under investigation. The positive effects of SrRan on osteoblast replication and survival may involve, in part, the seven-transmembrane-spanning extracellular calcium-sensing receptor that is expressed by osteoblastic cells (14) and that responds to strontium (15–17). The signaling pathways activated by SrRan via the calcium-sensing receptor in osteoblasts include mitogen-activated protein kinases (MAPK) ERK1/2, phosphatidylinositol 3-kinase, and phospholipase C (15, 17). However, we recently showed that SrRan increases osteoblastic cell replication and survival independently of the calcium-sensing receptor (17), indicating that other signaling mechanisms may contribute to the positive effects of SrRan on osteoblastogenesis.
Nuclear factor of activated Tc (NFATc) are transcription factors that are highly phosphorylated and remain in the cytoplasm in unstimulated cells. An increase in intracellular calcium level leads to activation of the heterodimeric serine/threonine phosphatase calcineurin (Cn) that is involved in the regulation of various physiological processes (18). This induces NFATc dephosphorylation, leading to its nuclear translocation and regulation of target genes (18, 19). In bone, Cn-NFATc signaling has recently emerged as an important activator of bone resorption, bone formation, and bone mass (20–26). Recent studies indicate that NFATc1 and NFATc2 may positively control bone formation through increased osteoblast replication and function (27–31), highlighting the importance of the Cn-NFATc signaling in the control of both bone resorption and bone formation.
Because strontium is a cation chemically close to calcium and Cn is a calcium-dependent phosphatase, we hypothesized that increasing strontium concentration in the osteoblast environment may activate Cn and subsequent NFATc-mediated signals in osteoblasts. In this study, we investigated the effects of SrRan on Cn-NFATc signaling in osteoblasts and determined the implication of this pathway on the osteoblast phenotype. We show here that SrRan activates Cn-NFATc1 signaling in osteoblasts, which results in increased osteoblastic cell replication and phenotypic gene expression. We further show that Cn-NFATc1 activation induced by SrRan results in induction of Wnt expression and activation of canonical and non-canonical Wnt signaling pathways that are involved in SrRan-induced osteoblastogenesis.
EXPERIMENTAL PROCEDURES
Cells and Reagents
MC3T3-E1 cells were obtained from ATCC. Primary mouse calvaria osteoblastic cells were dissected from normal newborn mice, as described previously (32). SrRan was a mix of strontium chloride (SrCl2, 6H2O) (Sigma) and sodium ranelate (Technologie Servier, Orléans, France) and was added to the culture medium at a molar ratio of 1:100 respectively to reflect the ratio found in blood of postmenopausal osteoporotic patients treated with strontium ranelate (2 g/day) (7, 8). SrRan concentrations were expressed as Sr2+ concentrations. The calcineurin inhibitors cyclosporin A (CSA) and FK506, PMA, ionomycin, calcium chloride, dexamethasone, and rabbit anti-β-actin antibody were from Sigma. Mouse anti-p84 antibody was from Abcam (Cambridge, UK). Mouse anti-NFATc was from BD Biosciences (Erembodegen, Belgium). Mouse anti-β-catenin was from Santa Cruz Biotechnology (Santa Cruz, CA). Secondary antibodies conjugated to CY2 or CY3 were from Jackson ImmunoResearch (Newmarket, UK). Recombinant human sFRP1 and BMP2 were from R&D Systems (Lille, France). Wnt3a-conditioned medium was prepared as described previously (33).
Transfection and Luciferase Activity
Transient transfections with pGL3-9xNFAT-luc plasmid (34) (kindly provided by Dr. Molkentin, Cincinnati Children's Hospital Medical Center, Cincinnati, OH), DKK1 encoding plasmid (kindly provided by Galapagos, Romainville, France), dominant-negative RhoA (DN-RhoA, kindly provided by Dr. Jacques Bertoglio, INSERM, Chatenay-Malabry, France), or control vectors were performed as described previously (33). For luciferase reporter assays, pCMV-β-galactosidase was added to the transfection mix (TCF and TopFlash or FopFlash). Transcriptional activity was evaluated by luciferase assay using a luciferase assay kit (Promega) and β-galactosidase gene reporter assay kit (Roche Diagnostics, Meylan, France) (33).
Immunofluorescence Microscopy
Subconfluent MC3T3-E1 osteoblastic cells and primary mouse calvaria osteoblasts were fixed in 4% PFA, permeabilized in 0.5% Triton X-100 for 15 min, saturated with 0.5% bovine serum albumin, incubated with primary antibody (1/100), and then incubated with corresponding secondary antibodies (donkey anti-mouse conjugated to CY3 or donkey anti-rabbit conjugated to CY2; Jackson ImmunoResearch Laboratories, West Grove, PA). 4′,6-Diamidino-2-phenylindole (300 nm) was used to localize the nuclei. The signal was visualized by confocal fluorescence microscopy (Apotome, Carl Zeiss SAS, Le Pecq, France).
Western Blot Analysis
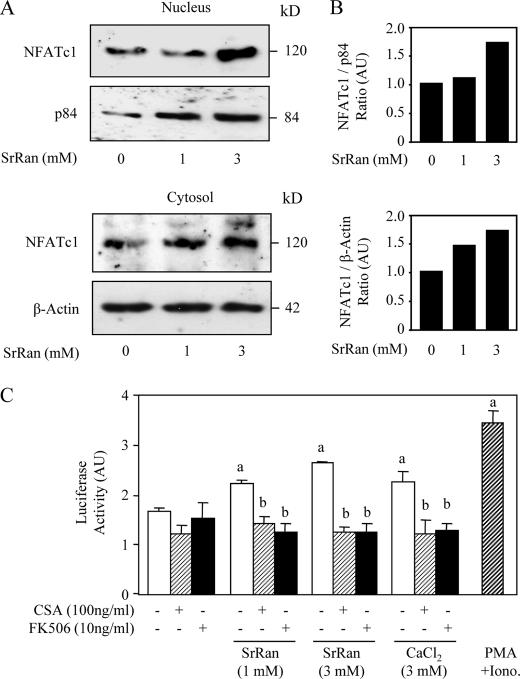
MC3T3-E1 osteoblastic cell lysates were prepared as described previously (32). Protein concentrations in both cytosolic and nuclear subcellular fractions were measured using the DC protein assay (Bio-Rad Laboratories). Equal aliquots (60 μg) of cell lysates were resolved on 10% SDS-PAGE. Western blot was performed as described previously (17) using specific primary antibodies raised against NFATc1 (1:1000) and against p84 (1/1000) for nuclear fraction or β-actin (1/2000) for cytosolic fraction. Representative images of immunoblots are shown in Fig. 2. Relative levels are expressed as a ratio of treated over control, after correction to the housekeeping protein. Results are representative of 2–3 independent experiments.
FIGURE 2.
NFATc1 nuclear translocation induced by SrRan results in NFATc transactivation in osteoblasts. A, MC3T3-E1 osteoblastic cells were treated with the Cn inhibitor CSA (100 ng/ml) for 10 min, rinsed, and incubated with or without SrRan (1–3 mm) for 30 min. Cytosolic and nuclear cell lysates were immunoreacted with NFATc1 and β-actin or with NFATc1 and p84 antibody, respectively. B, Western blots were scanned, quantified, and reported as ratios in arbitrary units (AU). C, MC3T3-E1 osteoblastic cells were transfected with a vector encoding luciferase sequence under NFAT-response elements linked to a basic promoter and then incubated with or without PMA (25 nm)/ionomycin (Iono, 1 μm) used as positive control, SrRan (1–3 mm) or CaCl2 (3 mm) for 24 h in the presence or absence of the Cn inhibitors CSA (100 ng/ml) or FK506 (10 ng/ml). Transcriptional activity was evaluated by luciferase assay. Data are the mean ± S.D. a, p < 0.05 versus control cells; b, p < 0.05 versus SrRan- or calcium-treated cells.
Cell Replication Analysis
Cells were plated at 6000 cells/cm2 in 96 wells and treated as indicated, and cell replication was determined using a bromodeoxyuridine enzyme-linked immunosorbent assay (Roche Diagnostics), according to the manufacturer's instructions. The experiments were repeated three times with at least six replicates per experiment.
Quantitative Reverse Transcription-PCR Analysis
Total RNAs were isolated using the TRIzol reagent (Invitrogen, Cergy Pontoise, France) according to the manufacturer's recommendations and stored at −80 °C in RNASecure (Applied Biosystems, Courtaboeuf, France) supplemented with H2O. cDNAs were generated from 3 μg of total RNA of each sample by reverse transcription using 300 units of Moloney murine leukemia virus reverse transcriptase, 15 μg of oligo(dT) primers, 1 mm dNTP, in 30 μl of total volume. The relative mRNA levels were evaluated by quantitative PCR using the SYBR Green master kit (ABGen, Courtaboeuf, France) supplemented with 0.5 μm of specific primers (Table 1). Signal was normalized to 18S as internal control. Reactions were performed in triplicate on the LightCycler 480 instrument (Roche Diagnostics) using the following thermal conditions: activation for one cycle at 95 °C for 15 min; 40 cycles of denaturation at 95 °C for 20 s; annealing at 58 °C for 15 s; and extension at 72 °C for 15 s. Melting curve analysis was included to assure that only one PCR product was formed. The relative amount of RNA was calculated by the 2−ΔΔCt method.
TABLE 1.
Sequences of primers used
| Housekeeping gene | ||
| 18S | Sense | 5′-TCAAGAACGAAAGTCGGAGG-3′ |
| Antisense | 5′-GGACATCTAAGGGCATCACA-3′ | |
| Histone | ||
| H4 | Sense | 5′-GCAAAGTCTTGCGTGACAAC-3′ |
| Antisense | 5′-CACGGGTCTCCTCGTAGATG-3′ | |
| Osteoblast markers | ||
| Runx2 | Sense | 5′-TTGACCTTTGTCCCAATGC-3′ |
| Antisense | 5′-AGGTTGGAGGCACACATAGG-3′ | |
| ALP | Sense | 5′-AAGGCTTCTTCTTGCTGGTG-3′ |
| Antisense | 5′-GCCTTACCCTCATGATGTCC-3′ | |
| Col1A1 | Sense | 5′-CTTGGTGGTTTTGTATTCGATGAC-3′ |
| Antisense | 5′-GCGAAGGCAACAGTCGCT-3′ | |
| Wnt family members | ||
| Wnt3a | Sense | 5′-CTTAGTGCTCTGCAGCCTGA-3′ |
| Antisense | 5′-AGTGCTCAGAGAGGAGTACTGG-3′ | |
| Wnt4 | Sense | 5′-CTGGACTCCCTCCCTGTCTT-3′ |
| Antisense | 5′-ATGCCCTTGTCACTGCAAA-3′ | |
| Wnt5a | Sense | 5′-ATGAAGCAGGCCGTAGAAC-3′ |
| Antisense | 5′-CTTCTCCTTGAGGGCATCG-3′ | |
| Wnt11 | Sense | 5′-GAGGATCCCAAGCCAATAAA-3′ |
| Antisense | 5′-TCCAGGGAGGCACGTAGA-3′ | |
| DKK2 | Sense | 5′-CTGGTACCCGCTGCAATAAT-3′ |
| Antisense | 5′-CATGGTTGCGATCTCTATGC-3′ | |
| sFRP2 | Sense | 5′-GACAACGACCTCTGCATCC-3′ |
| Antisense | 5′-TCACACACCTTGGGAGCTT-3′ | |
| Frz9 | Sense | 5′-AGACGGGAGGCACCAATAC-3′ |
| Antisense | 5′-TGGAAAAGACTCCGATTTTGA-3′ | |
Statistical Analysis
All values are presented as the mean ± S.D. Data were analyzed with the unpaired two-tailed Student's t test as appropriate for the data set. A p value <0.05 was considered statistically significant.
RESULTS
SrRan Induces NFATc1 Nuclear Translocation and Transcriptional Activity in Osteoblasts
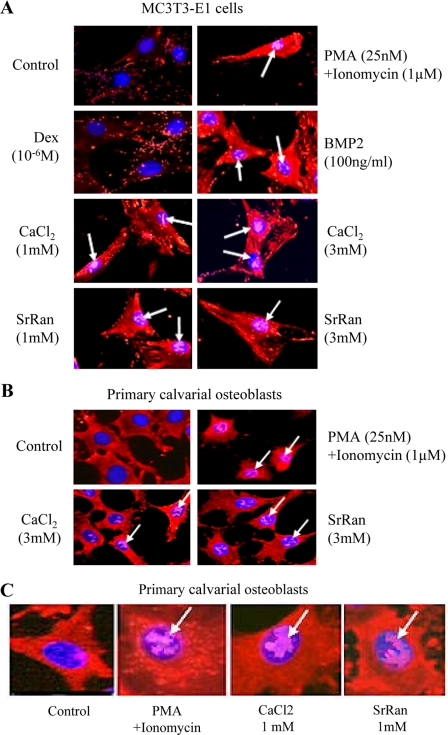
Activation of Cn is known to lead to dephosphorylation and nuclear translocation of NFATc1. We first determined the effect of SrRan on NFATc1 translocation. As expected, treatment of MC3T3-E1 cells with PMA and ionomycin, used as a positive control, led to NFATc1 nuclear translocation (Fig. 1A). We found that BMP2 induced NFATc1 translocation, whereas dexamethasone had no apparent effect. In these cells, both CaCl2 and SrRan induced NFATc1 translocation (Fig. 1A). Importantly, similar effects were observed in murine primary calvaria osteoblasts (Fig. 1, B and C). These results indicate that SrRan, like calcium, induces NFATc1 nuclear translocation in osteoblasts.
FIGURE 1.
SrRan induces NFATc1 nuclear translocation in osteoblasts. MC3T3-E1 osteoblastic cells (A) or mouse primary calvaria osteoblastic cells (B and C) were treated with CSA (100 ng/ml) for 10 min to fully phosphorylate NFATc1 and then rinsed and incubated with or without PMA (25 nm)/ionomycin (1 μm) for 4 h, or dexamethasone (Dex, 10−6 m), BMP2 (100 ng/ml), SrRan (1–3 mm), or CaCl2 (1–3 mm) for 30 min and immunostained for NFATc1 (red) and 4′,6-diamidino-2-phenylindole (blue). Arrows indicate nuclear localization of NFATc1 (violet spots). B, magnification, × 250; C, magnification, ×500.
To confirm this finding, we analyzed NFATc1 protein levels in MC3T3-E1 osteoblasts. As shown in Fig. 2A, Western blot analysis showed that SrRan increased NFATc1 levels in the nucleus, confirming the results obtained by immunocytochemical analysis. Quantification of the scanned blots confirmed the positive concentration-dependent effect of SrRan on NFATc1 nuclear translocation (Fig. 2B). We then determined whether NFATc1 translocation induced by SrRan had a functional impact on NFATc1-transactivating activity. As expected, PMA-ionomycin increased NFATc1 transcriptional activity by about 2-fold in MC3T3-E1 osteoblasts (Fig. 2C). We found that both CaCl2 and SrRan increased NFATc1 transcriptional activity in these cells. Moreover, this effect was fully abrogated in the presence of the Cn inhibitors CSA or FK506 (Fig. 2C). Taken together, these results indicate that SrRan, like calcium, induces NFATc1 translocation into the nucleus and activation of NFATc1 transcriptional activity.
SrRan-induced Osteoblastic Cell Replication and Differentiation Are Abrogated by Cn Inhibitors
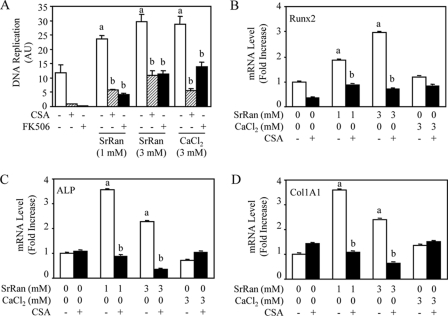
Having shown that SrRan activates NFATc1 signaling in osteoblasts, we then asked whether this effect may have significant impact on cell replication or differentiation. We found that SrRan increased MC3T3-E1 replication, as assessed by the bromodeoxyuridine assay (Fig. 3A). Remarkably, this effect was fully abolished by Cn inhibition using CSA or FK506. Similar effects were observed with CaCl2 (Fig. 3A). These data indicate that NFATc1 signaling mediates, at least in part, the increased cell replication induced by SrRan.
FIGURE 3.
SrRan-induced osteoblastic cell replication and differentiation are mediated by Cn/NFATc signaling. MC3T3-E1 osteoblastic cells were treated with the Cn inhibitor CSA (100 ng/ml) for 10 min, rinsed, and incubated with or without SrRan (1–3 mm) or CaCl2 (3 mm) in the presence or absence of the Cn inhibitors CSA (100 ng/ml) or FK506 (10 ng/ml). A, DNA replication was determined by bromodeoxyuridine incorporation assay after 24 h. AU, arbitrary units. B–D, mRNA expression of osteoblast markers (Runx2, ALP, Col1A1) was determined by quantitative PCR analysis after 48 h of incubation. Data are the mean ± S.D. a, p < 0.05 versus control cells; b, p < 0.05 versus SrRan- or calcium-treated cells.
We then investigated whether NFATc1 signaling may impact on osteoblast differentiation. As shown in Fig. 3, B–D, treatment with SrRan increased mRNA levels of the phenotypic osteoblast markers Runx2, ALP, and COL1A1 by 2–3-fold, as determined by quantitative reverse transcription-PCR, whereas CaCl2 had no significant effect. Importantly, the positive effect of SrRan on Runx2, ALP, and COL1A1 expression was abolished by the Cn inhibitor CSA (Fig. 3, B–D). Similar effects were observed in the presence of FK506 (data not shown). These results indicate that activation of NFATc1 signaling is involved in the increased osteoblast marker gene expression induced by SrRan in murine osteoblasts.
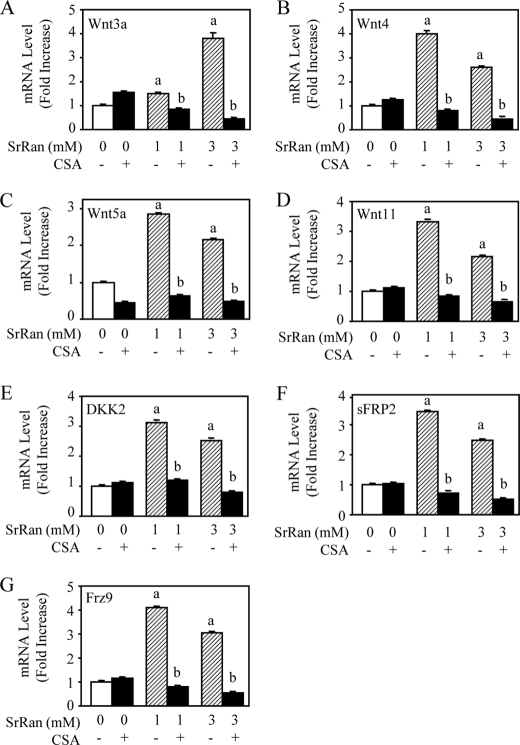
SrRan Induces Expression of Several Wnt-related Proteins
Calcineurin is known to affect β-catenin-associated pathways (35). Recent data indicate that activation of NFATc1 signaling in osteoblasts is associated with the production of Wnt-related proteins (28). Given that we found that SrRan activates NFATc1 transactivation and osteoblast differentiation (Fig. 3), we asked whether this effect may be mediated by the expression of Wnt proteins. Interestingly, we found that treatment of MC3T3-E1 osteoblastic cells with SrRan increased the mRNA expression of canonical and non-canonical Wnt partners (Wnt3a and Wnt4, Wnt5a, Wnt11, respectively). In addition, SrRan increased the expression of the Wnt inhibitor DKK2 and sFRP2 and receptor Frizzled9, suggesting a physiological balance in terms of stimulation of the Wnt pathway. The positive effect of SrRan on both Wnt activators and inhibitors was related to Cn-NFATc1 signaling because it was abrogated by CSA (Fig. 4, A–G) or FK506 (data not shown). These results indicate that multiple positive and negative regulators of Wnt pathway are targets of SrRan-mediated activation of Cn-NFATc1 signaling in osteoblastic cells.
FIGURE 4.
SrRan-mediated Cn/NFATc signaling induces expression of Wnt family molecules. MC3T3-E1 osteoblastic cells were treated with or without SrRan (1–3 mm) for 48 h in the presence or absence of the Cn inhibitor CSA (100 ng/ml). A–G, mRNA expression of Wnt molecules (A–D) and Wnt-related molecules (E–G) was determined by quantitative PCR analysis. Data are the mean ± S.D. a, p < 0.05 versus control cells; b, p < 0.05 versus SrRan-treated cells.
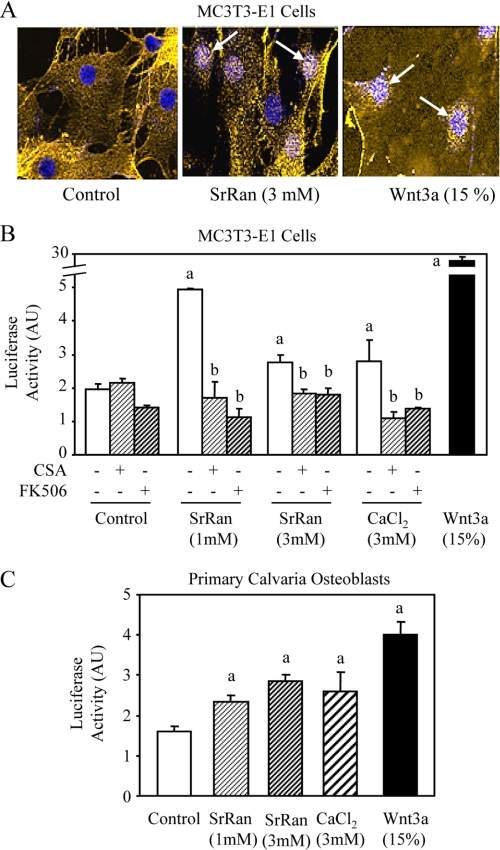
The above results, indicating that Wnt3a expression is increased by SrRan as a result of SrRan-induced Cn-NFATc1 activation, suggest that canonical Wnt signaling may be activated by SrRan in osteoblasts. To test this hypothesis, we determined whether SrRan activates β-catenin translocation into the nucleus. Confocal microscopy showed that SrRan increased β-catenin translocation into the nucleus in MC3T3-E1 osteoblasts, although to a lesser extent than Wnt3a, used as positive control (Fig. 5A). To confirm this finding, we determined whether SrRan promotes β-catenin transcriptional activity in osteoblasts using a reporter gene assay. As expected, Wnt3a markedly increased TCF-TOP transactivation (Fig. 5B). Interestingly, SrRan and CaCl2 also increased β-catenin transcriptional activity in MC3T3-E1 osteoblasts. This effect was not restricted to this cell line because both SrRan and CaCl2 increased TCF-TOP transactivation in primary murine calvaria osteoblasts, although the effect was limited (Fig. 5C). Importantly, the positive effects of SrRan and calcium on TCF-TOP transactivation were both related to Cn-NFATc1 activation because these effects were abolished by Cn inhibitors CSA and FK506 (Fig. 5B). These results indicate that SrRan promotes β-catenin transcriptional activity and that this effect is mediated by Cn/NFATc1 activation.
FIGURE 5.
SrRan-mediated Cn/NFATc signaling promotes β-catenin nuclear translocation and transcriptional activity. A, MC3T3-E1 osteoblastic cells were treated with 15% Wnt3a conditioned medium (used as positive control) or SrRan (1 mm) and then immunostained for β-catenin (yellow) and 4′,6-diamidino-2-phenylindole (blue), and β-catenin translocation in the nucleus (white spots, arrows) was determined by confocal microscopy. B and C, MC3T3-E1 cells or primary mouse calvaria cells were treated with 15% Wnt3a conditioned medium, SrRan (1–3 mm), or CaCl2 (3 mm) for 24 h in the presence or absence of the Cn inhibitors CSA (100 ng/ml) or FK506 (10 ng/ml), and β-catenin transcriptional activity was evaluated by a luciferase assay kit. Data are the mean ± S.D. a, p < 0.05 versus control cells; b, p < 0.05 versus SrRan or calcium-treated cells. AU, arbitrary units.
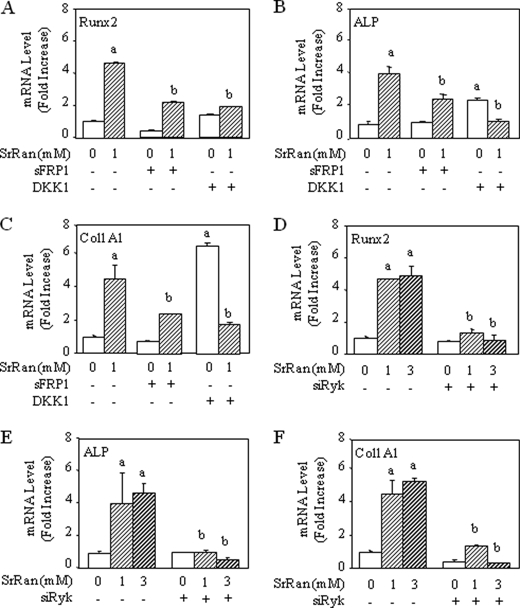
Because we found that SrRan increased the expression of non-canonical Wnt genes (Wnt4, Wnt5a, and Wnt11) (Fig. 4), we examined the possible role of non-canonical Wnt signaling in the response to SrRan in osteoblasts. To investigate the possible implication of the canonical and non-canonical Wnt pathways in SrRan-induced osteoblast gene expression, MC3T3-E1 osteoblasts were treated with SrRan in the presence of DKK1 that inhibits the Wnt-LRP5 interaction (36) or in the presence of sFRP1 that blocks Wnt proteins more effectively than sFRP2 (37). As shown in Fig. 6, A–C, both sFRP1 and DKK1 markedly reduced the expression of Runx2, ALP, and COL1A1 induced by SrRan. These results imply that both Wnt canonical and non-canonical pathways may be involved in osteoblast differentiation induced by SrRan.
FIGURE 6.
Inhibition of Wnt5a signaling blocks SrRan-induced osteoblast gene expression. A–C, MC3T3-E1 osteoblastic cells were transiently transfected with a vector encoding DKK1 or empty vector or incubated in the presence of sFRP1 and then treated with or without SrRan (1 mm) for 48 h. D–F, MC3T3-E1 osteoblastic cells were transiently transfected with a vector encoding a small interfering RNA raised against Ryk (siRyk) or an empty vector and then treated with or without SrRan (1–3 mm) for 48 h. mRNA expression of osteoblast markers (Runx2, ALP, and Col1A1) was determined by quantitative PCR analysis after an incubation of 48 h. Data are the mean ± S.D. a, p < 0.05 versus control cells; b, p < 0.05 versus SrRan-treated cells.
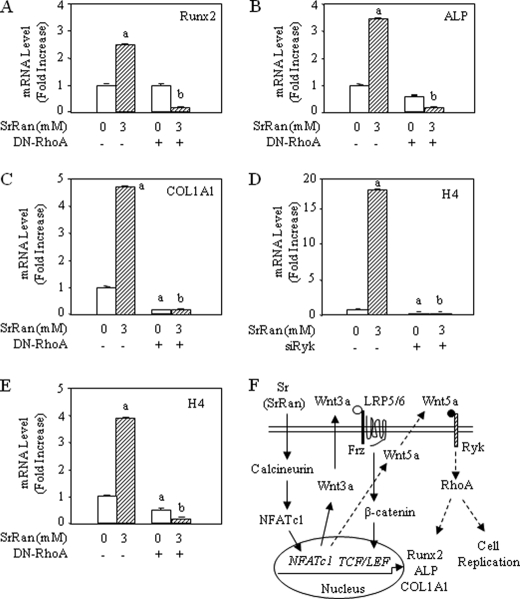
Non-canonical signals are transduced through frizzled receptors and co-receptors such as Ryk, which activate small G proteins including RhoA and c-Jun N-terminal kinase (JNK) and the calcium-dependent Nemo-like kinase (NLK) and NFAT signaling cascades (38). To analyze the implication of non-canonical Wnt signaling in the response to SrRan, MC3T3-E1 osteoblasts were transfected with a vector encoding a small interfering RNA targeting Ryk. As shown in Fig. 6, D–F, blunting the Wnt5a receptor Ryk using small interfering RNA abrogated the induction of Runx2, ALP, and COL1A1 induced by SrRan in MC3T3-E1 osteoblasts. To confirm this finding, MC3T3-E1 osteoblasts were transfected with a vector encoding a dominant-negative form of RhoA (DN-RhoA) to inhibit RhoA that acts downstream of Ryk (39). As shown in Fig. 7, A–C, transfection with DN-RhoA blunted the increased Runx2, ALP, and COL1A1 expression induced by SrRan. These data indicate that osteoblast gene expression induced by SrRan is at least in part dependent on Wnt5a-Ryk-RhoA activation.
FIGURE 7.
Inhibition of RhoA signaling blocks SrRan-induced osteoblast gene expression. A–E, MC3T3-E1 osteoblastic cells were transiently transfected with DN-RhoA, a small interfering RNA raised against Ryk (siRyk), or empty vector and then treated with or without SrRan (3 mm) for 48 h. mRNA expression of osteoblast markers (Runx2, ALP, Col1A1, histone H4) was determined by quantitative PCR analysis. Data are the mean ± S.D. a, p < 0.05 versus control cells; b, p < 0.05 versus SrRan-treated cells. F, schematic representation of the role of calcineurin-NFATc signaling in osteoblast replication and differentiation induced by SrRan. Treatment with SrRan activates Cn, leading to nuclear translocation of NFATc1 that acts with other transcription factors to up-regulate cell replication. Additionally, activation of Cn-NFATc1 by SrRan results in increased expression of several genes, including Wnt3a, which activates β-catenin transcription, and Wnt5a, which activates Ryk-RhoA to promote cell replication and osteoblast gene expression.
Finally, we investigated whether non-canonical Wnt signaling may be implicated in stimulating proliferation of osteoblastic cells induced by SrRan. Using measurement at histone H4 to monitor DNA synthesis, we found that transfection with a small interfering RNA raised against Ryk (siRyk) or DN-RhoA abrogated the increased DNA synthesis induced by SrRan (Fig. 7, D and E), suggesting that the non-canonical Wnt signaling is implicated in SrRan-induced stimulation of MC3T3-E1 osteoblastic cells. Taken together, the results indicate that SrRan activates Cn-mediated NFATc1 nuclear translocation and transcriptional activity, resulting in increased expression of Wnt proteins, activation of canonical and non-canonical Wnt signaling, cell proliferation, and osteoblast gene expression (Fig. 7F).
DISCUSSION
Previous studies indicated that the antiosteoporotic treatment SrRan increases osteoblast replication and differentiation in vitro (3). To determine the molecular mechanisms by which SrRan may act on cultured osteoblasts, we investigated the effect of SrRan on NFATc1 signaling, which is a positive regulator of osteoblastogenesis in vitro (40) and in vivo (28). In this study, we show that SrRan induces translocation and transcriptional activity of the Cn substrate NFATc1, indicating that SrRan activates Cn-NFATc signaling in these cells. Calcineurin is known to be activated by low, sustained increases in intracellular calcium (41). Consistently, increased intracellular calcium influx was shown to activate Cn and NFATc signaling in several cell types (42, 43). The activation of Cn-NFATc signaling by Sr may thus result indirectly from the increased intracellular calcium levels induced by SrRan via activation of the calcium-sensing receptor in osteoblasts (15).
Having shown that SrRan activates Cn-NFATc signaling in osteoblasts, we investigated the functional implication of NFATc signaling induced by SrRan in osteoblasts. We (11, 17) and others (10, 15) have previously shown that SrRan increases osteoblastic cell replication. The present finding that the increased osteoblastic cell replication induced by SrRan was blunted by Cn inhibitors indicates that this effect involves NFATc signaling. This mechanism is consistent with the recent finding that NFATc1 directs osteoblast replication in vivo (28). Previous evidence indicated that SrRan promotes osteoblastic cell replication in part via activation of ERK1/2 MAPK signaling (15, 17). The present data provide another mechanism by which SrRan promotes osteoblastic cell replication via NFATc1 signaling. In addition to this effect, our data reveal that SrRan increased Runx2 and other osteoblast phenotypic genes via activation of Cn-NFATc1 signaling. This indicates that activation of Cn-NFATc1 signaling is implicated in the increased osteoblast gene expression induced by SrRan. These results therefore identify Cn-NFAT signaling as an important mechanism that is involved in SrRan-induced osteoblastic cell replication and differentiation in vitro.
Several molecular mechanisms may be involved in the control of osteoblast differentiation by Cn-NFAT signaling. NFATc and Osterix (Osx) were found to cooperate in the control of osteoblast differentiation and bone formation (27, 45). However, we did not detect any changes in Osx gene expression by SrRan (data not shown). In the search for other NFATc target genes in osteoblasts, we found that SrRan increased the expression of several Wnt-related genes. This is consistent with the finding that NFATc1 signaling regulates components of Wnt signaling in other cell types (28, 35). Wnt proteins are known to play an important role in the control of cell replication, differentiation, and survival through signals involving β-catenin-dependent and -independent pathways. Binding of canonical Wnt proteins to Frizzled receptors and low density lipoprotein 5 and 6 (LRP5/6) co-receptors initiates a cascade of intracellular events that lead to inhibition of β-catenin phosphorylation and stabilization. This results in the translocation of β-catenin into the nucleus where it interacts with TCF-LEF transcription factors to activate the expression of Wnt-responsive genes (46). Here, we found that several genes that control osteoblast differentiation were up-regulated by SrRan. For example, Wnt11 promotes osteoblast maturation via β-catenin (47), and DKK2 is an early stimulator of osteoblast differentiation (48), suggesting that components of Wnt signaling may mediate the effects of SrRan on osteoblasts. Our findings that SrRan increased Wnt3a expression and β-catenin transcriptional activity, and that DKK1 blocked the effect of SrRan on osteoblast gene expression suggest that canonical Wnt signaling mediates in part the effects of SrRan on osteoblast differentiation. The fact that SrRan-induced activation of Wnt3a-β-catenin signaling was blunted by both Cn and Wnt inhibitors is suggestive of a regulatory loop in which activation of Cn-NFATc1 signaling by SrRan induces Wnt3a expression, which thereby activates canonical Wnt-β-catenin signaling and osteoblast gene expression (Fig. 7F). Recently, several molecules have been shown to exert a bone anabolic effect in part via canonical Wnt signaling (49, 50). The present data reveal that SrRan is another agent that may exert bone anabolism in part by enhancing Wnt signaling.
Our data suggest that SrRan-induced osteoblast gene expression may also be in part mediated by non-canonical Wnt signaling. We found that sFRP1 that effectively binds to and antagonizes all Wnt proteins blocked the effect of SrRan on osteoblast gene expression. Additionally, SrRan increased the expression of Wnt5a, a non-canonical Wnt family member that was found to enhance osteoblast differentiation (51–54). The mechanisms of action of Wnt5a are complex (38, 39, 55). Wnt5a signaling is transduced through the Frizzled family receptors and Ror2/Ryk co-receptors to the Dishevelled-dependent RhoA and JNK or Ca2+-dependent signaling (38). Here, we found that knocking down the transmembrane receptor tyrosine kinase Ryk or its downstream effector RhoA abolished SrRan-induced cell replication and osteoblast gene expression, indicating that Wnt5a-Ryk-RhoA signaling is involved in SrRan-induced osteoblastic cell proliferation and differentiation, which is consistent with the recent findings supporting a role of Wnt5a in osteoblastogenesis in vitro and in vivo (44, 53). These results reveal that besides canonical Wnt3a-β-catenin signaling, non-canonical Wnt signaling mediates osteoblastic cell proliferation and differentiation induced by SrRan. Thus, several cross-talks between Cn-NFATc signaling and Wnt signaling may contribute to the enhanced osteoblastogenesis induced by SrRan (Fig. 7F).
In summary, our data indicate that the stimulatory effects of SrRan on osteoblast replication and differentiation involve the activation of Cn-NFATc1 and components of canonical and non-canonical Wnt signaling, which provides novel molecular mechanisms by which SrRan may promote osteoblastogenesis.
Acknowledgments
We thank Galapagos Pharma (Romainville, France) for the DKK1 plasmid, Dr. Jacques Bertoglio (Inserm, Chatenay-Malabry, France) for the mutant RhoA plasmid, Dr. Molkentin (Cincinnati, OH) for the 9xNFAT-luc plasmid, and Dr. Zuzana Saidak for language corrections.
This work was supported by grants from Inserm, University Paris Diderot, and the Institut de Recherches Servier (IRIS, Courbevoie, France).
- SrRan
- strontium ranelate
- CSA
- cyclosporin A
- Cn
- calcineurin
- FK506
- Fujimycin/tacrolimus
- NFATc
- nuclear factor of activated T cells, cytoplasmic
- PMA
- phorbol 12-myristate 13-acetate
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- JNK
- c-Jun N-terminal kinase
- ALP
- alkaline phosphatase
- DN
- dominant-negative.
REFERENCES
- 1.Raisz L. G. (2005) J. Clin. Invest. 115, 3318–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riggs B. L., Parfitt A. M. (2005) J. Bone Miner. Res. 20, 177–184 [DOI] [PubMed] [Google Scholar]
- 3.Marie P. J. (2005) Curr. Opin. Pharmacol. 5, 633–636 [DOI] [PubMed] [Google Scholar]
- 4.Ammann P., Badoud I., Barraud S., Dayer R., Rizzoli R. (2007) J. Bone Miner. Res. 22, 1419–1425 [DOI] [PubMed] [Google Scholar]
- 5.Hott M., Deloffre P., Tsouderos Y., Marie P. J. (2003) Bone 33, 115–123 [DOI] [PubMed] [Google Scholar]
- 6.Arlot M. E., Jiang Y., Genant H. K., Zhao J., Burt-Pichat B., Roux J. P., Delmas P. D., Meunier P. J. (2008) J. Bone Miner. Res. 23, 215–222 [DOI] [PubMed] [Google Scholar]
- 7.Meunier P. J., Roux C., Seeman E., Ortolani S., Badurski J. E., Spector T. D., Cannata J., Balogh A., Lemmel E. M., Pors-Nielsen S., Rizzoli R., Genant H. K., Reginster J. Y. (2004) N. Engl. J. Med. 350, 459–468 [DOI] [PubMed] [Google Scholar]
- 8.Reginster J. Y., Seeman E., De Vernejoul M. C., Adami S., Compston J., Phenekos C., Devogelaer J. P., Curiel M. D., Sawicki A., Goemaere S., Sorensen O. H., Felsenberg D., Meunier P. J. (2005) J. Clin. Endocrinol. Metab. 90, 2816–2822 [DOI] [PubMed] [Google Scholar]
- 9.Barbara A., Delannoy P., Denis B. G., Marie P. J. (2004) Metabolism 53, 532–537 [DOI] [PubMed] [Google Scholar]
- 10.Bonnelye E., Chabadel A., Saltel F., Jurdic P. (2008) Bone 42, 129–138 [DOI] [PubMed] [Google Scholar]
- 11.Canalis E., Hott M., Deloffre P., Tsouderos Y., Marie P. J. (1996) Bone 18, 517–523 [DOI] [PubMed] [Google Scholar]
- 12.Choudhary S., Halbout P., Alander C., Raisz L., Pilbeam C. (2007) J. Bone Miner. Res. 22, 1002–1010 [DOI] [PubMed] [Google Scholar]
- 13.Zhu L. L., Zaidi S., Peng Y., Zhou H., Moonga B. S., Blesius A., Dupin-Roger I., Zaidi M., Sun L. (2007) Biochem. Biophys. Res. Commun. 355, 307–311 [DOI] [PubMed] [Google Scholar]
- 14.Brown E. M., MacLeod R. J. (2001) Physiol. Rev. 81, 239–297 [DOI] [PubMed] [Google Scholar]
- 15.Chattopadhyay N., Quinn S. J., Kifor O., Ye C., Brown E. M. (2007) Biochem. Pharmacol. 74, 438–447 [DOI] [PubMed] [Google Scholar]
- 16.Coulombe J., Faure H., Robin B., Ruat M. (2004) Biochem. Biophys. Res. Commun. 323, 1184–1190 [DOI] [PubMed] [Google Scholar]
- 17.Fromigué O., Haÿ E., Barbara A., Petrel C., Traiffort E., Ruat M., Marie P. J. (2009) J. Cell Mol. Med. 13, 8B, 2189–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hogan P. G., Chen L., Nardone J., Rao A. (2003) Genes Dev. 17, 2205–2232 [DOI] [PubMed] [Google Scholar]
- 19.Crabtree G. R., Olson E. N. (2002) Cell 109, (suppl.) S67–S79 [DOI] [PubMed] [Google Scholar]
- 20.Aliprantis A. O., Ueki Y., Sulyanto R., Park A., Sigrist K. S., Sharma S. M., Ostrowski M. C., Olsen B. R., Glimcher L. H. (2008) J. Clin. Invest. 118, 3775–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asagiri M., Sato K., Usami T., Ochi S., Nishina H., Yoshida H., Morita I., Wagner E. F., Mak T. W., Serfling E., Takayanagi H. (2005) J. Exp. Med. 202, 1261–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirotani H., Tuohy N. A., Woo J. T., Stern P. H., Clipstone N. A. (2004) J. Biol. Chem. 279, 13984–13992 [DOI] [PubMed] [Google Scholar]
- 23.Ikeda F., Nishimura R., Matsubara T., Hata K., Reddy S. V., Yoneda T. (2006) J. Immunol. 177, 2384–2390 [DOI] [PubMed] [Google Scholar]
- 24.Sun L., Peng Y., Zaidi N., Zhu L. L., Iqbal J., Yamoah K., Wang X., Liu P., Abe E., Moonga B. S., Epstein S., Zaidi M. (2007) Am. J. Physiol. Renal Physiol. 292, F285–F291 [DOI] [PubMed] [Google Scholar]
- 25.Takayanagi H. (2007) Ann. N.Y. Acad. Sci. 1116, 227–237 [DOI] [PubMed] [Google Scholar]
- 26.Takayanagi H., Kim S., Koga T., Nishina H., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., Wagner E. F., Mak T. W., Kodama T., Taniguchi T. (2002) Dev. Cell 3, 889–901 [DOI] [PubMed] [Google Scholar]
- 27.Koga T., Matsui Y., Asagiri M., Kodama T., de Crombrugghe B., Nakashima K., Takayanagi H. (2005) Nat. Med. 11, 880–885 [DOI] [PubMed] [Google Scholar]
- 28.Winslow M. M., Pan M., Starbuck M., Gallo E. M., Deng L., Karsenty G., Crabtree G. R. (2006) Dev. Cell 10, 771–782 [DOI] [PubMed] [Google Scholar]
- 29.Yeo H., Beck L. H., McDonald J. M., Zayzafoon M. (2007) Bone 40, 1502–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeo H., Beck L. H., Thompson S. R., Farach-Carson M. C., McDonald J. M., Clemens T. L., Zayzafoon M. (2007) J. Biol. Chem. 282, 35318–35327 [DOI] [PubMed] [Google Scholar]
- 31.Zayzafoon M. (2006) J. Cell. Biochem. 97, 56–70 [DOI] [PubMed] [Google Scholar]
- 32.Haÿ E., Laplantine E., Geoffroy V., Frain M., Kohler T., Müller R., Marie P. J. (2009) Mol. Cell. Biol. 29, 953–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haÿ E., Faucheu C., Suc-Royer I., Touitou R., Stiot V., Vayssière B., Baron R., Roman-Roman S., Rawadi G. (2005) J. Biol. Chem. 280, 13616–13623 [DOI] [PubMed] [Google Scholar]
- 34.Dai Y. S., Xu J., Molkentin J. D. (2005) Mol. Cell. Biol. 25, 9936–9948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fadel M. P., Szewczenko-Pawlikowski M., Leclerc P., Dziak E., Symonds J. M., Blaschuk O., Michalak M., Opas M. (2001) J. Biol. Chem. 276, 27083–27089 [DOI] [PubMed] [Google Scholar]
- 36.Mao B., Wu W., Davidson G., Marhold J., Li M., Mechler B. M., Delius H., Hoppe D., Stannek P., Walter C., Glinka A., Niehrs C. (2002) Nature 417, 664–667 [DOI] [PubMed] [Google Scholar]
- 37.Kawano Y., Kypta R. (2003) J. Cell Sci. 116, 2627–2634 [DOI] [PubMed] [Google Scholar]
- 38.Katoh M., Katoh M. (2007) Clin. Cancer Res. 13, 4042–4045 [DOI] [PubMed] [Google Scholar]
- 39.Angers S., Moon R. T. (2009) Nat. Rev. Mol. Cell Biol. 10, 468–477 [DOI] [PubMed] [Google Scholar]
- 40.Smerdel-Ramoya A., Zanotti S., Deregowski V., Canalis E. (2008) J. Biol. Chem. 283, 22690–22699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dolmetsch R. E., Lewis R. S., Goodnow C. C., Healy J. I. (1997) Nature 386, 855–858 [DOI] [PubMed] [Google Scholar]
- 42.Gooch J. L., Gorin Y., Zhang B. X., Abboud H. E. (2004) J. Biol. Chem. 279, 15561–15570 [DOI] [PubMed] [Google Scholar]
- 43.Gwack Y., Feske S., Srikanth S., Hogan P. G., Rao A. (2007) Cell Calcium 42, 145–156 [DOI] [PubMed] [Google Scholar]
- 44.Yang Y., Topol L., Lee H., Wu J. (2003) Development 130, 1003–1015 [DOI] [PubMed] [Google Scholar]
- 45.Ali M. M., Yoshizawa T., Ishibashi O., Matsuda A., Ikegame M., Shimomura J., Mera H., Nakashima K., Kawashima H. (2007) J. Cell Sci. 120, 2565–2573 [DOI] [PubMed] [Google Scholar]
- 46.Logan C. Y., Nusse R. (2004) Annu. Rev. Cell Dev. Biol. 20, 781–810 [DOI] [PubMed] [Google Scholar]
- 47.Friedman M. S., Oyserman S. M., Hankenson K. D. (2009) J. Biol. Chem. 284, 14117–14125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X., Liu P., Liu W., Maye P., Zhang J., Zhang Y., Hurley M., Guo C., Boskey A., Sun L., Harris S. E., Rowe D. W., Ke H. Z., Wu D. (2005) Nat. Genet. 37, 945–952 [DOI] [PubMed] [Google Scholar]
- 49.Cheng S. L., Shao J. S., Cai J., Sierra O. L., Towler D. A. (2008) J. Biol. Chem. 283, 20505–20522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wan M., Yang C., Li J., Wu X., Yuan H., Ma H., He X., Nie S., Chang C., Cao X. (2008) Genes Dev. 22, 2968–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arnsdorf E. J., Tummala P., Jacobs C. R. (2009) PLoS One 4, e5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baksh D., Boland G. M., Tuan R. S. (2007) J. Cell. Biochem. 101, 1109–1124 [DOI] [PubMed] [Google Scholar]
- 53.Guo J., Jin J., Cooper L. F. (2008) Bone 43, 961–971 [DOI] [PubMed] [Google Scholar]
- 54.Liu Y., Bhat R. A., Seestaller-Wehr L. M., Fukayama S., Mangine A., Moran R. A., Komm B. S., Bodine P. V., Billiard J. (2007) Mol. Endocrinol. 21, 376–387 [DOI] [PubMed] [Google Scholar]
- 55.Mikels A. J., Nusse R. (2006) PLoS Biol. 4, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]