Abstract
Curcumin activates diverse anticancer activities that lead to inhibition of cancer cell and tumor growth, induction of apoptosis, and antiangiogenic responses. In this study, we observed that curcumin inhibits Panc28 and L3.6pL pancreatic cancer cell and tumor growth in nude mice bearing L3.6pL cells as xenografts. In addition, curcumin decreased expression of p50 and p65 proteins and NFκB-dependent transactivation and also decreased Sp1, Sp3, and Sp4 transcription factors that are overexpressed in pancreatic cancer cells. Because both Sp transcription factors and NFκB regulate several common genes such as cyclin D1, survivin, and vascular endothelial growth factor that contribute to the cancer phenotype, we also investigated interactions between Sp and NFκB transcription factors. Results of Sp1, Sp3, and Sp4 knockdown by RNA interference demonstrate that both p50 and p65 are Sp-regulated genes and that inhibition of constitutive or tumor necrosis factor-induced NFκB by curcumin is dependent on down-regulation of Sp1, Sp3, and Sp4 proteins by this compound. Curcumin also decreased mitochondrial membrane potential and induced reactive oxygen species in pancreatic cancer cells, and this pathway is required for down-regulation of Sp proteins in these cells, demonstrating that the mitochondriotoxic effects of curcumin are important for its anticancer activities.
Keywords: Mitochondrial Transport, Pancreas, Reactive Oxygen Species (ROS), RNA Interference (RNAi), Sp1, NFκB, Sp Transcription Factors, Curcumin, Pancreatic Cancer
Introduction
Pancreatic ductal adenocarcinoma is a major cause of cancer-related deaths in developed countries and, in 2009, it is estimated that in excess of 34,000 new cases will be diagnosed in the United States (1). Pancreatic ductal adenocarcinoma is a highly aggressive disease that invariably evades early diagnosis (2, 3). The mean survival time for patients with metastatic disease is only 3–6 months, and only 20–30% of pancreatic cancer cases are alive after 12 months. Several factors are associated with increased risk for pancreatic cancer and these include chronic pancreatitis, prior gastric surgery, smoking, diabetes, exposure to certain classes of organic solvents, radiation, and specific gene polymorphisms (4, 5). In addition to heritable mutations, several acquired gene mutations have been identified in sporadic pancreatic tumors (6, 7). The K-Ras oncogene is primarily mutated in codon 12 in >90% of pancreatic tumors and the mutation results in a constitutively active form of ras that can lead to increased cell proliferation. Mutations in the cyclin-dependent kinase inhibitor p16, the tumor suppressor gene p53, and SMAD4, a downstream target of transforming growth factor β also exhibit high mutation frequencies in pancreatic tumors.
Because pancreatic cancers are frequently detected at an advanced stage, treatments have provided very limited improvements in tumor regression and overall survival times after diagnosis (8, 9). 5-Fluorouracil alone or in combination with other drugs has been extensively used for treatment of advanced pancreatic cancer, and gemcitabine, a deoxycytidine analog (or antimetabolite), has partially replaced 5-fluorouracil as a treatment for pancreatic cancer. Gemcitabine provides increased clinical benefits in terms of response rate, time to progression, and median survival and several other drugs for treatment of pancreatic cancer are also being investigated (10–13). Curcumin (diferuloylmethane) is a polyphenolic phytochemical that exhibits a broad spectrum of anticancer activities against multiple tumor types (14–16), including pancreatic cancer (17–21). Curcumin decreased survival and induced apoptosis in pancreatic cancer cells and, in the same cells, curcumin also decreased pro-survival nuclear factor κB (NFκB) DNA binding in a gel mobility shift assay (17). Treatment of athymic nude mice with orthotopically implanted tumors with 1 g/kg of curcumin daily did not inhibit tumor volume but in combination studies, curcumin enhanced the activity of gemcitabine as an inhibitor of pancreatic tumor growth (19). Curcumin also decreased several NFκB-regulated genes in tumors and these include cyclin D1, c-myc, bcl-2, cyclooxygenase-2 (COX-2),3 and vascular endothelial growth factor (VEGF) (19).
Recent studies in this laboratory demonstrated that the anticancer activity of curcumin in bladder cancer cells and tumors was associated with repression of specificity protein (Sp) transcription factors Sp1, Sp3, and Sp4, which was accompanied by decreased expression of Sp-regulated survival, angiogenic and growth promoting genes (22). In this study, we show that curcumin also decreased expression of Sp proteins and Sp-dependent gene products in pancreatic cancer cells and mouse tumors (xenograft). Moreover, in pancreatic cancer cells, the p65 and p50 subunits of NFκB are also Sp-regulated genes and inhibition of constitutive and induced NFκB expression by curcumin is also due, in part, to down-regulation of Sp transcription factors. Moreover, the mechanism of Sp down-regulation by curcumin is due to the mitochondriotoxicity of this compound and the subsequent induction of reactive oxygen species (ROS).
EXPERIMENTAL PROCEDURES
Cell Lines
The Panc28 cell line was a generous gift from Dr. Paul Chiao and L3.6pL cells were kindly provided by Dr. Isaiah Fidler (University of Texas M.D. Anderson Cancer Center, Houston, TX). Panc1 and PC3 cells were obtained from ATCC (Manassas, VA) and RKO cells were kindly provided by Dr. Stanley Hamilton (M.D. Anderson Cancer, Houston, TX).
Antibodies and Reagents
Both pancreatic cancer cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM)/F-12 supplemented with 5% FBS, 0.22% sodium bicarbonate, and 10 ml/liter of ×100 antibiotic/antimycotic mixture solution (Sigma). Cells were grown in 150-cm2 culture plates in an air/CO2 (95:5) atmosphere at 37 °C. Cyclin D1, Sp3, Sp4, VEGF, GKLF4, c-jun, and p50 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Cleaved PARP and COX-2 antibody were purchased from Cell Signaling Technology (Danvers, MA) and Sp1 antibody was purchased from Millipore (Billerica, MA). Survivin antibody was purchased from R&D Systems (Minneapolis, MN). NFκB-p65 antibody was from Abcam (Cambridge, MA). Monoclonal β-actin antibody was purchased from Sigma. Horseradish peroxidase substrate for Western blot analysis was obtained from Millipore. Dithiothreitol and γ-l-glutamyl-l-cysteinyl-glycine (GSH) were obtained from Sigma. TNFα was purchased from R&D Systems. Curcumin (98% pure) was purchased from Indofine Chemical Company, Inc. (Hillsborough, NJ). Lipofectamine and Lipofectamine 2000 was purchased from Invitrogen. Luciferase reagent was from Promega (Madison, WI). β-Galactosidase reagent was obtained from Tropix (Bedford, MA). The VEGF and survivin promoter constructs were provided by Drs. Gerhard Siemeister and Gunter Finkenzeller (Institute of Molecular Medicine, Tumor Biology Center, Freiburg, Germany) and Dr. M. Zhou (Emory University, Atlanta, GA) respectively. Sp1 and Sp3 promoter constructs were kindly provided by Drs. Carlos Cuidad and Veronique Noe (University of Barcelona, Barcelona, Spain). NFκB promoter construct was purchased from Stratagene (Cedar Creek, TX).
Cell Proliferation Assay
Pancreatic cancer cells (1 × 105 per well) were plated in 12-well plates and allowed to attach for 24 h. The medium was then changed to DMEM/Ham's F-12 medium containing 2.5% charcoal-stripped FBS, and vehicle (DMSO), GSH, DTT, and/or curcumin were added. Cells were then trypsinized and counted at the indicated times using a Coulter Z1 particle counter. Each experiment was done in triplicate and results are expressed as mean ± S.E. for each treatment group.
Transfection and Luciferase Assay
The pancreatic cancer cells (1 × 105 per well) were plated in 12-well plates in DMEM/Ham's F-12 medium supplemented with 2.5% charcoal-stripped FBS. After 24 h, various amounts of DNA (i.e. 0.4 μg of PGL2-Luc, 0.4 μg of PGL2-Luc, 0.04 μg of β-galactosidase, and 0.4 μg of pSp1 (4)-Luc, 0.4 μg of pSp3-Luc, 0.4 μg of VEGF (2068)-Luc, 0.4 μg of pSurvivin (269)-Luc) were transfected using Lipofectamine reagent according to the manufacturer's protocol. Five h post-transfection, the transfection mixture was replaced with complete medium containing either vehicle (DMSO) or the indicated compound in DMSO. After 22 h, cells were then lysed with 100 μl of 1× reporter lysis buffer, and cell extracts (30 ml) were used for luciferase and β-galactosidase assays. A Lumicount luminometer was used to quantitate luciferase and β-galactosidase activities, and the luciferase activities were normalized to β-galactosidase activity.
Western Blots
Pancreatic cancer cells were seeded in DMEM/Ham's F-12 medium containing 2.5% charcoal-stripped FBS and after 24 h, cells were treated with either vehicle (DMSO) or the indicated compounds. Cells were collected using high-salt buffer (50 mmol/liter of HEPES, 0.5 mol/liter of NaCl, 1.5 mmol/liter of MgCl2, 1 mmol/liter of EGTA, 10% glycerol, and 1% Triton X-100) and 10 μl/ml of Protease Inhibitor Mixture (Sigma). Protein lysates were incubated for 3 min at 100 °C before electrophoresis, and then separated on 10% SDS-PAGE at 120 V for 3–4 h. Proteins were transferred onto polyvinylidene difluoride membranes by wet electroblotting in a buffer containing 25 mmol/liter of Tris, 192 mmol/liter of glycine, and 20% methanol for 1.5 h at 180 mA. Membranes were blocked for 30 min with 5% TBST-Blotto (10 mmol/liter of Tris-HCl, 150 mmol/liter of NaCl (pH 8.0), 0.05% Triton X-100, and 5% nonfat dry milk) and incubated in fresh 5% TBST-Blotto with 1:500 primary antibody overnight with gentle shaking at 4 °C. After washing with TBST for 10 min, the polyvinylidene difluoride membrane was incubated with secondary antibody (1:5000) in 5% TBST-Blotto for 2 h by gentle shaking. The membrane was washed with TBST for 10 min, incubated with 6 ml of chemiluminescence substrate for 1 min, and exposed to Kodak image station 4000 mm Pro (Carestreamhealth, Woodbridge, CT).
Electrophoretic Mobility Shift Assay
Cells were rinsed in cold phosphate-buffered saline buffer and harvested in reporter lysis buffer (Promega). After a 15-min incubation on ice and 10-min centrifugation at 16,000 × g, 4 °C, the pellet was resuspended in 1× reporter lysis buffer (Promega) supplemented with 0.5 mol/liter of KCl and incubated on ice for 30 min. The supernatant containing nuclear proteins was collected after centrifugation for 10 min at 16,000 × g at 4 °C, and quantified for protein concentrations by the Bradford method. The NFκB probe was prepared by annealing the two complementary polynucleotides, the NFκB sense strand probe was 5′-AGT TGA GGG GAC TTT CCC AGG C-3′. The annealed probe was 5′-end labeled using T4 polynucleotide kinase (Invitrogen) and [γ-32P]ATP (PerkinElmer Life Sciences). The labeled probe was purified with the Chroma Spin TE-10 column (BD Biosciences, San Jose, CA). The electrophoretic mobility shift assay reaction was carried out in the reporter lysis buffer supplemented with 0.1 mol/liter of KCl. Each reaction contained 2 μg of nuclear protein, 1 μg of poly(dI-dC) (Roche Molecular Biochemicals) with or without unlabeled competitor oligonucleotides, and 10 fmol of labeled probe; the mixture was incubated for 15 min on ice. Protein-DNA complexes were resolved by 5% native PAGE at 160 V at room temperature for 1.5 h and visualized after exposing it to ImageTek-H autoradiography X-Ray film.
siRNA Interference Assays
siRNAs for Sp1, Sp3, Sp4, p65, p50, and LMN were purchased from Sigma. The siRNA complexes used in this study are indicated as follows: LMN, SASI_Hs02_00367643; Sp1, SASI_Hs02_00363664; Sp3, 5′-GCG GCA GGU GGA GCC UUC ACU TT; Sp4, 5′-GCA GUG ACA CAU UAG UGA GCT T; p65 (REL1096), 5′-GAT TGA GGA GAA ACG TAA ATT; and p50 (REL 1911), 5′-GTC ACT CTA ACG TAT GCA ATT.
The Panc28 and L3.6pL pancreatic cancer cell lines were seeded (6 × 104 per well) in 12-well plates in DMEM/Ham's F-12 medium supplemented with 2.5% charcoal-stripped FBS without antibiotic and left to attach for 1 day. The triple Sp siRNA knockdown (iSp1, iSp3, and iSp4 complex) along with iLamin as control was performed using Lipofectamine 2000 transfection reagent as per the manufacturer's instructions.
Xenograft Study
Female athymic nude mice, age 4 to 6 weeks, were purchased from Harlan. L3.6pL cells (3 × 105) in a 1:1 ratio of Matrigel (BD Biosciences) were injected into the either side of the flank area of nude mice. Seven days after tumor cell inoculation, mice were divided into two groups of 10 animals each. The first group received 100 μl of vehicle (corn oil) by intraperitoneal injection, and the second group of animals received 100 mg/kg/day injection of curcumin in corn oil every 2nd day for 18 days (9 doses) by intraperitoneal injection. The mice were weighed, and tumor areas were measured throughout the study. After 20 days, the animals were sacrificed; final body and tumor weights were determined and plotted.
GSH Estimation
GSH-Glo Glutathione assay kit (Promega) was used to estimate GSH levels according to the manufacturer's protocol using a 96-well cell culture plate and luminescence measured using a Lumicount luminometer.
ROS Estimation
Cellular ROS levels were evaluated with the cell permeant probe CM-H2DCFDA (5-(and-6)-chloromethyl-2′7′-dichlorodihydrofluorescein diacetate acetyl ester) from Invitrogen. Following 20–24 h treatment, cells plated on a 96-well cell culture plate were loaded with 10 μm CM-H2DCFDA for 30 min, washed once with serum-free medium, and analyzed for ROS levels using the BioTek Synergy 4 plate reader (Winooski, VT) set at 480 and 525 nm excitation and emission wavelengths, respectively. Following reading of ROS, cultures were then treated with Janus green and cell counts were determined with the plate reader set to an absorbance of 610 nm, and ROS intensities were then corrected accordingly. Each experiment was done in triplicate and results are expressed as mean ± S.E. for each treatment group.
Measurement of Mitochondrial Membrane Potential (MMP)
MMP was measured with Mitochondrial Membrane Potential Detection Kit (Stratagene) according to the manufacturer's protocol using JC-1 dye and mitochondrial membrane potential shift was measured using FACS Calibur flow cytometer using CellQuest acquisition software (BD Biosciences). J-aggregates are detected as red fluorescence and J-monomers are detected as green fluorescence.
Immunohistochemistry
Tissue sections were deparaffinized in xylene and treated with a graded series of alcohol and rehydrated in phosphate-buffered saline. Antigen retrieval was done using 10 mm sodium citrate (pH 6.0–6.2) and endogenous peroxidase was blocked by the 3% hydrogen peroxide in methanol for 6 min. Slides were then incubated with blocking serum (Vecstatin ABC Elite kit, Vector Laboratories, Burlingame, CA) for 45 min. Samples were then incubated overnight with Sp1, Sp3, Sp4, VEGF, cyclin D1, survivin, p50, and p65 antibodies at 4 °C. Sections were then washed in phosphate-buffered saline/Tween and then incubated with biotinylated secondary antibody followed by streptavidin. The brown staining specific for antibody binding was developed by exposing the avidin and biotinylated peroxidase complex to diaminobenzidine reagent (Vector Laboratories) and sections were then counterstained with hematoxylin (Vector Laboratories).
Quantitative Real Time PCR of mRNA
cDNA was prepared from Panc28 and L3.6pL cell lines using Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's protocol. Each PCR was carried out in triplicate in a 20-μl volume using SYBR GreenER (Invitrogen) at 95 °C for 10 min, then 40 cycles of 95 °C for 15 s and 60 °C for 1 min in the Applied Biosystems 7500 Fast Real-time PCR System. The following primers were used: p50 (forward), 5′-ACCCTGACCTTGCCTATTTG-3′ and (reverse). 5′-AGCTCTTTTTCCCGATCTCC-3′; p65 (forward), 5′-CGGGATGGCTTCTATGAGG-3′ and (reverse) 5′-CTCCAGGTCCCGCTTCTT-3′. The primers for TBP, Sp1, Sp3, and Sp4 genes have previously been described (23).
Fluorescence-activated Cell Sorting Assays
Both Panc28 and L3.6pL pancreatic cancer cells were treated with vehicle, curcumin, or siRNA for iLamin or Sp1/3/4. Cells were analyzed on a FACSCalibur flow cytometer using CellQuest acquisition software (BD Biosciences). Propidium iodide fluorescence was collected through a 585/42-nm band pass filter, and list mode data were acquired on a minimum of 20,000 single cells defined by a dot plot of propidium iodide width versus propidium iodide area. Data analysis was performed using Modfit LT software (Verity Software House, Topsham, ME).
Statistical Analysis
Statistical significance of differences between the treatment groups was determined by an analysis of variance and Student's t test, and levels of probability were noted. IC50 values were calculated using linear regression analysis and expressed in micromolar at 95% confidence intervals.
RESULTS
Curcumin Inhibits Constitutive NFκB
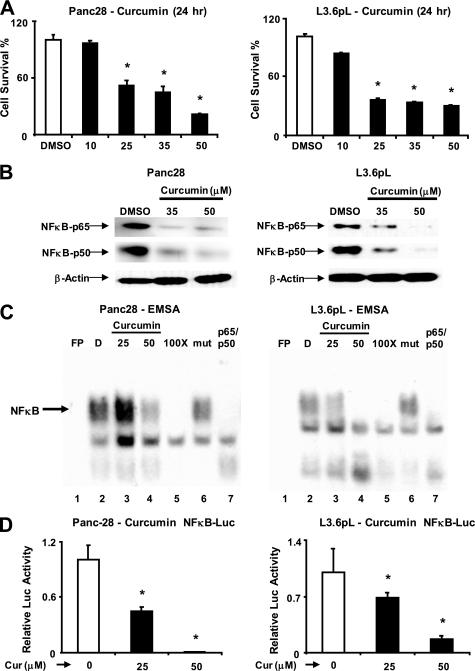
Fig. 1 illustrates the concentration-dependent inhibition of Panc28 and L3.6pL pancreatic cancer cell proliferation after treatment with 10–50 μm curcumin for 24 h, and IC50 values for this response were 34.0 and 28.8 μm, respectively. After prolonged treatment of these cells for 96 and 144 h, IC50 values were 12.4 and 11.2 μm and 11.8 and 9.9 μm in Panc28 and L3.6pL cells, respectively, and concentrations of curcumin required for growth inhibition decreased with increasing treatment times as observed for many anticancer drugs. Supplemental Fig. S1A shows that Panc28 and L3.6pL cells represent gemcitabine-resistant and non-resistant cell lines, respectively. The effects of curcumin on distribution of cells in Go/G1, S, and G2/M phases of the cell cycle were determined 0–24 h after treatment with 50 μm curcumin (supplemental Fig. S1B). In Panc28 cells, curcumin decreased G2/M and increased the percentage of cells in S phase over time and this effect was also observed in L3.6pL cells after treatment for 24 h. Curcumin-dependent inhibition of DNA replication as evidenced by inhibition of G0/G1 to S phase progression was not observed. Interactions between curcumin and NFκB signaling have been extensively reported (14–16). Fig. 1B illustrates Western blots of nuclear extracts from DMSO- and curcumin (35 and 50 μm)-treated L3.6pL and Panc28 cells; 35 μm decreased p65 and p50 protein levels in L3.6pL but not Panc28 cells, whereas 50 μm curcumin decreased expression of both proteins in both cell lines. It has also been reported that curcumin decreases NFκB binding to its cognate response element in gel mobility shift assays (17), and Fig. 1C summarizes binding nuclear extracts from L3.6pL cells treated with DMSO (solvent) or curcumin (25 and 50 μm) to an oligonucleotide containing a consensus NFκB site. The free probe in the absence of nuclear extract (lane 1) did not form a retarded band; however, nuclear extracts from solvent-treated cells formed an NFκB retarded band (lane 2, indicated by an arrow). The retarded band intensity was decreased when nuclear extracts from cells treated with 25 or 50 μm curcumin were used (lanes 3 and 4). The intensity of the NFκB-DNA complex (lane 2) was decreased after competition with 100-fold excess of unlabeled wild-type (lane 5) but not mutant (lane 6) NFκB oligonucleotide. In addition, the intensity of the NFκB-DNA complex was also decreased after incubation with p50/p65 (combined) antibodies due to immunodepletion (lane 7); however, we did not observe a supershift complex with these antibodies. We also investigated the effects of curcumin on NFκB-dependent transactivation in pancreatic cancer cells transfected with pNFκB-luc, a construct containing 5 tandem NFκB response elements linked to a luciferase reporter gene. The results show that curcumin decreased luciferase activity (Fig. 1D) and this was consistent with results in Fig. 1, A–C, showing that curcumin repressed constitutive NFκB primarily through down-regulation of p50 and p65 proteins.
FIGURE 1.
Curcumin inhibits pancreatic cancer cell growth, decreases expression of p65, p50 proteins, NFκB-DNA binding, and transactivation of the NFκB promoter. A, inhibition of Panc28 and L3.6pL cell growth. Cells were treated with DMSO (solvent control) or 10, 25, 35, or 50 μmol/liter of curcumin, and effects on cell growth were determined after treatment for 24 h as described under “Experimental Procedures.” B, effects of curcumin on p65 and p50 subunits of NFκB in Panc28 and L3.6pL cells. Cells were treated with DMSO (0), 35 or 50 μmol/liter of curcumin for 24 h, and p65 and p50 protein levels in nuclear extracts were determined as described under “Experimental Procedures.” β-Actin served as loading control. C, gel mobility shift assay. Panc28 and L3.6pL cells were treated with DMSO or 25 or 50 μmol/liter of curcumin for 24 h, and nuclear lysates were incubated with 32P-labeled GC-rich oligonucleotide alone or in the presence of other factors. Retarded bands were analyzed by electrophoretic mobility shift assay as described under “Experimental Procedures.” D, decrease in transactivation of NFκB promoter. Panc28 and L3.6pL cells were transfected with the NFκB-luc construct, then treated with DMSO or 25 and 50 μm curcumin, and luciferase activity was determined as described under “Experimental Procedures.” Results are expressed as mean ± S.E. for three replicate determinations for each treatment group, and significant (p < 0.05) compared with solvent (DMSO) control indicated by an asterisk. EMSA, electrophoretic mobility shift assay.
Curcumin Decreases Sp Transcription Factors and Sp-dependent Responses
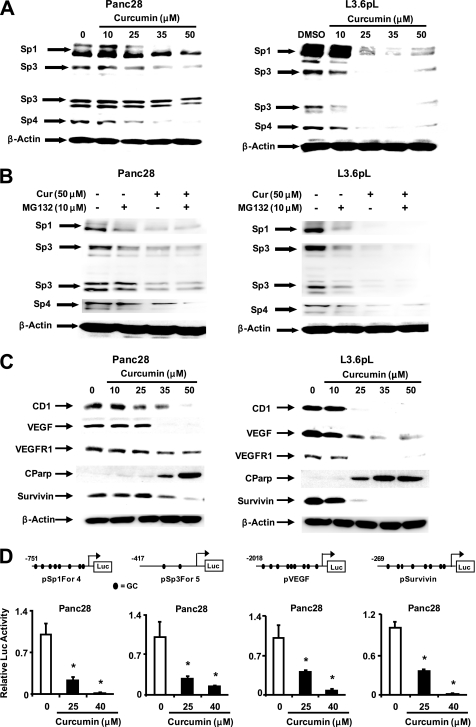
Recent studies in this laboratory showed that curcumin decreased expression of Sp transcription factors Sp1, Sp3, and Sp4 in bladder cancer cells (22). We also observed that these proteins were overexpressed in L3.6pL and Panc28 (Fig. 2A) cells, and curcumin induced a concentration-dependent decrease of these proteins in both cell lines. In bladder cancer cells, curcumin-induced Sp down-regulation was blocked by proteasome inhibitors; however, results in Fig. 2B show that the proteasome inhibitor MG132 did not alter the effects of curcumin on Sp1, Sp3, and Sp4 protein expression in pancreatic cancer cells. Curcumin also decreased Sp3 and Sp4 mRNA in both cell lines and decreased Sp1 mRNA in L3.6pL but not in Panc28 cells (supplemental Fig. S2). Curcumin also decreased expression of several Sp-dependent genes in Panc28 and L3.6pL cells and these included VEGF, VEGFR1, cyclin D1, and survivin (Fig. 2C). This was accompanied by increased PARP cleavage, a marker of apoptosis. Using Panc28 cells as a model, curcumin decreased luciferase activity in cells transfected with constructs containing GC-rich promoter inserts from the Sp1 (pSp1For4), Sp3 (pSp3For5), VEGF (pVEGF), and survivin (pSurvivin) genes linked to a luciferase reporter gene (Fig. 2D). Thus, curcumin decreased expression of Sp1, Sp3, Sp4, and several Sp-dependent gene products in pancreatic cancer cells. Previous studies showed that curcumin decreased growth and expression of Sp1, Sp3, and Sp4 in bladder cancer cells (22), and results in supplemental Fig. S3 show that similar results were observed after treatment of Panc1 pancreatic, PC3 prostate, and RKO colon cancer cells.
FIGURE 2.
Curcumin activates proteosome-independent down-regulation of Sp proteins, decreases cell growth, angiogenic and apoptotic proteins, and their promoters. A, decreased Sp proteins. Panc28 and L3.6pL cells were treated with DMSO or 10, 25, 35, and 50 μmol/liter of curcumin for 24 h and whole cell lysates were analyzed by Western blot analysis as described under “Experimental Procedures.” B, curcumin causes proteasome-independent Sp degradation. Cells were treated with DMSO or 50 μmol/liter of curcumin in the presence or absence of proteasome inhibitor MG132, and the effects on Sp protein degradation were determined after treatment for 24 h by Western blot as described under “Experimental Procedures.” C, curcumin decreases expression of Sp-dependent gene products. Panc28 and L3.6pL cells were treated with DMSO or 10, 25, 35, or 50 μmol/liter of curcumin for 24 h, and whole cell lysates were analyzed by Western blot analysis as described under “Experimental Procedures.” β-Actin served as a loading control. D, curcumin decreases transactivation in cells transfected with Sp1, Sp3, VEGF, and survivin promoter constructs. Cells were treated with DMSO (solvent control) or 25 or 40 μmol/liter of curcumin, and the effects on transactivation of promoters were determined after treatment for 24 h as described under “Experimental Procedures.” Results are expressed as mean ± S.E. for three replicate determinations for each treatment group, and significant (p < 0.05) decreases in luciferase activity compared with the solvent (DMSO) control are indicated (*).
Sp Transcription Factors Regulate NFκB
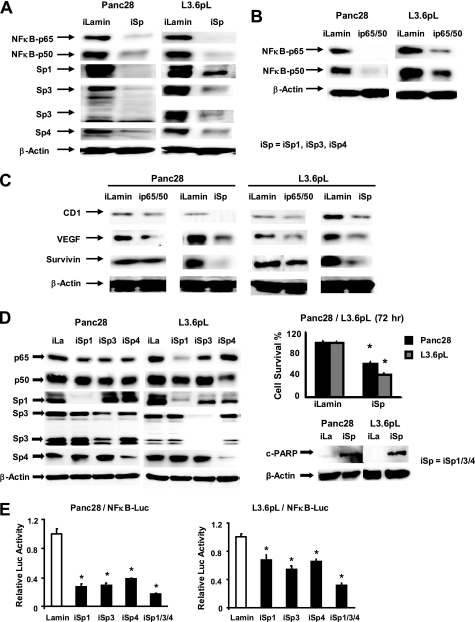
The role of Sp transcription factors in regulating p65 and p50 proteins that form the NFκB complex was investigated in Panc28 and L3.6pL cells transfected with a mixture (iSp) containing small inhibitory RNAs for Sp1 (iSp1), Sp3 (iSp3), and Sp4 (iSp4) as previously described (22). Fig. 3A illustrates that after transfection of these cells with iSp, there was a decrease in p65 and p50 expression and Sp proteins were also down-regulated. Similarly, transfection of cells with siRNA for p65 plus p50 (combined) (ip65–50) decreased expression of p65 and p50 proteins (Fig. 3B); however, Sp1, Sp3, and Sp4 protein expression was unchanged (data not shown). These results demonstrate that Sp transcription factors regulate expression of p65 and p50. Results in Fig. 3C compare the effects of transfection of iSp and ip65–50 on expression of putative NFκB- and Sp-regulated gene products, cyclin D1, VEGF, and survivin (19, 22, 24–32) and iSp > ip65–50 in decreasing expression of all three proteins. In these studies, cells were transfected with siRNAs and whole cell lysates were analyzed by Western blots and, therefore, the observed extent of down-regulation was limited not only by the effectiveness of the siRNAs but also by transfection efficiencies. The effects of individual knockdown of Sp1 (iSp1), Sp3 (iSp3), and Sp4 (iSp4) on expression of p65 and p50 were also examined in Panc28 and L3.6pL cells (Fig. 3D). Transfection of Panc28 cells with iSp1, iSp3, or iSp4 decreased expression of p65 and p50 proteins; however, the effects of combined knockdown (iSp) of all 3 transcription factors was much more effective than individual Sp knockdown. The effects of iSp1, iSp3, and iSp4 on p65 and p50 protein expression in L3.6pL cells was highly variable. iSp1 and iSp4 were the most effective oligonucleotides for decreasing expression of p65 and p50, respectively, and iSp4 had no effect on p65 expression. The combined knockdown of Sp1, Sp3, and Sp4 (iSp) was the most effective treatment for decreasing p65 and p50 protein expression in L3.6pL cells. The differential effect of Sp1, Sp3, and Sp4 on regulation of other genes has also previously been reported in different cancer cell lines (22, 33–35). In addition, Sp knockdown also inhibited Panc28 and L3.6pL cell growth and induced PARP cleavage, indicating that agents such as curcumin that decrease Sp transcription factors also decrease Sp-dependent cell growth and survival pathways. We also investigated the effects of iSp and individual siRNAs for Sp1, Sp3, and Sp4 on luciferase activity in Panc28 and L3.6pL cells transfected with pNFκB-luc, and luciferase activity was significantly decreased in all treatment groups (Fig. 3E). Results in Fig. 3, D and E, show that knockdown of individual Sp proteins is more effective as an inhibitor of NFκB-dependent transactivation than their individual effects on decreasing levels of p65 or p50 proteins. This may be due, in part, to a role for Sp transcription factors in cooperatively activating NFκB (36, 37) and this is being further investigated. In contrast to the effects of Sp transcription factor knockdown on Sp-regulated proteins and NFκB, the effects on distribution of Panc28 and L3.6pL cells in G0/G1, S, and G2/M phases as determined by FACS analysis were minimal (supplemental Fig. S4). As a control, we show that curcumin differentially affects KLF4 but does not induce c-jun protein expression in Panc28 and L3.6pL cells (supplemental Fig. S5).
FIGURE 3.
Sp and NFκB knockdown and effects on NFκB subunits, angiogenic and survival proteins. Sp (A) and NFκB (B) knockdown by RNA interference. Panc28 and L3.6pL were transfected with iSp (A) or ip65/p50 (B), and effects on Sp proteins, and p65 and p50 subunits of NFκB were determined by Western blot analysis as described under “Experimental Procedures.” C, effects of Sp and NFκB knockdown on expression of CD1, VEGF, and survivin proteins. Protein lysates from Panc28 and L3.6pL cells transfected with iSp or ip65/p50 were analyzed for CD1, VEGF, and survivin proteins by Western blot analysis as described under “Experimental Procedures.” D, effects of Sp knockdown on p65, p50, cell proliferation, and PARP cleavage. Cells were transfected with various oligonucleotides, and cell numbers were counted or cell lysates were analyzed by Western blots as described under “Experimental Procedures.” E, effects of iSp on transactivation of NFκB promoter. Cells were transfected iSp and NFκB-luc, and luciferase activity was estimated as described under “Experimental Procedures.” β-Actin and Lamin served as a loading control and similar results were observed in duplicate experiments (A–C). Cell numbers (D) or luciferase activity (E) in transfection experiments were expressed as mean ± S.E. for 3 replicate experiments and significant (p < 0.05) decreases are indicated (*).
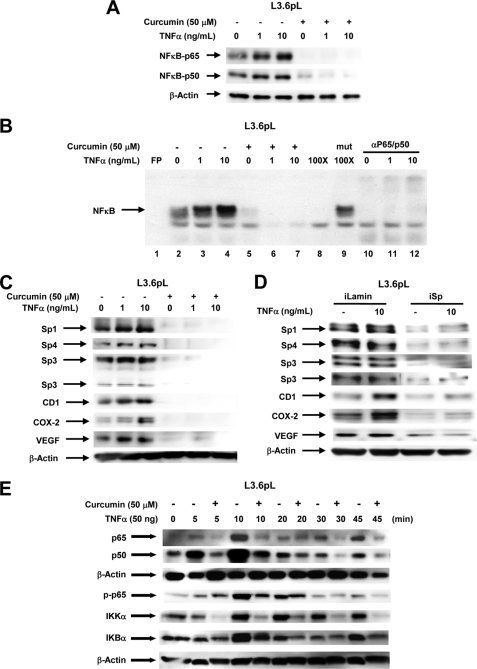
Stressors such as TNFα induce NFκB-dependent responses through increased formation of the nuclear NFκB complex and this was observed in L3.6pL cells, whereas minimal induction by TNFα was observed in Panc28 cells. Using L3.6pL cells as a model, we show that 1 and 10 ng/ml of TNFα increased nuclear p65/p50 levels and cotreatment with curcumin decreased TNFα-induced nuclear accumulation of p65/p50 (Fig. 4A). Similar results were observed in a gel mobility shift assay (Fig. 4B). Nuclear extracts from cells treated with solvent (control) or 1 and 10 ng/ml of TNFα formed an NFκB-DNA retarded band using a 32P-labeled consensus NFκB oligonucleotide and TNFα increased retarded band intensity, whereas band intensities were decreased using extracts from cells cotreated with TNFα plus curcumin. Retarded band intensities decreased after cotreatment with unlabeled wild-type but not with mutant NFκB oligonucleotide, and after coincubation with p65 plus p50 antibodies (combined). Fig. 4C shows that TNFα had a minimal effect on expression of Sp1, Sp3, Sp4, and Sp-dependent gene products VEGF; cyclin D1 was induced and COX-2 expression was the most highly induced by TNFα. Curcumin alone (50 μm) or in combination with TNFα resulted in decreased expression of all of these proteins. Results in Fig. 3A show that iSp inhibited basal expression of p65 and p50, and this is confirmed in Fig. 4D, which also shows that iSp inhibited TNFα-induced p65 and p50 expression in L3.6pL cells. TNFα had minimal effects on Sp expression but enhanced levels of cyclin D1 and COX-2; however, in cells cotransfected with iSp, there was a significant decrease in Sp1, Sp3, and Sp4, and TNFα-induced expression of COX-2 and cyclin D1 was also decreased. These results suggest that although TNFα-dependent induction of cyclin D1 and COX-2 correlated with induction of p65/p50, these responses were blocked after down-regulation of Sp transcription factors. TNFα and other stressors also rapidly induce several responses in cancer cell lines. Fig. 4E shows that after treatment of L3.6pL cells with TNFα, there was increased expression of p65, p50, phosphorylated p65, IKKα, and IKBα within 5–10 min. However, in cells cotreated with curcumin plus TNFα, all the stress-induced responses were inhibited and Sp transcription factors were not decreased over this short incubation period. These Sp-independent effects of curcumin also contribute to the anticancer activity of this compound.
FIGURE 4.
Role of Sp proteins in curcumin-dependent inhibition of TNFα inducible responses in L3.6pL pancreatic cancer cells. A, curcumin decreases TNFα-induced expression of p65 and p50 proteins. Cells were treated with TNFα in the presence or absence of 50 μm curcumin, and nuclear lysates were examined for expression of p65 and p50 proteins by Western blots as described under “Experimental Procedures.” B, curcumin decreased TNFα-induced NFκB oligonucleotide-protein binding. L3.6pL cells were treated with DMSO or 50 μmol/liter of curcumin in the presence or absence of TNFα for 24 h, and nuclear extracts were incubated with 32P-labeled NFκB oligonucleotide alone or in the presence of other factors. Retarded bands were analyzed by electrophoretic mobility shift assay as described under “Experimental Procedures.” Effects of curcumin (C) and iSp (D) on Sp/NFκB-dependent protein expression are shown. L3.6pL cells were treated with 50 μm curcumin (C) or transfected with iSp (D) in the presence or absence of TNFα, and whole cell and nuclear lysates were analyzed for Sp1, Sp3, Sp4, p65, p50, CD1, COX-2, and VEGF proteins by Western blot analysis as described under “Experimental Procedures.” The gels were typical of results of at least two replicate determinations per treatment group. E, effects of curcumin on TNFα-induced responses. L3.6pL cells were treated with curcumin or TNFα alone or in combination for up to 45 min, whole cell lysates were obtained and analyzed by Western blots as described under “Experimental Procedures.”
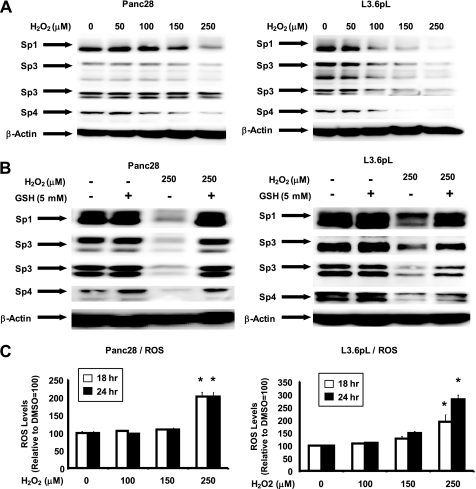
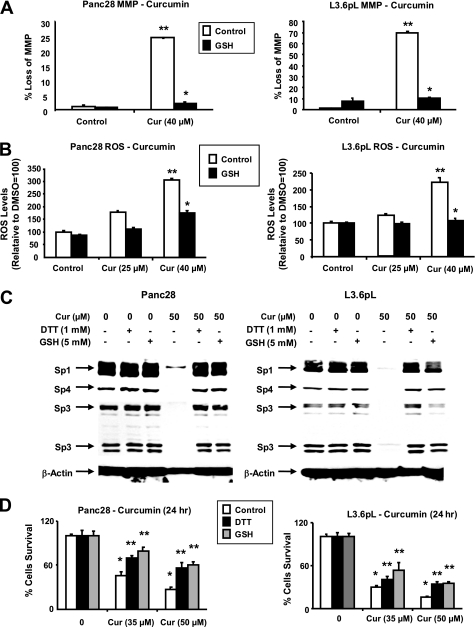
Ongoing studies in this laboratory indicate that drug-induced down-regulation of MMP and induction of ROS in cancer cell lines leads to decreased expression of Sp1, Sp3, and Sp4 and this is also observed after treatment with hydrogen peroxide (Fig. 7). Results in Fig. 5A show that after treatment of L3.6pL and Panc28 cells with 40 μm curcumin for 20 h, there was a significant loss of MMP, respectively (also see supplemental Fig. S6). Moreover, in these same cells, cotreatment with curcumin and the thiol antioxidants DTT and GSH, the curcumin-induced loss of MMP was significantly inhibited. In addition, the loss of MMP in L3.6pL and Panc28 cells treated with curcumin was accompanied by induction of ROS, which was also significantly attenuated after cotreatment with DTT or GSH (Fig. 5B). The role of antioxidants in protecting against curcumin-induced down-regulation of Sp1, Sp3, and Sp4 was also investigated in Panc28 and L3.6pL cells. Treatment with curcumin alone decreased expression of Sp1, Sp3, and Sp4 proteins in L3.6pL and Panc28 cells; however, cotreatment with the antioxidants GSH or DTT blocked down-regulation of these transcription factors (Fig. 5C). Curcumin-dependent inhibition of Panc28 and L3.6pL cell growth was also reversed in cells cotreated with GSH or DTT and the effects of the antioxidants were more pronounced in Panc28 than L3.6pL cells (Fig. 5D).
FIGURE 7.
Hydrogen peroxide decreases Sp proteins and induces ROS. Effects of hydrogen peroxide alone (A) and in combination with GSH (B) are shown. Cells were treated with hydrogen peroxide or GSH alone or in combination for 24 h and whole cell lysates were analyzed by Western blots as described under “Experimental Procedures.” C, induction of ROS. Panc28 and L3.6pL cells were treated with hydrogen peroxide for 18 or 24 h and ROS was determined as described under “Experimental Procedures.” Results are mean ± S.E. (3 replicates/group) and significant (p < 0.05) induction of ROS is indicated (*).
FIGURE 5.
Effects of curcumin on MMP and ROS and related responses. Induction of changes in loss of MMP (A) and ROS (B) by curcumin. Panc28 and L3.6pL cells were treated with DMSO or 25 or 40 μmol/liter of curcumin for 24 h, in the presence or absence of antioxidant GSH, and mitochondrial membrane potential and ROS were determined as described under “Experimental Procedures.” ROS mediated Sp degradation (C) cell growth inhibition (D) in the presence or absence of antioxidants. Cells were treated with DMSO or 35 or 50 μmol/liter of curcumin in the presence or absence of thiol antioxidants for 24 h, and cells were then counted or the whole cell lysates were analyzed by Western blots as described under “Experimental Procedures.” β-Actin served as a loading control. Results in A, B, and D are expressed as mean ± S.E. for three replicate determinations for each treatment group, and significant (p < 0.05). Curcumin-mediated decreases (*) or increases after cotreatment with antioxidants (**) compared with the solvent (DMSO) control are indicated. GSH levels in Panc28 (4.33 μm) and L3.6pL (2.64 μm) cells were also determined as described under “Experimental Procedures.”
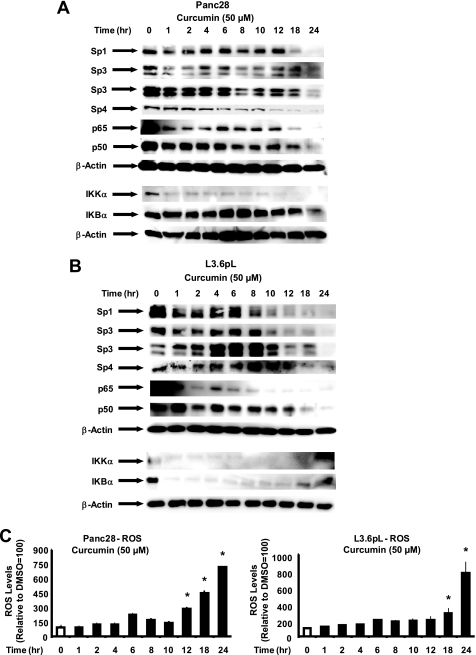
We also investigated the time course effects of curcumin on Sp1, Sp3, Sp4, p65, and p50 protein levels in Panc28 and L3.6pL (Fig. 6, A and B) and correlated these effects with induction of ROS (Fig. 6C). Treatment of Panc28 cells with 50 μm curcumin resulted in decreased levels of Sp1, Sp3, Sp4, p65, and p50 after 18, 8, 12, 8, and 6 h, respectively, whereas in L3.6pL cells decreased levels were observed after 8, 10, 18, 8, and 18 h, respectively. In addition, there was some variability in protein levels at earlier time points; however, with few exceptions, the major effects were observed after 18–24 and 12–24 h in Panc28 and L3.6pL cells, respectively. Maximal effects were observed after 24 h in both cell lines. In parallel studies, ROS was induced in Panc28 and L3.6pL cells 12–24 and 18–24 h, respectively, after treatment with curcumin. These results demonstrate a time-dependent overlap in curcumin-induced ROS and the down-regulation of Sp1, Sp3, Sp4, p65, and p50.
FIGURE 6.
Time course effects of curcumin on Sp1, Sp3, Sp4, p65 and p50, and ROS. Panc28 (A) and L3.6pL (B) cells were treated with DMSO (0 time) or 50 μm curcumin for different times over a 24-h period and whole cell lysates were analyzed by Western blots as described under “Experimental Procedures.” C, induction of ROS. The time course induction of ROS by curcumin in Panc28 and L3.6pL cells was determined as described under “Experimental Procedures.” Results are mean ± S.E. (3 replicates/group) and significant (p < 0.05) induction of ROS is indicated (*).
The direct effects of ROS on Sp proteins was investigated by treating cells with hydrogen peroxide (Fig. 7). Hydrogen peroxide (50–250 μm) decreased expression of Sp1, Sp3, and Sp4 proteins in Panc28 and L3.6pL cells (Fig. 7A), and in cells cotreated with hydrogen peroxide in combination with glutathione, the down-regulation of Sp transcription factors was reversed by the antioxidant (Fig. 7B). This interaction was similar to the effects of glutathione on curcumin-induced down-regulation of Sp proteins illustrated in Fig. 5C. Moreover, we also observed that, like curcumin, hydrogen peroxide induced ROS in the pancreatic cancer cell lines (Fig. 7C). These results further support that induction of ROS by curcumin results in down-regulation of Sp transcription factors, and activation of this pathway contributes to the anticancer activity of curcumin.
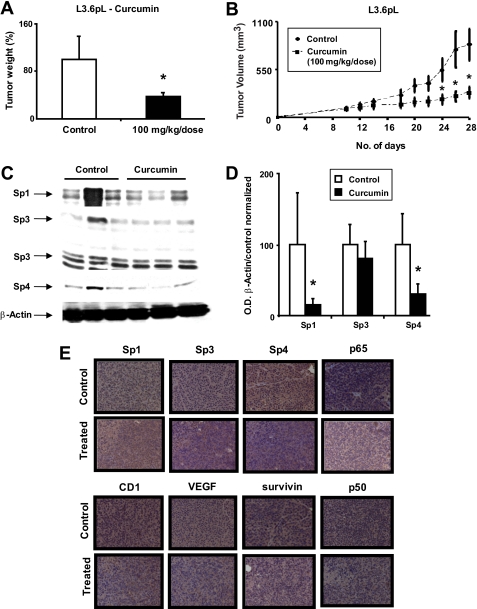
Curcumin Inhibits Tumor Growth and Down-regulates Sp Transcription Factors
The in vivo antitumorigenic activity of curcumin was investigated in athymic nude bearing L3.6pL cells as xenografts. At a dose of 100 mg/kg/days, curcumin inhibited tumor weights (Fig. 8A) and growth (Fig. 8B) over an 18-day treatment period. We also examined Sp1, Sp3, and Sp4 protein expression in tumors from the corn oil (control)- and curcumin-treated mice (Fig. 8, C and D). All three transcription factors were decreased after treatment with curcumin; however, only Sp1 and Sp4 were significantly (p < 0.05) lower due to the inter-animal variability. Immunohistochemical staining also showed that curcumin decreased expression of nuclear Sp proteins, p65, p50, VEGF, survivin, and cyclin D1 (Fig. 8E). Thus, curcumin-dependent down-regulation of Sp transcription factors correlated with the growth inhibitory effects of this compound in both in vitro and in vivo pancreatic cancer cells, suggesting that targeting these transcription factors play a role in the antitumorigenic activity of curcumin.
FIGURE 8.
Curcumin inhibits pancreatic cancer xenograft tumor growth. Tumor weights (A) and volume (B) are shown. Athymic nude mice bearing L3.6pL xenografts were treated with corn oil or curcumin (100 mg/kg/day), and tumor weights and volumes (mm3) were determined as described under “Experimental Procedures.” C, Western blot analysis of tumor lysates. Lysates from three mice in the treated and control groups were analyzed by Western blots as described under “Experimental Procedures.” β-Actin served as loading control and for standardizing quantitative protein determinations. D, Sp proteins levels of control animals were set at 100%. Columns, means for three separate determinations; bars, S.E.; *, significantly (p < 0.05) decreased protein levels. E, immunohistochemical staining. Tumor slides from treated and untreated animals were stained as described under “Experimental Procedures.”
DISCUSSION
The nuclear NFκB complex containing p65 (Rel A) and p50 (NFκB1) or closely related proteins is a multifunctional nuclear transcription factor that regulates expression of multiple genes that promote inflammation and carcinogenesis (30–32). The inactive cytosolic NFκB-IκB complex is activated and processed through phosphorylation and proteasome-dependent degradation of IκB and this results in enhanced accumulation of nuclear NFκB and modulation of NFκB-dependent gene expression. Upstream activators of nuclear NFκB include various cellular stressors such as cytokines, apoptosis inducers, carcinogens and tumor promoters, ROS, endotoxins, and bacterial and viral infections (30–32). Activation of NFκB in a cancer cell context results in the induction of cancer cell proliferation, survival, angiogenesis and metastasis, epithelial to mesenchymal transition, and inflammation, and this is accompanied by induction of genes such as cyclin D1, survivin, VEGF, bcl-2, and COX-2 that contribute to these responses (30–32). Curcumin has been extensively characterized as an anti-inflammatory and anticancer agent and these effects have been linked to modulation of several pathways and genes in different cancer cell lines (14–16). In addition, curcumin has been extensively investigated as an inhibitor of basal and induced NFκB-dependent responses and this plays an important role in the remarkable anticancer activities of this compound (14–16).
Studies in this laboratory have been focused on drugs such as tolfenamic acid, betulinic acid, and the synthetic triterpenoid methyl 2-cyano-3,11-dioxo-18β-olean-1,12-dien-30-oate (CDODA-Me) that also inhibit tumor growth and this is due, in part, to down-regulation of Sp1, Sp3, and Sp4 transcription factors (25–27, 38). These agents also decrease expression of several Sp-dependent genes including survivin, VEGF, and its receptors, cyclin D1, bcl-2, EGFR, and several other genes. Initial studies showed that Sp1 was highly expressed in many pancreatic cancer cell lines and was required for VEGF expression (39), and it has subsequently been shown that Sp1, Sp3, and Sp4 are highly expressed in pancreatic and other cancer cell lines (data not shown and see Refs. 23, 25–27, and 38). Moreover, a recent report showed that Sp1 was a negative prognostic factor for pancreatic cancer patient survival (40). Many Sp-dependent genes are also co-regulated in some cells by NFκB and not surprisingly, there is also a striking similarity between Sp- and NFκB-dependent growth inhibitory, angiogenic and survival responses, and genes. Moreover, our recent studies with curcumin in bladder cancer cells (22) showed that this compound also decreased expression of Sp transcription factors and Sp-dependent genes, and there was evidence in 253JB-V cells that p65 was also an Sp-regulated gene. Curcumin is currently in clinical trials for pancreatic cancer (21) and we used this tumor type as a model for investigating the effects of this compound on Sp1, Sp3, Sp4, and NFκB and also Sp-NFκB interactions.
Curcumin inhibited Panc28 and L3.6pL cell proliferation (Fig. 1A) and decreased expression of both p65 and p50 and their DNA binding activity (Fig. 1, B and C) and luciferase activity in cells transfected with an NFκB-luc construct (Fig. 1D). Thus, basal NFκB, which is overexpressed in many cancer cell lines and tumors including pancreatic cancer (17), is also inhibited in Panc28 and L3.6pL cells treated with curcumin and this is related, in part, to decreased expression of p65 and p50. In parallel experiments, we also demonstrated that curcumin decreased expression of Sp1, Sp3, and Sp4 (Fig. 2A) and several Sp-dependent genes (Fig. 2C), and similar results were previously observed in bladder cancer cells (22), and results in supplemental Fig. S3 show that curcumin inhibits growth and down-regulates Sp1, Sp3, and Sp4 in Panc1, RKO, and PC3 cells. In bladder cancer cells, Sp down-regulation after treatment with curcumin was blocked by the proteasome inhibitor MG-132, whereas in pancreatic cancer cells, MG-132 did not affect curcumin-dependent repression of Sp1, Sp3, and Sp4 (Fig. 2B). Curcumin decreased pancreatic tumor growth in athymic nude mice bearing L3.6pL cells as xenografts and this was also accompanied by Sp down-regulation (Fig. 8) and parallels the in vitro effects of curcumin in pancreatic cancer cells (Fig. 2). Thus, curcumin decreases both Sp and NFκB transcription factors in pancreatic cancer cells and this is accompanied by decreased expression of several genes that may be regulated by both NFκB and Sp transcription factors, depending on the cell context.
Because curcumin decreased p65 and p50 proteins in Panc28 and L3.6pL cells (Fig. 1B), we hypothesized that this response may be dependent, in part, on down-regulation of Sp1, Sp3, and Sp4 in pancreatic cancer cells (Fig. 2A). Direct evidence for the role of Sp transcription factors in regulating NFκB was obtained by RNA interference in which cells were transfected with iSp, which contained siRNAs for Sp1, Sp3, and Sp4 (in combination). The results showed that knockdown of Sp transcription factors decreased expression of both p65 and p50 proteins (Fig. 3A); combined knockdown of p65 and p50 (ip65-p50) by RNA interference decreased expression of both proteins (Fig. 3B). Moreover, a comparison of the effects of iSp versus ip65-p50 on several putative Sp- and NFκB-regulated genes (cyclin D1, VEGF, and survivin (Fig. 3C)) confirmed that expression of these genes was primarily dependent on Sp transcription factors, and luciferase activity in cells transfected with NFκB-luc was also decreased by Sp knockdown (Fig. 3E). However, comparison of the effects of iSp versus ip65/p50 on down-regulation of cyclin D1 (in L3.6pL cells) and VEGF in both cell lines (Fig. 3C) also suggests that Sp transcription factors and NFκB may coordinately regulate expression of these genes. Knockdown of individual Sp proteins in Panc28 and L3.6pL cells differentially affected expression of p65 and p50 (Fig. 3D) and this was more pronounced in L3.6pL cells where iSp1 and iSp4 were the most effective oligonucleotides for decreasing p65 and p50 proteins, respectively. Previous studies have reported similar differences in gene-specific regulation by Sp1, Sp3, and Sp4. For example, Sp1 (but not Sp3 or Sp4) regulates estrogen receptor α expression in breast cancer cells (33), whereas all 3 transcription factors contribute to basal expression of VEGF receptor 2 in pancreatic cancer cells (34). Knockdown of Sp proteins also inhibited pancreatic cancer cell growth and induced PARP cleavage (Fig. 3D). These responses were previously observed in L3.6pL and Panc1 cells4 and confirm that curcumin-induced effects on Sp proteins contributes to the growth inhibitory and proapoptotic responses induced by this compound.
TNFα induced levels of nuclear p65 and p50 proteins (Fig. 4, A and B) and this resulted in induction of some NFκB-dependent gene products such as COX-2 (Fig. 4C); both curcumin and iSp inhibited not only basal (Fig. 3) but TNFα-induced responses (Fig. 4) in pancreatic cancer cells. Thus, curcumin-dependent inhibition of NFκB is due, in part, to down-regulation of Sp transcription factors and these results are consistent with previous reports showing that the p65 and p50 promoters contain functional GC-rich Sp binding sites and both genes are regulated by Sp1 (41, 42). However, the role of Sp1, Sp3, and Sp4 in regulation of p65 and p50 will also be dependent on cell context because we previously observed that knockdown of Sp transcription factors in bladder cancer cells decreased p65 but not p50 proteins (22). The temporal effects of curcumin on decreased expression of Sp1, Sp3, Sp4, p65, and p50 are maximal after 24 h but are also decreased to a lesser extent after 8–12 h (Fig. 6, A and B). However, previous studies show that curcumin can rapidly decrease TNFα or stress-induced responses after short time periods, and TNFα-induced activation of p65 and p50, phospho-p65, IKKα, and IKBα were all inhibited by curcumin within a 45-min time period (Fig. 4E). Thus, curcumin-mediated short-term effects are independent of decreased Sp protein levels and these rapid effects of curcumin also contribute to the overall anticancer activity of this compound.
Ongoing studies in this laboratory have been investigating the mechanisms associated with drug-induced Sp down-regulation in cancer cells (data not shown and see Ref. 38), and recently we have shown that induction of ROS is a critical element for this response (43, 44). For example, arsenic trioxide decreased MMP and induced ROS and this resulted in down-regulation of Sp1, Sp3, and Sp4 in bladder and pancreatic cancer cells. Treatment with hydrogen peroxide also induced Sp down-regulation and in cells cotreated with arsenic trioxide or hydrogen peroxide and the thiol antioxidants DTT or GSH, the effects of Sp protein and cell growth inhibition were reversed (43, 44). Previous reports show that curcumin decreases MMP and induces ROS in some cancer cell lines (45–47), and this was also observed in pancreatic cancer cells (Fig. 5, A and B). Moreover, in cells cotreated with curcumin and GSH or DTT, the effects of curcumin on down-regulation of Sp1, Sp3, and Sp4 and growth inhibition were significantly reversed by the antioxidants (Fig. 5, C and D). Curcumin-mediated down-regulation of Sp proteins in Panc28 and L3.6pL cells and induction of ROS were observed primarily after prolonged treatment (8–12 h) and were maximal after 24 h (Fig. 6). In addition, hydrogen peroxide also decreased expression of Sp1, Sp3, and Sp4 proteins in Panc28 and L3.6pL cells and cotreatment with glutathione attenuated these effects (Fig. 7), and current studies are focused on analysis of the ROS species generated by treatment with curcumin and their individual role in repression of Sp proteins. Interaction of curcumin with pancreatic cancer cell mitochondria, induction of ROS, and the attenuation of curcumin-induced Sp down-regulation by antioxidants is also consistent with a role for ROS in regulating expression of Sp1, Sp3, and Sp4. Previous studies showed that among several cancer cell lines, their sensitivity to arsenic trioxide was dependent, in part, on constitutive glutathione levels (48), and the higher levels of glutathione in Panc28 (4.33 μm) versus L3.6pL (2.64 μm) cells may explain the increased resistance of the former cell line to curcumin-mediated repression of Sp1, Sp3, and Sp4 proteins (Fig. 2A). In contrast, we also observed that antioxidants were less effective in reversing curcumin-mediated inhibition of cell proliferation in L3.6pL compared with Panc28 cells (Fig. 5D) and this is an example of cell context-dependent differences in the contribution of the ROS-Sp degradation pathway to pancreatic cancer cell growth inhibition.
Thus, like arsenic trioxide (43) and other mitochondriotoxic drugs, curcumin induces ROS in pancreatic cancer cells and this results in down-regulation of Sp1, Sp3, and Sp4 proteins and Sp-dependent gene products, which includes NFκB. Moreover, curcumin inhibited pancreatic tumor growth and this was also accompanied by down-regulation of Sp1, Sp3, Sp4, and Sp-regulated genes (Fig. 8). These results highlight a novel mechanism of action for curcumin that includes ROS-Sp and Sp-NFκB interactions and further demonstrates that in pancreatic cancer cells, Sp transcription factors are an important drug target. The downstream targets of curcumin-induced ROS are also being investigated and these include microRNAs such as miR-27a that inhibit expression of the Sp repressor, ZBTB10 (38). Preliminary studies indicated that only minimal induction of ZBTB10 by curcumin is observed in pancreatic cancer cells and a search for other curcumin-induced Sp repressor genes is ongoing. Current studies are also focused on development of new agents that repress expression of Sp transcription factors in pancreatic cancer and their applications as stand-alone drugs or in combination with other agents such as gemcitabine for treatment of this devastating disease.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01CA108718 and R01CA136571 and a grant from Texas A&M AgriLife Research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
I. Jutooru, G. Chadalapaka, P. Lei, and S. Safe, unpublished results.
- COX-2
- cyclooxygenase 2
- Sp
- specificity protein
- ROS
- reactive oxygen species
- DMEM
- Dulbecco's modified Eagle's medium
- PARP
- poly(ADP-ribose) polymerase
- FACS
- fluorescence-activated cell sorter
- DMSO
- dimethyl sulfoxide
- VEGF
- vascular endothelial growth factor
- FBS
- fetal bovine serum
- TNFα
- tumor necrosis factor α
- DTT
- dithiothreitol
- siRNA
- small interfering RNA
- MMP
- mitochondrial membrane potential.
REFERENCES
- 1.Jemal A., Siegel R., Ward E., Hao Y., Xu J., Murray T., Thun M. J. (2008) CA Cancer J. Clin. 58, 71–96 [DOI] [PubMed] [Google Scholar]
- 2.Evans D. B., Abbruzzese J. L., Willett C. G. (1997) in Cancer: Principles and Practice of Oncology (DeVita V. T., Hellman S., Rosenberg S. A. eds) pp. 1126–1161, Lippincott, Williams & Wilkins, Philadelphia [Google Scholar]
- 3.Hruban R. H. (2001) J. Gastrointest. Surg. 5, 583–587 [DOI] [PubMed] [Google Scholar]
- 4.Li D. (2001) Cancer J. 7, 259–265 [PubMed] [Google Scholar]
- 5.Gold E. B., Goldin S. B. (1998) Surg. Oncol. Clin. N. Am. 7, 67–91 [PubMed] [Google Scholar]
- 6.Klein A. P., Hruban R. H., Brune K. A., Petersen G. M., Goggins M. (2001) Cancer J. 7, 266–273 [PubMed] [Google Scholar]
- 7.Jaffee E. M., Hruban R. H., Canto M., Kern S. E. (2002) Cancer Cell 2, 25–28 [DOI] [PubMed] [Google Scholar]
- 8.Haller D. G. (2003) Int. J. Radiat. Oncol. Biol. Phys. 56, 16–23 [DOI] [PubMed] [Google Scholar]
- 9.McKenna S., Eatock M. (2003) Oncologist 8, 149–160 [DOI] [PubMed] [Google Scholar]
- 10.Moore M. J., Hamm J., Dancey J., Eisenberg P. D., Dagenais M., Fields A., Hagan K., Greenberg B., Colwell B., Zee B., Tu D., Ottaway J., Humphrey R., Seymour L. (2003) J. Clin. Oncol. 21, 3296–3302 [DOI] [PubMed] [Google Scholar]
- 11.Bramhall S. R., Rosemurgy A., Brown P. D., Bowry C., Buckels J. A.Marimastat Pancreatic Cancer Study Group (2001) J. Clin. Oncol. 19, 3447–3455 [DOI] [PubMed] [Google Scholar]
- 12.Xiong H. Q., Rosenberg A., LoBuglio A., Schmidt W., Wolff R. A., Deutsch J., Needle M., Abbruzzese J. L. (2004) J. Clin. Oncol. 22, 2610–2616 [DOI] [PubMed] [Google Scholar]
- 13.Hotz H. G., Reber H. A., Hotz B., Sanghavi P. C., Yu T., Foitzik T., Buhr H. J., Hines O. J. (2001) J. Gastrointest. Surg. 5, 131–138 [DOI] [PubMed] [Google Scholar]
- 14.Sharma R. A., Gescher A. J., Steward W. P. (2005) Eur. J. Cancer 41, 1955–1968 [DOI] [PubMed] [Google Scholar]
- 15.Thangapazham R. L., Sharma A., Maheshwari R. K. (2006) Am. Assoc. Pharmacol. Sci. J. 8, E443–E449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aggarwal B. B., Kumar A., Bharti A. C. (2003) Anticancer Res. 23, 363–398 [PubMed] [Google Scholar]
- 17.Li L., Aggarwal B. B., Shishodia S., Abbruzzese J., Kurzrock R. (2004) Cancer 101, 2351–2362 [DOI] [PubMed] [Google Scholar]
- 18.Li L., Braiteh F. S., Kurzrock R. (2005) Cancer 104, 1322–1331 [DOI] [PubMed] [Google Scholar]
- 19.Kunnumakkara A. B., Guha S., Krishnan S., Diagaradjane P., Gelovani J., Aggarwal B. B. (2007) Cancer Res. 67, 3853–3861 [DOI] [PubMed] [Google Scholar]
- 20.Sun M., Estrov Z., Ji Y., Coombes K. R., Harris D. H., Kurzrock R. (2008) Mol. Cancer Ther. 7, 464–473 [DOI] [PubMed] [Google Scholar]
- 21.Dhillon N., Aggarwal B. B., Newman R. A., Wolff R. A., Kunnumakkara A. B., Abbruzzese J. L., Ng C. S., Badmaev V., Kurzrock R. (2008) Clin. Cancer Res. 14, 4491–4499 [DOI] [PubMed] [Google Scholar]
- 22.Chadalapaka G., Jutooru I., Chintharlapalli S., Papineni S., Smith R., 3rd, Li X., Safe S. (2008) Cancer Res. 68, 5345–5354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mertens-Talcott S. U., Chintharlapalli S., Li X., Safe S. (2007) Cancer Res. 67, 11001–11011 [DOI] [PubMed] [Google Scholar]
- 24.Abdelrahim M., Smith R., 3rd, Burghardt R., Safe S. (2004) Cancer Res. 64, 6740–6749 [DOI] [PubMed] [Google Scholar]
- 25.Chintharlapalli S., Papineni S., Ramaiah S. K., Safe S. (2007) Cancer Res. 67, 2816–2823 [DOI] [PubMed] [Google Scholar]
- 26.Abdelrahim M., Baker C. H., Abbruzzese J. L., Safe S. (2006) J. Natl. Cancer Inst. 98, 855–868 [DOI] [PubMed] [Google Scholar]
- 27.Abdelrahim M., Baker C. H., Abbruzzese J. L., Sheikh-Hamad D., Liu S., Cho S. D., Yoon K., Safe S. (2007) Cancer Res. 67, 3286–3294 [DOI] [PubMed] [Google Scholar]
- 28.Guttridge D. C., Albanese C., Reuther J. Y., Pestell R. G., Baldwin A. S., Jr. (1999) Mol. Cell. Biol. 19, 5785–5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arbiser J. L., Klauber N., Rohan R., van Leeuwen R., Huang M. T., Fisher C., Flynn E., Byers H. R. (1998) Mol. Med. 4, 376–383 [PMC free article] [PubMed] [Google Scholar]
- 30.Aggarwal B. B. (2004) Cancer Cell 6, 203–208 [DOI] [PubMed] [Google Scholar]
- 31.Karin M. (2006) Mol. Carcinog. 45, 355–361 [DOI] [PubMed] [Google Scholar]
- 32.Bassères D. S., Baldwin A. S. (2006) Oncogene 25, 6817–6830 [DOI] [PubMed] [Google Scholar]
- 33.Chadalapaka G., Jutooru I., Burghardt R., Safe S. (2010) Mol. Cancer Res. 8, 739–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Higgins K. J., Abdelrahim M., Liu S., Yoon K., Safe S. (2006) Biochem. Biophys. Res. Commun. 345, 292–301 [DOI] [PubMed] [Google Scholar]
- 35.Khan S., Wu F., Liu S., Wu Q., Safe S. (2007) J. Mol. Endocrinol. 39, 289–304 [DOI] [PubMed] [Google Scholar]
- 36.Hirano F., Tanaka H., Hirano Y., Hiramoto M., Handa H., Makino I., Scheidereit C. (1998) Mol. Cell. Biol. 18, 1266–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu A., Hoffman P. W., Lu W., Bai G. (2004) J. Biol. Chem. 279, 17449–17458 [DOI] [PubMed] [Google Scholar]
- 38.Chintharlapalli S., Papineni S., Abdelrahim M., Abudayyeh A., Jutooru I., Chadalapaka G., Wu F., Mertens-Talcott S., Vanderlaag K., Cho S. D., Smith R., 3rd, Safe S. (2009) Int. J. Cancer 125, 1965–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi Q., Le X., Abbruzzese J. L., Peng Z., Qian C. N., Tang H., Xiong Q., Wang B., Li X. C., Xie K. (2001) Cancer Res. 61, 4143–4154 [PubMed] [Google Scholar]
- 40.Jiang N. Y., Woda B. A., Banner B. F., Whalen G. F., Dresser K. A., Lu D. (2008) Cancer Epidemiol. Biomarkers Prev. 17, 1648–1652 [DOI] [PubMed] [Google Scholar]
- 41.Yurochko A. D., Mayo M. W., Poma E. E., Baldwin A. S., Jr., Huang E. S. (1997) J. Virol. 71, 4638–4648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yurochko A. D., Kowalik T. F., Huong S. M., Huang E. S. (1995) J. Virol. 69, 5391–5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jutooru I., Chadalapaka G., Sreevalsan S., Lei P., Barhoumi R., Burghardt R., Safe S. (2010) Exp. Cell Res. 316, 2174–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jutooru I., Chadalapaka G., Abdelrahim M., Basha M. R., Samudio I., Konopleva M., Andreef M., Safe S. H. (2010) Mol. Pharmacol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thayyullathil F., Chathoth S., Hago A., Patel M., Galadari S. (2008) Free Radic. Biol. Med. 45, 1403–1412 [DOI] [PubMed] [Google Scholar]
- 46.McNally S. J., Harrison E. M., Ross J. A., Garden O. J., Wigmore S. J. (2007) Int. J. Mol. Med. 19, 165–172 [PubMed] [Google Scholar]
- 47.Bhaumik S., Anjum R., Rangaraj N., Pardhasaradhi B. V., Khar A. (1999) FEBS Lett. 456, 311–314 [DOI] [PubMed] [Google Scholar]
- 48.Yang C. H., Kuo M. L., Chen J. C., Chen Y. C. (1999) Br. J. Cancer 81, 796–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.