Abstract
Certain bacteria synthesize glutathionylspermidine (Gsp), from GSH and spermidine. Escherichia coli Gsp synthetase/amidase (GspSA) catalyzes both the synthesis and hydrolysis of Gsp. Prior to the work reported herein, the physiological role(s) of Gsp or how the two opposing GspSA activities are regulated had not been elucidated. We report that Gsp-modified proteins from E. coli contain mixed disulfides of Gsp and protein thiols, representing a new type of post-translational modification formerly undocumented. The level of these proteins is increased by oxidative stress. We attribute the accumulation of such proteins to the selective inactivation of GspSA amidase activity. X-ray crystallography and a chemical modification study indicated that the catalytic cysteine thiol of the GspSA amidase domain is transiently inactivated by H2O2 oxidation to sulfenic acid, which is stabilized by a very short hydrogen bond with a water molecule. We propose a set of reactions that explains how the levels of Gsp and Gsp S-thiolated proteins are modulated in response to oxidative stress. The hypersensitivities of GspSA and GspSA/glutaredoxin null mutants to H2O2 support the idea that GspSA and glutaredoxin act synergistically to regulate the redox environment of E. coli.
Keywords: Glutathione, Oxidation-reduction, Oxidative Stress, Post-translational Modification, Sulfhydryl, Thiol, Glutathionylspermidine, Sulfenic Acid, Very Short Hydrogen Bond
Introduction
Protein thiols are readily oxidized and reduced to form sulfenates, sulfinates, sulfonates, and intra- and intermolecular disulfides. Most organisms have complex systems that regulate the intracellular redox states of thiols (1, 2). Small thiol-containing biomolecules (e.g. GSH and cysteine, form mixed-disulfides with protein thiols (S-thiolation). These post-translational modifications protect proteins from overoxidation and regulate certain protein functions (3, 4). For example, S-glutathionylation of Escherichia coli methionine synthase, which occurs when E. coli is oxidatively stressed, suppresses the synthase activity, thereby decreasing cellular methionine concentration (5). Because GSH is abundant in most organisms (often existing at 1–10 mm), protein S-glutathionylation (GSH S-thiolation) is considered to be a reversible and universal cellular process. In E. coli, GSH and spermidine (Spd)4 form N1-glutathionylspermidine (Gsp) via an ATP-dependent reaction catalyzed by the C-terminal Gsp synthetase domain (supplemental Fig. S1) (6–8). E. coli GspSA also hydrolyzes Gsp to GSH and Spd via the N-terminal amidase domain (supplemental Fig. S1) (6). Although GspSA was first found in E. coli more than 3 decades ago (9), it is not known what the physiological role of Gsp is or how the two opposing activities of GspSA are regulated in vivo.
Herein, we report that Gsp S-thiolated proteins (GspSSPs) have mixed disulfides of Gsp and protein thiols in E. coli. Intriguingly, we found that the level of GspSSPs increased when E. coli was treated with H2O2, indicating that this modification probably inhibits oxidation of protein thiols. The accumulation of GspSSPs probably occurred because, although Gsp amidase was inactivated by H2O2, Gsp synthetase was mostly unaffected. According to an x-ray crystallography study and a chemical modification study, we found that H2O2 oxidized the thiol of the amidase active-site nucleophile, Cys59, to a sulfenic acid and that the sulfenic acid was stabilized by several hydrogen bonds, one of which involved a water molecule and was unusually short. We propose a set of reactions that explain how the transient inactivation of Gsp amidase leads to an accumulation of Gsp and an increased level of GspSSPs after oxidative stress. With elimination of the oxidative threat, Gsp amidase, GSH reductase, and glutaredoxin act in concert to convert oxidized Gsp (as the disulfide of Gsp ((GspS)2), mixed disulfides of Gsp, and other small thiol-containing compounds and/or GspSSPs) to GSH. That GspSA and glutaredoxin act synergistically is supported by the hypersensitivity of E. coli mutants that lack both enzymes to H2O2.
EXPERIMENTAL PROCEDURES
The following experiments are described in the supplemental material due to space limitations: the construction of GspSA disrupted strain, protein crystallization and refinement, identification of sulfenic acid by dimedone, and the hydrolysis of GspSSPs by Gsp amidase.
Reagents and Chemicals
The chromogenic tripeptide γ-Glu-Ala-Gly-p-nitroanilide (γ-EAG-pNA), which was the amidase substrate, was synthesized as reported (10). The thiol-labeling reagent, monobromobimane (mBBr) and (GspS)2 were purchased from Bachem and Invitrogen, respectively. Other chemicals were purchased from Sigma and Merck unless specified otherwise.
Protein Expression and Purification of GspSA
The genes, GspSA, GspAF (residues 1–197 of full-length GspSA), GrxA (coding for Grx1 (glutaredoxin 1)), and GrxB (coding for Grx2 (glutaredoxin 2)), were each subcloned into a pET28a plasmid (Novagen). Protein expression and purification procedures followed a previous report (11).
Detection of E. coli Gsp S-Thiolated Proteins
NR754 (wild type) and HA61002 (ΔgspSA) were each cultured in 1 ml of M9 minimal medium at 37 °C until the A600 values of the cultures were 1.0. Then, 250 nmol of 14C-labeled spermidine (GE Healthcare) was added to each culture, and they were then incubated for an additional 10 h. Samples of the cells were treated with 0.5 mm H2O2 for 30 min, collected by centrifugation, mixed with SDS-loading buffer containing 0 or 50 mm 2-mercaptoethanol (2-ME), and then boiled for 15 min. After removing cell debris by centrifugation, the samples were subjected to SDS-PAGE. The separated proteins were stained with Coomassie Brilliant Blue or detected using a PhosphorImager system BAS-1500 (Fuji, Tokyo, Japan).
In Vitro and in Vivo Determination of Gsp Levels after H2O2 Treatment
For the in vitro assay, a 1.8 nm GspSA solution was first incubated with 500 μm H2O2 for 5 min to inactivate the GspSA amidase activity and then treated with catalase (final concentration 1 mg/ml) for 20 min to remove any remaining H2O2. Omission of H2O2 served as the control. The samples were added to 250 mm Tris-HCl (pH 7.3), 2 mm GSH, 2 mm Spd, 1 mm ATP, 4 mm phosphoenolpyruvate, 10 mm MgCl2, and 5 units/ml pyruvate kinase. Aliquots of 100 μl were withdrawn periodically and heated at 95 °C for 10 min to inactivate the enzyme activities. Samples were then derivatized with mBBr, and Gsp and GSH contents were analyzed by HPLC (12).
For the in vivo assay, E. coli BL21(DE3) was cultured in M9 minimal medium until its A600 was 1.5. Then the culture was treated with 1, 5, or 10 mm H2O2. After 10 min, the cells were centrifuged and lyophilized. Cells of suitable dry weight were lysed, and 800 μm 2-ME (final concentration) was added. The lysates were derivatized with mBBr and then centrifuged at 8000 × g for 30 min to remove cell debris. The resulting samples were subjected to HPLC to identify and quantify cellular thiol levels.
GspSA Activities Measured after H2O2 Treatment
To measure the degree to which Gsp amidase was inactivated by H2O2, γ-EAG-pNA was used as the substrate. The assay mixtures, at 25 °C, contained 0.45 nm GspSA, 125 mm Tris-HCl (pH 7.3), 1.25 mm γ-EAG-pNA, and various concentrations of H2O2. p-Nitroanilide formation was monitored as an increase in absorbance at 405 nm.
Gsp synthetase activity in the presence of H2O2 was assayed by a pyruvate kinase/lactate dehydrogenase-coupled assay that used ADP (a product of Gsp synthetase) and consumed NADH. NADH consumption was monitored by fluorescence emission at 460 nm (excitation at 340 nm). The assay mixture contained 125 mm Tris-HCl (pH 7.3), 0.2 mm NADH, 1 mm phosphoenolpyruvate, 10 units/ml pyruvate kinase, 10 units/ml lactate dehydrogenase, 2 mm ATP, 10 mm GSH, 10 mm Spd, 10 mm MgCl2, and various concentrations of H2O2. Control experiments indicated that H2O2 did not affect NADH consumption or the pyruvate kinase/lactate dehydrogenase couple.
The ability of GSH to restore amidase activity was assessed. GspSA (0.45 nm) was first inactivated with 500 μm H2O2 for 5 min. Then catalase (final concentration 1 mg/ml) was added, and 4 mm GSH was incubated for 20 min. Inactivated Gsp amidase was then incubated with the aforementioned reagents to ascertain if they could reactivate Gsp amidase. The levels of recovered activity were measured using the γ-EAG-pNA assay. The control experiment used H2O instead of H2O2, and its activity level was normalized to 100%.
The Viabilities of GspSA and Glutaredoxin Null Mutants after H2O2 Treatment
Overnight cultures (ΔgrxA, ΔgrxB, ΔgrxAΔgspSA, and ΔgrxBΔgspSA) were diluted with fresh M9 minimal medium containing 0.4% glucose and cultivated until A600 of 1.0 were reached. Then different concentrations of H2O2 were added to the cultures, and they were incubated for an additional 1 h. The cultures were serially diluted with PBS buffer and plated onto LB agar plates. The survival percentage (survivability) was calculated as ((CFU in the presence of H2O2)/(CFU in the absence of H2O2))/100, where CFU represents colony-forming units. The survivability values (%) for each H2O2 concentration were calculated using those of at least three replicates and are presented as the means ± S.D.
Conversion of Gsp-disulfide to GSH by a Gsp Amidase/GSH Reductase Couple
To determine if Gsp amidase could catalyze the hydrolysis of (GspS)2, 5 mm Gsp-disulfide and 10 μm GspSA (1 μg) were incubated at 37 °C and then subjected to MALDI-TOF-MS. Similarly, to determine if (GspS)2 could be converted to GSH by the Gsp amidase/GSH reductase couple, 0.23 unit/ml of GSH reductase (final concentration) was added to mixtures of 100 mm Tris-HCl (pH 7.3), 0.5 mm NADPH, 600 μm (GspS)2 at 25 °C with or without 28.7 μm GspSA. NADPH consumption was measured by monitoring fluorescence emission at 460 nm (excitation at 340 nm). Replacement of (GspS)2 with oxidized GSH (GSSG, the native substrate of GSH reductase) served as the positive control, whereas the negative control did not include (GspS)2 and GSSG.
RESULTS
Identification of E. coli Gsp S-Thiolated Proteins
To investigate if Gsp S-thiolates E. coli proteins, 14C-labeled spermidine (Spd*) was added to cultures of NR754 (wild type) and HA61002 (ΔgspSA). NR754 has an endogenous GspSA that can conjugate GSH and Spd* to form 14C-labeled Gsp (Gsp*; Fig. 1A). After uptake of Spd*, Gsp*-labeled proteins were detected in lysates of NR754 by phosphorimaging after SDS-PAGE (Fig. 1B, right). The proteins were Gsp S-thiolated was confirmed by treatment of the Gsp*-labeled cell lysates with 2-ME (Fig. 1B, right), which decreased the amount of radiolabeled proteins observed and was attributed to disulfide cleavage by 2-ME. Proteins of HA61002 (ΔgspSA) were not radiolabeled because Gsp could not be synthesized in HA61002 (Fig. 1B, right). Interestingly, the enhanced level of GspSSPs after H2O2 treatment was reminiscent of the protective effects offered by GSH S-thiolation (13), which implied that Gsp might also protect protein thiols against irreversible oxidation. Various Gsp-protein conjugates were formed (Fig. 1B, right), suggesting that Gsp S-thiolation of proteins may be involved in diverse types of biological processes.
FIGURE 1.
Detection of GspSSPs in E. coli. A, a schematic diagramming how Gsp S-thiolation of E. coli proteins was detected. 14C-Labeled spermidine (Spd*) was added into an E. coli culture and was then conjugated with GSH by Gsp synthetase activity in GspSA to form radiolabeled Gsp (Gsp*). Gsp*SSPs were detected by phosphorimaging after SDS-PAGE. The levels of radiolabeled proteins were reduced when extracts were treated with 2-ME. B, SDS-PAGE analysis of E. coli proteins in NR754 (wild type) and HA61002 (ΔgspSA). Left, Coomassie Brilliant Blue-stained proteins. Right, phosphorimaging of radiolabeled proteins.
Rapid Accumulation of Gsp in Vivo in the Presence of H2O2
To measure the intracellular Gsp level after treatment with H2O2, M9 minimal medium cultures of E. coli BL21(DE3) that were in stationary phase (A600 = 1.2) were treated with 1, 5, or 10 mm H2O2 and then incubated for an additional 10 min. The cells were collected by centrifugation and then lysed. Monobromobimane (mBBr) was added into the lysates to derivatize thiol-containing compounds (12), and after HPLC analysis, mBBr-modified Gsp in the eluates was quantified (fluorescence emission at 480 nm, excitation at 375 nm). The addition of 1 or 10 mm H2O2 led to a 2.2- or 3.6-fold increase in Gsp concentration, respectively, within 10 min (Fig. 2A). Expression of GspSA was not altered by H2O2 treatment (Fig. 2B), indicating that the observed increase in Gsp was not linked to the level of GspSA.
FIGURE 2.
Gsp accumulation in E. coli after H2O2 treatment. The levels of Gsp and GspSA in E. coli were measured after the bacteria had been cultured in M9 minimal medium, treated with H2O2, lysed, and treated with mBBr. A, HPLC chromatograms of mBBr-derivatized thiol compounds detected using fluorescence spectroscopy. The arrows indicate the elution position of Gsp. The numbers above the arrows are the amounts (μmol) of Gsp obtained from 1 mg, dry weight, of cells. B, SDS-PAGE of GspSA shows that its expression level was unaffected by H2O2.
Selective Inactivation of Gsp Amidase by H2O2
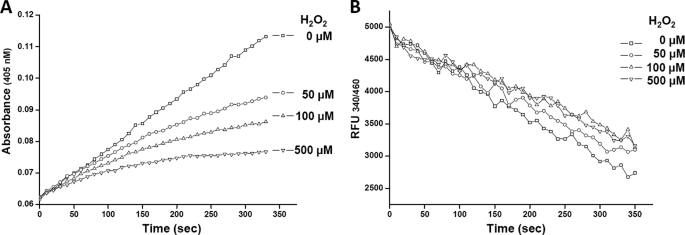
To determine how the GspSA in E. coli is affected by oxidative damage, the enzyme was treated with 50, 100, and 500 μm H2O2, and the synthetase and amidase activities were determined. In all cases, the Gsp amidase activity decreased as functions of time and dose (Fig. 3A) with 50% inhibitory concentration of 250 μm (data not shown). Inactivation was >95% when GspSA was incubated with 500 μm H2O2 for 5 min. Conversely, the synthetase activity remained almost completely active under the same conditions (Fig. 3B). The finding that H2O2 affects the two activities differently is supported by the structural information described below.
FIGURE 3.
Effects of H2O2 on GspSA amidase and synthetase activities. A, plots of Gsp amidase activity that remained after treatment with four different concentrations of H2O2. Activity was measured using the chromogenic substrate γ-EAG-pNA. B, plots of Gsp synthetase activity that remained after treatment with four different concentrations of H2O2. At worst, there was only a 10% reduction in synthetase activity compared with that of the negative control (0 m H2O2). Synthetase activity was measured as the consumption of NADH by the pyruvate kinase/lactate dehydrogenase couple, which also used ADP generated by Gsp synthetase.
To determine whether H2O2-mediated amidase inactivation could be rescued by a biological reducing agent, inactivated GspSA (treated with 500 μm H2O2 for 5 min and then incubated with catalase for 20 min to remove excess H2O2) was exposed to GSH. The addition of 4 mm GSH (physiological concentration) recovered ∼80% of the amidase activity (supplemental Fig. S2). To examine how selectively inactivated Gsp amidase affected the level of Gsp, GspSA was treated with GSH, Spd, ATP, phosphoenolpyruvate, and pyruvate kinase with or without prior exposure to H2O2. The mixtures were then treated with mBBr and analyzed by HPLC. Whether H2O2-treated or not, the Gsp concentrations initially increased and then declined (supplemental Fig. S3); therefore, the level of Gsp was tightly regulated. After pretreatment with H2O2, Gsp accumulated to a greater extent than it did otherwise (no treatment). Additionally, less GSH was present at all times longer than the initial stage (∼25 min). Both of these observations suggested that H2O2 selectively inactivates Gsp amidase, but the amidase activity recovers at a later stage. More Gsp was produced as a consequence of oxidative damage.
Identification of Cys59-Sulfenic Acid by X-ray Crystallography and a Chemical Modification Study
The structure of the H2O2-treated Gsp amidase fragment (GspAF_H2O2; supplemental Table S1) was obtained by soaking a GspAF crystal in crystallization buffer containing 1 mm H2O2 for 3 days before x-ray crystallography. The electron density map of GspAF_H2O2 (Protein Data Bank entry 3A2Z) at 1.5 Å resolution has a density not seen in untreated GspAF that extends from Cys59 Sγ and corresponds to two adjacent oxygen atoms (Fig. 4A). The distance between the S and O1 is 2.1 Å, the C-S–O1 angle is 96.0°, and the S-O1-O2 angle is 94.2°. The second oxygen atom (O2) is unusually close to O1 (2.2 Å). Using the structural and other information (see below), we suggest that the Cys59 thiol was oxidized to sulfenic acid and that its oxygen atom forms a very short hydrogen bond (<2.3 Å) (14) with a water molecule (H2Otriad; Fig. 4B and supplemental Table S1); therefore, the extra electron density probably arose as a consequence of an S-O1···H···O2-H hydrogen bond. Partial negative charges could be present at O1 and O2 because such hydrogen bonds have partial ionic character. Conversely, we do not believe that a hydroperoxide-derivatized cysteine (–S-O–O-H) was present because the S-O and O-O distances are not compatible with such a compound, and it would be less stable than a sulfenic acid (15).
FIGURE 4.
Identification of the Cys59 sulfenic acid by x-ray crystallography and chemical modification/mass spectrometry. A, stereo view of the 2Fo − Fc electron density map at the active site of GspAF_H2O2. The electron density map, drawn at a contour level of 1σ, shows continuous density (in red) connected to the Sγ atom of Cys59. This density was fitted with the oxygen atom of a sulfenic acid (-SOH) and the oxygen of a tightly hydrogen-bonded water molecule. Limited by the structure resolution, it is possible that the Asn149 side chain can be flipped. B, schematic of the hydrogen bond network associated with the Cys59 sulfenic acid in GspAF_H2O2. The sulfenic acid/water hydrogen bond is shown in red. Two other water molecules that contribute to the stability of the sulfenic acid are shown in blue. C, MALDI-MS-MS spectrum of the tryptic peptides that contained the Cys59-dimedone adduct. Gsp amidase, after oxidation by H2O2, was treated with dimedone, digested with trypsin, and subjected to MALDI-MS. The dimedone-modified Cys59-containing peptide (57WQCVEFAR64) has a molecular weight of 1176.6. That covalent dimedone modification of Cys59 had occurred is evidenced by the y- and b-ions identified with asterisks.
The sulfenic acid that was produced when a GspAF crystal was soaked with 1 mm H2O2 for 20 min was reduced to a free thiol when subsequently treated with 1 mm GSH (data not shown). Interestingly, the sulfenic acid was not further oxidized by either prolonged incubation with H2O2 or by a greater concentration of H2O2 (10 mm). To our surprise, Cys59 was covalently bonded to an acetate oxygen in the GspAF_acetate crystal structure (Protein Data Bank code 3A30; supplemental Fig. S4). This S-modification was probably caused by a nucleophilic attack of an acetate ion on the sulfenic acid, which was favorable because of the large acetate concentration (100 mm) and the prolonged exposure.
Although Cys59, which is the catalytic nucleophile for GspSA amidase, was oxidized by H2O2, other GspSA cysteines, including Cys338 and Cys539, which form part of the synthetase binding site (9), were not oxidized when crystals of GspSA were treated with H2O2 (data not shown).
To further corroborate that the Cys59 thiol was oxidized to a sulfenic acid (16), GspAF was first treated with 100 μm H2O2 for 10 min, which fully inactivated GspAF, and then reacted with 100 μm 5,5-dimethyl-1,3-cyclohexanedione (dimedone), which specifically reacts with sulfenic acid (16), at 25 °C for 12 h. After removing excess reagents, derivatized GspAF was digested with trypsin, and its hydrolytic products were analyzed by MALDI-TOF-MS (Fig. 4C). The signal (M + H)+ at m/z 1176.8 is expected for the adduct formed by the GspSA sequence 57WQCVEFAR64 (Mr = 1037) and dimedone (Mr = 140). The presence of b- and y-series ions confirmed that dimedone modified the Cys59 sulfenic acid. Therefore, the thiol of Cys59 was oxidized to the sulfenic acid by H2O2.
Hydrolysis of Gsp-disulfide and Gsp S-Thiolated Proteins and Peptides by Gsp Amidase
E. coli lacks an enzyme that can reduce (GspS)2 (17). If (GspS)2 is involved in E. coli redox regulation, then there must be a way to convert (GspS)2 to Gsp. We next showed that it is possible to convert (GspS)2 to GSH by coupling Gsp amidase and GSH reductase activities. (GspS)2 could be hydrolyzed by Gsp amidase, as shown by MALDI-TOF-MS analysis. After a 10-min or 4-h incubation (GspS)2 with GspSA, Spd, Gsp-GSH mixed disulfide (Gsp-SG), and GSH disulfide (GSSG) were generated, as indicated by the presence of MS peaks at m/z 740.6 ((M + H)+, Gsp-SG), at m/z 613.3 ((M + H)+, GSSG), and at m/z 635.3, ((M + Na)+, GSSG) (Fig. 5A). A second assay was performed to determine if (GspS)2 was a substrate for GSH reductase. GSH reductase activity was detected as NADPH consumption, which was monitored by NADPH absorption at 340 nm (Fig. 5B). GSH reductase alone (without the addition of GSSG) served as the negative control (open circles in Fig. 5B). No activity was observed when (GspS)2 was incubated with only GSH reductase and NADPH (filled squares), in contrast to the result found for GSSG (filled circles). Activity was also observed when (GspS)2 was treated with GspSA and GSH reductase (open triangles). The experimental results presented in Fig. 5B suggested that both (GspS)2 and Gsp-SG were converted to GSH and Spd via consecutive reactions by Gsp amidase and GSH reductase (Fig. 5C).
FIGURE 5.
Conversion of (GspS)2 to GSH and Spd by the Gsp amidase/GSH reductase couple. A, a MALDI-TOF spectrum that shows the molecular weights of the hydrolysis products after (GspS)2 was treated with GspSA for 0 min (top), 10 min (middle), or 4 h (bottom). GSSG was the end product. B, the Gsp amidase/GSH reductase coupled assay. (GspS)2 cannot be reduced by GSH reductase alone (■), but it is reduced when both GspSA and GSH reductase are present (▵). GSSG plus GSH reductase served as the positive control (●). GSH reductase alone served as the negative control (○). C, reaction scheme for the enzymatic conversion of (GspS)2 to GSH and Spd by the Gsp amidase/GSH reductase couple.
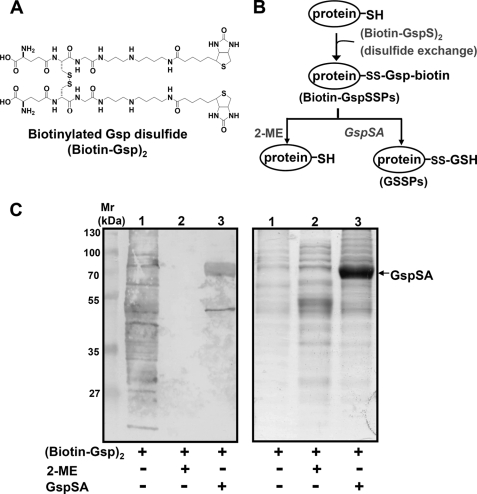
We next tested whether Gsp S-thiolated peptides and proteins were substrates for Gsp amidase. Lysates of E. coli proteins were treated with disulfide of biotinated Gsp ((Biotin-GspS)2) (Fig. 6A) to generate biotin-labeled GspSSPs (biotin-GspSSPs), which could be detected with an anti-biotin antibody (Fig. 6B). Treatment of these proteins with GspSA removed Spd-biotin moieties because biotin-GspSSPs were no longer observed after staining with the anti-biotin antibody (Fig. 6C, left panel, lane 3). Treatment with 2-ME also produced the same result, presumably because 2-ME cleaved Gsp-protein disulfides (Fig. 6C, left panel, lane 2). A synthetic Gsp-containing peptide (T28-Gsp) was also incubated with GspSA, and a MALDI-TOF-MS spectrum of that incubation mixture indicated that T28-Gsp was a substrate for Gsp amidase (supplemental Fig. S5). Therefore, Gsp amidase can hydrolyze a variety of Gsp-derivatized substrates, yielding Spd and GSH S-thiolated proteins/peptides.
FIGURE 6.
Conversion of GspSSPs to GSSPs by Gsp amidase-catalyzed hydrolysis. A, the (biotin-Gsp)2 structure. B, schematic showing the preparation of biotinylated Gsp S-thiolated proteins (biotin-GspSSPs). Treatment of biotin-GspSSPs with GspSA generated GSH S-thiolated proteins. Treatment of biotin-GspSSPs with 2-ME generated unlabeled proteins. C, SDS-PAGE of biotin-GspSSPs. Right, Coomassie Brilliant Blue-stained proteins. Left, Western blot using an alkaline phosphatase-conjugated anti-biotin antibody. Mr, molecular weight markers. Lane 1, biotin-GspSSPs from an E. coli lysate. Lane 2, biotin-GspSSPs as in lane 1 that were treated with 20 mm 2-ME before SDS-PAGE. Lane 3, biotin-GspSSPs as in lane 1 that were treated with 5 μg of GspSA before SDS-PAGE.
Sensitivities of Different E. coli Strains to Oxidative Stress
To characterize how Gsp and GspSA protect against oxidative stress, cultures of NR754 (wild type) and HA61002 (ΔgspSA) were treated with various concentrations of H2O2, and the viabilities of the cells were examined after treatment. The strains were cultured in M9 minimal medium until the cells reached mid-exponential phases, at which point they were treated with H2O2, incubated for 1 h, serially diluted with PBS buffer, and then transferred to LB plates. The viabilities of NR754 and HA61002 under oxidative stress were similar (supplemental Fig. S6), which is consistent with a previous report that an E. coli strain lacking the ability to synthesize GSH was as resistant to H2O2 as the corresponding wild-type strain was (18).
Because glutaredoxins can reduce GSH S-thiolated proteins (GSSPs) (19), we wanted to determine if these enzymes are part of a redox cycle involving Gsp S-thiolation. The viabilities of two glutaredoxin null strains (ΔgrxA and ΔgrxB) and two double mutant strains (ΔgrxAΔgspSA and ΔgrxBΔgspSA) were examined after they were treated with H2O2. When preincubated with 1 or 5 mm H2O2 for 1 h, ΔgrxA and ΔgrxB had survivability values of 60 and 30%, respectively (Fig. 7). The double mutant strains were even more susceptible to oxidative damage because their survivabilities were reduced by 70 and 99% after exposure to 1 and 5 mm H2O2, respectively. Therefore, there is a synergistic protective effect by GspSA and Grx against oxidative damage. Moreover, in a separate experiment, the levels of GspSSPs were substantially decreased when recombinant glutaredoxin (Grx1 or Grx2) was added to E. coli lysates that contained Gsp*SSPs (supplemental Fig. S7). These results strengthen the idea that Grxs participate in the reduction of Gsp S-thiolated proteins.
FIGURE 7.
Glutaredoxin- and GspSA/glutaredoxin-null mutants exhibit hypersensitivities to exogenous H2O2. All strains were grown in M9 minimal medium until the cell densities reached 1.0 A600. Then the cultures were treated with various concentrations of H2O2 for 1 h, and then 0.1 ml of each culture was transferred onto LB plates. The numbers of viable cells were then determined. Survival percentages (survivabilities) of ΔgrxA (▵, blue), ΔgrxB (▴, black), ΔgrxAΔgspSA (○, brown), and ΔgrxBΔgspSA (●, red) were calculated as ((CFU in the presence of H2O2)/(CFU in the absence of H2O2))/100. Survivability values (%) are the means of three replicates ± S.D. (error bars).
DISCUSSION
Redox Regulation of Gsp and Gsp S-Thiolated Proteins
Most organisms use GSH to regulate their intracellular thiol and disulfide levels. Gsp is a GSH derivative found only in some bacteria and parasitic protozoa. As reported herein, we showed that Gsp forms mixed disulfides with the thiols of E. coli proteins in vivo and that the amounts of these Gsp derivatives are linked to intracellular redox conditions. In vivo Gsp S-thiolation of E. coli proteins is affected by several factors, including the intracellular Gsp concentration. We demonstrated that the Gsp level increases when E. coli is oxidatively stressed. The selective inactivation of Gsp amidase provides a rational explanation for this event. When E. coli is exposed to reactive oxygen species, the thiol of Cys59 of the active-site amidase domain is oxidized to sulfenic acid, which inactivates Gsp amidase and consequently causes Gsp to accumulate (Fig. 8). Accumulated Gsp possibly scavenges oxidants directly or forms mixed disulfides with protein thiols. Once the source of the oxidative stress has been eliminated, Gsp amidase activity can be rescued by reaction of the sulfenic acid with a reducing reagent (e.g. GSH or Gsp). Sulfenic acid is a reactive electrophile and reacts with thiol reagents, such as GSH, to generate mixed disulfides. The amidase active site is solvent-exposed, which should allow GSH or other small thiol-containing molecules to react with the sulfenic acid. After formation of a mixed disulfide, an additional thiol exchange may continue regenerating the thiol of Cys59 and thereby recovering amidase activity (supplemental Fig. S8). The reactivated amidase may then hydrolyze either excessive Gsp to GSH and Spd and/or GspSSPs to GSSPs and Spd. Gsp could also be oxidized to produce (GspS)2 and/or other mixed disulfides upon reaction with reactive oxygens. We showed that GSH reductase alone cannot reduce (GspS)2 but that Gsp amidase must first remove the spermidine moiety. The conversion of (GspS)2 to Spd and GSH by the Gsp amidase/GSH reductase couple suggests that Gsp may be part of an oxidative defense mechanism that does not require a Gsp-specific reductase. Such a reductase has not been found in E. coli (17). The results allow us to propose a reaction scheme (Fig. 8) that explains how Gsp amidase activity is modulated in vivo so that the Gsp level reflects the intracellular redox condition.
FIGURE 8.
The roles of Gsp and glutathionylspermidine synthetase/amidase in the intracellular redox regulation of E. coli. When exposed to reactive oxygen species (ROS), the active-site Cys59 thiol of Gsp amidase is oxidized to sulfenic acid (1), which causes the inactivation of Gsp amidase (×) (2). Because Gsp synthetase is not affected by ROS, intracellular Gsp accumulates (3). Gsp may then scavenge harmful oxidants by forming Gsp-disulfides and other small molecule disulfide compounds (4A) and/or protecting protein thiols from oxidation by Gsp S-thiolation (4B). With the removal of the oxidative stresses, intracellular GSH and/or Gsp may rescue oxidized Gsp amidase, restoring amidase activity (4C). Reactivated Gsp amidase may hydrolyze Gsp to GSH and Spd or hydrolytically remove Spd from Gsp-disulfide or Gsp-modified proteins. Finally, the amount of Gsp returns to its basal level (5).
Glutaredoxins reduce mixed disulfide bonds between GSH and GSSPs by forming a GrxSSG intermediate that can be subsequently reduced by a molecule of GSH (20). The protein expression level of Grx1 is enhanced under oxidative stress in E. coli, whereas Grx2 is constitutively produced (21). We found that the amounts of GspSSPs decreased in the presence of Grx1 or Grx2 (supplemental Fig. S7), suggesting that Grxs can reduce mixed disulfides. As mentioned, Gsp amidase catalyzes the hydrolytic removal of Spd from GspSSPs, leading to the formation of GSSPs. A Grx can then reduce the GSSPs to generate free protein thiol(s) (supplemental Fig. S9). However, we cannot exclude the possibility that glutaredoxin directly reduces GspSSPs, with the resulting Grx-Gsp mixed disulfide intermediate subsequently reduced via thiol-disulfide exchange (supplemental Fig. S9). Because ΔgrxAΔgspSA and ΔgrxBΔgspSA were more easily killed by H2O2 than were ΔgrxA and ΔgrxB, it appears that the activities of Grx and GspSA can provide a coordinated defense against oxidative damage in E. coli.
To date, most studies concerning protein S-thiolation have focused on GSH, because GSH is abundant in cells (4). However, protein S-thiolation by other thiol-containing molecules, such as cysteine or Gsp, may have different effects. For example, a previous study demonstrated that GSH S-thiolation of Ca2+-dependent protein kinase Cα inhibits the activity of Ca2+-dependent protein kinase Cα and its isozymes, whereas cysteine S-thiolation does not (22). Both trypanothione (TSH) and Gsp are more efficient reducing agents than is GSH. The non-enzymatic reductions of dehydroascorbate and H2O2 by Gsp and TSH are several times faster than those by GSH (23, 24). It has also been found that Gsp was a more efficient S-thiolating agent than was GSH (25). GSH is negatively charged (−1) at physiological pH, whereas Gsp is positively charged (+2). Gsp and GSH therefore introduce opposite charges into the proteins that they thiolate. Protein thiols are deprotonated to form thiolates that interact more readily with (GspS)2 at pH >6, rather than GSSG (25). Although GSH and Gsp have distinct physical and chemical properties, it is still difficult to elucidate how GSH and Gsp function in vivo because of the involvement of the GspSA amidase-catalyzed conversion of GspSSPs to GSSPs.
Dissimilar Amidase Active Sites Linked to Differential Redox Regulation
Parasitic protozoa use TSH, which is synthesized by TSH synthetase/amidase to defend against oxidative damage. Like GspSA, TSH synthetase/amidase has an amidase activity that can hydrolyze the amide bond of Gsp. Therefore, it would be interesting to know if TSH amidase can be selectively and reversibly inactivated. In the x-ray structure of Leishmania major TSH synthetase/amidase, the C-terminal segment partially obstructs accessibility to the catalytic Cys59 nucleophile (26). Consequently, the active site of TSH amidase is substantially blocked, in contrast with that of GspSA, which is part of a large, solvent-accessible cleft. Even if the TSH amidase Cys59 thiol could be oxidized to sulfenic acid and/or form a mixed disulfide with GSH or TSH, the restricted access almost certainly would impede subsequent thiol exchange (supplemental Figs. S8and S10). Because access to the TSH amidase active site is partially obscured, TSH reductase and TSH tryparedoxin may compensate as a defense mechanism against reactive oxygen in parasitic protozoa. It is also possible that, with the evolutionary emergence of TSH reductase and TSH tryparedoxin, TSH amidase degenerates and no longer participates in redox regulation.
Cys59 Sulfenic Acid of GspSA Is Primarily Stabilized by a Very Short Hydrogen Bond
Sulfenic acid is an unstable compound and is highly reactive; therefore, it usually acts as an intermediate en route to other more stable oxidized sulfur compounds (e.g. sulfinic and sulfonic acids) (27). Nature has used different strategies to stabilize sulfenic acids. For example, the sulfur of sulfenic acid and an amide nitrogen can form a cyclic sulfonamide, which has been found in the crystal structure of protein-tyrosine phosphatase (Protein Data Bank entry 1OEM) (28). The sulfenic acid of an archaeal 2-Cys peroxiredoxin (Protein Data Bank entry 2ZCT) is hypervalent with an S–N covalent bond (2.2 Å) between Sγ of Cys50 and Nδ1 of His42 (29). Other sulfenic acids derived from cysteines exist in solvent-inaccessible and nonpolar microenvironments. Their limited solvent accessibilities prevent further oxidation to a sulfinic or sulfonic acid, which are not reducible. Conversely, Cys59 of Gsp amidase is solvent-accessible because it is located on the surface. We did not observe formation of Cys59 sulfinic acid or sulfonic acid even when GspAF was exposed to 10 mm H2O2. The sulfenic acid of GspAF_H2O2 is most likely stabilized by hydrogen bonds formed with water oxygens. In the GspAF_H2O2 structure, the S–O1 bond (2.1 Å) is longer than those S–O bonds commonly observed in proteins (∼1.7 Å) (e.g. NAD(P)H oxidase (Protein Data Bank entry 2CDU)) (30). The S-O1 of GspAF_H2O2 is thus polarized, which probably causes the sulfur to be partially positively charged and thereby prevents further oxidation. The very short hydrogen bond between O1 and O2 (O1···H···O2) also stabilizes the sulfenic acid (Fig. 4, A and B). Because both O1 and O2 should be partially negatively charged, the repulsion between them might be compensated for by three hydrogen bonds (hydrogen bonds of a, b, and e in Fig. 4B) found in the GspAF crystal structure. Two of the three hydrogen bonds involving O1 are the hydrogen bonds with a water molecule (H2Ooxy) and with the side-chain O of Asn149, which is an oxyanion binding site residue, with distances of 2.9 and 3.5 Å, respectively. Interestingly, the Nϵ of Gln58, which is also the other residue in the oxyanion hole, forms a hydrogen bond network with two water molecules (H2Ooxy and H2Odistal) (Fig. 4, A and B). The third hydrogen bond is formed between a hydrogen of O2 and an imidazole nitrogen of His131. These observations are consistent with the suggestion of Salsbury et al. (31) that surrounding hydrogen bond networks stabilize the sulfenic acids found in proteins.
In summary, Gsp formed mixed disulfides with the thiols of a variety of E. coli proteins. These mixed disulfides represent a previously uncharacterized type of post-translational modification. The E. coli GspSA amidase activity was selectively inhibited in vivo by oxidation of its Cys59 thiol to sulfenic acid. This inhibition led to a rapid increase in the amounts of intracellular Gsp and GspSSPs. Basal levels of Gsp and GspSSPs were recovered once the oxidative threat was eliminated, and at that time, the amidase activity was restored. Moreover, the hypersensitivities of the GspSA/Grx null mutants to H2O2 support the idea that GspSA and Grx synergistically defend against oxidative damage.
Supplementary Material
Acknowledgments
Some E. coli strains were obtained from the National Bioresource Project, Japan. We thank the National Synchrotron Radiation Research Center of Taiwan and the National Core Facility of Proteomics for mass spectrometry analysis.
This work was supported by National Science Council of Taiwan Grant NSC97-2628-M-001-016-MY3.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S10.
- Spd
- spermidine
- (GspS)2
- disulfide of glutathionylspermidine
- GSSG
- disulfide form of GSH
- GSSP
- GSH S-thiolated protein
- Gsp
- glutathionylspermidine
- GspSA
- glutathionylspermidine synthetase/amidase
- Gsp-SG
- Gsp glutathione mixed disulfide
- GspSSPs
- Gsp S-thiolated proteins
- Grx
- glutaredoxin
- mBBr
- monobromobimane
- 2-ME
- β-mercaptoethanol
- TSH
- trypanothione
- pNA
- p-nitroanilide
- MALDI
- matrix-assisted laser desorption ionization
- TOF
- time-of-flight
- MS
- mass spectrometry.
REFERENCES
- 1.Winterbourn C. C., Hampton M. B. (2008) Free Radic. Biol. Med. 45, 549–561 [DOI] [PubMed] [Google Scholar]
- 2.Ritz D., Beckwith J. (2001) Annu. Rev. Microbiol. 55, 21–48 [DOI] [PubMed] [Google Scholar]
- 3.Dalle-Donne I., Rossi R., Giustarini D., Colombo R., Milzani A. (2007) Free Radic. Biol. Med. 43, 883–898 [DOI] [PubMed] [Google Scholar]
- 4.Dalle-Donne I., Rossi R., Colombo G., Giustarini D., Milzani A. (2009) Trends Biochem. Sci. 34, 85–96 [DOI] [PubMed] [Google Scholar]
- 5.Hondorp E. R., Matthews R. G. (2004) PLoS Biol. 2, e336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bollinger J. M., Jr., Kwon D. S., Huisman G. W., Kolter R., Walsh C. T. (1995) J. Biol. Chem. 270, 14031–14041 [DOI] [PubMed] [Google Scholar]
- 7.Koenig K., Menge U., Kiess M., Wray V., Flohé L. (1997) J. Biol. Chem. 272, 11908–11915 [DOI] [PubMed] [Google Scholar]
- 8.Oza S. L., Tetaud E., Ariyanayagam M. R., Warnon S. S., Fairlamb A. H. (2002) J. Biol. Chem. 277, 35853–35861 [DOI] [PubMed] [Google Scholar]
- 9.Tabor H., Tabor C. W. (1975) J. Biol. Chem. 250, 2648–2654 [PubMed] [Google Scholar]
- 10.Lin C. H., Kwon D. S., Bollinger J. M., Jr., Walsh C. T. (1997) Biochemistry 36, 14930–14938 [DOI] [PubMed] [Google Scholar]
- 11.Pai C. H., Chiang B. Y., Ko T. P., Chou C. C., Chong C. M., Yen F. J., Chen S., Coward J. K., Wang A. H., Lin C. H. (2006) EMBO J. 25, 5970–5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shim H., Fairlamb A. H. (1988) J. Gen. Microbiol. 134, 807–817 [DOI] [PubMed] [Google Scholar]
- 13.Aracena P., Tang W., Hamilton S. L., Hidalgo C. (2005) Antioxid. Redox Signal. 7, 870–881 [DOI] [PubMed] [Google Scholar]
- 14.Katz B. A., Elrod K., Luong C., Rice M. J., Mackman R. L., Sprengeler P. A., Spencer J., Hataye J., Janc J., Link J., Litvak J., Rai R., Rice K., Sideris S., Verner E., Young W. (2001) J. Mol. Biol. 307, 1451–1486 [DOI] [PubMed] [Google Scholar]
- 15.Dierks T., Dickmanns A., Preusser-Kunze A., Schmidt B., Mariappan M., von Figura K., Ficner R., Rudolph M. G. (2005) Cell 121, 541–552 [DOI] [PubMed] [Google Scholar]
- 16.Ellis H. R., Poole L. B. (1997) Biochemistry 36, 15013–15018 [DOI] [PubMed] [Google Scholar]
- 17.Smith K., Borges A., Ariyanayagam M. R., Fairlamb A. H. (1995) Biochem. J. 312, 465–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenberg J. T., Demple B. (1986) J. Bacteriol. 168, 1026–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lundström-Ljung J., Vlamis-Gardikas A., Aslund F., Holmgren A. (1999) FEBS Lett. 443, 85–88 [DOI] [PubMed] [Google Scholar]
- 20.Gallogly M. M., Starke D. W., Mieyal J. J. (2009) Antioxid. Redox Signal. 11, 1059–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Potamitou A., Neubauer P., Holmgren A., Vlamis-Gardikas A. (2002) J. Biol. Chem. 277, 17775–17780 [DOI] [PubMed] [Google Scholar]
- 22.Chu F., Ward N. E., O'Brian C. A. (2001) Carcinogenesis 22, 1221–1229 [DOI] [PubMed] [Google Scholar]
- 23.Krauth-Siegel R. L., Lüdemann H. (1996) Mol. Biochem. Parasitol. 80, 203–208 [DOI] [PubMed] [Google Scholar]
- 24.Ariyanayagam M. R., Fairlamb A. H. (2001) Mol. Biochem. Parasitol. 115, 189–198 [DOI] [PubMed] [Google Scholar]
- 25.Melchers J., Dirdjaja N., Ruppert T., Krauth-Siegel R. L. (2007) J. Biol. Chem. 282, 8678–8694 [DOI] [PubMed] [Google Scholar]
- 26.Fyfe P. K., Oza S. L., Fairlamb A. H., Hunter W. N. (2008) J. Biol. Chem. 283, 17672–17680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poole L. B., Karplus P. A., Claiborne A. (2004) Annu. Rev. Pharmacol. Toxicol. 44, 325–347 [DOI] [PubMed] [Google Scholar]
- 28.Salmeen A., Andersen J. N., Myers M. P., Meng T. C., Hinks J. A., Tonks N. K., Barford D. (2003) Nature 423, 769–773 [DOI] [PubMed] [Google Scholar]
- 29.Nakamura T., Yamamoto T., Abe M., Matsumura H., Hagihara Y., Goto T., Yamaguchi T., Inoue T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6238–6242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lountos G. T., Jiang R., Wellborn W. B., Thaler T. L., Bommarius A. S., Orville A. M. (2006) Biochemistry 45, 9648–9659 [DOI] [PubMed] [Google Scholar]
- 31.Salsbury F. R., Jr., Knutson S. T., Poole L. B., Fetrow J. S. (2008) Protein Sci. 17, 299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.