Abstract
RNA helicases are proteins essential to almost every facet of RNA metabolism, including the gene-silencing pathways that employ small RNAs. A phylogenetically related group of helicases is required for the RNA-silencing mechanism in Caenorhabditis elegans. Dicer-related helicase 3 (DRH-3) is a Dicer-RIG-I family protein that is essential for RNA silencing and germline development in nematodes. Here we performed a biochemical characterization of the ligand binding and catalytic activities of DRH-3 in vitro. We identify signature motifs specific to this family of RNA helicases. We find that DRH-3 binds both single-stranded and double-stranded RNAs with high affinity. However, the ATPase activity of DRH-3 is stimulated only by double-stranded RNA. DRH-3 is a robust RNA-stimulated ATPase with a kcat value of 500/min when stimulated with short RNA duplexes. The DRH-3 ATPase may have allosteric regulation in cis that is controlled by the stoichiometry of double-stranded RNA to enzyme. We observe that the DRH-3 ATPase is stimulated only by duplexes containing RNA, suggesting a role for DRH-3 during or after transcription. Our findings provide clues to the role of DRH-3 during the RNA interference response in vivo.
Keywords: ATPases, Double-stranded RNA, RNA Helicase, RNA Interference (RNAi), siRNA
Introduction
Diverse groups of small RNAs regulate gene expression through a set of conserved pathways, of which RNA interference (RNAi)2 is the best characterized (1). The RNAi response is initiated by small interfering RNAs (siRNA) that are cut (“diced”) from long, double-stranded RNA that has been introduced to the organism by an exogenous source (i.e. viral genomic RNA) (2). One strand of the siRNA duplex is assembled into an effector complex, and this single-stranded RNA acts as a guide sequence for seeking out complementary RNA targets (3). In nematodes, fungi, and plants, a second set of siRNAs is produced from these RNA targets, which function as templates for RNA-dependent RNA polymerases (RdRPs) (4) that produce secondary siRNAs. In the nematode Caenorhabditis elegans, secondary siRNAs are produced directly by RdRP transcription, without a double-stranded RNA intermediate or dicing (5, 6).
A group of conserved helicases is essential to all gene-silencing pathways that employ small RNAs, including RNAi (7–9). Dicer-related helicase-3 (DRH-3) is an essential RNA helicase that functions in germline development and the RNAi pathway in C. elegans (10–13). Biochemical data show that DRH-3 interacts with members of the C. elegans RNAi machinery, including Dicer (DCR-1) and the RdRP, RRF-1 (11, 14). Mutations in the helicase motif of DRH-3 cause defects in RNAi, including the loss of RdRP-dependent small RNAs, suggesting that the helicase is essential for producing small silencing RNAs (10, 12). Recently, DRH-3 was also shown to be required for heterochromatin formation and for euchromatic chromosome segregation in worms (10, 15).
DRH-3 is an ortholog of the Dicer- and RIG-I-like family of RNA helicases that are essential for RNAi and the innate immune response to viruses in mammals, respectively. Multiple groups have conducted phylogenetic analyses of the Dicer-RIG-I helicase family (11, 13, 16, 17) and found that DRH-3 sequence identity and structure are more closely related to RIG-I than Dicer (17). Moreover, the central helicase motif of DRH-3 is enveloped by N-terminal and C-terminal appendages that suggest a protein structure more similar to RIG-I than Dicer, which has an N-terminal helicase motif and C-terminal domains of specific function. RIG-I initiates the antiviral response by recognizing multiple motifs on viral RNA including short double-stranded RNA and single-stranded RNA with 5′-triphosphates (18–20), features similar to those of small RNAs in nematode RNAi pathway.
We sought to explore the function of DRH-3 by conducting a quantitative, biochemical analysis of this RNA helicase in vitro. Based on an alignment of helicase motifs, we confirm that DRH-3 is an ortholog of the Dicer-RIG-I family of RNA helicases, and we identify two sequence motifs that distinguish this family from other helicases. As a preliminary step toward defining how DRH-3 contributes to the RNAi pathway, we examined its interactions with ligands that are implicated in siRNA production. In vitro, DRH-3 binds both single-stranded and double-stranded RNA; however, its ATPase activity is stimulated only by double-stranded RNA (dsRNA). DRH-3 ATPase activity is stimulated by nucleic acid duplexes that contain at least one RNA strand, implicating a role in either transcriptional or post-transcriptional regulation. Kinetic analysis of the ATPase function indicates that DRH-3 is a strictly RNA-dependent ATPase with a kcat of ∼500 min−1 and a Km of 36 μm. Although these parameters are remarkably similar to those reported for RIG-I, there are important differences in enzyme specificity as DRH-3 does not bind 5′-triphosphates. In addition, DRH-3 appears to have some form of allosteric regulation mechanism, potentially through oligomerization. We find that DRH-3 is inhibited by high concentrations of duplex RNA supporting an autoregulatory mechanism that may limit DRH-3 function during siRNA production.
EXPERIMENTAL PROCEDURES
General Methods
All binding and ATPase reactions were performed at 25 °C unless otherwise noted. RNA was prepared by in vitro transcription by T7 RNA polymerase using DNA oligomers as templates (supplemental Table 1), resolved through a polyacrylamide gel, UV-shadowed, excised from the gel, eluted in 1 m NaCl, and purified on a MegaClear filter (Ambion). Double-stranded RNA was annealed by heating to 95 °C for 2 min and slow cooling to 25 °C for 1 h in 1× annealing buffer (25 mm MOPS [pH 6.3], 30 mm NaCl, 0.1 mm EDTA). When needed, RNA was treated with calf intestine alkaline phosphatase (New England Biolabs) and 5′-32P-radiolabeled using T4 polynucleotide kinase (New England Biolabs).
Protein Expression and Purification
DRH-3 open reading frame was reverse transcribed from N2 worm total RNA using a SuperScript II reverse transcriptase (Invitrogen) and cloned into the Champion pET-SUMO protein expression vector (Invitrogen). Full-length protein was overexpressed in bacteria in the Rosetta II cell line (Novagen) and purified by batch method over nickel-nitrilotriacetic acid-agarose (Qiagen). After eluting from nickel-nitrilotriacetic acid with 160 mm imidazole, the protein was bound to a heparin column (GE healthcare) and eluted over a 100–600 mm salt gradient. The SUMO tag was then cleaved overnight using SUMO site-specific protease. Cleaved protein was further purified by gel filtration using HiLoad Superdex 200 column (GE healthcare). Peak fractions were concentrated using an Amicon molecular weight cutoff column (Millipore). Protein concentration was determined by absorbance at 280 nm of protein denatured in 6 m guanidium hydrochloride. The extinction coefficient at 280 nm of DRH-3 is 126,060 cm−1 m−1.
RNA Binding
Double filter binding assays were done as described (21). Briefly, 25-μl reactions containing 25 mm MOPS [pH 6.3], 30 mm NaCl, 3 mm MgCl2, 2 mm DTT, 0.1 unit of RNase inhibitor (Roche Applied Science), 1% glycerol, 0.5 nm radiolabeled RNA, and varying concentrations of DRH-3 were incubated for 60 min at 25 °C. Incubation times were varied to ensure that the binding reaction was at equilibrium. The entire reaction was loaded onto a dot blot assembly under vacuum with nitrocellulose (top) and nylon (bottom) membranes stacked in the assembly. The blots were washed twice with 1× reaction buffer and then dried, exposed to an imaging plate, and analyzed by phosphorimaging (Storm, GE Healthcare). Data were analyzed using the IGOR Pro software (Softzymics) and best fit to a hyperbolic
 |
or sigmoidal (variation of the Hill equation)
curves using the VisualEnzymics package.
RNA binding by native gel analysis was performed as outlined for filter binding experiments with slight modifications. 25-μl reactions containing 25 mm HEPES [pH 7.4], 100 mm NaCl, 3 mm MgCl2, 2 mm DTT, 0.1 of unit RNase inhibitor (Roche Applied Science), 10% glycerol, 0.5 nm radiolabeled RNA, and varying concentrations of protein were incubated for 15 min at 25 °C. One-half of the reaction was loaded onto 6 or 8% native polyacrylamide gels pre-run for 2 h at 5 watts at room temperature. Samples were resolved on the pre-run gel at 5 watts for 1 h and then dried, exposed to an imaging plate, and analyzed by phosphorimaging.
ATPase Assays
ATP hydrolysis by DRH-3 was measured using thin layer chromatography (22). A typical reaction consisted of 25 mm MOPS [pH 6.3], 30 mm NaCl, 3 mm MgCl2, 2 mm DTT, 0.1 unit of RNase inhibitor (Roche Applied Science), 1% glycerol, 1 mm cold ATP, and 7.5 nm [α-32P]ATP (PerkinElmer Life Sciences). All reactions were done at 25 °C. At each time point, the reaction was quenched by spotting on a cellulose polyethyleneimine flexible TLC plate (Selecto Scientific). The plate was subjected to 0.75 m KH2PO4 [pH 4] buffer in a glass chamber. After migration of buffer to the top of the plate, the plates were then dried, and ATP hydrolysis (ADP production) was analyzed by phosphorimaging. An ATP hydrolysis ladder was prepared using a reaction containing 1:1000-fold dilution of alkaline phosphatase (New England Biolabs) incubated at 37 °C for 20 min. In the kinetic analysis of DRH-3 ATPase, initial rates of ATP hydrolysis were calculated (IGOR Pro) and plotted as a function of substrate (ATP) or activator (RNA) concentration. Data were fit to either a hyperbolic (Michaelis-Menten) equation
 |
or a sigmoidal (Hill) equation
 |
NTPase Assays
GTPase, CTPase, and UTPase activity was measured using a TLC-based assay. Reactions consisted of 25 mm MOPS [pH 6.3], 30 mm NaCl, 3 mm MgCl2, 2 mm DTT, 0.1 unit of RNase inhibitor (Roche Applied Science), 1% glycerol, 200 μm cold NTP, and 7.5 nm [α-32P]NTP (PerkinElmer Life Sciences). All reactions were done at 25 °C. After 90 min, the reaction was quenched by spotting on a cellulose polyethyleneimine flexible TLC plate (Selecto Scientific). The plate was subjected to 0.75 m KH2PO4 [pH 4] (ATP and GTP) or 0.5 m LiCl2/0.5 M formic acid (CTP and UTP) in a glass chamber. After migration of buffer to the top of the plate, the plates were then dried, and NTP hydrolysis (NDP production) was analyzed by phosphorimaging. An NTP hydrolysis ladder was prepared using a reaction containing 1:1000-fold dilution of alkaline phosphatase (New England Biolabs) incubated at 37 °C for 20 min.
Unwinding Assays
RNA-unwinding assays were done as described previously (32). Briefly, 100 nm DRH-3 and 0.1 nm 5′-32P-radiolabeled RNA duplexes were preincubated for 5 min in a reaction mix containing 25 mm MOPS [pH 6.3], 30 mm NaCl, 3 mm MgCl2, 2 mm DTT, 0.1 unit of RNase inhibitor (Roche Applied Science), 1% glycerol. After 5 min, 4 mm ATP and varying concentrations of poly[ribo(I-C)] (Midland) were added to the reactions. Reactions were incubated at 25 °C for 1 h and stopped with 1 volume of 2× quench buffer (30 mm EDTA, 25% sucrose, 1% SDS, 0.015% xylene cyanol, and 0.015% bromphenol blue). Samples were resolved through a 14% semi-native polyacrylamide gel (3 m urea, 0.5× Tris-borate-EDTA), dried, and analyzed by phosphorimaging.
Analytical Ultracentrifugation
Analytical ultracentrifugation experiments were done on a Beckman Xl-I ultracentrifuge using an An-60 Ti analytical rotor. Sedimentation velocity experiments containing 1 or 5 μm DRH-3 in 25 mm HEPES (pH 7.5), 100 mm NaCl, 5% glycerol, 2 mm DTT were run at 35,000 rpm at 16 °C for 12 h. Velocity data were analyzed using SedFit with a frictional coefficient of 1.7 and density of 1.0175 to predict the sedimentation coefficient of 4.5 s for 130-kDa DRH-3.
NADH-coupled ATPase Assay
The NADH-coupled assay was done as described (27). Briefly, reactions contained the reagents of a typical ATPase assay with 0.5 mm phosphoenolpyruvate, 100 units of pyruvate kinase, 200 μm NADH, and 20 units of lactic dehydrogenase. ATP hydrolysis is coupled to NADH oxidation, detected by the change in absorbance at 340 nm.
RESULTS
DRH-3 Is a Member of the Dicer-RIG-I Family of DEXH Helicases
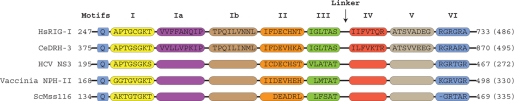
We conducted a comparative analysis of conserved motifs from diverse members of helicase superfamily II (SF2), including DRH-3 (Fig. 1). Based on its primary sequence, the helicase motif of DRH-3 is more similar to RIG-I than Dicer, based on ClustalW scoring (supplemental Fig. S1), and therefore, RIG-I was used as a prototypical Dicer-RIG-I family member during the sequence analysis. The primary sequences of five diverse RNA helicases were aligned based on helicase motif sequences that begin at the conserved glutamine upstream of motif I and extend through motif VI. Motifs I (Walker A), II (Walker B), III, and VI are well conserved among the diverse set of eukaryotic and viral helicases analyzed (Fig. 1). However, significant sequence variations in motifs Ia, Ib, and V distinguish DRH-3 from other SF2 helicases (based on an alignment of 21 sequences within this specific family). These same motifs are well conserved within the DRH-3-Dicer-RIG-I subfamily, and they can be used as markers to differentiate this group (Fig. 1, supplemental Fig. S1). Motifs 1a and 1b, which are almost identical between RIG-I and DRH-3, are intriguing because they contain amino acids that interact with the backbone of duplex nucleic acid, perhaps explaining their divergence from motifs within other SF2 helicase families (23).
FIGURE 1.
DRH-3 is a member of the Dicer-RIG-I family of superfamily II RNA helicases. A helicase motif alignment comparing five SF2 RNA helicase proteins including human RIG-I, nematode DRH-3, viral proteins NS3 and NPH-II, and yeast DEAD box helicase Mss116 is shown. Although the sequences of the highlighted motifs were well established for NS3, NPH-II, and Mss116, only sequences that aligned with RIG-I are shown. HCV, hepatitis C virus.
A second feature that distinguishes the RIG-I-DRH-3 subfamily is that conserved motifs I-VI are distributed over a larger span of the protein. For example, the conserved motifs span ∼500 residues in DRH-3 and RIG-I, whereas the same motifs span ∼300 residues in viral DEXH proteins NS3 and NPH-II and the DEAD box helicase Mss116 (Fig. 1). Most of this difference in sequence length can be attributed to a linker region between motifs III and IV in the RIG-I/DRH-3 family.
DRH-3 Binds Both Single-stranded and Double-stranded RNA in Vitro
DRH-3 is essential for the production of RdRP-dependent small silencing RNAs in C. elegans. However, the mechanism by which DRH-3 facilitates small RNA production remains unclear. We sought to understand the function of C. elegans DRH-3 in the RNA interference pathway by first studying its biochemical properties in vitro. To this end, we cloned and expressed the 129-kDa recombinant DRH-3 protein (supplemental Fig. S2), and we began our analysis by testing the RNA binding properties of DRH-3. As a homolog of RIG-I, it was expected that DRH-3 might bind RNA with double-stranded character or RNA transcripts bearing 5′-triphosphates (18–20).
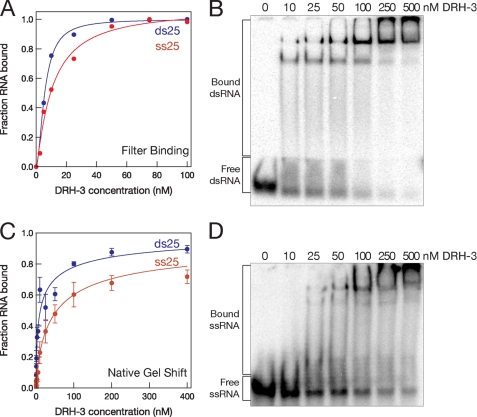
DRH-3 was observed to bind both single-stranded RNA (ssRNA) and double-stranded RNA (dsRNA) in filter binding experiments, with equilibrium binding constants of 10 nm (± 3 nm) and 6 nm (± 0.3 nm) respectively (Fig. 2A). To test whether 5′-triphosphates are essential for DRH-3 affinity (supplemental Fig. S3), we examined the binding of an 18-nt RNA that had been prepared by in vitro transcription. DRH-3 readily binds both untreated (5′-triphosphate) and phosphatase-treated (5′-hydroxyl) RNA with equal efficiency, indicating a possible difference in the substrate specificity for DRH-3 and RIG-I (18, 19). Using the native gel shift assay, we also monitored the oligomeric state of DRH-3 upon binding RNA (Fig. 2, C and D). DRH-3 forms a predominantly monomeric complex with both ss25 and ds25 RNA at DRH-3 concentrations less than 25 nm and a multimeric complex at higher concentrations of protein.
FIGURE 2.
DRH-3 binds both single-stranded and duplex RNA. A, by filter binding, DRH-3 binds single-stranded RNA (25 nt) with a binding constant of 10 ± 3 nm and duplex RNA (25 bp) of the same sequence with a binding constant of 5.6 ± 0.3 nm. Error for each was based on the best fit of the binding data. C, using native gel analysis, apparent binding constants were calculated to be 47 nm for single-stranded RNA and 11 nm for duplex. These parameters were calculated based on binding reactions that were incubated for 15 min. Error bars indicate S.E. B and D, native gel analysis showing DRH-3 binding both single-stranded (25 nt)(D, 6% polyacrylamide) and double-stranded (25 bp)(B, 8% polyacrylamide) RNA.
ATPase Activity of DRH-3 and Cofactor Requirements
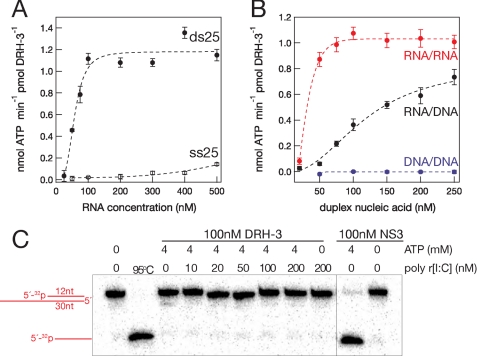
Most DEXH/D helicases require nucleic acid cofactors to stimulate their ATPase and unwinding activities (24). We therefore tested the ATPase activity of DRH-3 in the absence and presence of various RNA cofactors, monitoring phosphate release by thin layer chromatography (TLC, supplemental Fig. S4) (22). In the absence of RNA and in the presence of ribonucleases to remove residual nucleic acid in the protein preparation, DRH-3 does not exhibit any ATPase activity (supplemental Fig. S4). In the presence of RNA, robust ATPase activity was observed, indicating that DRH-3 is a tightly coupled RNA-dependent ATPase. However, ATPase activity was dependent on the type of RNA ligand employed in the assay. For example, in the presence of ss25 RNA, DRH-3 displayed low levels of ATPase activity (0.1 nmol of ATP hydrolyzed/min/pmol of DRH-3 at 500 nm RNA). Detectable ss25-activated ATPase activity required RNA concentrations exceeding 500 nm (Fig. 3A, supplemental Fig. S5) despite the fact that DRH-3 binds ssRNA at nanomolar concentrations (Fig. 2). By contrast, DRH-3 displayed robust ATPase in the presence of duplex RNA of the same sequence, even at low RNA concentrations (ds25 RNA, a maximum of 1.2 nmol of ATP hydrolyzed/min/pmol of DRH-3 at 100 nm RNA) (Fig. 3B). These data suggest that functional complexes of DRH-3 require the binding of duplex RNA.
FIGURE 3.
DRH-3 ATPase is stimulated by double-stranded RNA. A, analysis showing RNA-stimulated ATPase rates of DRH-3 by short (25-mer) single- and double-stranded RNA. B, DRH-3 ATPase activity required at least one strand of the duplex nucleic acid (34 bp) to contain RNA. Errors for each of the ATPase rates were based on the fit of the data. Error bars in A and B indicate S.E. C, DRH-3 cannot unwind RNA duplexes in vitro. No unwinding activity was seen for 12-bp RNA duplexes in the presence or absence of dsRNA in trans. Hepatitis C virus NS3 was used as a positive control for unwinding (right).
Based on our comparative sequence analysis of SF2 RNA helicases, we observed potential similarities in DRH-3, a DEXH protein, to the DEAD box proteins including a putative Q-motif that is believed to confer specificity for ATP (Fig. 1) (24). Therefore, we asked whether DRH-3 is a general NTPase similar to other DEXH proteins or whether DRH-3 hydrolyzes only ATP-like DEAD box proteins. Using TLC to test NTPase activity, we observed that DRH-3 was able to hydrolyze all four NTPs in the presence of dsRNA (supplemental Fig. S4). Although ATP is the hydrolyzed the best, these data suggest that DRH-3 behaves more like a DEXH protein with relatively non-selective NTPase activity (24).
To test whether RNA 5′-triphosphates influence the rate constant for ATP hydrolysis by DRH-3, protein activity was monitored in the presence of phosphatase-treated ds25 (supplemental Fig. S3). Equivalent rate constants for ATP hydrolysis were observed in reactions stimulated by either untreated or CIP-treated blunt duplexes at three different RNA concentrations. These data indicate, along with the direct binding experiments, that DRH-3 binds functionally to dsRNA and that it does not require 5′-triphosphate moieties.
Although DRH-3 is essential for the production of small silencing RNAs in C. elegans (10, 12), it is also required for chromosomal maintenance in these animals (10, 13, 15), suggesting possible activity in the presence of DNA. To evaluate the polymer specificity of DRH-3, we compared the RNA-stimulated ATPase activity with activity measured in the presence of hybrid duplexes and DNA duplexes. The ATPase activity of DRH-3 was measured in the presence of 34-bp duplexes composed of all RNA (RNA/RNA), a hybrid (RNA/DNA), or all DNA (DNA/DNA) (Fig. 3C, supplemental Fig. S5). DRH-3 ATPase activity was observed in the presence of duplexes containing at least one RNA strand (Fig. 3C, supplemental Fig. S5), but activity was not observed in the presence of DNA duplexes. RNA/DNA duplexes stimulated the ATPase activity of DRH-3 to an intermediate level when compared with the all RNA or all DNA duplex, activating DRH-3 to a maximum of 0.8 nmol of ATP hydrolyzed/min/pmol of DRH-3 at 500 nm hybrid duplex (Fig. 3B). These data suggest that RNA is essential for functional binding and for stimulating ATPase activity. Moreover, the results imply that DRH-3 plays a role at the transcriptional or posttranscriptional level but not at the genomic level.
Unwinding Activity of DRH-3
To test whether DRH-3 is truly a helicase with duplex-unwinding capabilities, we examined DRH-3-dependent strand separation with different RNA duplexes. A variety of potential substrates were generated by annealing a 12-nt top strand RNA (5′-32P-radiolabeled) with one of three different bottom strands: 1) a 12-nt RNA, thereby forming a blunt duplex; 2) a 30-nt RNA, resulting in an 18-nt 3′ overhang; 3) a 30-nt RNA, resulting in an 18-nt 5′ overhang (Fig. 3C, supplemental Fig. S6). When these potential unwinding substrates were incubated with DRH-3 and ATP and the reaction products were subjected to electrophoresis, no strand separation was observed. Unwinding was also not observed in the presence of a nucleic acid trap that prevents reannealing of unwound strands.
Because the ATPase activity of DRH-3 is activated by dsRNA, we then tested unwinding activity by incubating DRH-3 with a longer duplex of 18 bp. Despite the increase in duplex length, no unwinding was observed (supplemental Fig. S6). We then asked whether DRH-3 might be stimulated by an RNA duplex that is presented in trans. To test this idea, we added increasing concentrations of cold poly[ribo(I-C)] to reactions that contained one of the three unwinding substrates described above. However, unwinding activity by DRH-3 was not observed upon supplementation with RNA duplex in trans (supplemental Fig. S6). Taken together, the results show that DRH-3 does not act as an RNA helicase in vitro, and it is unlikely to do so in vivo.
The RNA Binding Stoichiometry of DRH-3
The native gel analysis suggests that DRH-3 can bind a 25-bp RNA duplex as both a monomer and a dimer at non-limiting protein concentrations (Fig. 2A). Before analyzing the stoichiometry of RNA binding by DRH-3, we first evaluated the oligomeric state of free DRH-3 in the absence of RNA. To test this, DRH-3 was subjected to analytical ultracentrifugation (supplemental Fig. S7). The sedimentation velocity of DRH-3 was measured at 1 μm (data not shown) and 5 μm DRH-3. At both concentrations, DRH-3 was a monomer in solution, having a Svedberg coefficient of 4.5 (120 kDa). Less than 1% of the population behaved as a multimeric species in solution (> 8 S).
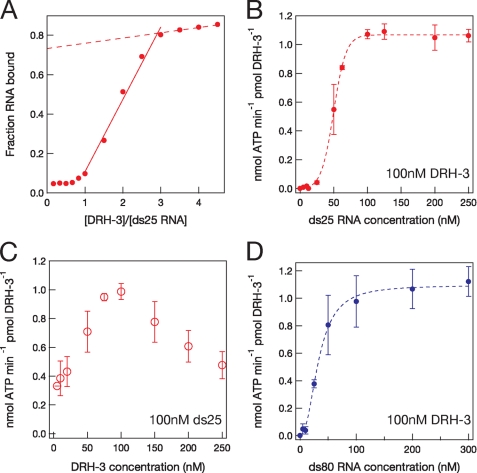
DRH-3-RNA stoichiometry was first monitored using a direct binding assay in which DRH-3 (5–300 nm) was mixed with dsRNA (66 nm, which is 11-fold above the Kd), and the extent of binding was monitored using native gel electrophoresis. The fraction of RNA bound was plotted as a function of the ratio DRH-3 to ds25 (Fig. 4A). The linear increase in the fraction of RNA bound saturated at a ratio of 3, suggesting that no more than three DRH-3 molecules bind per one molecule short dsRNA.
FIGURE 4.
RNA binding stoichiometry of DRH-3. A, results of a direct RNA binding assay showing a plateau at a ratio of three DRH-3 molecules per molecule of ds25. B, the functional binding stoichiometry of DRH-3 and ds25 RNA. 100 nm DRH-3 was incubated with varying concentrations of 25-bp RNA and ATP, and ATP hydrolysis was measured by TLC. ATPase activity as a function of DRH-3 concentration was then plotted versus RNA concentration. C, determining the peak ATPase activity of DRH-3 (75 nm) by keeping the RNA concentration constant (100 nm) and varying the DRH-3 concentration. D, functional binding of DRH-3 to a longer, 80-bp RNA duplex. Error bars in B–D indicate S.E.
To explore the stoichiometry of functional DRH-3-RNA binding, we monitored the RNA concentration dependence of ATPase activity (25) using a DRH-3 concentration (100 nm) that exceeds Kd by 16-fold. Initial rates of ATPase activity were measured with increasing concentrations of ds25 RNA (5–250 nm), and the ATPase activity of DRH-3 was then plotted as a function of RNA concentration (Fig. 4B). The DRH-3 ATPase activity peaked at 100 nm of ds25 RNA, which suggests a functional complex of 1:1 DRH-3 to ds25 (Fig. 4B). However, the sigmoidal shape of the data implies that DRH-3 may function as a cooperative oligomer or that it may have two RNA binding sites. To explore the functional stoichiometry further, the conditions of the experiment were inverted; the dsRNA concentration was held constant at a level (100 nm, which is 16-fold above Kd), and the ATPase activity of DRH-3 was measured at varying enzyme concentrations (5–250 nm). When the ATPase activity of DRH-3 was plotted versus enzyme concentration, activity peaked between 75 and 100 nm DRH-3, which is approximately equivalent to the total concentration of RNA in the reaction (Fig. 4C). Taken together, these data indicate that DRH-3 binds functionally to one short dsRNA per molecule protein; however, they also suggest that the behavior of DRH-3 is somewhat more complex than simple, non-cooperative binding.
One way to test whether DRH-3 has multiple RNA binding sites is to measure ATPase activity in the presence of longer RNA duplexes that might provide more than one binding site for DRH-3. If one molecule of DRH-3 can bind one RNA molecule at distinct sites, then fewer molecules of RNA would be needed to occupy all the sites on DRH-3. To this end, we measured the ATPase activity of DRH-3 (100 nm) in the presence of an 80-bp duplex (5–300 nm, Fig. 4D). Although the final stoichiometry appears to be similar, the ds80-stimulated ATPase activity displayed less sigmoidal behavior than the ds25-stimulated activity, and the apparent stoichiometry appears to be slightly less than one. These findings suggest that DRH-3 may contain two RNA binding sites, both of which may be occupied by a single long dsRNA in cis.
Kinetic Analysis of DRH-3 ATPase Activity
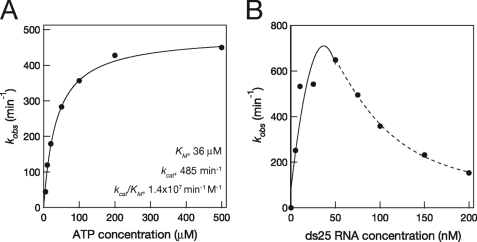
To characterize DRH-3 as an enzyme, we set out to establish the kinetic parameters for ATPase activity under conditions where DRH-3 was fully stimulated by RNA (at RNA saturation). In these experiments, the short 25-bp (ds25, 66 nm) RNA was employed because 1) DRH-3 likely binds small RNA duplexes in vivo (12, 14) and 2) short duplexes have few potential binding sites for DRH-3. To obtain an apparent binding constant for ATP (the Km(ATP)), we measured the rate of ATP hydrolysis as a function of ATP concentration (Fig. 5A). The resulting hyperbolic curve shows that DRH-3 has robust ATPase activity when stimulated with dsRNA, with a Vmax of 2.43 pmol of ATP hydrolyzed/min and a kcat of ∼500 min−1. Interestingly, the Km value for DRH-3 (36 μm) is almost identical to that calculated for RIG-I by single molecule methods (37 μm) (26), further underscoring the relatedness of these proteins.
FIGURE 5.
ATP hydrolysis kinetics of DRH-3. A, kobs values (initial velocity divided by the DRH-3 concentration) were plotted as a function of ATP concentration and fit to the Michaelis-Menten equation (Equation 3) with a Km = 36 ± 12 μm. ATPase reactions contained 5 nm DRH-3 and varying ATP concentrations. Error for each was based on the fit of the data. B, ATP hydrolysis by DRH-3 plotted versus ds25 concentration. DRH-3 (5 nm) ATPase activity peaked at 50 nm dsRNA and was inhibited at higher RNA concentrations. The initial peak in rate constant (0–50 nm RNA) was best fit to a polynomial (n = 3), and the inhibition data (50–200 nm) were best fit to a parameter sigmoidal equation (y = A + (B − A)/(1 + (x/Kd)n)).
To evaluate the influence of RNA concentration on ATPase activity, the concentrations of DRH-3 (5 nm, which is <Kd) and ATP (200 μm, which is ≫Kd) were held constant while the concentration of ds25 was varied (5–200 nm). Although we expected that the DRH-3 ATPase activity would display a hyperbolic dependence upon increasing dsRNA concentration, we observed more complicated behavior. ATP hydrolysis peaked at 50 nm activator RNA (kcat, 700 min−1) and then declined at higher concentrations of RNA (Fig. 5B). As a control for our TLC ATPase assay, an indirect NADH-coupled ATPase assay was employed (27). Using the coupled assay, we found identical results at high concentration of RNA as observed in the direct assay where ATPase activity declined after a peak at 50 nm ds25 (supplemental Fig. S8). These results are consistent with a mechanism in which high concentrations of dsRNA inhibit the ATPase activity of DRH-3.
DISCUSSION
DRH-3 Is a Bona Fide Member of the Dicer-RIG-I Family of RNA Helicases
The data shown here indicate that DRH-3 behaves like its Dicer and RIG-I orthologs in that DRH-3 binds and is activated by dsRNA. Thus, dsRNA activation appears to be a function conserved by this family of RNA enzymes. DRH-3 is more closely related to RIG-I at the primary sequence level than its Dicer homologs (supplemental Fig. S1). It is feasible that the C. elegans Dicer-related helicase proteins are prototypes of the RIG-I family of helicases (17) and that the mammalian proteins apply the machinery associated with dsRNA ATPase activation to newer, more advanced innate immune responses to viral infection (28, 29). Importantly, the RNA binding motifs Ia, Ib, and V are well conserved within the Dicer-RIG-I family. Structural studies of helicases bound specifically to double-stranded nucleic acids, such as the chromatin-remodeling enzyme SNF/SWI, indicate that amino acids within motifs 1a, 1b, and V are essential for recognition of the double-stranded lattice (23). Based on all these lines of evidence, together with the observed duplex specificity of the protein family, it is likely that specialized motifs Ia, Ib, and V have adapted to provide a platform for binding dsRNA.
Characterization of an RNAi Pathway Helicase
A conserved class of RNA-dependent ATPases is essential to all gene-silencing pathways that act through small RNAs. Our study of DRH-3 is the first biochemical analysis of one of these motors essential to RNA silencing, and the behaviors we have observed are likely to inform our understanding of their biological function. In terms of ligand and cofactor specificity, we have shown that DRH-3 binds both single-stranded and double-stranded RNA with nanomolar affinity (Fig. 2). However, DRH-3 binds dsRNA four times more tightly than its ssRNA counterpart. This binding could be mediated by the motifs within the conserved “helicase core,” or it may also be mediated by appended domains of DRH-3, as suspected for RIG-I (30, 31). Perhaps most importantly, we show that DRH-3 requires at least one of the two duplex strands to be composed of RNA, thereby suggesting that DRH-3 is involved in post-transcriptional processes rather than the reorganization of chromatin DNA. In addition, we observe that DRH-3 ssRNA binding does not require 5′-triphosphates (supplemental Fig. S3), a property different from its mammalian ortholog, RIG-I (18, 19). Taken together, the dsRNA-stimulated ATPase activity of DRH-3 suggests that dsRNA is the natural ligand for DRH-3 (Fig. 3).
A kinetic analysis of DRH-3 ATPase activity has revealed important attributes of DRH-3 as an enzyme and shed light on its relatedness to other SF2 proteins. DRH-3 is a robust and fully RNA-dependent ATPase, hydrolyzing 500–700 ATPs/min, which is a kcat similar to the ATPase activity of the processive hepatitis C virus NS3 RNA helicase (Fig. 5A) (32). Of note, the ATP Km of 36 μm for DRH-3 is the same as the parameters determined for RIG-I ATP binding under single molecule conditions (26). Curiously, high concentrations (10-fold > [E]) of dsRNA inhibit the ATPase activity of DRH-3 (Fig. 5B), resembling a case in which free substrate inhibits the activity of an enzyme that requires an essential activator (33). In our case, free activator causes inhibition of DRH-3 ATPase rate. Moreover, activator inhibition occurs at multiple DRH-3 concentrations (data not shown), supporting an autoinhibition mechanism that may limit DRH-3 ATPase function (see below).
The RNA Binding Stoichiometry of DRH-3 and Small RNA Duplexes, Monomer or Dimer?
The RNA binding behavior of DRH-3 suggests that there may be interesting modes of allosteric regulation for the protein. Several observations may be particularly relevant. We find an apparent difference between the functional (active) and non-functional (passive) dsRNA binding activities of DRH-3. In direct binding assays (the absence of ATP), DRH-3 binds RNA as a monomer, dimer, or multimer (∼3), and binding in multimeric form is dependent on protein concentration (Fig. 4A), as observed for almost every other SF2 family member (34–36). SF2 proteins appear to be “sticky,” having strong and often nonspecific electrostatic interactions with negatively charged polymers. In the case of DRH-3, oligomerization is a direct result of RNA binding because DRH-3 is a monomer in solution according to sedimentation velocity experiments (supplemental Fig. S7). Native gel analysis shows that DRH-3 binds as a multimer to both ssRNA and dsRNA (Fig. 2, C and D). However, DRH-3 is activated only by dsRNA, suggesting that oligomerization results from nonspecific binding to RNA. Therefore, direct binding of RNA may or may not reflect functional binding.
DRH-3 requires dsRNA to hydrolyze ATP, and therefore, ATPase activity can provide a metric of functional dsRNA binding. Using this approach, we determined the stoichiometry of functional dsRNA binding and observed its sensitivity to variance in dsRNA concentration. Based on this analysis, we observe that DRH-3 binds one short RNA duplex per molecule of protein (Fig. 4B). However, the reduced ATPase activity at low ds25 RNA concentrations suggests some form of allosteric behavior. We can envision two models to fit the sigmoidal functional binding data. First, DRH-3 may have two binding sites for RNA (as observed for other RNA helicases and polymerases) (30, 31, 37), and these distinct sites may have different functions. For example, one site may behave as the activator site (site A), and the other site may behave as an inhibitor site (site I). The sigmoidal curve shown in Fig. 4B would require that site I has a greater affinity for dsRNA at low concentrations. At higher concentrations of dsRNA, binding of site I is displaced, and there is a shift in dsRNA binding from site I to site A. ATPase activity is then restored at a 1:1 stoichiometry of short RNA duplex to DRH-3.
A second model to explain the functional binding of short RNA duplexes is that DRH-3 may be active only as an obligate dimer. In this model, a combination of fewer than two molecules of either ds25 RNA or DRH-3 represents an inactive complex, thus explaining the sigmoidal shape of the data. When DRH-3 is a dimer, two RNA molecules must be present for DRH-3 ATPase to function, equaling an apparent stoichiometry of 1:1 (Fig. 4B). When DRH-3 ATPase is activated by an 80-bp RNA duplex, outlined in Fig. 4C, ATPase activity plateaus at slightly less than 100 nm RNA, suggesting that DRH-3 binds functionally 1:1 to ds80. However, these data show less sigmoidal shape than the analysis of ds25-ATPase activity, suggesting that a longer duplex may occupy more than one RNA binding site on DRH-3 at a given time or at least part of the time.
Insight into the Role of DRH-3 in the RNAi Pathway
If the helicase motif of DRH-3 is essential for the production of small silencing RNAs (12), then DRH-3 function involves a nucleic acid component. Here we show the ATPase activity of DRH-3 requires at least one strand of the nucleic acid duplex to be RNA (Fig. 3C). Therefore, DRH-3 likely operates at the transcriptional (RNA/DNA) or post-transcriptional (RNA/RNA) level but not at the genomic (DNA/DNA) level. Moreover, DRH-3 may sense the A-form helices of RNA/RNA and RNA/DNA duplexes as part of its mechanism of ATPase function. Along with a recent study of DRH-3 (12), the data here support the idea that DRH-3 has some translocase/ATPase function during the production of small silencing RNAs, a subset of which may direct proper chromosome maintenance during development (10, 11, 13, 15).
In DRH-3 helicase motif mutants, a reduced number of RdRP-dependent siRNAs are produced when compared with wild-type worms. Of the few small RNAs found in the absence of intact DRH-3, all are transcribed from the 3′-most end of RdRP templates (12). It has been hypothesized that the RdRP alone initiates small RNA production at the 3′ end and then extends small RNA production along template RNA with the aid of DRH-3 helicase activity (12). One idea is that DRH-3 processively unwinds newly formed RNA duplexes, after they are produced by RdRPs, and allows efficient turnover (i.e. release) of small RNAs from the template. However, using a variety of unwinding substrates, we do not observe unwinding activity by DRH-3 (Fig. 3C, supplemental Fig. S6). These data are consistent with previous results showing that Drosophila Dicer-2, an ortholog of DRH-3, cannot unwind siRNAs in vivo or in vitro (38, 39). Although the Dicer-RIG-I-DRH family is a member of the processive DEXH RNA helicase group, it does not seem likely that these helicases function as “unwindases” (26, 38, 39). Moreover, the ATPase activity of dsRNA binding suggests that DRH-3 is inhibited by high concentrations of dsRNA (Fig. 5A). This autoinhibition of DRH-3 may function to perhaps limit the number of small RNAs produced by an RdRP from a single template (12, 14).
In single molecule experiments, the DRH-3 ortholog RIG-1 translocates along duplex RNA (26). It is plausible that DRH-3 has some ATP-dependent motor activity on double-stranded RNA and may apply this activity to some function in the RNAi pathway. Because DRH-3 interacts with both C. elegans Dicer, DCR-1, and RdRP, RRF-1 (11, 14), it is unclear whether DRH-3 is required for primary (Dicer-dependent) or secondary (RdRP-dependent) siRNA production, and it may play a possible role in the production of either siRNA species. Because C. elegans encode only one Dicer, required for processing both miRNAs and siRNAs, DRH proteins may function by helping Dicer or Dicer-containing complexes release themselves from RNA duplexes. It is unclear how DRH-3 could function in the production of secondary siRNAs given its preference for duplex RNA. A possible dsRNA ligand for DRH-3 could arise from the base-pairing of initial RdRP transcripts and its template RNA during secondary siRNA production.
Of note, our study of DRH-3 is the first biochemical analysis of an RNA helicase implicated in the RNA-silencing mechanism. There are many types of the helicases essential for gene silencing by small RNAs. A closer look at these important proteins is needed to further our understanding of the biological role they play in regulatory RNA pathways.
Supplementary Material
Acknowledgments
We thank Victor Serebrov for helpful discussion during the preparation of the manuscript. We thank Phil Zamore, Enrique De La Cruz, and members of the Pyle Laboratory for helpful comments on the manuscript. We thank Steve Ding, Andrew Kohlway, and Gabriele Drews for technical help. We thank Frank Slack and members of the laboratory for help with collecting total RNA from C. elegans for cloning experiments.
This work was supported, in whole or in part, by a National Institutes of Health National Research Service Award fellowship (F32GM086949) (to C. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8 and Table 1.
- RNAi
- RNA interference
- siRNA
- small interfering RNA
- DRH
- Dicer-related helicase
- dsRNA
- double-stranded RNA
- ssRNA
- single-stranded RNA
- RdRP
- RNA-dependent RNA polymerase
- DTT
- dithiothreitol
- SUMO
- small ubiquitin-like modifier
- nt
- nucleotide
- MOPS
- 4-morpholinepropanesulfonic acid.
REFERENCES
- 1.Zamore P. D., Haley B. (2005) Science 309, 1519–1524 [DOI] [PubMed] [Google Scholar]
- 2.Ding S. W., Voinnet O. (2007) Cell 130, 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomari Y., Zamore P. D. (2005) Genes Dev. 19, 517–529 [DOI] [PubMed] [Google Scholar]
- 4.Girard A., Hannon G. J. (2008) Trends Cell Biol. 18, 136–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pak J., Fire A. (2007) Science 315, 241–244 [DOI] [PubMed] [Google Scholar]
- 6.Sijen T., Steiner F. A., Thijssen K. L., Plasterk R. H. (2007) Science 315, 244–247 [DOI] [PubMed] [Google Scholar]
- 7.Tabara H., Yigit E., Siomi H., Mello C. C. (2002) Cell 109, 861–871 [DOI] [PubMed] [Google Scholar]
- 8.Tomari Y., Du T., Haley B., Schwarz D. S., Bennett R., Cook H. A., Koppetsch B. S., Theurkauf W. E., Zamore P. D. (2004) Cell 116, 831–841 [DOI] [PubMed] [Google Scholar]
- 9.Zhou R., Hotta I., Denli A. M., Hong P., Perrimon N., Hannon G. J. (2008) Mol Cell 32, 592–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claycomb J. M., Batista P. J., Pang K. M., Gu W., Vasale J. J., van Wolfswinkel J. C., Chaves D. A., Shirayama M., Mitani S., Ketting R. F., Conte D., Jr., Mello C. C. (2009) Cell 139, 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duchaine T. F., Wohlschlegel J. A., Kennedy S., Bei Y., Conte D., Jr., Pang K., Brownell D. R., Harding S., Mitani S., Ruvkun G., Yates J. R., 3rd, Mello C. C. (2006) Cell 124, 343–354 [DOI] [PubMed] [Google Scholar]
- 12.Gu W., Shirayama M., Conte D., Jr., Vasale J., Batista P. J., Claycomb J. M., Moresco J. J., Youngman E. M., Keys J., Stoltz M. J., Chen C. C., Chaves D. A., Duan S., Kasschau K. D., Fahlgren N., Yates J. R., 3rd, Mitani S., Carrington J. C., Mello C. C. (2009) Mol. Cell 36, 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura M., Ando R., Nakazawa T., Yudazono T., Tsutsumi N., Hatanaka N., Ohgake T., Hanaoka F., Eki T. (2007) Genes Cells 12, 997–1010 [DOI] [PubMed] [Google Scholar]
- 14.Aoki K., Moriguchi H., Yoshioka T., Okawa K., Tabara H. (2007) EMBO J. 26, 5007–5019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.She X., Xu X., Fedotov A., Kelly W. G., Maine E. M. (2009) PLoS Genet. 5, e1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deddouche S., Matt N., Budd A., Mueller S., Kemp C., Galiana-Arnoux D., Dostert C., Antoniewski C., Hoffmann J. A., Imler J. L. (2008) Nat. Immunol. 9, 1425–1432 [DOI] [PubMed] [Google Scholar]
- 17.Zou J., Chang M., Nie P., Secombes C. J. (2009) BMC Evol. Biol. 9, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K. K., Schlee M., Endres S., Hartmann G. (2006) Science 314, 994–997 [DOI] [PubMed] [Google Scholar]
- 19.Pichlmair A., Schulz O., Tan C. P., Näslund T. I., Liljeström P., Weber F., Reis e Sousa C. (2006) Science 314, 997–1001 [DOI] [PubMed] [Google Scholar]
- 20.Schlee M., Roth A., Hornung V., Hagmann C. A., Wimmenauer V., Barchet W., Coch C., Janke M., Mihailovic A., Wardle G., Juranek S., Kato H., Kawai T., Poeck H., Fitzgerald K. A., Takeuchi O., Akira S., Tuschl T., Latz E., Ludwig J., Hartmann G. (2009) Immunity 31, 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong I., Lohman T. M. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 5428–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laggerbauer B., Lauber J., Lührmann R. (1996) Nucleic Acids Res. 24, 868–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dürr H., Körner C., Müller M., Hickmann V., Hopfner K. P. (2005) Cell 121, 363–373 [DOI] [PubMed] [Google Scholar]
- 24.Pyle A. M. (2008) Annu Rev. Biophys. 37, 317–336 [DOI] [PubMed] [Google Scholar]
- 25.Tsu C. A., Uhlenbeck O. C. (1998) Biochemistry 37, 16989–16996 [DOI] [PubMed] [Google Scholar]
- 26.Myong S., Cui S., Cornish P. V., Kirchhofer A., Gack M. U., Jung J. U., Hopfner K. P., Ha T. (2009) Science 323, 1070–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De La Cruz E. M., Sweeney H. L., Ostap E. M. (2000) Biophys. J. 79, 1524–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowie A. G., Fitzgerald K. A. (2007) Trends Immunol. 28, 147–150 [DOI] [PubMed] [Google Scholar]
- 29.Rehwinkel J., Reis e Sousa C. (2010) Science 327, 284–286 [DOI] [PubMed] [Google Scholar]
- 30.Cui S., Eisenächer K., Kirchhofer A., Brzózka K., Lammens A., Lammens K., Fujita T., Conzelmann K. K., Krug A., Hopfner K. P. (2008) Mol. Cell 29, 169–179 [DOI] [PubMed] [Google Scholar]
- 31.Takahasi K., Yoneyama M., Nishihori T., Hirai R., Kumeta H., Narita R., Gale M., Jr., Inagaki F., Fujita T. (2008) Mol. Cell 29, 428–440 [DOI] [PubMed] [Google Scholar]
- 32.Beran R. K., Serebrov V., Pyle A. M. (2007) J. Biol. Chem. 282, 34913–34920 [DOI] [PubMed] [Google Scholar]
- 33.Segel I., H. (1993) Enzyme Kinetics, 2nd Ed., pp. 242–267, John Wiley & Sons, Inc., New York [Google Scholar]
- 34.Banroques J., Cordin O., Doère M., Linder P., Tanner N. K. (2008) Mol. Cell. Biol. 28, 3359–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider S., Campodonico E., Schwer B. (2004) J. Biol. Chem. 279, 8617–8626 [DOI] [PubMed] [Google Scholar]
- 36.Sikora B., Chen Y., Lichti C. F., Harrison M. K., Jennings T. A., Tang Y., Tackett A. J., Jordan J. B., Sakon J., Cameron C. E., Raney K. D. (2008) J. Biol. Chem. 283, 11516–11525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng L., Gershon P. D. (1997) EMBO J. 16, 1103–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee Y. S., Nakahara K., Pham J. W., Kim K., He Z., Sontheimer E. J., Carthew R. W. (2004) Cell 117, 69–81 [DOI] [PubMed] [Google Scholar]
- 39.Tomari Y., Matranga C., Haley B., Martinez N., Zamore P. D. (2004) Science 306, 1377–1380 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.