Abstract
Group 5 allergens from house dust mites elicit strong IgE antibody binding in mite-allergic patients. The structure of Der p 5 was determined by x-ray crystallography to better understand the IgE epitopes, to investigate the biologic function in mites, and to compare with the conflicting published Blo t 5 structures, designated 2JMH and 2JRK in the Protein Data Bank. Der p 5 is a three-helical bundle similar to Blo t 5, but the interactions of the helices are more similar to 2JMH than 2JRK. The crystallographic asymmetric unit contains three dimers of Der p 5 that are not exactly alike. Solution scattering techniques were used to assess the multimeric state of Der p 5 in vitro and showed that the predominant state was monomeric, similar to Blo t 5, but larger multimeric species are also present. In the crystal, the formation of the Der p 5 dimer creates a large hydrophobic cavity of ∼3000 Å3 that could be a ligand-binding site. Many allergens are known to bind hydrophobic ligands, which are thought to stimulate the innate immune system and have adjuvant-like effects on IgE-mediated inflammatory responses.
Keywords: Immunochemistry, Immunology, Innate Immunity, X-ray Crystallography, X-ray Scattering, Der p 5, Allergens, Allergy, Asthma, Dust Mites
Introduction
Allergic diseases are estimated to afflict 20% of the world population. House dust mites are a significant source of indoor allergens, which are associated strongly with extrinsic asthma (1). More than 20 allergen groups have been characterized from dust mites with varying levels of patient response (2). An important group is the group 5 allergens from dust mites, which elicit strong IgE binding in about 30% of mite-allergic patients (3).
Two dust mite species Dermatophagoides farinae and Dermatophagoides pteronyssinus are found worldwide and predominate in climates with less humidity (D. farinae) or with seasonal fluctuations in humidity (D. pteronyssinus) (4, 5). In tropical climates, Blomia tropicalis is found in addition to Dermatophagoides spp. Indeed, asthmatic patients from Florida, Puerto Rico, and Brazil were found to be sensitized to both D. pteronyssinus and B. tropicalis (6). Patients from the United Kingdom who were sensitized only to D. pteronyssinus gave positive skin tests to B. tropicalis extract, but this apparent cross-reactivity was not due to the group 5 allergens (7). The group 5 allergens Der p 5 and Blo t 5 share 42% sequence identity and have been studied extensively for their cross-reactivity in patients. Nearly all studies with recombinant allergens have concluded that the IgE antibody responses to the group 5 allergens are species-specific (6–9). Furthermore, recent studies using polyclonal rabbit IgG antibodies raised against Der p 5 detected no binding with group 5 allergens from storage mites (i.e. Glycyphagus domesticus or Lepidoglyphus destructor) or B. tropicalis (10).
Structural characterization of allergens can provide important insights into the biologic function of allergens, which may influence their ability to cause IgE responses. Among dust mite allergens, many are proteases, and this enzymatic property may be related to the chronic inflammation of the lung in asthmatic patients (11–13). Other studies suggested that the proteolytic activity enhanced IgE synthesis and/or skewed the immune response away from tolerance and toward an allergic response (14, 15). However, it was reported that Der p 5 activated human airway-derived epithelial cells via a protease-independent mechanism (16). Recently, crystallographic studies revealed that Der p 7 was distantly related to hydrophobic ligand-binding proteins of the human innate immune system (17); this relationship was not previously identified from sequence information alone. It was suggested that Der p 7 may co-opt the innate immune response into promoting allergenicity through the binding and delivery of hydrophobic ligands (17), as has been demonstrated for the mite allergen Der p 2 (18).
Previous structural studies of the Blo t 5 and related Der p 21 allergens have not identified a natural function in mites (19–21). Despite a preliminary x-ray report of Der p 5 crystals in 1998 (22), a definitive structure of Der p 5 has not been obtained. The structure of Blo t 5 was determined in two studies using NMR (19, 20). Both studies confirmed the results of CD spectroscopy and bioinformatics predictions of other group 5 and the highly similar group 21 mite allergens, which suggested that the proteins were largely helical in nature (10, 21–23). A superficial comparison of the two protein structures of Blo t 5 (2JMH and 2JRK in the Protein Data Bank) shows two elongated proteins, both with three α-helices all in a parallel orientation, and a disordered N terminus. A more careful comparison indicates that the folding topology of the two structures is different. Because of the degree of sequence identity between Der p 5 and Blo t 5, it was anticipated that the Der p 5 structure might shed light on the unexpected inconsistency between the two previously determined structures of Blo t 5. In this study, we have solved the first crystal structure of Der p 5 at 2.8 Å resolution. The Der p 5 structure was found to adopt a space group in which the asymmetric unit contains six Der p 5 molecules in an apparent trimer of dimers. The structure presented here resolves the ambiguity between the two existing Blo t 5 structures and provides insights into the function of the group 5 and group 21 mite allergens.
EXPERIMENTAL PROCEDURES
Expression and Purification
The construct designed for Der p 5 sought to reduce the number of disordered residues for crystallization based on previous NMR studies that indicated significant flexibility of the N-terminal residues (19, 20). A construct of Der p 5 encoding residues Leu34–Val132 was inserted into a pGEX4T3 vector that had been modified to include a tobacco etch virus restriction site between the BamHI and EcoRI site. Der p 5 was inserted using the EcoRI and XhoI sites in the multicloning site. This vector was transformed into BL21(DE3)RIL cells. For protein expression of the GST-Der p 5 construct, 20 ml of an overnight culture grown in LB + 100 μg/ml ampicillin at 37 °C and 275 rpm was added to each of 6 1.8-liter Fernbach flasks containing 1 liter of LB media and 100 μg/ml ampicillin. These were placed on shakers at 37 °C and 275 rpm until an A600 of 0.8 was reached. The temperature was set to 18 °C. When the temperature reached 22 °C, isopropyl 1-thio-β-d-galactopyranoside was added to a final concentration of 500 μm, and cells were allowed to shake overnight at 18 °C. Cells were pelleted at 4000 × g. The cell pellets from 6 liters of culture were resuspended in sonication buffer consisting of 25 mm Tris, pH 7.5, and 500 mm NaCl to a final volume of 120 ml. Cells were sonicated on an ice-water bath for three bursts at 20 s each and then spun down for 35 min at 48,000 × g. The supernatant was mixed with 12 ml of glutathione-Sepharose 4B resin and gently rocked at 4 °C for 1 h. The resin was pelleted at 500 × g for 5 min and washed extensively with sonication buffer. Resin was transferred to the elution buffer consisting of 25 mm Tris, pH 7.5, and 75 mm NaCl in a 50-ml Falcon tube. Der p 5 was eluted from the resin by adding 400 μl of 1.5 mg/ml tobacco etch virus protease to the Falcon tube and gently rocked at 4 °C overnight. The eluted protein was concentrated down to 40 mg/ml and loaded onto a 16/60 Superdex 200 column, which was equilibrated in the elution buffer. Fractions containing purified Der p 5 were pooled and concentrated to 30.7 mg/ml and used for crystallization trials. For selenomethionine-labeled protein, the GST-Der p 5 fusion protein was expressed in B834 (DE3) cells using minimal media supplemented with amino acids where selenomethionine was substituted for methionine. Expression and purification procedures were similar to above except 2 mm dithiothreitol was present in all buffers.
IgE Binding
IgE antibody binding to the shorter Der p 5 construct was assessed by chimeric enzyme-linked immunosorbent assay. Microtiter plates were coated at 4 °C overnight with 10 μg/ml of the short recombinant Der p 5 construct. Plates were blocked for 30 min with 1% bovine serum albumin/phosphate-buffered saline, 0.05% Tween 20, pH 7.4. A 2-h incubation with sera (dilutions 1:2 and 1:10) was performed. All other incubations were for 1 h, and plates were washed three times between steps with phosphate-buffered saline, 0.05% Tween 20. Bound IgE was detected using biotin-labeled goat anti-human IgE (Kirkegaard and Perry Laboratories, Gaithersburg, MD) at a 1:4,000 dilution. Streptavidin peroxidase was added at 250 μg/ml, followed by development with 1 mm 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) and 0.03% hydrogen peroxide as substrate. Plates were quantitated by measuring the absorbance at 405 nm, and an absorbance of >0.2 was considered positive. The sera from allergic patients were obtained from PlasmaLab International (Everett, WA), which operates in full compliance of Food and Drug Administration regulations. An informed donor consent was obtained from each individual prior to the first donation.
Crystallization, Data Collection, and Refinement
Crystals of Der p 5 Leu34–Val132 were obtained using the sitting and hanging drop vapor diffusion technique at room temperature by mixing 1 μl of protein with 1 μl of the reservoir solution consisting of 31.5% methylpentanediol and 90 mm sodium/potassium phosphate buffer, pH 6.2. For data collection, a crystal was taken directly from the sitting drop tray and frozen in liquid nitrogen. Data were collected at −180 °C on an in-house Saturn92 CCD detector mounted on a 007HFmicromax generator equipped with VarimaxHF mirrors. Data were indexed and integrated and scaled in HKL2000 (24) and also scaled in XDS (25). Selenomethionine-labeled protein crystals were grown from similar conditions. Data were collected at wavelengths 0.97243 (peak) and 0.97943 (edge) at the SER-CAT beamline ID-22 at Argonne National Laboratory. All data were processed and scaled using HKL2000. Initial phases were obtained with a figure of merit of 0.29 by using the Auto_Sol routines in Phenix (26), which found 23 of the possible 36 selenomethionine sites. The Auto_build routine in Phenix built 231 amino acids (polyalanines) of the 608 present in the asymmetric unit producing a map with an figure of merit of 0.63. Manual model building and real space refinement in Coot (27) followed by refinement in CNS (28) and Phenix produced a model with six monomers in the asymmetric unit and an Rwork of 25.8% and an Rfree of 35.7%. The model was refined against the native data sets scaled in either HKL2000 or XDS using CNS and Phenix along with manual building in Coot. It was determined that there was no benefit to the XDS refined data so the final reported structure was refined against HKL2000 processed data in Phenix using noncrystallographic symmetry restraints along with TLS refinement to obtain a model that includes all residues of the six Der p 5 molecules in the asymmetric unit with an Rwork of 23.1% and an Rfree of 28.7% (Table 1). The structure was deposited in the Protein Data Bank, PDB code 3MQ1.
TABLE 1.
Crystallographic data statistics
| Data set | Native | Selenomethionine (peak) | Selenomethionine (edge) |
| Wavelength | 1.5418 Å | 0.97243 Å | 0.97943 Å |
| Unit cell (a, b, c) | 46.78, 96.57, 167.89 Å | 46.78, 96.70, 168.14 Å | 46.77, 96.64, 168.13 Å |
| Space group | P212121 | P212121 | P212121 |
| Resolution | 50 to 2.8 Å | 50 to 2.7 Å | 50 to 2.7 Å |
| No. of observations | 124,140 | 22,515 | 22,461 |
| Unique reflections | 19,292 | 254,289 | 271,926 |
| Rsym(%)a,b | 6.9 (51.8) | 14.7 (50.4) | 13.7 (55.0) |
| I/σI | 15.7 (2.5) | 15.1 (2.2) | 12.1 (2.1) |
| Mosaicity range | 1.3–1.6 | 0.4–1.9 | 0.4–1.3 |
| Completeness | 98.8% (98.5%) | 99.0% (90.6%) | 98.7% (89.2%) |
| Refinement statistics | |||
| Rcryst,cRfreed | 22.6, 27.9% | ||
| No. of waters | 45 | ||
| Overall Wilson B value | 62.5 Å2 | ||
| Average B for | |||
| Molecules A, B | 67.1 | ||
| Molecules C, D | 86.2 | ||
| Molecules E, F | 101.4 | ||
| Water | 69.0 | ||
| Root mean square deviation from ideal values | |||
| Bond length | 0.001 Å | ||
| Bond angle | 0.307° | ||
| Dihedral angle | 8.78° | ||
| Ramachandran statisticse | |||
| Favored (98%) regions | 98.0% | ||
| Allowed (>99.8%) regions | 99.8% | ||
a Rsym = Σ (|Ii − 〈I〉|)/Σ(Ii), where Ii is the intensity of the ith observation, and 〈I〉 is the mean intensity of the reflection. Selenomethionine data were scaled by merging Friedel pairs, but I+ and I− were written out separately.
b The last resolution shell is in parentheses.
c Rcryst = Σ‖Fo| − |Fo ‖/Σ|Fo| calculated from the working data set.
d Rfree was calculated from 5% of data randomly chosen not to be included in refinement.
e Ramachandran results were determined by MolProbity.
SAXS4 Data Acquisition and Analysis
SAXS data were collected at room temperature on the X9 beam line at the National Synchrotron Light Source (Brookhaven National Laboratory) using the small Der p 5 construct. The wavelength of the beam was 0.855 Å, and the sample to detector distance was 2 m. Der p 5 sample concentrations were 10, 5, and 2.5 mg/ml. Scattering data were circularly averaged and scaled to obtain a relative scattering intensity (I) as a function of momentum transfer vector, q (q = (4πsinθ)/λ), after subtraction of buffer-scattering contributions. SAXS data on a standard protein, hen egg white lysozyme (14.2 kDa; Acros Organics, Morris Plains, NJ), were collected over a concentration series in the same capillary and used for I0 analyses.
All scattering data were analyzed using the Primus software package (29); the GNOM45 software package (30) was used for all P(r) and I0 analyses. The radius of gyration, Rg, and forward scattering, I0, were calculated from the second moment and the start of P(r), respectively, where Rg is the root mean square of all elemental volumes from the center-of-mass of the particle, weighted by their scattering densities, and I0 is directly proportional to the molar particle concentration multiplied by the square of the scattering particle molecular weight for particles with the same mean scattering density. Guinier plots for 10.0, 5.0, and 2.5 mg/ml Der p 5 samples were linear over a q range of 0.010–0.0385, 0.0095–0.044, and 0.010–0.0495 Å−1, respectively.
Initial results on Der p 5 indicated a particle size larger than a monomer; hence the data were analyzed for the presence of multiple species with the program OLIGOMER (29). Molecular models of possible solution conformers were generated from the crystallographic results, the Blo t 5 structure 2JMH, and the SAXS model of Der p 21 (21).
RESULTS
IgE Binding to Der p 5
A total of 67 sera from allergic patients were tested for binding to rDer p 5. Five negative control sera from allergic patients with no IgE reactivity to natural Der p 1 and Der p 2 (measured by multiplex array technology) were tested. The remaining 62 sera were from patients sensitized to either Der p 1 or Der p 2 or both. In data not shown, 22 of the 62 sera (35.5%) had IgE antibody binding to rDer p 5, in agreement with the range of reported prevalences of IgE antibody to this allergen (7, 8, 32, 33), indicating that this construct is a good model of the natural allergen.
Crystal Structure
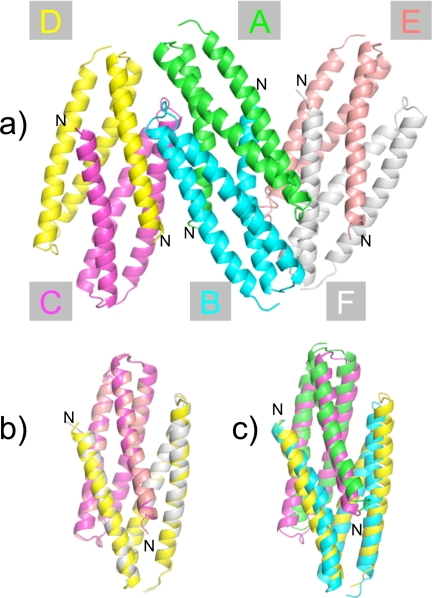
The structure of Der p 5 contains three helices connected by short loops. In the crystal structure determined here, the helices are splayed to interact with another Der p 5 molecule to form dimers. Fig. 1a shows the six Der p 5 molecules in the asymmetric unit, arranged as a trimer of imperfectly matched dimers. The chains in the central dimer are designated A and B, and the peripheral dimers contain chains C paired with D and E paired with F. The N terminus in the crystal form is exposed to the solvent channels suggesting these trimers of dimers are still possible in the full-length construct (Fig. 1a). The A-B dimer is the best ordered in the structure (average B factor 67.1) based on overall B factor analysis followed by C-D (〈B〉 = 86.2) and E-F (〈B〉 = 101.4). The two chains that make up the A-B dimer align very well, with a root mean square deviation <0.5 Å, but the same is not true of the C-D and E-F dimers. This indicates that the C-D and E-F dimers are not as symmetric as the A-B dimer. The C-D and E-F dimers are very similar as can be seen in Fig. 1b (Cα root mean square deviation = 0.56 Å over 191 atoms). Fig. 1c shows the alignment of A-B with C-D, which do not align as well (Cα root mean square deviation = 3.2 Å over 201 atoms). The N-terminal helix is significantly more bent in A-B, whereas all the helices in C-D and E-F appear straighter. The kink in the N-terminal helix occurs at Gly45, which probably provides flexibility and confers upon Der p 5 a significant degree of conformational plasticity.
FIGURE 1.
Asymmetric unit of the Der p 5 crystal structures. a, hexamer of Der p 5. There are six molecules of Der p 5 in the asymmetric unit of the crystal forming a trimer of dimers. The N terminus is designated, as well as the chain name of the molecules. b, alignment of the C-D and E-F dimers. c, alignment of the A-B and C-D dimers. The PDB code for the Der p 5 structure is 3MQ1.
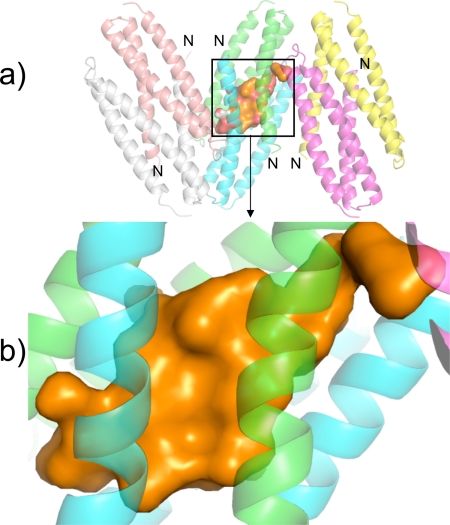
A particularly interesting feature of the Der p 5 dimer is the formation of a large hydrophobic pocket (Fig. 2). The volume of the pocket ranges from 2960 to 3109 Å3 in the three dimers, evaluated by the program CastP (34). The values given should be considered approximate because it is unclear exactly where the pocket begins and ends. The pocket runs almost half the length of the dimers with openings to solvent at both ends of the cavity. The pockets contain ordered density, which was fit with water, phosphate, or methylpentanediol molecules, because all were present during crystallization. The greatest amount of ligand density was observed in the A-B dimer, and poorer density and/or fewer molecules were found in the C-D or E-F dimers. This result suggests that Der p 5 has the potential to bind hydrophobic ligands, which is a common feature of many allergens (35).
FIGURE 2.
Cavity in the Der p 5 dimer. a, cavity in the A-B dimer is rendered with a solid surface, and the helices in the asymmetric unit are rendered as semi-transparent ribbons. b, zoomed region of a. Ordered density in the cavity was assumed to be either water or methylpentanediol; however, the electron density is poorly determined so it is rendered with a solid surface just to give an impression of the size of the cavity.
Relation to Blo t 5 Structures
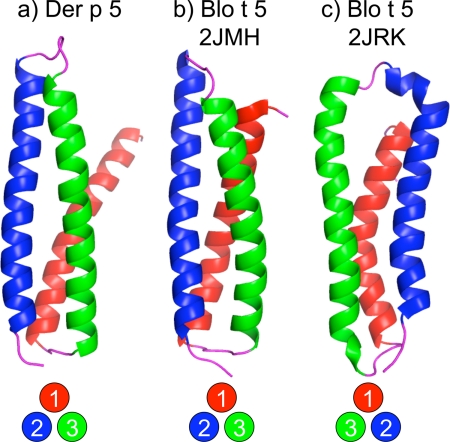
Fig. 3 compares the folding topology of the Der p 5 monomer of chain A with the Blo t 5 structures 2JMH and 2JRK (19, 20). Viewed along an axis parallel to the main helical axes, the helices of 2JMH are arranged in a counterclockwise orientation, whereas the helices of 2JRK are arranged in a clockwise orientation. The two topologies thus approximate mirror images of each other. As a result of the 41% sequence identity between Der p 5 and Blo t 5, it was anticipated that the Der p 5 structure might shed light on this unexpected inconsistency between the two previously determined Blo t 5 structures. The fold topology of the Der p 5 monomer is counterclockwise like the 2JMH structure supporting this as the correct fold for group 5 proteins.
FIGURE 3.
Comparisons of Der p 5 with published Blo t 5 structures. The monomeric form of Der p 5 from chain A is displayed in a. b shows the first structure from 2JMH, and c is the first structure from 2JRK. Helix 1 is red; helix 2 is blue, and helix 3 is green. Circles at the bottom indicate the “handedness” of the structures, looking “down” the helical axis.
Analysis of Interfaces in the Crystal
A detailed analysis of the important residues for the various interfaces was provided using the program Protein Interfaces Surfaces and Assemblies (PISA) (36). The A-B dimer interface contains 11 hydrogen bonds and 6 salt bridges. With only one exception, all of these interactions are side chain to side chain as might be expected for interactions between helices. Similarly, the C-D and E-F dimer interfaces contain 14 and 6 hydrogen bonds and 6 and 2 salt bridges, respectively. In all three dimers, the important residues for hydrophobic interactions include Met35–Phe50, Leu34–Ile115, and Val88–Gly84.
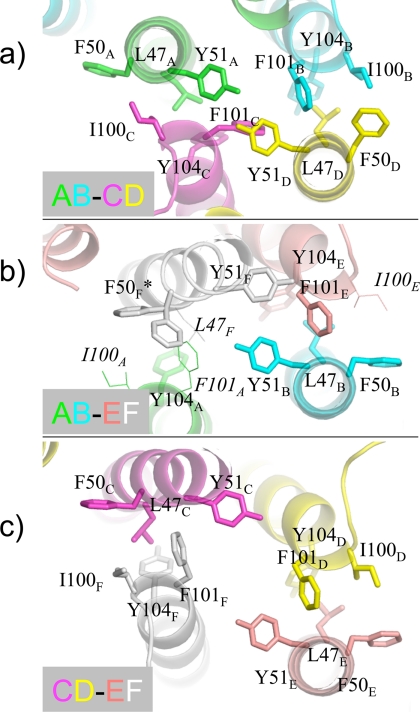
The inter-dimer interfaces also contain a number of hydrophobic contacts. Similar residues can be found at each interface, but in slightly different orientations, as can be seen in Fig. 4. For example, common hydrophobic residues at each interface include Leu47, Phe50, Tyr51, Ile100, Phe101, and Tyr104 from different monomers as denoted with a subscript letter. Fig. 4, a and b, compare the two dimer interfaces within the unit cell. There was very little electron density for several residues at the AB-EF interface, so in Fig. 4b, these residues were modeled based on the best available rotamer to give perspective in the figure. Fig. 4c shows that at the crystallographic interface between dimers C-D and E-F, similar hydrophobic residues can be found again.
FIGURE 4.
Important residues for various inter-dimer interfaces. a shows the interface between A-B and C-D; b shows the interface between A-B and E-F, and c shows a crystallographic interface between C-D and E-F. Note that similar residues appear at each interface but in different conformations. Residues rendered with a thin line lack electron density and are modeled based on best allowed rotamer. F50F has two conformations in the crystal structure.
The various interfaces were examined to evaluate the buried surface area and shape complementarity (37). Table 2 lists the intra-dimer interfaces, the inter-dimer interfaces, and the crystallographic interfaces for which there is significant contact within 4 Å. The average surface area reported is the average from the two partners. In general, the largest values of buried surface area exist between the intra-chain dimer surfaces, which buries between 507 and 775 Å2 with shape complementarity scores between 0.55 and 0.64. For comparison, antibody-antigen interactions typically involve 600–900 Å2 and have shape complementarity scores of 0.6 (37). The inter-dimer interfaces that are noncrystallographic bury between 149 and 247 Å2 per molecule, which is between a half and a third of that found for the intra-dimer interactions. Similarly, the crystallographic interface that would continue the hexameric pattern to the left or right in Fig. 1 buries 199 Å2. For each of these inter-dimer interactions, the buried surface area is low, but the shape complementarity is relatively high. Finally, there are AB-AB dimer contacts in the direction perpendicular to the page, which buries only 107 Å2.
TABLE 2.
Interface summary
| Interface | Chains | 〈Surface area〉 | Shape complementarity |
|---|---|---|---|
| Å2 | |||
| Intra-chain | AB | 755 | 0.63 |
| CD | 715 | 0.64 | |
| EF | 509 | 0.55 | |
| Inter-chain | AB-CD | 246 | 0.69 |
| AB-EF | 150 | 0.64 | |
| Crystallographic | CD-EF | 199 | 0.56 |
| AB-AB | 107 | 0.66 |
Characterization with SAXS
Small angle x-ray scattering was employed to determine the solution stoichiometry of Der p 5, using I0 analysis. Table 3 shows the calculated molecular mass based on the average particle size in solution. The data suggest that a large fraction of Der p 5 is monomeric at low concentrations. However, the average particle size of Der p 5 increases with concentration indicating a tendency to form oligomers. Consistent with this observation, multiangle light scattering confirms that Der p 5 is predominantly monomeric in solution but exhibits a concentration-dependent increase in apparent molecular mass from 14 to 19 kDa, where the actual mass is 12.0 kDa for the construct used here (data not shown).
TABLE 3.
SAXS analysis
| Der p 5 Concentration (mg/ml) | 2.5 | 5 | 10 |
|---|---|---|---|
| I0 analysis | 43.5 kDa | 59.9 kDa | 82.6 kDa |
| Oligomer analysis of relative populationsa | |||
| 1. Der p 5-like monomer | 0.0% | 0.0% | 0.0% |
| 2. Blo t 5-like monomer | 74.0% | 65.0% | 54.1% |
| 3. Der p 5, AB dimer | 0.0% | 0.0% | 0.0% |
| 4. Der p 21-like dimer | 4.1% | 0.0% | 0.0% |
| 5. Der p 5 tetramer (AB and CD) | 0.0% | 0.0% | 0.0% |
| 6. Der p 5 hexamer | 20.3% | 30.7% | 32.6% |
| 7. Der p 5 dodecamer | 1.5% | 4.3% | 13.3% |
| 8. Der p 5 “18-mer” | 0.0% | 0.0% | 0.0% |
| Weighted average of populations | 44.1 kDa | 63.9 kDa | 85.6 kDa |
| χ2 oligomer fit | 1.5 | 3.3 | 11.1 |
a See text for more description and Fig. 5a for visual depictions of models.
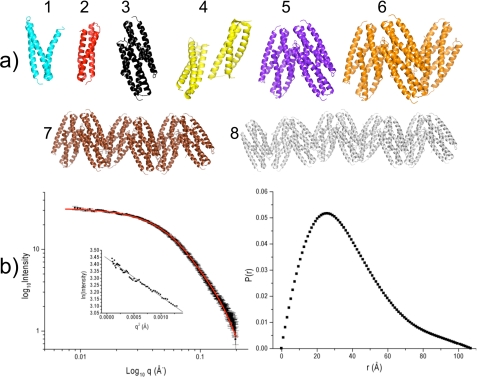
SAXS data also contain information about molecular shapes, and given the crystal structure with a trimer of dimers, we assessed the relative populations of different multimeric structures that might be in solution. Fig. 5a shows diagrams of the structures that were considered for evaluation as follows: 1) a monomer with the structure of Der p 5 chain A; 2) a monomer like Blo t 5 (2JMH); 3) a Der p 5 dimer using chains A and B; 4) a dimer like Der p 21; 5) a tetramer using chains A–D; 6) a hexamer of chains A–F; 7) a dodecamer using two symmetry-related hexamers; and 8) an 18-mer using three symmetry-related hexamers. Model 4 was based on ab initio modeling of SAXS data for Der p 21 (21), which resembles a dimer between chains B and D. The experimental scattering curve was analyzed as a weighted contribution of the theoretical scattering from the eight structure possibilities listed above using the program OLIGOMER (29). Fig. 5b shows the fit to the data obtained at a concentration of 2.5 mg/ml, and Table 3 lists the results in conjunction with the average particle size. The analysis indicates that the primary solution structure is possibility 2, a monomer like Blo t 5, which represents 54–74% of the species present depending on the concentration. At low concentrations, dimers shaped like the low resolution SAXS structure of Der p 21 constitute 4% of the population. Additionally, there are hexamers and dodecamers that increase in relative population as the concentration of Der p 5 increases. At each concentration, a weighted average of molecular mass based on the relative populations very closely matches the predicted particle size based on I0 analysis (Table 3). As a control, we also attempted to simultaneously fit other structure possibilities like trimers and pentamers, but these shapes were rejected by the OLIGOMER analysis (data not shown).
FIGURE 5.
SAXS analysis. a shows the eight tested structures for fit to the SAXS data as described in the text. b, left panel, shows the theoretical fit of the SAXS data (red line) to the acquired scattering data (black squares with error bars) for the 2.5 mg/ml sample of Der p 5 and inset with the linear region of the Guinier fit. b, right panel, shows the P(r) plot for the same sample.
DISCUSSION
This paper presents the first crystal structure of a group 5 mite allergen, Der p 5, from D. pteronyssinus. A short construct was designed based on the available NMR Blo t 5 structures to decrease the number of disordered residues. The prevalence of IgE antibody binding to the shorter Der p 5 construct used here is the same as reported for the full length allergen, indicating that this construct is physiologically relevant. A previous study using peptides to map patient IgE epitopes on Blo t 5, found very few patients with high IgE reactivity to the N-terminal peptides (19). This suggests that IgE antibodies are not directed against the flexible N-terminal residues of Der p 5, which are absent in the shorter construct.
The folding topology of the helices in Der p 5 is similar to that reported for the Blo t 5 solution structure (2JMH) and differs from the reported structure (2JRK). The most likely interpretation of the discrepancy is that the predicted nuclear Overhauser effect differences were sufficiently small so that an incorrect structure was obtained in one of the studies. Indeed, when we examined the reported distance restraints for 2JMH, ∼90% were satisfied in the 2JRK structure (data not shown). Generally, there is enough information in the nuclear Overhauser effect data to differentiate between these possibilities, but a 3-helical bundle represents a challenging situation where errors have been known to occur (38, 39). Given the degree of sequence identity between Blo t 5 and Der p 5, we suggest that the structure 2JMH is most likely to be correct.
The results of the SAXS data show a large population of Der p 5 to be monomeric in solution, although Der p 5 appears to show concentration-dependent oligomerization (Table 3). The asymmetric unit of the Der p 5 crystal is a hexamer, which consists of an imperfect trimer of dimers, whereas Blo t 5 is reported to be a monomer (20). The discovery of a multimeric form of Der p 5 is not unprecedented in the literature. In a previous crystallization report, the Matthew's coefficient predicted that the unit cell could accommodate 12 molecules per asymmetric unit (22). The crystals were grown at low pH, and subsequent studies of Der p 5 reported a strong tendency to polymerize in acidic conditions (23). At neutral pH (7.4) and low concentrations, the protein was largely monomeric, but some dimers could be detected in cross-linking studies. Small angle x-ray studies of Der p 21 (34% identity to Der p 5; see Fig. 6) indicated that the Der p 21 allergen was dimeric (21). Studies of immuno-stained mites noted that Der p 5 appeared on fibrous structures in the food ball (10). In combination with the present results, these observations suggest that Der p 5 may be an integral structural component of these fibers given the apparent proclivity to polymerize. It is also conceivable that given the similarities between Der p 5 and Der p 21, both could be involved in forming polymers.
FIGURE 6.
Sequence comparisons. ClustalW alignments of Der p 21, Blo t 21, Blo t 5, and Der p 5 are shown. Color red are residues important for the Der p 5 intra-dimer interface; blue are residues important for inter-dimer interfaces, and green are residues involved in both types of interfaces. Asterisks indicate identity; colons indicate strong similarity, and periods indicate weaker similarity among the four proteins.
An interesting question is whether or not other group 5 and group 21 allergens could polymerize similarly to what the crystal structure and past biochemical analysis of Der p 5 supports, bearing in mind that biophysical data indicate Blo t 5 to be monomeric (20). Fig. 6 presents a multiple sequence alignment of Der p 5, Blo t 5, Der p 21, and Blo t 21. Residues that are strongly conserved in the intra-dimer and inter-dimer interfaces are shown in color. Residue Gly45, which appears important for the flexibility of the N-terminal helix, is conserved in Blo t 5 and Der p 5 but not in the group 21 allergens. Many of the other residues that are important for intra- and inter-dimer interactions are highly conserved. The most compelling distinction that might provide a basis for the preference of Blo t 5 to not polymerize appears to be related to the sequence 84–88, which is GVRGV in Der p 5 and is GAQGA in Blo t 5. In the dimeric structure of Der p 5, this helical sequence interacts in an antiparallel manner with itself. Residue Val88 from one chain fits into a notch created by Gly84 of the other chain, with Val85 contributing hydrophobic interactions to create a “valine zipper” (analogous to a leucine zipper) at the intra-dimer interface. In Blo t 5, these residues are alanines, which may not contribute enough hydrophobic surface to stabilize a dimer. However, the group 21 allergens have a bulky side chain at the comparative position 88 and a smaller side chain at 85 (Ala or Ser), so one could imagine the intra-dimer interactions forming. A monomer of Der p 5 like that found in the crystal is not found in the SAXS analysis possibly because helix 1 in the molecules are “splayed” in the crystal dimer and not straight like the more compact monomeric Blo t 5 structure.
The cavity observed in the Der p 5 dimer is particularly interesting, considering the existence of many allergens with hydrophobic cavities that bind lipid-like ligands. For example, the cat allergen Fel d 1 binds steroid-like molecules (40); the horse allergen Equ c 1 and the mouse allergen Mus m 1 are lipocalins (41, 42), and Der f 2 was demonstrated to bind lipopolysaccharide (43). Interestingly, the crystal structures of both Der p 2 and Bet v 1 contained apparent ligands in a hydrophobic environment with difficult to interpret electron density (44, 45). The connection of hydrophobic ligand binding to allergy is thought to be related to hydrophobic compounds from bacteria that are commonly recognized by the innate immune system (18). It has been suggested that allergens carrying hydrophobic compounds shift the immune response from tolerance to Th2-type immune responses that are associated with allergic inflammation (31). The function of several dust mite allergens supports this contention. For example, Der p 2 functionally substituted for murine MD-2, an innate immune protein known to bind a lipopolysaccharide-binding protein from Gram-negative bacteria (18). Similarly, the structure of Der p 7 resembles lipopolysaccharide-binding protein, and Der p 7 was shown to bind a lipopeptide from Gram-positive bacteria (17). The structure of Der p 5 presented here suggests that a Der p 5 dimer may also have a propensity to bind hydrophobic compounds. Future studies are planned to investigate ligand binding in an effort to better understand the natural function of this important allergen and to determine whether this function is related to its allergenicity. We hypothesize that hydrophobic ligands or the natural ligand could induce a structural change to encourage dimer formation as seen in the crystal.
Acknowledgments
We thank Doan-Thu Nguyen, Cynthia Holley, and Andrea Moon for helpful contributions, and Dr. Lin Yang (National Synchrotron Light Source, Brookhaven National Laboratory) for assistance with data collection. We are grateful to Dr. Wayne Thomas (Western Australian Research Institute for Child Health, Perth, Western Australia) for providing the pGEX Der p 5 construct. Use of the X9 beamline at National Synchrotron Light Source at Brookhaven National Laboratory is supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-98CH10886.
This work was supported, in whole or in part, by National Institutes of Health Grant R01AI077653 from NIAID (to A. P. and M. D. C.), by the Intramural Research Program, and by NIEHS.
The atomic coordinates and structure factors (code 3MQ1) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- SAXS
- small angle x-ray scattering.
REFERENCES
- 1.Sporik R., Chapman M. D., Platts-Mills T. A. (1992) Clin. Exp. Allergy 22, 897–906 [DOI] [PubMed] [Google Scholar]
- 2.Chapman M. D., Pomés A., Breiteneder H., Ferreira F. (2007) J. Allergy Clin. Immunol. 119, 414–420 [DOI] [PubMed] [Google Scholar]
- 3.Thomas W. R., Hales B. J. (2007) Immunol. Res. 37, 187–199 [DOI] [PubMed] [Google Scholar]
- 4.Arruda L. K., Rizzo M. C., Chapman M. D., Fernandez-Caldas E., Baggio D., Platts-Mills T. A., Naspitz C. K. (1991) Clin. Exp. Allergy 21, 433–439 [DOI] [PubMed] [Google Scholar]
- 5.Caraballo L., Puerta L., Fernández-Caldas E., Lockey R. F., Martínez B. (1998) J. Investig. Allergol. Clin. Immunol. 8, 281–284 [PubMed] [Google Scholar]
- 6.Arruda L. K., Vailes L. D., Platts-Mills T. A., Fernandez-Caldas E., Montealegre F., Lin K. L., Chua K. Y., Rizzo M. C., Naspitz C. K., Chapman M. D. (1997) Am. J. Respir. Crit. Care Med. 155, 343–350 [DOI] [PubMed] [Google Scholar]
- 7.Simpson A., Green R., Custovic A., Woodcock A., Arruda L. K., Chapman M. D. (2003) Allergy 58, 53–56 [DOI] [PubMed] [Google Scholar]
- 8.Chew F. T., Yi F. C., Chua K. Y., Fernandez-Caldas E., Arruda L. K., Chapman M. D., Lee B. W. (1999) Clin. Exp. Allergy 29, 982–988 [DOI] [PubMed] [Google Scholar]
- 9.Kuo I. C., Cheong N., Trakultivakorn M., Lee B. W., Chua K. Y. (2003) J. Allergy Clin. Immunol. 111, 603–609 [DOI] [PubMed] [Google Scholar]
- 10.Weghofer M., Grote M., Dall'Antonia Y., Fernández-Caldas E., Krauth M. T., van Hage M., Horak F., Thomas W. R., Valent P., Keller W., Valenta R., Vrtala S. (2008) Int. Arch. Allergy Immunol. 147, 101–109 [DOI] [PubMed] [Google Scholar]
- 11.Chapman M. D., Wünschmann S., Pomés A. (2007) Curr. Allergy Asthma Rep. 7, 363–367 [DOI] [PubMed] [Google Scholar]
- 12.King C., Brennan S., Thompson P. J., Stewart G. A. (1998) J. Immunol. 161, 3645–3651 [PubMed] [Google Scholar]
- 13.Tomee J. F., van Weissenbruch R., de Monchy J. G., Kauffman H. F. (1998) J. Allergy Clin. Immunol. 102, 75–85 [DOI] [PubMed] [Google Scholar]
- 14.Hewitt C. R., Brown A. P., Hart B. J., Pritchard D. I. (1995) J. Exp. Med. 182, 1537–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulz O., Sewell H. F., Shakib F. (1998) J. Exp. Med. 187, 271–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kauffman H. F., Tamm M., Timmerman J. A., Borger P. (2006) Clin. Mol. Allergy 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mueller G. A., Edwards L. L., Aloor J. J., Fessler M. B., Glesner J., Pomés A., Chapman M. D., London R. E., Pedersen L. C. (2010) J. Allergy Clin. Immunol. 125, 909–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trompette A., Divanovic S., Visintin A., Blanchard C., Hegde R. S., Madan R., Thorne P. S., Wills-Karp M., Gioannini T. L., Weiss J. P., Karp C. L. (2009) Nature 457, 585–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan S. L., Ong T. C., Gao Y. F., Tiong Y. S., Wang de Y., Chew F. T., Mok Y. K. (2008) J. Immunol. 181, 2586–2596 [DOI] [PubMed] [Google Scholar]
- 20.Naik M. T., Chang C. F., Kuo I. C., Kung C. C., Yi F. C., Chua K. Y., Huang T. H. (2008) Structure 16, 125–136 [DOI] [PubMed] [Google Scholar]
- 21.Weghofer M., Dall'Antonia Y., Grote M., Stöcklinger A., Kneidinger M., Balic N., Krauth M. T., Fernández-Caldas E., Thomas W. R., van Hage M., Vieths S., Spitzauer S., Horak F., Svergun D. I., Konarev P. V., Valent P., Thalhamer J., Keller W., Valenta R., Vrtala S. (2008) Allergy 63, 758–767 [DOI] [PubMed] [Google Scholar]
- 22.Liaw S. H., Chen H. Z., Kuo I. C., Chua K. Y. (1998) J. Struct. Biol. 123, 265–268 [DOI] [PubMed] [Google Scholar]
- 23.Liaw S. H., Chen H. Z., Liu G. G., Chua K. Y. (2001) Biochem. Biophys. Res. Commun. 285, 308–312 [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 25.Kabsch W. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zwart P. H., Afonine P. V., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., McKee E., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Storoni L. C., Terwilliger T. C., Adams P. D. (2008) Methods Mol. Biol. 426, 419–435 [DOI] [PubMed] [Google Scholar]
- 27.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 28.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 29.Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H., Svergun D. I. (2003) J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 30.Svergun D. I. (1992) J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 31.Thomas W. R., Hales B. J., Smith W. A. (2005) Curr. Allergy Asthma Rep. 5, 388–393 [DOI] [PubMed] [Google Scholar]
- 32.Caraballo L., Mercado D., Jiménez S., Moreno L., Puerta L., Chua K. Y. (1998) Int. Arch. Allergy Immunol. 117, 38–45 [DOI] [PubMed] [Google Scholar]
- 33.Lin K. L., Hsieh K. H., Thomas W. R., Chiang B. L., Chua K. Y. (1994) J. Allergy Clin. Immunol. 94, 989–996 [DOI] [PubMed] [Google Scholar]
- 34.Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., Liang J. (2006) Nucleic Acids Res. 34, W116–W118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Radauer C., Bublin M., Wagner S., Mari A., Breiteneder H. (2008) J. Allergy Clin. Immunol. 121, 847–852 [DOI] [PubMed] [Google Scholar]
- 36.Xu Q., Canutescu A. A., Wang G., Shapovalov M., Obradovic Z., Dunbrack R. L., Jr. (2008) J. Mol. Biol. 381, 487–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrence M. C., Colman P. M. (1993) J. Mol. Biol. 234, 946–950 [DOI] [PubMed] [Google Scholar]
- 38.Derose E. F., Kirby T. W., Mueller G. A., Chikova A. K., Schaaper R. M., London R. E. (2004) Structure 12, 2221–2231 [DOI] [PubMed] [Google Scholar]
- 39.Mueller G. A., Kirby T. W., DeRose E. F., Li D., Schaaper R. M., London R. E. (2005) J. Bacteriol. 187, 7081–7089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaiser L., Grönlund H., Sandalova T., Ljunggren H. G., van Hage-Hamsten M., Achour A., Schneider G. (2003) J. Biol. Chem. 278, 37730–37735 [DOI] [PubMed] [Google Scholar]
- 41.Lascombe M. B., Grégoire C., Poncet P., Tavares G. A., Rosinski-Chupin I., Rabillon J., Goubran-Botros H., Mazié J. C., David B., Alzari P. M. (2000) J. Biol. Chem. 275, 21572–21577 [DOI] [PubMed] [Google Scholar]
- 42.Böcskei Z., Groom C. R., Flower D. R., Wright C. E., Phillips S. E., Cavaggioni A., Findlay J. B., North A. C. (1992) Nature 360, 186–188 [DOI] [PubMed] [Google Scholar]
- 43.Ichikawa S., Takai T., Yashiki T., Takahashi S., Okumura K., Ogawa H., Kohda D., Hatanaka H. (2009) Genes Cells 14, 1055–1065 [DOI] [PubMed] [Google Scholar]
- 44.Derewenda U., Li J., Derewenda Z., Dauter Z., Mueller G. A., Rule G. S., Benjamin D. C. (2002) J. Mol. Biol. 318, 189–197 [DOI] [PubMed] [Google Scholar]
- 45.Gajhede M., Osmark P., Poulsen F. M., Ipsen H., Larsen J. N., Joost van Neerven R. J., Schou C., Løwenstein H., Spangfort M. D. (1996) Nat. Struct. Biol. 3, 1040–1045 [DOI] [PubMed] [Google Scholar]