Abstract
Caldecrin/chymotrypsin C is a novel secretory-type serine protease that was originally isolated as a serum calcium-decreasing factor from the pancreas. Previously, we reported that caldecrin suppressed the bone-resorbing activity of rabbit mature osteoclasts (Tomomura, A., Yamada, H., Fujimoto, K., Inaba, A., and Katoh, S. (2001) FEBS Lett. 508, 454–458). Here, we investigated the effects of caldecrin on mouse osteoclast differentiation induced by macrophage-colony stimulating factor and the receptor activator of NF-κB ligand (RANKL) from the monocyte/macrophage cell lineage of bone marrow cells. Wild-type and protease-deficient mutant caldecrin dose-dependently inhibited RANKL-stimulated tartrate-resistant acid phosphatase-positive osteoclast formation from bone marrow cells. Caldecrin did not affect macrophage colony formation from monocyte/macrophage lineage cells or osteoclast progenitor generation in cultures of bone marrow cells. Caldecrin inhibited accumulation of the RANKL-stimulated nuclear factor of activated T-cells, cytoplasmic 1 (NFATc1) mRNA in bone marrow cells, which is a key transcription factor for the differentiation of osteoclasts. Caldecrin also suppressed RANKL-induced differentiation of the RAW264.7 monocyte/macrophage cell line into osteoclasts. Caldecrin reduced the transcriptional activity of NFATc1 in RAW264.7 cells, whereas those of NF-κB and c-Fos, which are also transcription factors involved in osteoclast differentiation, were unaffected. Caldecrin inhibited RANKL-stimulated nuclear translocation of NFATc1 and the activity of the calcium/calmodulin-dependent phosphatase, calcineurin. Caldecrin inhibited phospholipase Cγ1-mediated Ca2+ oscillation evoked by RANKL stimulation. RANKL-stimulated phosphorylation of spleen tyrosine kinase (Syk) was also attenuated by caldecrin. Taken together, these results indicate that caldecrin inhibits osteoclastogenesis, without its protease activity, by preventing a phospholipase Cγ1-mediated Ca2+oscillation-calcineurin-NFATc1 pathway.
Keywords: Calcium/Bone, Calcium/Calcineurin, Calcium/Cellular Regulation, Cell/Blood, Cell/Differentiation, Proteases/Serine Protease, Transcription/Regulation, Calcium, Phospholipase C
Introduction
Calcium ions have numerous cellular functions, and serum calcium homeostasis is tightly regulated by the intestines, kidneys, and bones, which are governed by the key systemic hormones, parathyroid hormone, 1α,25-dihydroxy vitamin D3, and calcitonin. Bone is a dynamic serum calcium-regulating tissue. Bone formation by osteoblasts involves deposition of minerals, including calcium, and bone resorption by osteoclasts releases calcium from bone (1).
We have purified and cloned the serum calcium-decreasing factor caldecrin from the pancreas (2–6). Caldecrin is a secretory-type serine protease that has chymotryptic activity and is also known as chymotrypsin C (EC 3.4.21.2) (8). We originally reported that the administration of caldecrin decreases mouse serum calcium concentration in a dose-dependent manner, and serum calcium decreasing activity is correlated with a decrease in serum hydroxyproline, which is included as a component of collagen and is a marker of bone resorption (2). Interestingly, this does not depend on its protease activity. Furthermore, caldecrin inhibits not only parathyroid hormone-stimulated bone destruction in mouse long bone organ culture but also bone resorption by rabbit mature osteoclasts in vitro (7). Taken together with our previous results, these observations suggest that caldecrin may suppress osteoclast differentiation and/or its activity through as yet unknown mechanisms.
Osteoclasts, multinucleated giant cells that have a unique bone resorption activity, are differentiated from hematopoietic cells of the monocyte/macrophage lineage (9–11). Osteoclast differentiation (osteoclastogenesis) from bone marrow cells (BMCs)4 requires the cell-to-cell contact of osteoclast progenitor cells with osteoblasts or bone marrow stromal cells. The key molecule for osteoclastogenesis is a member of the tumor necrosis factor family (receptor activator of NF-κB ligand (RANKL) (12), also called TRANCE (13)/ODF (14)/OPGL (15)) and is expressed in the osteoblast and bone marrow stromal cells. Osteoclast precursors express RANK, which is a member of the tumor necrosis factor receptor family, bind with RANKL, and stimulate osteoclast formation in the presence of M-CSF (11). Parathyroid hormone induces expression of RANKL on the cell surface of osteoblasts, resulting in osteoclast formation (14, 16). Furthermore, RANKL activates mature osteoclasts in vitro, and administration of RANKL to mice induces an increase in blood calcium concentration (17). Binding of RANKL to RANK induces receptor oligomerization and recruitment of signaling adaptor molecules, such as the tumor necrosis factor receptor-associated factor 6 (TRAF6) (18). TRAF6 can activate downstream signaling pathways, NF-κB and AP-1 and finally induces expression of the NFAT family member, NFATc1, which is the master regulator of osteoclast differentiation (11, 19). The RANKL-RANK signal also activates Ca2+ signaling through the activation of phospholipase Cγ (PLCγ). Takayanagi et al. (19) reported that RANKL evokes Ca2+ oscillation and accelerates Ca2+-dependent NFAT translocation. Nuclear-translocated NFATc1 forms a complex with a transcription factor, such as NF-κB and/or c-Fos, and activates the transcription of NFAT target genes, including those encoding TRAP and calcitonin receptor. The immunosuppressant drugs cyclosporin A (CsA) and FK506 inhibited RANKL-induced differentiation of cells into osteoclasts by the inhibition of calcineurin (19, 20). Calcineurin, which is a calcium/calmodulin-dependent serine/threonine phosphatase that is activated by cellular calcium concentration, causes dephosphorylation of NFATc1 proteins and induces their translocation into the nucleus (21, 22). Therefore, the activation of NFATc1 requires assembly of the RANKL-RANK-TRAF6 axis and PLCγ-Ca2+-calcineurin signaling.
Recently, immunoreceptor tyrosine-based activation motifs (ITAM)-harboring adaptor molecules DAP12 (DNAX-activating protein 12) and FcRγ (Fc receptor γ-chain) have been shown to cooperate with the RANKL-induced signaling pathways, leading to NFATc1 up-regulation and osteoclast-specific gene expression (23). ITAM signaling is coupled to the downstream pathway through Syk-tyrosine kinase (45). On the other hand, ITIM-harboring receptors LILR B (leukocyte Ig-like receptor B) and PIRB (paired Ig-like receptor B) are also expressed in preosteoclasts and inhibit osteoclastogenesis (46). RANKL-stimulated osteoclast differentiation is negatively controlled by osteoprotegerin, a tumor necrosis factor receptor-like soluble decoy receptor for RANKL (24, 14), and interferon-β, a negative regulator of c-Fos (25). Interferon-γ degrades TRAF6 and inhibits osteoclast formation (25). Interleukin-4 inhibits NF-κB through the STAT (signal transducers and activators of transcription)-dependent pathway and osteoclast formation (26–28). Interleukin-10 inhibits osteoclast formation by inhibition of Ca2+ mobilization (29, 30). Thus, osteoclast formation and bone homeostasis are controlled by many signaling pathways, cooperating with the RANKL-RANK signal. The molecular mechanisms remain unclear, especially the negative regulation of osteoclast formation.
In this study we investigated the effects of caldecrin on RANKL-induced osteoclast differentiation in vitro and characterized the signaling pathway of caldecrin in osteoclast differentiation. We provide the first evidence that caldecrin inhibits RANKL-stimulated osteoclastogenesis by suppression of NFATc1 activity through PLCγ1-mediated Ca2+ oscillation.
EXPERIMENTAL PROCEDURES
Preparation of Caldecrin
Recombinant wild-type (WT) and protease-deficient (S187A, Sm) mutant caldecrin were prepared as described previously (4). Briefly, cultured insect Sf9 cells were infected with baculovirus harboring WT or Sm caldecrin cDNA. On day 3 of culture, the secreted recombinant proteins were purified from culture media. The recombinant caldecrin was freshly activated by treatment with trypsin (50:1) for 30 min at room temperature followed by treatment with 0.1 mm α-amidinophenyl methanesulfonyl fluoride hydrochloride (Sigma) to terminate activation. The protease activity of caldecrin using N-succinyl-Ala-Ala-Pro-Phe p-nitroanilide (Sigma) as a substrate was measured at 405 nm with a plate reader (Dynatech MR 5000).
Preparation of Mouse BMCs and in Vitro Osteoclast Formation
4–6-Week-old male ddY mice (Saitama Experimental Animals) were sacrificed, and the femur and tibia were dissected out. After cutting both ends of the bone, BMCs were collected from the bone cavities by injection of α-minimal essential medium (α-MEM; Sigma) containing 10% fetal bovine serum (FBS; Sigma). The cell suspension was then passed through a 40-μm nylon cell strainer (BD Biosciences) and centrifuged at 450 × g for 5 min. The obtained cell pellet was resuspended and seeded on culture dishes or plates. All procedures were approved by the Animal Care and Use Committee of the Meikai University School of Dentistry (Saitama, Japan). BMCs (1–2 × 105 cells/ml) were cultured with α-MEM containing 10% FBS and M-CSF (10 ng/ml; R&D Systems) on 96-well plates (0.1 ml/well) for 3 days. The cells were cultured for a further 3–4 days in culture media containing M-CSF (10 ng/ml) and RANKL (10 ng/ml; R&D Systems) in the presence or absence of caldecrin (3 μg/ml). In some experiments with various inhibitors, the PLC inhibitor U73122 (10 μm; Cayman Chemicals), calcineurin inhibitor FK506 (2 μm; Alexis), CsA (1 μm; Merck), calcium chelator BAPTA-AM (20 μm; Merck), Syk inhibitor ER-27319 (15 μm; Sigma), and protein kinase C (PKC) inhibitor calphostin (100 nm; Merck) were used. In some experiments BMCs were cultured with M-CSF plus PKC activator 12-O-tetradecanoylphorbol-13-acetate (TPA, 20 nm; Merck) with or without RANKL.
For coculture with the stromal cell line ST2, BMCs (2 × 104 cells) were seeded on precultured ST2 cells (2 × 104 cells/96-well plate) and cultured for 7 days with 1α,25-dihydroxy vitamin D3 (10−8 m; Wako Pure Chemicals) and dexamethasone (10−8 m; Sigma) in the presence or absence of WT caldecrin (3 μg/ml) or inhibitors including FK506 and CsA.
For osteoclast formation experiments using RAW264.7 cells, the cells (104 cells/ml) with α-MEM containing 10% FBS were cultured on 96-well plates (0.1 ml/well) in the presence or absence of RANKL (10 ng/ml) with or without caldecrin or inhibitors. After 3–4 days in culture, TRAP activity of the medium and TRAP staining were performed.
Measurement of TRAP Activity and TRAP Staining
TRAP activities of osteoclasts were measured with culture media (30 μl) incubated for 15–30 min at 37 °C with 30 μl of 600 mm sodium acetate buffer, pH 5.5, containing l-ascorbic acid (17.6 mg/ml), sodium tartrate dehydrate (9.2 mg/ml), 4-nitrophenylphosphate-sodium (3.6 mg/ml), Triton X-100 (0.3%), EDTA (6 mm), and NaCl (600 mm). The reaction was terminated by the addition of 30 μl of NaOH (300 mm) and measured at 405 nm.
TRAP histochemical staining of the cells was performed using a leukocyte acid phosphatase kit (Sigma). Cultured cells were fixed with 100% methanol for 1 min at room temperature and air-dried. After TRAP-staining, TRAP-positive mono- and multinucleated (more than 3 nuclei) cells were counted under phase-contrast microscopy.
In Vitro Colony Formation
In vitro colony formation was measured according to the method described by Sato et al. (31). BMCs (1 × 104 cells/ml), which were suspended in α-MEM containing 30% FBS, 1.2% methylcellulose (Stem Cell Technologies), 1% BSA, and M-CSF with or without WT caldecrin (3 μg/ml) were cultured on 35-mm culture dishes for 1 week. At the end of culture, the macrophage colonies were counted by phase-contrast microscopy.
Frequency Analysis for Osteoclast Progenitor Cells
Frequency analysis for the osteoclast progenitor cells was performed according to a method described elsewhere (31, 32). BMCs (1, 15, 50, and 300 cells/well in 96-well culture plates) were cultured with α-MEM, 10% FBS containing M-CSF with or without WT caldecrin (3 μg/ml) on two 96-well culture plates. After culture for 3 days, the cells were washed and cultured for a further 3 days with medium containing M-CSF and RANKL and then stained for TRAP activity. The plates with appropriate numbers of osteoclast-positive wells (6–20 wells in 96) from each experimental group were selected for frequency analysis. The wells containing TRAP-positive multinucleated cells were counted. Frequency analysis was performed using the formula 1/frequency = N/ln(96/96 − P), where N is cell density in the well, and P is number of TRAP-positive wells.
Measurement of Cell Viability of BMCs
BMCs were cultured in medium containing M-CSF in the absence or presence of caldecrin (0.3, 1, 3 μg/ml) for 3 days. The cells were incubated with 3-(4,5-dimethyl-2-thiazol)-2,5-diphenyl-2H-tetrazolium bromide (0.2 mg/ml) for 4 h. The formazan produced was resolved with DMSO and measured at 600 nm.
Real-time Reverse Transcription-PCR
BMCs were cultured with M-CSF and RANKL with or without caldecrin as described above. Total RNA was prepared from BMCs using an RNeasy mini kit (Qiagen) on the indicated days. RNAs were treated with RNase-free DNase-1 at 37 °C for 90 min and repurified using the same kit. Total RNA (5.0 μg) was used to produce cDNA with random hexamers and SuperScript-II reverse transcriptase (Invitrogen). Aliquots (1/50 volume) of cDNA were used for real-time PCR amplification with TaqMan probe primer sets for NFATc1 and rRNA and Platinum Quantitative PCR SuperMix-UDG. NFATc1 and rRNA mRNAs were measured using a GeneAmp 5700 sequence detection system (Applied Biosystems).
Preparation of Cell Fraction
The cultured RAW264.7 cells at 70–80% confluence were incubated in medium (α-MEM, 5% FBS) containing RANKL with or without WT caldecrin for the indicated times, washed, and scraped in cold PBS. Cytosolic fractions and nuclear extracts were prepared as follows. The cells were harvested and collected by centrifugation at 450 × g for 5 min, and cell pellets were treated with hypotonic buffer (10 mm Tris-HCl, pH 7.4, 1 mm EDTA, and 1 mm EGTA) containing protease inhibitor mixture (Sigma) and phosphatase inhibitors (10 mm NaF and 2.5 mm Na2VO4) for 30 min on ice, then disrupted by passing through a 27-Gauge needle. After centrifugation at 10,000 × g for 20 min, the supernatant was used as the cytosolic fraction. The cell pellets were resuspended in extraction buffer (20 mm HEPES, pH 7.6, 1.5 mm MgCl2, 0.2 mm EDTA, 0.42 m NaCl, 10% glycerol) containing protease inhibitor mixture, incubated for 30 min on ice, and centrifuged at 20,000 × g for 10 min. The obtained supernatant was used as the nuclear extract.
Transcriptional Activity of NFATc1, NF-κB, and c-Fos
Activated transcription factors, NFATc1, NF-κB, and c-Fos were measured using a TransAM transcription factor enzyme-linked immunosorbent assay kit (Active Motif) according to the manufacturer's instructions. The nuclear extracts (10 μg) were incubated on NFATc1, NF-κB, and c-Fos consensus oligonucleotide-coated enzyme-linked immunosorbent assay plates for 30 min at room temperature. Activated transcription factors bound to consensus oligo DNA were detected with a specific antibody and measured at 450 nm.
Immunoblotting and Immunoprecipitation
Whole-cell lysates were prepared by extraction with radioimmune precipitation assay buffer (20 mm Tris, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 10 mm β-glycerophosphate, 10 mm NaF, 5 mm NaVO4, and protease inhibitor mixture; Roche Applied Science). The clarified lysates were separated by SDS-PAGE, transferred onto nitrocellulose membranes, and then immunoblotted with antibody against PLCγ1 (Cell Signaling), phospho-PLCγ1 (Tyr-783; Cell Signaling), NFATc1 (Santa Cruz Biotechnology), phospho-NFATc1 (Ser259; Santa Cruz Biotechnology), Syk (N-19; Santa Cruz Biotechnology), phosphotyrosine (4G10; Millipore), calcineurin (Sigma), histone H1 (Millipore), or β-actin (Sigma). The blots were incubated with horseradish peroxidase-conjugated secondary antibodies (Pierce) for 1 h, and chemiluminescence was detected with an ECL system (GE Healthcare). To assay phosphorylation of Syk, the clarified lysates were incubated with anti-Syk antibody for 3 h followed by a 1-h incubation with protein G-Sepharose beads (GE Healthcare). Immunoprecipitates were resolved by SDS-PAGE and immunoblotted with anti-phosphotyrosine 4G10.
Nuclear Translocation of NFATc1
For Western blotting to detect NFATc1 nuclear translocation, RAW264.7 cells were incubated in the presence or absence of RANKL (10 ng/ml) with or without WT caldecrin (5 μg/ml) for 30–40 min. The cell fractions were subjected to SDS-PAGE and immunoblotted with anti-NFATc1 antibody. For immunohistochemical detection of NFATc1, RAW264.7 cells grown on 2-mm glass coverslips in 24-well plates were treated for 30–40 min with RANKL in the presence or absence of caldecrin. The coverslips were then removed, washed in PBS, fixed in 4% paraformaldehyde, PBS, pH 7.2, for 1 h on ice, and washed in PBS, and cells were permeabilized with 0.2% Triton X-100 for 15 min. After blocking nonspecific binding with 5% goat serum for 30 min, cells were incubated with anti-NFATc1 antibody diluted 1:100 in PBS overnight. The cells were washed in PBS then incubated for 1 h with Alexa Fluor 488-conjugated anti-mouse secondary antibody (Invitrogen). After immunostaining, nuclei were stained using ethidium homodimer-1 (1:2000; Invitrogen). The fluorescence images were then visualized and photographed by laser scanning confocal microscopy (LSM 510; Carl Zeiss).
Calcineurin Activity
Cellular calcineurin phosphatase activity was measured in cell extracts using a Calcineurin Cellular Activity assay kit (Calbiochem) according to the manufacturer's instructions. Briefly, cultured RAW 264.7 cells at semi-confluence were further cultured for 30 min with α-MEM, 10% FBS and RANKL alone or in medium containing caldecrin. Cell extracts from cultured cells were prepared with lysis buffer containing EDTA and EGTA (50 μm each) and centrifuged. The fraction of total phosphatase activity due to calcineurin was determined by detection of free phosphate released from cAMP-dependent protein kinase regulatory subunit type II phosphopeptide, the best-known substrate for PP-2B, by treatment with okadaic acid in the absence or presence of EGTA buffer. Colorimetric measurements (assay with Malachite Green) were performed at 620 nm.
Intracellular Ca2+ Measurement
Intracellular Ca2+ was measured using a Fluo-4NW calcium assay kit (Invitrogen) according to the manufacturer's instructions as described elsewhere (19). Briefly, BMCs, which were cultured for 1 day with RANKL (10 ng/ml) and M-CSF (10 ng/ml), or RAW264.7 cells cultured with growth media only were loaded with 4 μm Fluo-4NW and 2.3 μm Fura Red AM “cell-permeant” (Invitrogen) in loading solution for 30 min. After washing, cells were incubated with α-MEM containing 10% FBS and stimulated with RANKL. Cells were excited at 488 nm, and the fluorescence images with emission at 505–530 nm for fluo-4 and 600–680 nm for Fura Red were monitored by laser scanning confocal microscopy at 1-s intervals. For inhibition assay, each of WT caldecrin or various inhibitors was subsequently added 10 min after Ca2+ oscillation by RANKL treatment. For ratiometric measurement of Ca2+ in a single cell, the fluorescence intensity of fluo-4 to Fura Red was calculated and expressed as the percent maximum ratio increase, which was obtained by the addition of 10 μm ionomycin (Sigma) at 30 min to terminate the experiment.
Statistics
Data were analyzed by analysis of variance and represent the means ± S.D. from at least three independent experiments. p < 0.05 was considered statistically significant.
RESULTS
Caldecrin Inhibits Osteoclast Formation but Not Macrophage and Osteoclast Progenitor Formation from Mouse BMCs
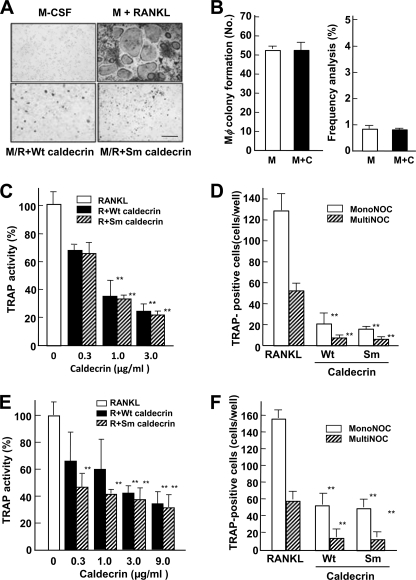
To determine whether the serum calcium-decreasing factor caldecrin inhibits osteoclastogenesis, we first evaluated the effects of caldecrin treatment on mouse osteoclast formation from BMCs in vitro. As shown in Fig. 1A, TRAP-positive multinucleated cells were observed in culture with M-CSF and RANKL, whereas TRAP-positive cells were not observed in M-CSF alone. Treatment with WT caldecrin in the presence of RANKL and M-CSF inhibited RANKL stimulated-osteoclast formation completely. Protease-deficient mutant (Sm) caldecrin also inhibited RANKL-stimulated osteoclast differentiation, suggesting that the inhibition of osteoclast formation by caldecrin is independent of its protease activity.
FIGURE 1.
Caldecrin inhibits RANKL-stimulated OC formation from BMCs and RAW264.7 cells without protease activity. A, BMCs were cultured for 3 days with M-CSF (M, 10 ng/ml) and further cultured for 3–4 days with M-CSF alone or M-CSF plus RANKL (R, 10 ng/ml) (M/R) with or without WT or Sm caldecrin (C, 3 μg/ml). The cells were stained for TRAP activity. The scale bar shows 200 μm. B, macrophage colony formation assay (left) and frequency analysis (right) were performed as described under “Experimental Procedures.” C, BMCs were cultured as shown in A except various concentrations of WT or Sm caldecrin were added, and TRAP activities of the culture media were compared (mean ± S.D. of three experiments; **, p < 0.01 versus RANKL-treated group). D, BMCs were cultured as shown in A and stained for TRAP activity. TRAP-positive mononucleated osteoclasts (MonoNOC) and multinucleated osteoclasts (MultiNOC) were counted (mean ± S.D. of three experiments; **, p < 0.01 versus the RANKL-treated mononucleated or multinucleated osteoclast control group, respectively). E, RAW264.7 cells were cultured for 3–4 days with RANKL (10 ng/ml) or RANKL plus various concentrations of WT or Sm caldecrin, and TRAP activities were measured. F, RAW264.7 cells were cultured as shown in E and stained for TRAP activity. TRAP-positive mononucleated and multinucleated osteoclasts were counted and compared with respective controls (mean ± S.D. of three experiments; **, p < 0.01 versus the RANKL-treated mononucleated or multinucleated osteoclast control group, respectively).
Osteoclasts were differentiated from the monocyte-macrophage lineage. Therefore, we examined whether caldecrin stimulates macrophage formation from BMCs. BMCs were cultured in the presence of M-CSF with or without WT caldecrin for 1 week, and macrophage colonies were estimated by colony formation assay. The macrophage-type colonies formed from BMCs in the absence or presence of WT caldecrin were not different (52.8 ± 1.3 versus 52.5 ± 3.7; Fig. 1B, left panel), suggesting that caldecrin does not stimulate macrophage formation. Next, we examined whether caldecrin inhibits osteoclast progenitor formation by frequency analysis. BMCs were cultured with M-CSF alone or in the presence of WT caldecrin to make the osteoclast progenitor. After washing, the progenitor cells were further cultured with M-CSF and RANKL to differentiate into osteoclasts. The frequency of osteoclast progenitor formation in the presence of M-CSF alone, which was estimated by RANKL-stimulated osteoclast formation, was not different from that in the presence of M-CSF and WT caldecrin (0.82 ± 0.13% versus 0.85 ± 0.05%; Fig. 1B, right panel). Thus, caldecrin does not affect osteoclast progenitor formation. Furthermore, BMC survival rates estimated by 3-(4,5-dimethyl-2-thiazol)-2,5-diphenyl-2H-tetrazolium bromide assay in WT or Sm caldecrin treatment were not different from the control culture (data not shown). These results suggest that caldecrin does not affect the microenvironment suitable for hematopoietic cell development, osteoclast progenitor formation, and macrophage differentiation.
The TRAP activities induced by RANKL treatment were inhibited in a dose-dependent manner by WT and Sm caldecrin (Fig. 1C). Osteoclast progenitor cells differentiate to TRAP-positive mononucleated osteoclasts at the early stage, and TRAP-positive multinucleated osteoclasts appear at the late stage of osteoclastogenesis. To determine which stage of osteoclastogenesis is inhibited by caldecrin, we compared the populations of mononucleated cells with those of multinucleated cells. RANKL-stimulated TRAP-positive multinucleated cell formation was about 40% that of TRAP-positive mononucleated cells (Fig. 1D). Treatment with WT or Sm caldecrin inhibited mononucleated cell differentiation to about 20% that stimulated by RANKL. However, there were no significant differences in the ratio of multinucleated/mononucleated cell formation between the control and WT or Sm caldecrin treatment, suggesting that caldecrin inhibits mononucleated cell formation at the early stage but not multinucleated cell formation at the late stage in osteoclastogenesis, and its inhibitory effect does not depend on protease activity.
The mouse macrophage-like cell line RAW 264.7 has been shown to readily differentiate into osteoclasts upon exposure to RANKL. This system has advantages because of the absence of osteoblast/stromal cells and M-CSF. As shown in Fig. 1E, caldecrin with or without protease activity dose-dependently suppressed RANKL-stimulated TRAP activities of RAW264.7 cells. Caldecrin also suppressed the TRAP-positive mononucleated cell formation from RAW264.7 cells (Fig. 1F). Comparing the results of BMC-derived osteoclast formation, inhibition of caldecrin on the osteoclast formation from RAW264.7 cells was slightly lower, suggesting that monocyte/macrophage-like RAW264.7 cells may proceed at the osteoclast differentiation step compared with M-CSF-treated BMCs.
Caldecrin Inhibits NFATc1 mRNA Accumulation and Its Transcriptional Activity
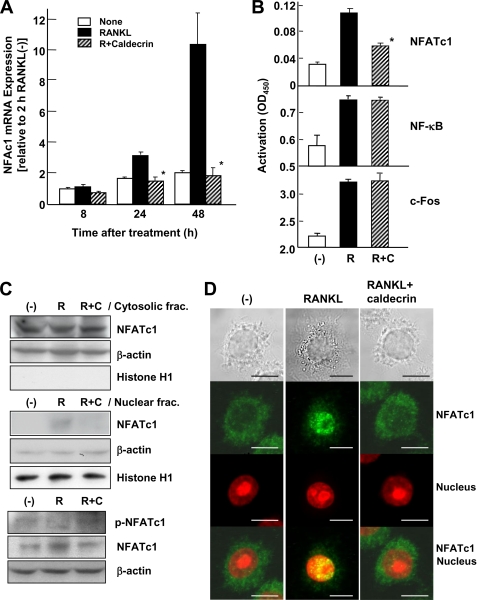
Induction and activation of NFATc1 is important for the RANKL-stimulated osteoclastogenesis (19). Therefore, we first investigated NFATc1 mRNA accumulation, which is related to the autoamplification of NFATc1. RANKL induced NFATc1 mRNA expression in a time-dependent manner (Fig. 2A). However, WT caldecrin completely inhibited the RANKL-stimulated NFATc1 accumulation (autoamplification) in osteoclasts derived from BMCs.
FIGURE 2.
Caldecrin inhibits RANKL-stimulated NFATc1 activity. A, BMCs were cultured as shown in Fig. 1A. At 8, 24, and 48 h after treatment with RANKL with or without WT caldecrin (3 μg/ml), total RNAs were isolated, and NFATc1 mRNA was evaluated by TaqMan real-time PCR (mean ± S.D. of three experiments; *, p < 0.05 versus RANKL-treated group with the same incubation time). B, RAW264.7 cells were incubated with RANKL alone (R) or in combination with WT caldecrin (9 μg/ml, shaded) for 30 min (R+C). Nuclear extracts were prepared, and the transcriptional activities of NFATc1, NF-κΒ, and c-Fos were measured (mean ± S.D. of three experiments: *, p < 0.05 versus RANKL-treated group). C, RAW264.7 cells were stimulated for 40 min with RANKL or RANKL plus WT caldecrin (5 μg/ml). Cytosolic and nuclear fractions were prepared from untreated (−), RANKL-stimulated (R), or RANKL plus WT caldecrin-treated (R+C) cells and analyzed by Western blotting with anti-NFATc1 antibody. The purity of the subcellular fractions and equal sample loading were controlled by analyzing β-actin and histone H1. Phosphorylation of NFATc1 in the whole-cell lysates from the same cells were evaluated by Western blotting with anti-phospho-NFATc1. D, RAW264.7 cells were incubated and stimulated for 40 min with RANKL or RANKL plus WT caldecrin (5 μg/ml). Cells were fixed and stained for anti-NFATc1 (green) and nucleus (red). The scale bar shows 10 μm.
Next, we determined the effect of caldecrin on the transcriptional activities of NFATc1. As shown in Fig. 2B, the transcriptional activity of NFATc1 was induced in RAW264.7 cells after a 30-min exposure to RANKL. However, WT caldecrin inhibited the RANKL-induced transcriptional activity of NFATc1. The transcriptional activities of NF-κB and c-Fos were also induced by RANKL treatment but not suppressed by caldecrin.
Caldecrin Inhibits NFATc1 Translocation to the Nucleus
NFATc1, which is phosphorylated in the cell cytoplasm, is translocated from the cytoplasm to the nucleus by calcineurin-mediated dephosphorylation. Therefore, we investigated whether RANKL-stimulated translocation of NFATc1 is suppressed by caldecrin treatment. RAW264.7 cells were incubated in the absence or presence of RANKL or RANKL plus WT caldecrin for 40 min, and the cytosolic and nuclear fractions were prepared. Western blotting analysis showed that NFATc1 in the nuclear fraction was increased in RANKL-stimulated cells (Fig. 2C). However, caldecrin reduced NFATc1 accumulation in the nucleus. NFATc1 phosphorylation states were examined in whole-cell lysates. The inactive form of NFATc1 (phospho-NFATc1) was decreased in RANKL-treated cells compared with untreated controls (Fig. 2C, lower panels). Caldecrin treatment prevented the disappearance of phospho-NFATc1 in RANKL-treated cells. In addition, immunofluorescence studies revealed more obvious nuclear translocation of NFATc1 in RANKL-treated cells. Caldecrin treatment suppressed the RANKL-stimulated translocation of NFATc1 to the nucleus in RAW264.7 cells (Fig. 2D). These results indicated that RANKL induces the translocation of NFATc1 into the nucleus and that caldecrin inhibits the translocation of NFATc1 into the nucleus.
Caldecrin Inhibits Calcineurin Activity
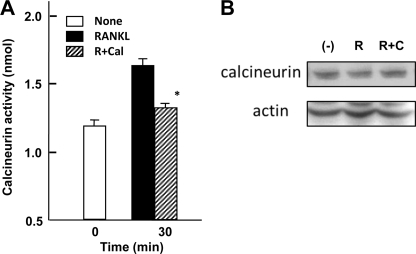
To determine how caldecrin inhibits osteoclast differentiation, we examined the upstream pathway(s) of NFATc1 in osteoclastogenesis using various inhibitors. The calcineurin inhibitors FK506 (2 μm) and CsA (1 μm), the PLCγ inhibitor U73122 (10 μm), and the intracellular calcium chelator BAPTA-AM (20 μm) inhibited RANKL-stimulated osteoclast formation (supplemental Fig. S1). In contrast, calphostin C (100 nm), an inhibitor of PKC, did not suppress RANKL-stimulated osteoclast formation in BMC. TPA alone did not induce osteoclast formation (supplemental Fig. S1). Therefore, in our in vitro system, the Ca2+-calcineurin axis but not PKC downstream of PLC may be involved in the activation of NFATc1. These results prompted us to examine the role of caldecrin in Ca2+-calcineurin signaling in osteoclasts. As we expected lower activity of calcineurin with caldecrin treatment, we determined the calcineurin activity of RAW264.7 cells treated with RANKL in the presence or absence of caldecrin. The calcineurin activity of RANKL-treated cells was increased (Fig. 3A). Caldecrin significantly reduced the RANKL-stimulated calcineurin activity. It was reported that RANKL treatment did not change the expression level of calcineurin subunits in RAW264.7 cells (41). Also, RANKL with or without caldecrin treatment did not change the protein level of calcineurin (Fig. 3B).
FIGURE 3.
Caldecrin inhibits RANKL-stimulated calcineurin activity in (RANKL) RAW 264.7 cells. A, RAW264.7 cells were cultured and stimulated for 30 min by RANKL with (R+Cal) or without (RANKL) WT caldecrin (9 μg/ml). Cell homogenates were prepared, and calcineurin activities were compared (mean ± S.D. of three experiments; *, p < 0.05 versus RANKL-treated group). B, RAW264.7 cells were cultured as shown in A, and calcineurin contents in the cells prepared from unstimulated (−), RANKL-stimulated (R), or RANKL plus WT caldecrin (R+C) were compared by Western blotting with anti-calcineurin antibody.
Caldecrin Inhibits PLCγ-mediated Ca2+ Oscillation Evoked by RANKL in BMC and RAW264.7 Cells
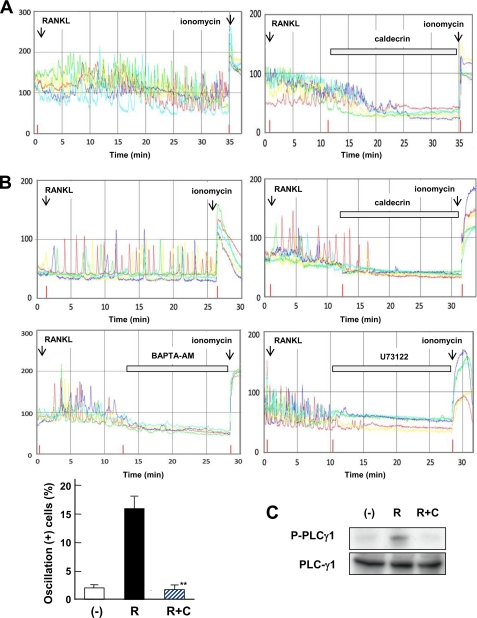
RANKL stimulates osteoclastogenesis by induction of long-lasting Ca2+ oscillation 24–48 h post-stimulation in BMC (19). We analyzed Ca2+ oscillation using 1-day-cultured BMCs in the presence of M-CSF and RANKL. As shown in Fig. 4A, RANKL evoked Ca2+ oscillation (left panel), whereas no oscillation was evoked by caldecrin treatment (right panel). Furthermore, we analyzed the effects of caldecrin on RANKL-evoked Ca2+ oscillation in RAW264.7 cells. As shown in Fig. 4B, RANKL treatment immediately produced Ca2+ oscillation in stock-cultured RAW264.7 cells (left panel). Caldecrin also completely abolished RANKL-evoked Ca2+ oscillation (right panel). The intracellular calcium chelator BAPTA-AM and PLC inhibitor U73122 abolished osteoclast formation (supplemental Fig. S1) and inhibited the RANKL-stimulated Ca2+ oscillation (middle panels), indicating that RANKL stimulated Ca2+ oscillation is driven by PLC activation and Ca2+ oscillation is required for osteoclastogenesis (Fig. 4B, lower panel).
FIGURE 4.
Caldecrin inhibits PLCγ1 mediated Ca2+ oscillation evoked by RANKL in BMCs and RAW 264.7 cells. A, BMCs were cultured for 1 day with M-CSF (10 ng/ml) plus RANKL (10 ng/ml). Intracellular [Ca2+]i in single cells was measured as described under “Experimental Procedures.” After a 10- min observation of Ca2+ oscillation with the addition of RANKL, WT caldecrin (10 nm) was further added. Each color indicates an individual cell in the same field. At the end of the experiment, ionomycin was added. B, [Ca2+]i in RAW264.7 cells was monitored as shown in A. Effects of BAPTA-AM (100 nm) and U73122 (100 nm) were evaluated (middle panels). The cells showed Ca2+ oscillation without RANKL (−), RANKL-triggered Ca2+ oscillation-acquired cells (R), and caldecrin-responsive cells in which RANKL-triggered Ca2+ oscillation was inhibited by WT caldecrin (R+C) were scored in the same field (mean ± S.D. of three experiments: *, p < 0.01 versus RANKL-treated group). C, RAW264.7 cells were incubated for 30 min with or without RANKL (10 ng/ml, R) or RANKL plus WT caldecrin (5 μg/ml, R+C). Cell lysates were subjected to Western blotting with anti-PLCγ1 or anti-phospho-PLCγ1 antibody.
It has been reported that RANKL-stimulated Ca2+ oscillation is evoked by PLCγ1 activation (23). Next, we analyzed activation of PLCγ1 by Western blotting with anti-phospho PLCγ1 antibody. As shown in Fig. 4C, phosphorylation of PLCγ1 induced by RANKL was reduced in caldecrin-treated cells. Taken together, these results suggest that caldecrin inhibits the RANKL-induced PLCγ1 activation followed by suppression of Ca2+ oscillation.
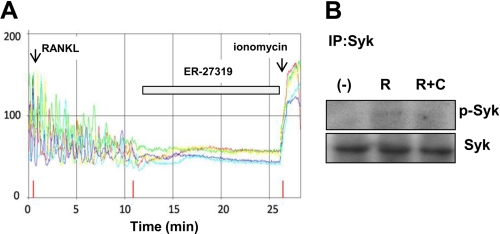
Caldecrin Suppresses RANKL-induced Syk Activation in RAW264.7 Cells
During osteoclastogenesis, ITAM-harboring adaptor protein and activation of PLCγ1 are involved in RANKL-stimulated Ca2+ oscillation (23). Syk is critical for osteoclastogenesis, which is activated by ITAM (49). Syk phosphorylates and activates the downstream target PLCγ1 (23). Therefore, we examined BMC osteoclastogenesis with the selective Syk inhibitor ER-27319, which resulted in impairment of RANKL-induced osteoclastogenesis (supplemental Fig. S1). We examined whether caldecrin suppressed RANKL-induced Syk activation in RAW264.7 cells. As shown in Fig. 5A, RANKL-evoked Ca2+ oscillation was inhibited by the addition of ER-27319, suggesting that RANKL-stimulated Syk activation is upstream of PLCγ1. On Western blotting analysis, phosphorylation of Syk was increased after 6 min of RANKL stimulation. Caldecrin also suppressed the RANKL-stimulated phosphorylation of Syk, suggesting that WT caldecrin suppressed RANKL-stimulated Syk activation, which was followed by activation of PLCγ1.
FIGURE 5.
Caldecrin suppresses RANKL-stimulated Syk phosphorylation in RAW264.7 cells. A, RANKL-evoked Ca2+ oscillation was monitored as shown in Fig. 4B. The Syk inhibitor ER-27319 (100 nm) was added at 10 min after RANKL treatment. B, RAW264.7 cells were incubated for 6 min with RANKL (R, 10 ng/ml) or RANKL plus WT caldecrin (R+C, 5 μg/ml). Total cell lysates were subjected to immunoprecipitation (IP) with anti-Syk antibody. The immunoprecipitates were separated by SDS-PAGE and immunoblotted with anti-phosphotyrosine antibody.
Coculture with Stromal/Osteoblast Partially Restores the Suppression of Osteoclast Formation by Caldecrin
ITAM-harboring adaptor DAP12 or Syk-deficient mouse impaired RANKL-stimulated osteoclastogenesis (23, 45). In contrast, bone marrow cells from these deficient mice cocultured with osteoblasts showed partial restoration of osteoclast formation, suggesting that the interaction between osteoclast precursors and osteoblasts may light up the existence of other signals for osteoclastogenesis. Moreover, osteoclast precursors from inositol 1,4,5-triphosphate receptor 2/3 double knock-out mice impaired Ca2+ oscillation and osteoclast formation but was partially restored in coculture with osteoblasts, suggesting that there is a Ca2+ oscillation-independent mechanism for NFATc1 activation in the coculture system (51). To investigate whether the suppression of caldecrin is restored by the interaction with osteoblasts, BMCs were cocultured with stromal ST2 cells. Compared with RANKL- and M-CSF-treated osteoclast culture, TRAP-positive multinucleated osteoclasts in the coculture system were partially restored in the presence of FK506 (supplemental Fig. S2). The inhibition of osteoclastogenesis by caldecrin was also partially restored, suggesting that osteoblast-induced osteoclastogenesis has Ca2+ oscillation/calcineurin-dependent (FK-sensitive) and -insensitive (FK-insensitive) pathways, and caldecrin is likely involved in the Ca2+ oscillation/calcineurin-dependent pathway.
DISCUSSION
Bone marrow cells develop into functionally active mature osteoclasts in various stages of differentiation; in the early stage, the osteoclast progenitors become mononucleated preosteoclasts expressing TRAP activity; in the late stage, the TRAP-positive mononucleated preosteoclasts fuse together to become TRAP-positive multinucleated osteoclasts; in the final stage, the multinucleated osteoclasts are activated by stimulants to absorb the bone matrix. We showed here that the serum calcium-decreasing factor, caldecrin, inhibits the early stage of preosteoclast formation stimulated by RANKL from BMCs. The fusion rates of TRAP-positive mononucleated preosteoclasts to become multinucleated osteoclasts were unaffected by treatment with caldecrin, indicating that caldecrin does not affect the fusion process of osteoclast formation (Fig. 1D). Furthermore, caldecrin treatment did not affect M-CSF-stimulated macrophage precursor generation or macrophage colony formation from BMCs or osteoclast progenitor generation in BMCs (Fig. 1B). Therefore, we focused on RANKL signaling. To clarify the inhibitory effects of caldecrin on the signaling pathway of RANKL-stimulated osteoclast differentiation, we studied the use of the macrophage cell line RAW264.7, which is sufficient to mimic the TRAP-positive multinucleated osteoclast-like phenotype by the sole addition of RANKL (Fig. 1, E and F). That is, caldecrin antagonized RANKL signaling.
Many cellular events including osteoclast formation are regulated by the Ca2+ oscillation frequency (33–37). NFAT is one of the cellular responses regulated by the Ca2+ oscillation frequency (35, 36). Ca2+ oscillation builds up dephosphorylation of NFAT by the Ca2+/calmodulin-dependent serine/threonine phosphatase calcineurin and increases NFAT nuclear translocation (38, 39). Takayanagi et al. (19) reported that RANKL-evoked Ca2+ oscillation triggers osteoclast differentiation through NAFTc1 activation, which induces osteoclast-specific gene expression. They observed that RANKL-evoked Ca2+ oscillation begins 24 h after RANKL stimulation and is sustained in BMCs. We observed RANKL-evoked Ca2+ oscillation and enhanced activation of NFATc1 expression in 24-h-cultured BMCs (Figs. 2A and 4A). On the other hand, Ca2+ oscillation occurred rapidly after RANKL-stimulation in RAW264.7 cells (Fig. 4B). The component(s) necessary for the Ca2+ oscillation may be absent or reduced in number and expressed during the first 24 h of differentiation in BMCs. Interestingly, caldecrin immediately and completely inhibited the RANKL-evoked Ca2+ oscillation in BMCs and RAW264.7 cells (Fig. 4, A and B). Therefore, the primary effect of caldecrin for the suppression of osteoclastogenesis is blocking the Ca2+ oscillation. The Ca2+ oscillation requires Ca2+ release from internal stores and extracellular Ca2+ influx. Activation of PLCγ stimulates Ca2+ release from the endoplasmic reticulum via inositol 1,4,5-triphosphate receptor. It was reported that RANKL activated PLCγ (40, 23, 55), and knock-down of PLCγ1 inhibited RANKL-induced Ca2+ oscillations (55), suggesting that PLCγ1 may cause RANKL-stimulated Ca2+ oscillation. Indeed, we found that the PLCγ inhibitor U73122 inhibited not only Ca2+ oscillation (Fig. 4B) but also osteoclast formation (supplemental Fig. S1). Interestingly, caldecrin suppressed RANKL-induced PLCγ1 activation (Fig. 4C).
There is another downstream pathway of PLCγ in addition to the Ca2+-calcineurin axis. PLCγ hydrolyzes phosphatidylinositol 4,5-bisphosphate to diacylglycerol, which activates PKC. Early studies indicated that NFAT integrated Ca2+ signals with those transduced by mitogen-activated protein kinase and/or PKC (52, 53). Thus, NFAT activation requires both Ras/PKC and Ca2+/calcineurin signaling (54). On the other hand, TPA, an activator of PKC, was reported to inhibit osteoclastogenesis by suppressing RANKL-induced NF-κB activation (50). We examined the effects of PKC activity on osteoclastogenesis, which is downstream of PLC (supplemental Fig. S1). TPA alone did not stimulate osteoclast formation, and TPA or PKC inhibitor calphostin A also did not affect RANKL-induced osteoclast formation, suggesting that PKC activity may not be involved in our osteoclastogenesis system. Therefore, the negative regulation of caldecrin in osteoclastogenesis is likely to be the blocking of RANKL-stimulated PLCγ/Ca2+-calcineurin-NFATc1 axis.
The series of signals after RANK stimulation of PLCγ activation is still unclear, although Syk has been implicated in the regulation of PLCγ and osteoclastogenesis. Syk-deficient BMCs showed impairment of normal osteoclastogenesis induced by RANKL (49). The immunoreceptor system was reported as the RANKL-RANK axis modifying signal (42). ITAM-harboring adaptor proteins, DAP12 and FcRγ, accelerate the RANKL stimulation in osteoclast differentiation (43, 45). Osteoclast-associated receptor and triggering receptor expressed on myeloid cell 2 (TREM2) recruit Fcγ and DAP12 adaptor protein, respectively, resulting in the activation of PLCγ and initiation of Ca2+ oscillation through Syk activation (45). DAP12−/−FcRγ−/− cells showed impaired RANKL-induced phosphorylation of PLCγ1 and Ca2+ oscillation (23). In addition, piceatannol, an inhibitor of Syk that is recruited to ITAM in the immune system (44), has an inhibitory effect on osteoclastogenesis (23). We demonstrated that the selective Syk inhibitor ER-27319 abolished not only osteoclastogenesis but also RANKL-evoked Ca2+ oscillation (supplemental Fig. S1 and Fig. 5A). Interestingly, caldecrin suppresses RANKL-induced Syk activation (Fig. 5B). Thus, caldecrin may be involved, at least in part, in ITAM-Syk-PLCγ1-mediated Ca2+ oscillation.
In osteoclast-osteoblast coculture, the formation of multinucleated osteoclasts was partially restored from DAP12−/− and to a lesser extent from Syk−/−, suggesting that FcRγ, adapting to osteoclast-associated receptor and binding to the osteoclast-associated receptor ligand on the osteoblast in coculture, is able to compensate for the lack of osteoclastogenesis in DAP12−/− cells and/or links to some Syk-independent pathway(s). It is likely that osteoblasts induce Ca2+oscillation-independent osteoclast formation (51). The suppressive effect of caldecrin on osteoclast formation in the coculture system is weak similarly to those of FK506 and CsA, suggesting that caldecrin may be involved in a Ca2+ oscillation-sensitive pathway. At present, we do not know how caldecrin suppresses Syk kinase. Caldecrin may associate with ITAM-harboring adaptor-associated receptor or ITIM-harboring receptor and modulate its kinase activity.
Recently, Kim et al. (55) reported a novel redox pathway that is upstream of PLCγ1 based on observations in peroxiredoxin (PrxII) knock-out mice. They demonstrated that RANKL-generated reactive oxygen species induces long-lasting Ca2+ oscillation and osteoclastogenesis. Whether caldecrin regulates this redox pathway and cross-talk to the Syk signal pathway is an intriguing question.
The pro-form of caldecrin requires trypsin treatment for activation of its protease activity (3). Serum Ca2+ decreasing activity and the inhibitory effects of caldecrin on the bone-resorbing activity of mature osteoclasts require trypsin activation even though protease-deficient mutants are used (4, 7). The inhibitory effects of caldecrin on osteoclastogenesis also require pretreatment with trypsin (data not shown). The inhibitory effect of caldecrin in osteoclastogenesis may be caused by the proteolytic cleavage of the propeptide, which needs to be exposed to the intramolecular responsive region after activation by trypsin treatment. The precise mechanism of the inhibitory effect of caldecrin on Ca2+ oscillation should be elucidated in future studies.
Considering the selective effects of caldecrin on Ca2+-calcineurin-NFATc1 inhibition and its nature as a secretory molecule that does not require protease activity in osteoclastogenesis, caldecrin may be a good therapeutic agent for suppressing osteoporosis and arthritis caused by enhanced NFATc1 activation (47, 48). Indeed, we have found that ovariectomized-induced bone loss was prevented by caldecrin treatment.5 In conclusion, caldecrin completely inhibits RANKL-triggered Ca2+ oscillation through the suppression of Syk-induced PLCγ activation in RAW264.7 cells, resulting in the inhibition of calcineurin activity, NFATc1 translocation into the nucleus, and osteoclast differentiation.
Supplementary Material
Acknowledgment
We thank Dr. Tatsuya Satoh (Meikai University) for the generous gift of RAW264.7 cell and ST2 cell and for advice and discussion for the osteoclast culture.
This work was supported in part by Grants-in-aid for Scientific Research C14571773 (to M. K.) and C16591864 (to A. T.) from the Ministry of Education, Culture, Sports, Science, and Technology of JAPAN.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
M. Ohi, S. Kido, H. Hasegawa, K. Fujimoto, M. Tomomura, H. Kanegae, A. Inaba, and A. Tomomura, manuscript in preparation.
- BMC
- bone marrow cell
- AP-1
- activator protein-1
- CsA
- cyclosporin A
- DAP
- DNAX-activating protein
- FcRγ
- Fc receptor γ-chain
- ITAM
- immunoreceptor tyrosine-based activation motif
- M-CSF
- monocyte/macrophage-colony stimulating factor
- NFAT
- nuclear factor of activated T cells
- NFATc1
- NFAT, cytoplasmic 1
- NF-κB
- nuclear factor κB
- PLC
- phospholipase C
- PKC
- protein kinase C
- RANK
- receptor activator of NF-κB
- RAKL
- RANK ligand
- Sm
- serine mutant
- Syk
- spleen-tyrosine kinase
- TRAF
- tumor necrosis factor receptor-associated factor
- TRAP
- tartrate-resistant acid phosphatase
- TPA
- 12-O-tetradecanoylphorbol-13-acetate
- WT
- wild type
- α-MEM
- α-minimum Eagle's medium
- FBS
- fetal bovine serum
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Manolagas S. C. (2000) Endocr. Rev. 21, 115–137 [DOI] [PubMed] [Google Scholar]
- 2.Tomomura A., Fukushige T., Noda T., Noikura T., Saheki T. (1992) FEBS Lett. 301, 277–281 [DOI] [PubMed] [Google Scholar]
- 3.Tomomura A., Fukushige T., Tomomura M., Noikura T., Nishii Y., Saheki T. (1993) FEBS Lett. 335, 213–216 [DOI] [PubMed] [Google Scholar]
- 4.Tomomura A., Tomomura M., Fukushige T., Akiyama M., Kubota N., Kumaki K., Nishii Y., Noikura T., Saheki T. (1995) J. Biol. Chem. 270, 30315–30321 [DOI] [PubMed] [Google Scholar]
- 5.Tomomura A., Akiyama M., Itoh H., Yoshino I., Tomomura M., Nishii Y., Noikura T., Saheki T. (1996) FEBS Lett. 386, 26–28 [DOI] [PubMed] [Google Scholar]
- 6.Yoshino-Yasuda I., Kobayashi K., Akiyama M., Itoh H., Tomomura A., Saheki T. (1998) J. Biochem. 123, 546–554 [DOI] [PubMed] [Google Scholar]
- 7.Tomomura A., Yamada H., Fujimoto K., Inaba A., Katoh S. (2001) FEBS Lett. 508, 454–458 [DOI] [PubMed] [Google Scholar]
- 8.Szilágyi L. (1998) Handbook of Proteolitic Enzymes (Barrett A. J., Rawlings N. D., Woessner J. F. eds) pp. 38–40, Academic Press, Inc., London [Google Scholar]
- 9.Chambers T. J., Revell P. A., Fuller K., Athanasou N. A. (1984) J. Cell Sci. 66, 383–399 [DOI] [PubMed] [Google Scholar]
- 10.Suda T., Takahashi N., Martin T. J. (1992) Endocr. Rev. 13, 66–80 [DOI] [PubMed] [Google Scholar]
- 11.Teitelbaum S. L. (2000) Science 289, 1504–1508 [DOI] [PubMed] [Google Scholar]
- 12.Anderson D. M., Maraskovsky E., Billingsley W. L., Dougall W. C., Tometsko M. E., Roux E. R., Teepe M. C., DuBose R. F., Cosman D., Galibert L. (1997) Nature 390, 175–179 [DOI] [PubMed] [Google Scholar]
- 13.Wong B. R., Rho J., Arron J., Robinson E., Orlinick J., Chao M., Kalachikov S., Cayani E., Bartlett F. S., 3rd, Frankel W. N., Lee S. Y., Choi Y. (1997) J. Biol. Chem. 272, 25190–25194 [DOI] [PubMed] [Google Scholar]
- 14.Yasuda H., Shima N., Nakagawa N., Yamaguchi K., Kinosaki M., Mochizuki S., Tomoyasu A., Yano K., Goto M., Murakami A., Tsuda E., Morinaga T., Higashio K., Udagawa N., Takahashi N., Suda T. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lacey D. L., Timms E., Tan H. L., Kelley M. J., Dunstan C. R., Burgess T., Elliott R., Colombero A., Elliott G., Scully S., Hsu H., Sullivan J., Hawkins N., Davy E., Capparelli C., Eli A., Qian Y. X., Kaufman S., Sarosi I., Shalhoub V., Senaldi G., Guo J., Delaney J., Boyle W. J. (1998) Cell 93, 165–176 [DOI] [PubMed] [Google Scholar]
- 16.Takahashi N., Udagawa N., Takami M., Suda T. (2005) in Principles of Bone Biology (Bilezikian J. P., Raiz L. G., Rodan G. A. eds) pp.109–126, Academic Press, Inc., New York [Google Scholar]
- 17.Burgess T. L., Qian Y., Kaufman S., Ring B. D., Van G., Capparelli C., Kelley M., Hsu H., Boyle W. J., Dunstan C. R., Hu S., Lacey D. L. (1999) J. Cell Biol. 145, 527–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong B. R., Josien R., Lee S. Y., Vologodskaia M., Steinman R. M., Choi Y. (1998) J. Biol. Chem. 273, 28355–28359 [DOI] [PubMed] [Google Scholar]
- 19.Takayanagi H., Kim S., Koga T., Nishina H., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., Wagner E. F., Mak T. W., Kodama T., Taniguchi T. (2002) Dev. Cell 3, 889–901 [DOI] [PubMed] [Google Scholar]
- 20.Hirotani H., Tuohy N. A., Woo J. T., Stern P. H., Clipstone N. A. (2004) J. Biol. Chem. 279, 13984–13992 [DOI] [PubMed] [Google Scholar]
- 21.Asagiri M., Sato K., Usami T., Ochi S., Nishina H., Yoshida H., Morita I., Wagner E. F., Mak T. W., Serfling E., Takayanagi H. (2005) J. Exp. Med. 202, 1261–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asagiri M., Takayanagi H. (2007) Bone 40, 251–264 [DOI] [PubMed] [Google Scholar]
- 23.Koga T., Inui M., Inoue K., Kim S., Suematsu A., Kobayashi E., Iwata T., Ohnishi H., Matozaki T., Kodama T., Taniguchi T., Takayanagi H., Takai T. (2004) Nature 428, 758–763 [DOI] [PubMed] [Google Scholar]
- 24.Simonet W. S., Lacey D. L., Dunstan C. R., Kelley M., Chang M. S., Lüthy R., Nguyen H. Q., Wooden S., Bennett L., Boone T., Shimamoto G., DeRose M., Elliott R., Colombero A., Tan H. L., Trail G., Sullivan J., Davy E., Bucay N., Renshaw-Gegg L., Hughes T. M., Hill D., Pattison W., Campbell P., Sander S., Van G., Tarpley J., Derby P., Lee R., Boyle W. J. (1997) Cell 89, 309–319 [DOI] [PubMed] [Google Scholar]
- 25.Takayanagi H., Kim S., Matsuo K., Suzuki H., Suzuki T., Sato K., Yokochi T., Oda H., Nakamura K., Ida N., Wagner E. F., Taniguchi T. (2002) Nature 416, 744–749 [DOI] [PubMed] [Google Scholar]
- 26.Abu-Amer Y. (2001) J. Clin. Invest. 107, 1375–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei S., Wang M. W., Teitelbaum S. L., Ross F. P. (2002) J. Biol. Chem. 277, 6622–6630 [DOI] [PubMed] [Google Scholar]
- 28.Moreno J. L., Kaczmarek M., Keegan A. D., Tondravi M. (2003) Blood 102, 1078–1086 [DOI] [PubMed] [Google Scholar]
- 29.Xu L. X., Kukita T., Kukita A., Otsuka T., Niho Y., Iijima T. (1995) J. Cell Physiol. 165, 624–629 [DOI] [PubMed] [Google Scholar]
- 30.Evans K. E., Fox S. W. (2007) BMC Cell Biol. 8, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato T., Shibata T., Ikeda K., Watanabe K. (2001) J. Bone Miner. Res. 16, 2215–2221 [DOI] [PubMed] [Google Scholar]
- 32.Hayashi S., Miyamoto A., Yamane T., Kataoka H., Ogawa M., Sugawara S., Nishikawa S., Nishikawa S., Sudo T., Yamazaki H., Kunisada T. (1997) J. Cell Physiol. 170, 241–247 [DOI] [PubMed] [Google Scholar]
- 33.Negulescu P. A., Shastri N., Cahalan M. D. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 2873–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Koninck P., Schulman H. (1998) Science 279, 227–230 [DOI] [PubMed] [Google Scholar]
- 35.Dolmetsch R. E., Xu K., Lewis R. S. (1998) Nature 392, 933–936 [DOI] [PubMed] [Google Scholar]
- 36.Li W., Llopis J., Whitney M., Zlokarnik G., Tsien R. Y. (1998) Nature 392, 936–941 [DOI] [PubMed] [Google Scholar]
- 37.Oancea E., Meyer T. (1998) Cell 95, 307–318 [DOI] [PubMed] [Google Scholar]
- 38.Rusnak F., Mertz P. (2000) Physiol. Rev. 80, 1483–1521 [DOI] [PubMed] [Google Scholar]
- 39.Tomida T., Hirose K., Takizawa A., Shibasaki F., Iino M. (2003) EMBO J. 22, 3825–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harris M. L., Schiller H. J., Reilly P. M., Donowitz M., Grisham M. B., Bulkley G. B. (1992) Pharmacol. Ther. 53, 375–408 [DOI] [PubMed] [Google Scholar]
- 41.Zhu L. L., Zaidi S., Moonga B. S., Troen B. R., Sun L. (2005) Biochem. Biophys. Res. Commun. 326, 131–135 [DOI] [PubMed] [Google Scholar]
- 42.Kim N., Takami M., Rho J., Josien R., Choi Y. (2002) J. Exp. Med. 195, 201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paloneva J., Mandelin J., Kiialainen A., Bohling T., Prudlo J., Hakola P., Haltia M., Konttinen Y. T., Peltonen L. (2003) J. Exp. Med. 198, 669–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raeder E. M., Mansfield P. J., Hinkovska-Galcheva V., Shayman J. A., Boxer L. A. (1999) J. Immunol. 163, 6785–6793 [PubMed] [Google Scholar]
- 45.Mócsai A., Humphrey M. B., Van Ziffle J. A., Hu Y., Burghardt A., Spusta S. C., Majumdar S., Lanier L. L., Lowell C. A., Nakamura M. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6158–6163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mori Y., Tsuji S., Inui M., Sakamoto Y., Endo S., Ito Y., Fujimura S., Koga T., Nakamura A., Takayanagi H., Itoi E., Takai T. (2008) J. Immunol. 181, 4742–4751 [DOI] [PubMed] [Google Scholar]
- 47.Takayanagi H., Iizuka H., Juji T., Nakagawa T., Yamamoto A., Miyazaki T., Koshihara Y., Oda H., Nakamura K., Tanaka S. (2000) Arthritis Rheum. 43, 259–269 [DOI] [PubMed] [Google Scholar]
- 48.Cenci S., Weitzmann M. N., Roggia C., Namba N., Novack D., Woodring J., Pacifici R. (2000) J. Clin. Invest. 106, 1229–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zou W., Kitaura H., Reeve J., Long F., Tybulewicz V. L., Shattil S. J., Ginsberg M. H., Ross F. P., Teitelbaum S. L. (2007) J. Cell Biol. 176, 877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang C., Steer J. H., Joyce D. A., Yip K. H., Zheng M. H., Xu J. (2003) J. Bone Miner. Res. 18, 2159–2168 [DOI] [PubMed] [Google Scholar]
- 51.Kuroda Y., Hisatsune C., Nakamura T., Matsuo K., Mikoshiba K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8643–8648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crabtree G. R. (1989) Science 243, 355–361 [DOI] [PubMed] [Google Scholar]
- 53.Woodrow M., Clipstone N. A., Cantrell D. (1993) J. Exp. Med. 178, 1517–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Timmerman L. A., Clipstone N. A., Ho S. N., Northrop J. P., Crabtree G. R. (1996) Nature 383, 837–840 [DOI] [PubMed] [Google Scholar]
- 55.Kim M. S., Yang Y. M., Son A., Tian Y. S., Lee S. I., Kang S. W., Muallem S., Shin D. M. (2010) J. Biol. Chem. 285, 6913–6921 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.