Abstract
Pasteurella multocida toxin (PMT) is a virulence factor responsible for the pathogenesis of some forms of pasteurellosis. The toxin activates Gq- and G12/13-dependent pathways through the deamidation of a glutamine residue in the α-subunit of heterotrimeric GTPases. We recently reported the crystal structure of the C terminus (residues 575–1285) of PMT (C-PMT), which is composed of three domains (C1, C2, and C3), and that the C1 domain is involved in the localization of C-PMT to the plasma membrane, and the C3 domain possesses a cysteine protease-like catalytic triad. In this study, we analyzed the membrane-targeting function of the C1 domain in detail. The C1 domain consists of seven helices of which the first four (residues 590–670), showing structural similarity to the N terminus of Clostridium difficile toxin B, were found to be involved in the recruitment of C-PMT to the plasma membrane. C-PMT lacking these helices (C-PMT ΔC1(4H)) neither localized to the plasma membrane nor stimulated the Gq/12/13-dependent signaling pathways. When the membrane-targeting property was complemented by a peptide tag with an N-myristoylation motif, C-PMT ΔC1(4H) recovered the PMT activity. Direct binding between the C1 domain and liposomes containing phospholipids was evidenced by surface plasmon resonance analyses. These results indicate that the C1 domain of C-PMT functions as a targeting signal for the plasma membrane.
Keywords: Bacterial Toxins, Crystal Structure, Heterotrimeric G Proteins, Lipid-binding Protein, Plasma Membrane
Introduction
Pasteurella multocida toxin (PMT)2 is a highly potent mitogen for various types of cultured cells including fibroblasts and osteoblastic cells (1–3). Because of this, PMT is also referred to as cyclomodulin, which promotes or interferes with the cell cycle progression of target cells (4). PMT exerts its effects by up-regulating various signaling cascades in which Rho, phospholipase Cβ, and mitogen-activated protein kinases (MAPKs) such as c-Jun N-terminal kinase (JNK) and extracellular signal-regulated kinase (ERK) are involved (5–10). The up-regulation is considered to arise from the activation of heterotrimeric GTPase (Gq- and G12/13)-dependent pathways (3, 10, 11–13). More recently, it was reported that PMT activated Gi2 through the deamidation of Gln205 of Gαi2 (14, 15). However, the mechanism of action of PMT is not completely understood. Previous studies implied that the toxin is internalized by endocytosis after binding to a putative receptor on target cells and escapes from endosomes into the cytoplasm (7, 16) where it activates heterotrimeric GTPases through deamidation. PMT consists of a single polypeptide chain of 1285 amino acids. Several lines of evidence indicate that the N-terminal region of the toxin binds to target cells and that the C-terminal region carries the intracellularly active moiety (17–20). The N-terminal region is partly homologous to Escherichia coli cytotoxic necrotizing factors CNF1 and CNF2 (21, 22). In contrast, in the C-terminal region of PMT, there is neither a known catalytic motif nor a region homologous with any other toxin or enzyme. Recently, we clarified the crystal structure of the C-terminal region (residues 575–1285) of PMT (C-PMT), which is toxic when directly introduced into cells (23), and found that C-PMT was composed of three domains (C1, C2, and C3). In addition, we clearly showed that the C1 domain is involved in the recruitment of C-PMT to the plasma membrane, and the C3 domain possesses a cysteine protease-like catalytic triad.
Here we analyzed the function of the C1 domain in detail. The results revealed that the C1 domain, essential to the toxicity of PMT, was exchangeable with a different signal motif for membrane targeting. The direct binding of the C1 domain with liposomes including phospholipids was also demonstrated. From these results, we conclude that the C1 domain functions to recruit PMT to the plasma membrane where the heterotrimeric GTPases, the substrates for the toxin, reside.
EXPERIMENTAL PROCEDURES
Plasmid Construction
Plasmids for Expression in Mammalian Cells
All the derivatives of pcDNA3-C1(4H)-EGFP and pcDNA3-TcdB(N4H)-EGFP were constructed by PCR cloning as follows. PCR was performed with a sense primer with a HindIII site for pcDNA3-C1(4H)-EGFP derivatives or a BamHI site for pcDNA3-TcdB(N4H)-EGFP and an antisense primer with a HindIII site (see supplemental Table S1). pPROEX-1-PMT (23) was used as the template DNA for construction of all the pcDNA3-C1(4H)-EGFP derivatives, and the genomic DNA of Clostridium difficile ATCC 17589 was used as the template for the construction of pcDNA3-TcdB(N4H)-EGFP. Consequently, each amplified DNA fragment was cloned into pCR2.1-TOPO TA (Invitrogen) and sequenced. The fragment containing the sequence of the PMT or tcd B gene was excised with HindIII and cloned into the HindIII site of pcDNA3-EGFP (24). The resultant plasmids were designated as shown in supplemental Table S1.
pcDNA3-C-PMT and pcDNA3-C-PMT ΔC1(4H) were constructed previously (23). pcDNA3-NM-C-PMT, pcDNA3-G2A-C-PMT, pcDNA3-NM-C-PMT ΔC1(4H), and pcDNA3-G2A-C-PMT ΔC1(4H) used in the reporter assay described later were also constructed by PCR cloning. PCR was performed with a sense primer with a BamHI site and an antisense primer (see supplemental Table S1) and with pPROEX-1-PMT (23) as the template DNA. Consequently, the amplified DNA fragment was cloned into pCR2.1-TOPO TA and sequenced. The DNA fragment with the sequence of C-PMT was excised with BamHI and XhoI and inserted into the BamHI-XhoI sites of pcDNA3 (Invitrogen).
Plasmids for Expression in E. coli
pPROEX-1-C-PMT was constructed previously (25). pPROEX-1-C-PMT ΔC1(4H) was constructed by PCR using the primers listed in supplemental Table S1 as follows. PCR was performed with a sense primer with a BamHI site and an antisense primer with an XhoI site for pPROEX-1-C-PMT (25). Consequently, the amplified DNA fragment was cloned into pCR2.1-TOPO TA and sequenced. The C-PMT ΔC1(4H) fragment with the sequence of the PMT gene was excised with BamHI and XhoI and inserted into the BamHI-XhoI sites of pPROEX-1 (Invitrogen). pPROEX-1-C-PMT for the C-PMT preparation used in the surface plasmon resonance analyses was newly constructed by inserting the BamHI-XhoI DNA fragment of C-PMT from pcDNA3-C-PMT (23) into the BamHI-XhoI site of pPROEX-1 so that the recombinant C-PMT expressed by the consequent plasmid had a spacer that exactly matched that of pPROEX-1-C-PMT ΔC1(4H) between the histidine tag and the N terminus of C-PMT. pGEX-C1(4H), which was used for expression of the GST-C1(4H) fusion protein, was constructed by PCR using the primers listed in supplemental Table S1. PCR was performed with a sense primer with a BamHI site and an antisense primer for pPROEX-1-PMT (23) as the template DNA. Consequently, the amplified DNA fragment was cloned into pCR2.1-TOPO TA and sequenced. The C1(4H) fragment with the correct sequence of the PMT gene was excised with BamHI and NotI and inserted into the BamHI-NotI sites of pGEX-4T3 (GE Healthcare).
Production of Recombinant Proteins in E. coli
All the recombinant proteins were produced by E. coli BL21-CodonPlus (DE3)-RIL (Stratagene, La Jolla, CA). pPROEX-1-C-PMT (25), the newly constructed pPROEX-1-C-PMT, and pPROEX-1-C-PMT ΔC1(4H) were used for the expression of C-PMTs and C-PMT ΔC1(4H). Recombinant C-PMT and C-PMT ΔC1(4H) were purified by affinity chromatography with nickel-nitrilotriacetic acid-agarose (Sigma-Aldrich) according to the manufacturer's instructions followed by DEAE-Sephacel (GE Healthcare) chromatography and Sephacryl S-200 (GE Healthcare) gel filtration. GST-C1(4H) from pGEX-C1(4H) and GST from pGEX-4T3 were purified by affinity chromatography with glutathione-Sepharose 4B (GE Healthcare) according to the manufacturer's instructions.
Cell Culture
293, 293T, and Swiss3T3 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and maintained at 37 °C under an atmosphere of 95% air, 5% CO2. Clones expressing C1(4H)-EGFP derivatives were selected from 293 cells transfected with each plasmid (shown in supplemental Table S1) with a selection marker (neomycin resistance gene) in the presence of G418 (Wako Chemicals). CHO-K1 cells were cultured in minimal essential medium-α supplemented with 10% fetal bovine serum and maintained at 37 °C under an atmosphere of 95% air, 5% CO2.
Fluorescence Microscopy
293, 293T, or CHO-K1 cells were seeded into 24-well plates containing glass coverslips (Matsunami). After incubation overnight, the cells were transfected with plasmids for the expression of C-PMT or C-PMT ΔC1(4H) (23) with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After 24 h, the cells were fixed with 3.0% paraformaldehyde in phosphate-buffered saline for 10 min, treated with 10% fetal bovine serum in phosphate-buffered saline for 10 min, and stained with a polyclonal anti-human CD46 rabbit antibody (H294, Santa Cruz Biotechnology, Santa Cruz, CA) and an Alexa Fluor® 546-conjugated anti-rabbit IgG antibody (Invitrogen). After permeabilization with 0.1% saponin in phosphate-buffered saline for 30 min, the cells were sequentially stained with the anti-PMT mouse monoclonal antibody 1B10 (23), an Alexa Fluor 488-conjugated anti-mouse IgG antibody (Invitrogen), and 4′,6-diamidino-2-phenylindole (Invitrogen). For the preparation of 293 cells constitutively expressing C1(4H)-EGFP variants, the cells were exposed to medium containing G418 after the transfection of each derivative plasmid. The immunolabeled cells and C1(4H)-EGFP variant-expressing cells were subjected to microscopy with an epifluorescence microscope (BX50, Olympus). All the images were captured and analyzed by SlideBook 4.0 (Roper Industries, Inc., Sarasota, FL) to control the fluorescence deconvolution microscopy.
Cell Fractionation
293T cells were seeded into 10-cm dishes. After an overnight incubation, the cells were transfected with plasmids for the expression of C-PMT or C-PMT ΔC1(4H) with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After a 48-h incubation, the cells were harvested, washed with ice-cold buffer (20 mm Hepes-NaOH, pH 7.4) containing a protease inhibitor mixture (Nacalai Tesque), suspended in and homogenized with the same buffer, and centrifuged at 1000 × g for 30 min at 4 °C. The supernatant was centrifuged again at 100,000 × g for 30 min at 4 °C, and the resultant supernatant and pellet suspended in the buffer were used as the cytosolic fraction and membranous fraction, respectively.
Reporter Assay
293T cells were seeded into 96-well plates and transfected with the aid of Lipofectamine 2000 (Invitrogen) with pcDNA3-C-PMT (23), pcDNA3-C-PMT ΔC1(4H) (23), pcDNA3-NM-C-PMT, pcDNA3-G2A-C-PMT, pcDNA3-NM-C-PMT ΔC1(4H), pcDNA3-G2A-C-PMT ΔC1(4H), or pcDNA3; pCDM8-β-gal, a β-galactosidase gene expression vector driven by the cytomegalovirus promoter (26); and p15xNFAT(MCIP1)-luc (27) or pSRE-luc (23) according to the manufacturer's instructions. After a 48-h incubation, the culture medium was discarded, and the cells were lysed with 100 μl of 1× Reporter Lysis Buffer (Promega, Madison, WI). The cell lysates were divided into two aliquots. One aliquot was used for the determination of luciferase activity with the Steady-Glo luciferase assay system (Promega). The relative light units of luciferase activity were measured with a GloMax 96 microplate luminometer (Promega). The other aliquot was used for the determination of β-galactosidase activity with the β-Galactosidase Enzyme Assay System with Reporter Lysis Buffer (Promega). The absorbance at 414 nm of β-galactosidase activity was measured with an enzyme-linked immunosorbent assay microtiter plate reader (Multiskan, Titertek). The value for the luciferase activity was divided by that for the β-galactosidase activity and normalized.
Measurement of Phospholipase C Activity
Swiss3T3 cells were seeded at 5 × 104 cells/well into a 24-well plate and incubated at 37 °C for 2 days. The cells were washed with inositol-free Dulbecco's modified Eagle's medium twice and labeled with myo-[3H]inositol (Moravek Biochemicals Inc.) at 37 kBq/ml in inositol-free Dulbecco's modified Eagle's medium containing 0.3% bovine serum albumin for 48 h. Cells were then washed twice with inositol-free Dulbecco's modified Eagle's medium containing 0.3% bovine serum albumin and 5 mm LiCl and treated with the HVJ (Hemagglutinating Virus of Japan) envelope vector (Ishihara Co. Ltd, Osaka, Japan) loaded with C-PMT or C-PMT ΔC1(4H) according to the manufacturer's instructions. The cells were incubated at 37 °C for 4 h after treatment with the toxins and lysed by incubation in 200 μl/well 0.1 m formic acid for 20 min at room temperature. The amount of total [3H]inositol phosphates was determined by the yttrium silicate scintillation proximity assay (28). Twenty microliters of cell extract was mixed with 80 μl of yttrium silicate scintillation proximity assay beads (GE healthcare) in water to give a final concentration of 1.0 mg of beads/ml in each well of white 96-well plates (Picoplate-96, PerkinElmer Life Sciences), and the plates were sealed with adhesive and clear plastic cover sheets (Topseal-A, PerkinElmer Life Sciences). The contents were mixed by shaking for 1 h. After the beads had settled for 2 h, the radioactivity of each well was measured by using a TopCount microplate scintillation counter (PerkinElmer Life Sciences).
Liposome Preparation
Egg phosphatidylcholine, egg phosphatidic acid (PA), egg phosphatidylethanolamine, brain phosphatidylserine (PS), liver phosphatidylinositol (PI), brain phosphatidylinositol 4,5-bisphosphate, egg phosphatidylglycerol (PG), and cholesterol were purchased from Sigma-Aldrich. A stock solution of lipids in chloroform was mixed in a 20-ml pear-shaped glass flask. The solvent was removed using a rotary evaporator or nitrogen purging. The lipid film was resuspended in 500 μl of phosphate-buffered saline and sonicated for 10 min with a water bath type sonicator. The lipid suspension was frozen in liquid nitrogen, thawed in a water bath at 30 °C, and vortexed for 30 s, a process that was repeated five times. The final suspension was extruded through a 50-nm pore-sized polycarbonate filter with a miniextruder (Avanti Polar Lipids, Alabaster, AL) 21 times. The liposome was stored at 4 °C and diluted with 10 mm Hepes pH 7.4 containing 150 mm NaCl to 0.5 mm just prior to use. The liposome composition was based on a background neutral lipid consisting of phosphatidylcholine (30–60%), phosphatidylethanolamine (20%), and cholesterol (20%). When indicated, anionic lipids such as PS, PG, PA, and PI were included at 30% at the expense of phosphatidylcholine.
Surface Plasmon Resonance Analysis
Surface plasmon resonance was measured with a Biacore2000 instrument and Pioneer Sensor L1 chip (GE Healthcare). The L1 chip was cleaned by a 5-min injection with 20 mm CHAPS and by a 5-min injection with 40 mm n-octyl β-glucoside and then coated with liposomes at 0.5 mm for 12 min in 10 mm Hepes, pH 7.4 containing 150 mm NaCl. These procedures were carried out at a flow rate of 5 μl/min. The unattached liposomes were washed out of the chip with 50 mm NaOH at a flow rate of 100 μl/min for 0.2 min. Nonspecific binding was probed by a 5-min injection of 0.1 mg/ml bovine serum albumin at a flow rate of 10 μl/min. Purified proteins were diluted with a running buffer of 10 mm Hepes, pH 7.4 containing 10 mm NaCl to the indicated concentration and perfused at a flow rate of 30 μl/min. The sample was allowed to associate for 3 min and then was dissociated for 4 min. The sensor surface was regenerated by 50 mm NaOH at a flow rate of 100 μl/min for 0.1 min.
Other Methods
The protein concentration in each sample was measured with a Micro BCA Protein Assay kit (Pierce). SDS-PAGE was carried out by the method of Laemmli (29) in a 7.5% polyacrylamide gel. For Western blotting, the samples in a gel after SDS-PAGE were electrophoretically transferred onto polyvinylidene difluoride membranes (Bio-Rad). The membranes were then treated with 5% skim milk, and the transferred proteins were probed with proper antibodies and visualized on Fuji Medical film (Fujifilm) with an enhanced chemiluminescence system according to the manufacturer's instructions (ECL Plus, GE Healthcare). Band intensity for images of Western blotting was determined by using NIH ImageJ 1.43r.3
RESULTS
C1 Domain of C-PMT Functions as Targeting Signal for Plasma Membrane
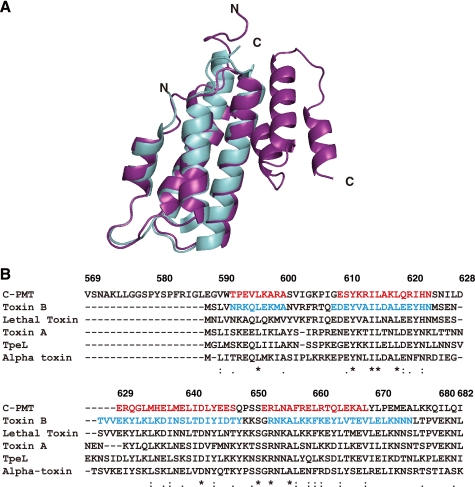
We have recently determined the crystal structure of C-PMT, which exhibits intracellular toxicity (23), and found that it was composed of three distinct domains, respectively named C1, C2, and C3. A DALI search of the database of protein three-dimensional structures (30) indicated significant structural homology between the N-terminal four helices (residues 590–670) of the C1 domain in C-PMT (C1(4H)) and the N-terminal first four helices (residues 1–100) of C. difficile toxin B (Fig. 1A). Furthermore, a PSI-BLAST search revealed that C1(4H) shares homology with the N-terminal regions of large clostridial cytotoxins including C. difficile toxin B with about 25% identity and about 40% similarity (Fig. 1B and Table 1).
FIGURE 1.
Homology of C1 domain in C-PMT to N termini of large clostridial cytotoxins. A, three-dimensional homology of the C1 domain to the N-terminal region of C. difficile toxin B. The C1 domain of C-PMT shown as a blue ribbon was superimposed on the N terminus of toxin B shown as a purple ribbon. B, alignment of amino acid sequences of the C1 domain of C-PMT and the N-terminal regions of large clostridial cytotoxins by ClustalW. Sequences for helices proved by the crystal structure are shown in red letters for the C1 domain of C-PMT and blue letters for toxin B. Toxin A (accession number A37052) and toxin B (accession number CAA63562) from C. difficile, lethal toxin (accession number CAA57959) from C. sordellii, α-toxin (accession number CAA88565) from Clostridium novyi, and TpeL (accession number BAF46125) from Clostridium perfringens are shown. The numbers above the alignment indicate the amino acid position of C-PMT. Identical amino acid residues are denoted by asterisks, highly conserved residues are denoted by double dots, and modestly conserved residues are denoted by dots. The accession numbers of amino acid sequences were obtained from NCBI.
TABLE 1.
Homology between C1 domain of C-PMT and large clostridial cytotoxins
The accession numbers of amino acid sequences were obtained from NCBI.
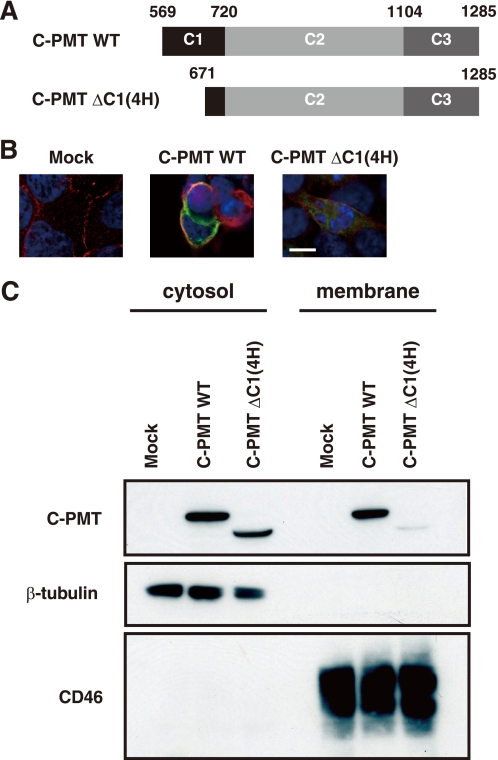
Among large clostridial cytotoxins, Clostridium sordellii lethal toxin and C. difficile toxin B were reported to have the ability to associate with anionic phospholipids through their N-terminal regions (31). This feature facilitated the enzymatic reactions of the toxins with the target molecule, hydrophobic geranylgeranylated Rho GTPases on the liposomes (31, 32). To examine whether the C1 domain, which is homologous to toxin B, has a similar ability, we constructed expression plasmids for C-PMT and a mutant C-PMT lacking the first four helices of the C1 domain (C-PMT ΔC1(4H), residues 672–1285) and localized the expressed proteins in 293T cells (Fig. 2). C-PMT but not C-PMT ΔC1(4H) was localized to the plasma membrane, which was labeled by the antibody against CD46, a typical plasma membrane protein. The same results were obtained in CHO-K1 cell-based experiments (supplement Fig. S2). Furthermore, Western blotting clearly showed the recruitment to the plasma membrane of C-PMT but not C-PMT ΔC1(4H) (Fig. 2C and supplemental Fig. S3). We further fractionated the plasma membrane and checked the distribution of C-PMT. C-PMT was not found in the fraction containing flotillin, a typical protein in the raft microdomain (data not shown). These results indicate that the C1 domain possesses a signal to lead the toxin molecule to the cell membrane excluding the raft microdomain.
FIGURE 2.
C1 domain of C-PMT targets plasma membrane. A, schematic representations of wild-type C-PMT (C-PMT WT) and the C-PMT variant deficient in the first four helices of the C1 domain (C-PMT ΔC1(4H)). B, intracellular localization of C-PMT wild type and C-PMT ΔC1(4H). C-PMT and C-PMT ΔC1(4H) were expressed in 293T cells and located by fluorescence microscopy (green). The plasma membrane region was visualized with anti-human CD-46 antibody (red), and the nucleus was visualized with 4′,6-diamidino-2-phenylindole (blue), respectively. Images are presented at the same magnification (scale bar, 10 μm). C, Western blot analysis for C-PMT wild type and C-PMT ΔC1(4H) in the cytoplasmic and membranous fractions of 293T cells. Equivalent amounts (10 μg protein/lane) of fractionated samples were subjected to 10% SDS-PAGE.
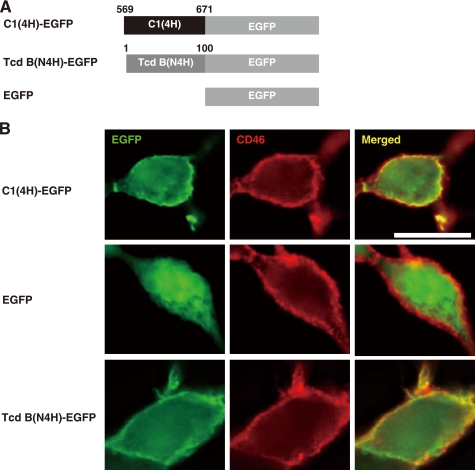
Next we examined whether the first four helices of C-PMT and the N terminus of large clostridial toxins lead a protein other than C-PMT to the plasma membrane. We prepared cells expressing EGFP tagged with C1(4H) (C1(4H)-EGFP) or the corresponding region of toxin B (TcdB(N4H)-EGFP) (Fig. 3A). Both proteins were co-localized with CD46 at the plasma membrane (Fig. 3B). The measurements of fluorescence intensity for C1(4H)-EGFP and TcdB(N4H)-EGFP showed maximum signals at cell-cell junctions compared with EGFP alone (supplemental Fig. S4).
FIGURE 3.
C1 domain and N terminus of toxin B function as membrane-targeting signals. A, schematic representations of C1(4H)-EGFP and TcdB(N4H)-EGFP. B, intracellular localization of C1(4H)-EGFP and TcdB(N4H)-EGFP. C1(4H)-EGFP and TcdB(N4H)-EGFP were expressed in 293 cells and located by fluorescence microscopy (green). The plasma membrane region was visualized with anti-human CD46 antibody (red). Images are presented at the same magnification (scale bar, 10 μm).
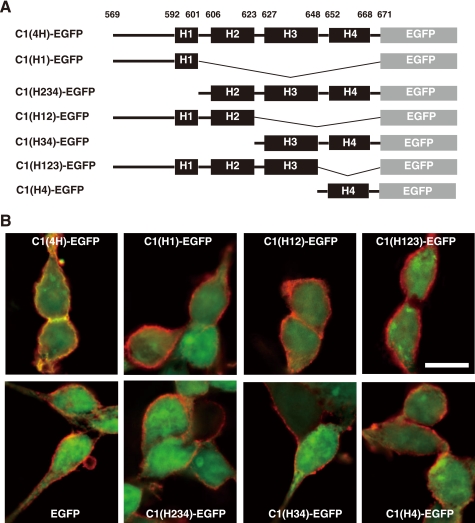
To know which of the four helices in C1(4H) is responsible for the membrane localization, we constructed six variants of C1(4H)-EGFP as shown in Fig. 4A and examined their distribution. When expressed in 293 cells, C1(4H)-EGFP but no other variant was localized to the membrane (Fig. 4B and supplemental Figs. S5 and S6). These results indicate that the plasma membrane-targeting function is not attributable to a specific helix in the C1 domain, and the overall structure of the four helices may be needed. Moreover, respective substitutions of eight of nine amino acid residues conserved between the C1 domain and the large clostridial cytotoxin family (Fig. 1B, asterisks) with other amino acids (alanine for Leu596, Tyr611, Ile614, Leu615, Leu618, Ser651, and Arg653 and glycine or valine for Ala656) abolished the membrane-targeting property (data not shown). These results also indicate that the overall structure of C1(4H) is presumably critical for the targeting of the plasma membrane.
FIGURE 4.
Structure of C1 domain in C-PMT is important to target plasma membrane. A, schematic representations of C1(4H)-EGFP variants. B, intracellular localization of the C1(4H)-EGFP variants. The C1(4H)-EGFP variants were expressed in 293 cells and located by fluorescence microscopy (green). The plasma membrane region was visualized with anti-human CD46 antibody (red). Images are presented at the same magnification (scale bar, 10 μm).
Toxicity of PMT Is Dependent on Plasma Membrane-targeting Signal of C1 Domain
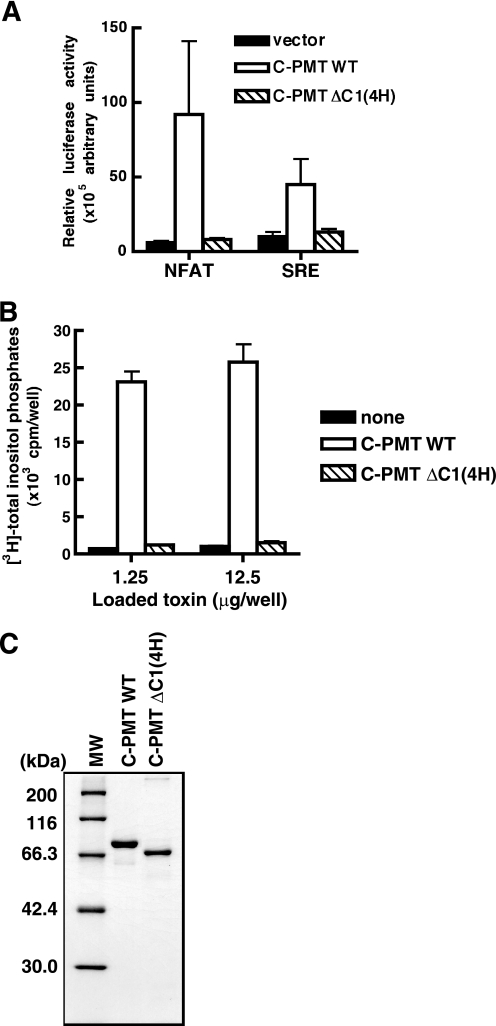
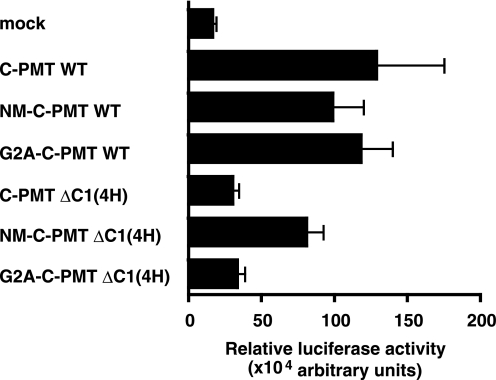
To understand the role of the C1 domain in the actions of PMT, we compared toxic activities of C-PMT and C-PMT ΔC1(4H). For this purpose, we performed a luciferase reporter assay with serum response factor and NFAT, transcription factors that are known to be activated by PMT through the heteromeric GTPases Gq and G12/13. C-PMT was active when ectopically expressed in the cells, consistent with our previous finding that recombinant C-PMT stimulated transcription of the reporters when introduced into the cytosol by HVJ liposomes (23). However, C-PMT ΔC1(4H) did not show activity (Fig. 5A). Phospholipase assays also revealed the inactivity of C-PMT ΔC1(4H) (Fig. 5, B and C, and supplemental Fig. S1B). These results indicate that C1(4H) plays some role in the toxicity probably through its membrane-targeting activity. To elucidate this, we constructed a variant of C-PMT ΔC1(4H) tagged with an N-myristoylation peptide, which functions as a membrane-targeting signal, and examined whether it is substitutable for C1(4H). As expected, the N-myristoylation peptide tag conferred the membrane-targeting characteristics on the tagged proteins (supplemental Fig. S7). In addition, the N-myristoylation peptide tag, but not its unmyristoylated analogue, G2A, made C-PMT ΔC1(4H) active when produced in the cells (Fig. 6 and supplemental Fig. S8). Thus, C1(4H) is considered to function as the membrane-targeting signal for PMT.
FIGURE 5.
C1 domain is required for PMT activity. A, NFAT- and SRE-luciferase reporter assays in 293T cells expressing either C-PMT wild type (WT) or C-PMT ΔC1(4H). 293T cells were co-transfected with the plasmids for C-PMTs and either NFAT- or SRE-luciferase. The luciferase activity in the cells was determined 48 h after the transfection as described under “Experimental Procedures.” B, stimulation of phospholipase C activity by C-PMT. Swiss3T3 cells, which had been labeled with myo-[3H]inositol for 48 h, were treated with the HVJ envelope vector containing 1.25 or 12.5 μg of C-PMT (residues 569–1285), or C-PMT ΔC1(4H) (residues 671–1285), and intracellular [3H]inositol phosphates, which are the products of the enzymatic action of phospholipase C, were measured as described under “Experimental Procedures.” Each data point represents the mean of at least triplicate measurements; the error bar shows S.D. Representative results from three independent experiments are shown. C, SDS-PAGE of purified recombinant C-PMT and C-PMT ΔC1(4H). One microgram of each recombinant protein was applied to a lane and visualized by Coomassie Brilliant Blue staining.
FIGURE 6.
N-Myristoylation peptide tag can functionally compensate for deletion of C1(4H). The effect of the N-myristoylation peptide-tagged C-PMT ΔC1(4H) on the NFAT-luciferase assay of 293T cells is shown. 293T cells were co-transfected with C-PMTs and NFAT-luciferase. The luciferase activity in the cells was determined 48 h after the transfection as described under “Experimental Procedures.” Each data point represents the mean of at least triplicate measurements; the error bar shows S.D. Representative results from three independent experiments are shown. NM, the N-myristoylated peptide tag (MGSSKSKPKVSN); G2A, an unmyristoylated analogue of the N-myristoylated peptide tag in which the second residue glycine is replaced by alanine. WT, wild type.
Direct Binding between C1 domain and Liposomes Containing Phospholipids Detected by Surface Plasmon Resonance
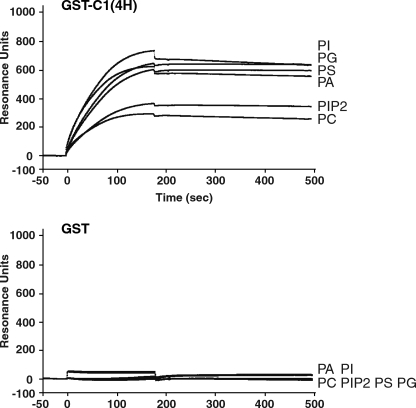
To detect the direct binding of the C1 domain to phospholipids, which compose the plasma membrane, we applied the recombinant C-PMT and C-PMT ΔC1(4H) to surface plasmon resonance analyses with the liposome-coated sensor chips. It was shown that C-PMT directly bound to various phospholipids, whereas C-PMT ΔC1(4H) did not (supplemental Fig. S9). GST-tagged C1(4H) (GST-C1(4H)) but not GST also bound to the liposomes as well as C-PMT. Both C-PMT and GST-C1(4H) bound to the liposomes containing PS, PG, PA, or PI more apparently than those containing phosphatidylinositol 4,5-bisphosphate and phosphatidylcholine without PS, PG, PA, or PI (Fig. 7, upper panel, and supplemental Fig. S9).
FIGURE 7.
C1 domain interacts with phospholipids. Binding of recombinant GST-C1(4H) (upper panel) to phospholipids was compared with binding of GST alone (lower panel) by surface plasmon resonance analyses. Purified recombinant GST-C1(4H) (100 nm) or GST (100 nm) was injected over the chip coated with the liposomes containing 30% PS, PG, PI, phosphatidylinositol 4,5-bisphosphate (PIP2), or PA. The composition of the liposomes was described under “Experimental Procedures.” PC, phosphatidylcholine.
DISCUSSION
In this study, we found that C1(4H) of the C1 domain functions as a membrane-targeting signal to recruit the PMT molecule to the cytoplasmic face of the plasma membrane. C1(4H) was also essential for intoxicating target cells. These results indicate that PMT, after entering the cytoplasm, needs to be delivered by C1(4H) beneath the plasma membrane where its target molecules should reside. Recently, it was reported that the toxin directly targets and deamidates Gαi and Gαq, the α subunits of heterotrimeric GTPases (15). Because the α subunits are known to be located on the cell membrane and transduce signals from the heptahelical transmembrane receptors to downstream pathways, it is conceivable that C1(4H) recruitment of PMT to the plasma membrane is essential for the actions of the toxin. This is supported by the finding that the N-myristoylation peptide tag could substitute for C1(4H), a result that also suggests that C1(4H) is not involved in the intoxication process other than in the membrane targeting.
C1(4H) shows homology with the N termini of C. difficile toxin B and related large clostridial cytotoxins in amino acid sequence (Fig. 1). Notably, the tertiary structures of C1(4H) and the N terminus of toxin B are quite similar. The N-terminal four helices of toxin B, like C1(4H), serve as the plasma membrane-targeting signal. Substitutions of amino acid residues conserved among C1(4H) and large clostridial cytotoxins abolished the membrane-targeting property of C1(4H). These results imply that C1(4H) and the large clostridial cytotoxins exert their membrane-targeting ability through the same mechanism. Of the large clostridial cytotoxins, C. sordellii lethal toxin (LT) was intensively analyzed for its ability to bind phospholipids, the major components of the biomembrane (31). Mesmin et al. (31) reported that the N-terminal region of LT bound to anionic phospholipids such as phosphatidylserine better than to neutral lipids. They presumed that the specificity for phosphatidylserine, which is more abundant in the cytoplasmic leaflet of cellular membranes, could be advantageous to LT for targeting its substrates, the membrane-associated small GTPases. In addition, they suggested that the basic residues in the N-terminal 18-amino acid region of LT were important for interaction with the anionic phospholipids. However, toxin B, which also has small GTPases as substrates, did not show a clear preference for phosphatidylserine probably because its N terminus is less basic than that of LT. In this study, C1(4H), like toxin B, was found to bind almost equally to any phospholipid and to be essential for the intracellular toxicity of PMT. Thus, in the case of PMT and toxin B, specificity in binding phosphatidylserine does not seem to be important.
The N-terminal 18 amino acids of LT that were reported to be essential for the interaction with phospholipids correspond to the first helix of the N termini of the large clostridial cytotoxins and the C1 domain of PMT. We demonstrated that the other three helices were also necessary for the membrane targeting: deletion mutants in which any helices of C1(4H) were missing lost the membrane-targeting property. Therefore, it is likely that the overall structure of C1(4H) is necessary for the membrane targeting. This may be the case for the large clostridial cytotoxins. C1(4H) and the N terminus of toxin B form the typical four-helix bundle. This characteristic structure per se may function to associate with phospholipids.
Previously, we demonstrated that the crystal structure of C-PMT is intracellularly active and revealed that it consists of three distinct domains, C1, C2, and C3. We evidenced that the C3 domain with a thiol protease-like catalytic triad organizes the enzymatic core, the nature of which was recently found to be a deamidase for the α subunits of heterotrimeric GTPases (15). This study demonstrated that the C1 domain functions as the membrane-targeting signal for PMT. However, the function of the C2 domain remains unknown. The C2 domain is composed of two α/β-folds, the structure of which is known to provide a biochemically active site in various functional proteins. Because C-PMT is considered to represent the unit necessary and sufficient for the intracellular action of PMT, the C2 domain should play some role in the target cells. In general, it is known that toxins modifying intracellular target molecules bind to their receptors on the cell surface, are internalized into the cells by receptor-mediated endocytosis, enter the cytoplasm crossing the membrane of the endocytotic vesicles, and eventually reach substrate molecules. During these procedures, the C2 domain may exert its function. For example, it was reported that Vibrio cholerae RTX (repeats-in-toxin) and C. difficile toxins A and B underwent autoproteolytic cleavage to deliver their catalytic domains into the cytosol of target cells (33–35). These autocleavages were accomplished by specific domains in the toxin molecules. Although it is unknown whether this is the case for PMT, it may be that any autocleavage of PMT is conducted by the C2 domain. To elucidate the true function of the C2 domain, we are now dissecting its tertiary structure in detail.
The N-terminal region of PMT is believed to be responsible for binding to an unidentified cell surface receptor. The domain that translocates at least the C-terminal catalytic domain from the endocytotic vesicles to the cytosol was reportedly located in the region spanning residues 402–457 (16). However, direct evidence demonstrating the role of each domain of the N-terminal region of PMT is not sufficient. We are also trying to determine the crystal structure of the full-length PMT, which should provide a complete understanding of the structure-function relationship of the toxin.
Supplementary Material
Acknowledgments
We greatly appreciate the gift of p15xNFAT(MCIP1)-luc plasmids from Dr. Eric Olson (University of Texas Southwestern Medical Center, Dallas, TX). We thank Tomoko Suzuki for secretarial assistance.
Addendum
After this manuscript had been submitted for publication, Geissler et al. (36) reported C1-like membrane-targeting domains that were conserved among many bacterial toxins. Although both their data and ours demonstrate the membrane-targeting function of the domains, its mechanism remains unknown. Analyses of the molecular interaction between the domains and phospholipid membranes may be required to clarify this issue.
This work was supported in part by grants-in-aid for Scientific Research from the Ministry of Education, Culture, Science, and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S9 and Table S1.
W. S. Rasband (1997–2009), ImageJ, National Institutes of Health.
- PMT
- P. multocida toxin
- SRE
- serum response element
- NFAT
- nuclear factor of activated T-cells
- GST
- glutathione S-transferase
- PA
- phosphatidic acid
- PS
- phosphatidylserine
- PI
- phosphatidylinositol
- PG
- phosphatidylglycerol
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- EGFP
- enhanced green fluorescent protein
- LT
- lethal toxin.
REFERENCES
- 1.Higgins T. E., Murphy A. C., Staddon J. M., Lax A. J., Rozengurt E. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4240–4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullan P. B., Lax A. J. (1996) Infect. Immun. 64, 959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rozengurt E., Higgins T., Chanter N., Lax A. J., Staddon J. M. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nougayrède J. P., Taieb F., De Rycke J., Oswald E. (2005) Trends Microbiol. 13, 103–110 [DOI] [PubMed] [Google Scholar]
- 5.Essler M., Hermann K., Amano M., Kaibuchi K., Heesemann J., Weber P. C., Aepfelbacher M. (1998) J. Immunol. 161, 5640–5646 [PubMed] [Google Scholar]
- 6.Lacerda H. M., Lax A. J., Rozengurt E. (1996) J. Biol. Chem. 271, 439–445 [DOI] [PubMed] [Google Scholar]
- 7.Murphy A. C., Rozengurt E. (1992) J. Biol. Chem. 267, 25296–25303 [PubMed] [Google Scholar]
- 8.Seo B., Choy E. W., Maudsley S., Miller W. E., Wilson B. A., Luttrell L. M. (2000) J. Biol. Chem. 275, 2239–2245 [DOI] [PubMed] [Google Scholar]
- 9.Staddon J. M., Barker C. J., Murphy A. C., Chanter N., Lax A. J., Michell R. H., Rozengurt E. (1991) J. Biol. Chem. 266, 4840–4847 [PubMed] [Google Scholar]
- 10.Wilson B. A., Zhu X., Ho M., Lu L. (1997) J. Biol. Chem. 272, 1268–1275 [DOI] [PubMed] [Google Scholar]
- 11.Orth J. H., Lang S., Aktories K. (2004) J. Biol. Chem. 279, 34150–34155 [DOI] [PubMed] [Google Scholar]
- 12.Orth J. H., Lang S., Taniguchi M., Aktories K. (2005) J. Biol. Chem. 280, 36701–36707 [DOI] [PubMed] [Google Scholar]
- 13.Zywietz A., Gohla A., Schmelz M., Schultz G., Offermanns S. (2001) J. Biol. Chem. 276, 3840–3845 [DOI] [PubMed] [Google Scholar]
- 14.Orth J. H., Fester I., Preuss I., Agnoletto L., Wilson B. A., Aktories K. (2008) J. Biol. Chem. 283, 23288–23294 [DOI] [PubMed] [Google Scholar]
- 15.Orth J. H., Preuss I., Fester I., Schlosser A., Wilson B. A., Aktories K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7179–7184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baldwin M. R., Lakey J. H., Lax A. J. (2004) Mol. Microbiol. 54, 239–250 [DOI] [PubMed] [Google Scholar]
- 17.Busch C., Orth J., Djouder N., Aktories K. (2001) Infect. Immun. 69, 3628–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orth J. H., Blöcker D., Aktories K. (2003) Biochemistry 42, 4971–4977 [DOI] [PubMed] [Google Scholar]
- 19.Pullinger G. D., Sowdhamini R., Lax A. J. (2001) Infect. Immun. 69, 7839–7850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward P. N., Miles A. J., Sumner I. G., Thomas L. H., Lax A. J. (1998) Infect. Immun. 66, 5636–5642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falbo V., Pace T., Picci L., Pizzi E., Caprioli A. (1993) Infect. Immun. 61, 4909–4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oswald E., Sugai M., Labigne A., Wu H. C., Fiorentini C., Boquet P., O'Brien A. D. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 3814–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitadokoro K., Kamitani S., Miyazawa M., Hanajima-Ozawa M., Fukui A., Miyake M., Horiguchi Y. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5139–5144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanajima-Ozawa M., Matsuzawa T., Fukui A., Kamitani S., Ohnishi H., Abe A., Horiguchi Y., Miyake M. (2007) Infect. Immun. 75, 565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyazawa M., Kitadokoro K., Kamitani S., Shime H., Horiguchi Y. (2006) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 906–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasai Y., Kageyama R., Tagawa Y., Shigemoto R., Nakanishi S. (1992) Genes Dev. 6, 2620–2634 [DOI] [PubMed] [Google Scholar]
- 27.Yang J., Rothermel B., Vega R. B., Frey N., McKinsey T. A., Olson E. N., Bassel-Duby R., Williams R. S. (2000) Circ. Res. 87, E61–E68 [DOI] [PubMed] [Google Scholar]
- 28.Brandish P. E., Hill L. A., Zheng W., Scolnick E. M. (2003) Anal. Biochem. 313, 311–318 [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 30.Holm L., Sander C. (1993) J. Mol. Biol. 233, 123–138 [DOI] [PubMed] [Google Scholar]
- 31.Mesmin B., Robbe K., Geny B., Luton F., Brandolin G., Popoff M. R., Antonny B. (2004) J. Biol. Chem. 279, 49876–49882 [DOI] [PubMed] [Google Scholar]
- 32.Reinert D. J., Jank T., Aktories K., Schulz G. E. (2005) J. Mol. Biol. 351, 973–981 [DOI] [PubMed] [Google Scholar]
- 33.Sheahan K. L., Cordero C. L., Satchell K. J. (2007) EMBO J. 26, 2552–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reineke J., Tenzer S., Rupnik M., Koschinski A., Hasselmayer O., Schrattenholz A., Schild H., von Eichel-Streiber C. (2007) Nature 446, 415–419 [DOI] [PubMed] [Google Scholar]
- 35.Egerer M., Giesemann T., Jank T., Satchell K. J., Aktories K. (2007) J. Biol. Chem. 282, 25314–25321 [DOI] [PubMed] [Google Scholar]
- 36.Geissler B., Tungekar R., Satchell K. J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 5581–5586 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.