Abstract
The superoxide-generating NADPH oxidase complex of resting phagocytes includes cytochrome b559, a membrane-associated heterodimer composed of two subunits (Nox2 and p22phox), and four cytosolic proteins (p47phox, p67phox, Rac, and p40phox). Upon stimulation, the cytosolic components translocate to the membrane, as the result of a series of interactions among the cytosolic components and among the cytosolic components and cytochrome b559 and its phospholipid environment. We described the construction of a tripartite chimera (trimera) consisting of strategic domains of p47phox, p67phox, and Rac1, in which interactions among cytosolic components were replaced by fusion (Berdichevsky, Y., Mizrahi, A., Ugolev, Y., Molshanski-Mor, S., and Pick, E. (2007) J. Biol. Chem. 282, 22122–22139). We now fused green fluorescent protein (GFP) to the N terminus of the trimera and found the following. 1) The GFP-p47phox-p67phox-Rac1 trimera activates the oxidase in amphiphile-dependent and -independent (anionic phospholipid-enriched membrane) cell-free systems. 2) Geranylgeranylation of the GFP-trimera makes it a potent oxidase activator in unmodified (native) membranes and in the absence of amphiphile. 3) Prenylated GFP-trimera binds spontaneously to native membranes (as assessed by gel filtration and in-line fluorometry), forming a tight complex capable of NADPH-dependent, activator-independent superoxide production at rates similar to those measured in canonical cell-free systems. 4) Prenylation of the GFP-trimera supersedes completely the dependence of oxidase activation on the p47phox phox homology domain and, partially, on the Rac1 polybasic domain, but the requirement for Trp193 in p47phox persists. Prenylated GFP-p47phox-p67phox-Rac1 trimera acts as a quintessential single molecule oxidase activator of potential use in high throughput screening of inhibitors.
Keywords: Cytochrome b, Oxidase, Oxygen Radicals, Phagocytosis, Phosphatidylcholine, Phosphatidylglycerol, Superoxide Ion, isoprenylation, p47phox, p67phox
Introduction
One of the most effective mechanisms for the destruction of pathogenic microorganisms is the production of reactive oxygen species, acting independently or in cooperation with granule-derived stored enzymes (reviewed in Ref. 1). All oxygen radicals are derived from superoxide (O2⨪),2 which is produced in response to stimuli typically represented by contact of membrane receptors with microorganisms and/or their opsonins (reviewed in Ref. 2). O2⨪ is generated by the NADPH-derived one-electron reduction of molecular oxygen catalyzed by a membrane-associated 91-kDa protein, known as gp91phox or Nox2, associated with a second membrane-bound subunit, p22phox, to form the cytochrome b559 heterodimer. Nox2 contains the NADPH-binding site and two redox stations (bound FAD and two hemes), generating a redox gradient along which electrons are carried from NADPH to oxygen. Despite the availability of both NADPH and oxygen, there is no electron flow in Nox2 in resting phagocytes; this is elicited as a consequence of the above-mentioned stimuli by signal transduction from membrane receptors to Nox2. Nox2 “activation” is mediated by four cytosolic components, p47phox, p67phox, Rac (1 or 2), and p40phox, which translocate to the membrane environment of Nox2 to form the active O2⨪ producing NADPH oxidase complex (briefly “oxidase”), a process known as oxidase assembly (reviewed in Refs. 3–6). Oxidase assembly can be divided into three stages as follows. The first stage consists of protein-protein interactions among the cytosolic components, in preparation for membrane translocation. The second stage includes cytosolic protein-membrane phospholipids and protein interactions, not involving Nox2, in preparation for the third and final stage of cytosolic components-Nox2 interaction.
The first stage involves interactions between p67phox and p47phox, based on a tail to tail interaction between a proline-rich region (PRR), at the C terminus of p47phox, and the C-terminal Src homology 3 (SH3) domain of p67phox (7), and between p67phox and Rac in the GTP-bound form, based on the interaction between the tetratricopeptide repeat domain in p67phox and pre-switch I and switch I regions in Rac (8, 9). There is evidence for Rac-GTP serving not only as a carrier for p67phox but also inducing a conformational change in p67phox, essential for its interaction with Nox2 (10). The second stage interactions include that between p47phox and p22phox, consequent to the substitution of the auto-inhibitory intramolecular bond between the SH3 tandem and a polybasic region, within p47PHOX, for an intermolecular bond between the same SH3 tandem and a PRR, at the C terminus of p22phox (11–13). Trp193, in the N-terminal SH3 domain of p47phox, is an essential residue in the mediation of this interaction (13). Yet another second stage interaction is that between the phox homology (PX) domain at the N terminus of p47phox (residues 4–121) and specific phosphoinositides in the membrane (14). A key second stage mediator is Rac, which contains two potent membrane-localizing elements, represented by the polybasic region at its C terminus (residues 183–188), and the geranylgeranyl tail. The first has affinity for anionic phospholipids on the cytosolic face of the membrane, based on electrostatic interaction, whereas the second is inserted into the lipid bilayer and is attached by hydrophobic forces (15). Prenylation compensates quite successfully for membrane attachment of Rac under conditions of reduced electrostatic attraction, but optimal attachment requires the participation of both mechanisms (10, 15, 16). In the view championed in this study, the third stage is represented predominantly by the interaction of p67phox with Nox2, with p47phox and Rac, seen as accessory molecules, acting as carriers of p67phox to the membrane vicinity of Nox2 or as stabilizers of the Nox2-p67phox bond. The main alternative to this is the proposal of a direct involvement of Rac in the induction of electron flow in Nox2, based on the evidence for binding of Rac2 to Nox2, involving the insert region of Rac (residues 124–135) (17–19). Contradictory data on the role of the insert region of Rac in oxidase activation left this issue unsettled (16, 20, 21).
A novel approach to the study of protein-protein interactions in oxidase assembly is the design of chimeric constructs, consisting of selected segments from two or three cytosolic components (22). Fusion of cytosolic components into a single molecule supersedes the need for first stage interactions and should allow a direct approach to second and third stage processes. The first such chimera was a p47phox-p67phox fusion protein (23), and this was followed by the independent description by two groups of p67phox-Rac1 chimeras (20, 24), which could be prenylated at the C terminus (16). We recently described the design and bacterial expression of a tripartite chimera consisting of selected segments of p47phox, p67phox, and Rac1, which we called “trimera” (25). Trimeras were found to act as potent amphiphile-dependent oxidase activators upon addition to native phagocyte membrane or purified cytochrome b559. Supplementation of the membrane with anionic phospholipids enabled oxidase activation by the trimera in the absence of amphiphile.
We now describe the design of a p47phox-p67phox-Rac1 trimera to which enhanced green fluorescent protein (GFP) was fused, to allow assessment of its binding to membrane liposomes. In the nonprenylated form, amphiphile-independent oxidase activation by the trimera required supplementation of the membrane with anionic phospholipid and was correlated with membrane association based on electrostatic forces. Upon prenylation, the trimera became a potent spontaneous (amphiphile-independent) oxidase activator in native membranes, an ability correlated with a high degree of membrane association, predominantly hydrophobic in nature.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
The synthetic phospholipids 1,2-dioleoyl-sn-glycero-3-(phospho-rac-(1-glycerol)) (sodium salt, synthetic, >99%, product 840475P) and 1,2-dioleoyl-sn-glycero-3-phosphocholine (synthetic, >99%, product 850375P) were obtained from Avanti Polar Lipids. Common laboratory chemicals were purchased from Sigma.
Construction of Recombinant Fluorescent Chimeric Proteins
We designed and expressed six recombinant chimeric proteins in which functionally important parts of the cytosolic NADPH oxidase components p47phox, p67phox, and Rac1 were joined in a tetrameric construct with “enhanced GFP” from Aequorea Victoria, the latter serving as a fluorescent tag to allow detection and quantification of the chimera. The present chimeras were based on those we recently described, which lacked a fluorescent tag (25). The chimeras include full-length GFP (residues 1–238; Clontech); residues 1–286 of p47phox, or parts of it; residues 1–212 of p67phox; and full-length Rac1 (residues 1–192), or parts of it. The composition of the six chimeras, which we named “GFP-trimeras,” is illustrated schematically in Fig. 1. As an initial step toward the construction of GFP-trimeras, we introduced the gfp gene into plasmid pET-30a (Novagen), creating suitable restriction sites for subsequent subcloning of GFP-trimera cDNAs C-terminal to the gfp gene. The gfp gene was generated by PCR with a sense primer 5′-NdeI-His6-NcoI-GFP 5′-GCAATCCATATGCACCATCATCATCATCATGGTTCCATGGTGAGCAAGGGCGAGGAGCTG-3′ and an antisense primer 3′-GFP-PstI-thrombin site-BamHI 5′- GCTCTCGGATCCACGCGGAACCAGACCAGACTGCAGCTTGTACAGCTCGTCCATGCCGAG-3′. Plasmid pgfpN1 (Clontech) was used as a template. This procedure introduced NdeI-NcoI restriction sites, separated by a stretch of six histidines, to the 5′ terminus of the gfp gene and PstI and BamHI restriction sites, separated by a thrombin cleavage site, to its 3′ terminus and resulted in the construction of plasmid pET-30a-His6 tag-gfp. The His6 tag-GFP-trimera Rac1 Q61L, in which Gln61 in Rac1 was substituted by leucine, was constructed by the subcloning of a BamHI-EcoRI fragment from plasmid pGEX-2T-trimera RAC1 Q61L into plasmid pET-30a-His6 tag-gfp. The plasmid pGEX-2T-trimera RAC1 Q61L was constructed by subcloning the AgeI-EcoRI fragment from plasmid pGEX-2T-chimera 3 (20), bearing a Q61L mutation in the RAC1 moiety, into plasmid pGEX-2T-prototype trimera (25). The His6 tag-GFP-trimera Rac1 Q61L was called “GFP-trimera” and the plasmid pET-30a-gfp-trimera was used as a platform for the construction of mutant GFP-trimeras that contained substitutions or deletions in the p47phox and Rac1 segments. The cDNA of His6 tag-gfp-trimera p47PHOX ΔPX domain (lacking the PX domain of p47PHOX) was constructed by subcloning of a 175-bp BamHI-digested fragment of a p47PHOX-(151–286) PCR fragment, amplified from plasmid pGEX-2T-trimera p47PHOX ΔPX domain (25), by applying a sense primer 5′-pGEX 5′-GGGCTGGCAAGCCACGTTTGGTG-3′ and an antisense primer 3′-p47PHOX-(280–286)-Stop-EcoRI 5′-GGGAGGGAAAGAATTCTTAGTCTTGCCCCGACTTTTGCAGG-3′ into the BamHI site of plasmid pET-30a-GFP-trimera. The cDNA of His6 tag-gfp-trimera p47PHOX W193R (in which Trp193 in p47PHOX was substituted by Arg) was generated by PCR with a sense primer 5′-BamHI-NcoI-p47PHOX (1–9) 5′-TACTACGGATCCATGGGTGACACCTTCATCCGTCACATCG-3′ and antisense primer 3′- p47PHOX-(280–286)-Stop-EcoRI. Plasmid pET-28a-trimera p47PHOX W193R (25) was used as a template. The PCR fragment was digested with BamHI, and a 622-bp DNA fragment was subcloned into the BamHI site of plasmid pET-30a-gfp-trimera. The cDNA of His6 tag-gfp-trimera RAC1 Δinsert region, in which the full-length Rac1 segment had the insert region (residues 124–135) deleted, was generated by subcloning of a 248-bp Bsu36I- and EcoRI-digested DNA fragment from plasmid pET-28a-trimera RAC1 Δinsert domain (25) into plasmid pET-30a-gfp-trimera digested with Bsu36I and EcoRI. The His6 tag-gfp-trimera RAC1 → RAC2 cDNA, in which the Rac1 polybasic region was substituted by that of Rac2, was constructed by subcloning of a 284-bp Bsu36I- and EcoRI-digested DNA fragment from plasmid pGEX-2T-trimera RAC1 → RAC2 (25) into plasmid pET-30a-GFP-trimera, digested with Bsu36I and EcoRI. The cDNA for the His6-gfp-trimera RAC1 Gln183–Gln188, in which all six amino acids comprising the Rac1 polybasic domain were substituted by glutamine, was constructed by subcloning of a 284-bp Bsu36I and EcoRI-digested DNA fragment from plasmid pGEX-2T-trimera RAC1 Gln183–Gln188 (25) into plasmid pET-30a-gfp-trimera, digested with Bsu36I and EcoRI. The PfuUltra® II Fusion HS DNA polymerase (Stratagene) was used in all PCRs. The reaction conditions were as follows: 94 °C for 2 min, followed by 30 cycles each of 95 °C for 20 s, 55 °C for 20 s, and 72 °C for 30 s, and finally 72 °C for 3 min. Digestion by restriction endonucleases (New England Biolabs), DNA purification from agarose gel, plasmid, and DNA fragment purifications after enzymatic reactions (using HiYield Gel/PCR DNA fragments extraction kit, and HiYield plasmid mini kit, respectively, both from RBC Real Biotech Corp.), and molecular cloning, were performed according to the standard procedures described in Ref. 26 or as recommended by the suppliers. Throughout the entire work, the correctness and integrity of p47PHOX, p67PHOX, and RAC1 genes were confirmed by DNA sequencing. It should be noted that all gfp-trimera cDNAs contained the Q61L mutation in the RAC1 segment, thus ensuring that Rac1 was in its GTP-bound, active form, eliminating the need for GDP to GTP exchange.
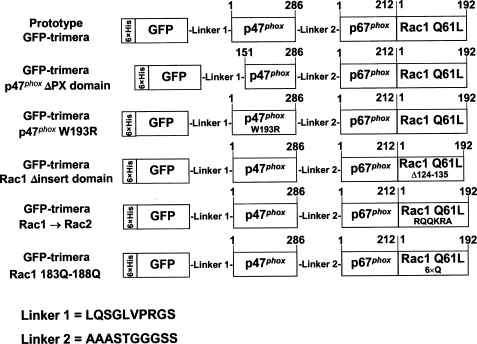
FIGURE 1.
Schematic representation of the prototype GFP-trimera and mutant GFP-trimeras. The component of origin of the segment incorporated in the GFP-trimeras appears within the rectangles. The fluorescent segment (GFP) is preceded by a polyhistidine tag. Above the rectangles are indicated the residue numbers in the native sequences of p47phox, p67phox, and Rac1, representing the N- and C-terminal limits of the segments forming the trimeras. Mutations within the different segments are indicated inside the rectangles. LQSGLVPRGS and AAASTGGGSS are 10-amino acid linkers between the GFP and p47phox segments (linker 1) and between the p47phox and p67phox segments (linker 2), respectively.
Expression and Purification of Recombinant Proteins
The recombinant proteins used in this work were expressed in and isolated from E. coli Rosetta (DE3, pLysS) (Novagen). All were expressed as N-terminal His6-tagged fusion proteins and purified by metal chelate affinity chromatography. Briefly, the expression vector pET-30a (Novagen) carrying cDNAs encoding the above-listed gfp-trimeras or gfp only was introduced into E. coli cells, and bacteria were induced with 0.4 mm isopropyl β-d-thiogalactopyranoside at 18 °C for 14–16 h. The induced cells were suspended in lysis buffer (20 mm sodium phosphate, 0.5 m NaCl, and 20 mm imidazole, pH 7.4) supplemented with “Complete” EDTA-free protease inhibitor (Roche Applied Science) and 1% Triton X-100 (Sigma). The bacteria were disrupted by stirring the suspension in an ice-cooled beaker with a magnetic bar for 30 min at room temperature followed by sonication, using a 500-watt sonic disruptor (Vibra Cell, Sonics and Materials) at 20% amplitude for 5 min in the 50% pulse mode in an ice-cooled beaker. The bacterial lysate was subjected to centrifugation at 27,000 × g for 25 min at 4 °C, and the cleared cell-free extract was filtered through a 0.45-μm cellulose acetate filter (Whatman). The material was applied to nickel-Sepharose 6 Fast Flow beads (GE Healthcare), and binding was allowed to proceed in the batch mode for 60 min on a rotary mixer at room temperature. The beads were washed with lysis buffer without Triton X-100, followed by two washes with washing buffer (20 mm sodium phosphate, 0.5 m NaCl, and 40 mm imidazole, pH 7.4). The protein bound to the beads was eluted from the resin with elution buffer (20 mm sodium phosphate, 0.5 m NaCl, and 300 mm imidazole, pH 7.4) at a volume corresponding to five times the volume of the beads. The purified proteins were supplemented with 30% glycerol and stored at −75 °C.
Further Purification of GFP-Trimeras
To improve the level of purity of the proteins, all GFP-trimeras purified on nickel-Sepharose were subjected to further purification by gel filtration on a Superdex 75 Hiload 16/60 fast protein liquid chromatography (FPLC) column. 10-mg amounts of GFP-trimera were injected into the column, and chromatography was performed with a buffer consisting of 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 5 mm MgCl2, on a high pressure liquid chromatography (HPLC) system (Waters) at a flow rate of 1 ml/min. 1-ml fractions were collected. Both absorbance (at 280 nm) and the fluorescent signal derived from GFP (excitation = 488 nm; emission = 511 nm) were measured continuously by passing the column eluate through a diode array detector (MD-1510, Jasco) and a spectrofluorometer (model FP-750, Jasco) fitted with an HPLC flow cell (MFC-132, Jasco). In the vast majority of separations, three absorbance peaks were found, all of which also exhibited a fluorescent signal (results not shown). Fractions making up the first peak were analyzed by SDS-PAGE, and those in which the GFP-trimera band (corresponding to a molecular mass of close to 108 kDa, for all GFP-trimeras with the exception of GFP-trimera p47phox ΔPX domain, which was 90.8 kDa in size) was present and represented 90% or more of all the detected proteins were pooled, supplemented with 30% glycerol and protease inhibitor mixture (Complete, EDTA-free), and stored in small aliquots at −75 °C until use.
Enzymatic Prenylation of GFP-Trimeras
Nonprenylated GFP-trimeras were geranylgeranylated in vitro by recombinant mammalian geranylgeranyltransferase type I. Recombinant human geranylgeranyltransferase type I was produced in Sf9 cells co-infected with baculoviruses encoding the two subunits of the enzyme and purified as described previously (27). Enzymatic prenylation was performed as described before (16), with minor changes. A typical reaction mixture contained 5 nmol of recombinant GFP-trimera, 10 nmol of geranylgeranyl pyrophosphate (Sigma), and 5 units of geranylgeranyltransferase type I (1 unit transfers 1 nmol of geranylgeranyl to protein/h) in 1 ml of 50 mm Tris-HCl, pH 7.7, 5 mm MgCl2, 50 μm ZnCl2, 2 mm dithiothreitol, and 10 μm GDP. The mixture was supplemented with octyl β-d-glucopyranoside (octyl glucoside) to a final concentration of 7 mm and incubated for 6 h at 25 °C in an orbital mixer (Thermomix Comfort, Eppendorf), at 500 rpm. The mixture was then cooled on ice and subjected to sonication by 5 cycles of 10 s each in an ultrasonic processor (Vibra Cell, 400-watt, fitted with a cup horn; Sonics and Materials), at 60% amplitude, to break up protein aggregates, which appeared occasionally. Aggregates, resistant to sonication, were removed by centrifugation at 12,000 × g for 30 min at 4 °C. Finally, glycerol was added to a final concentration of 20%, and the geranylgeranylated GFP-trimeras were stored at −75 °C until use. Prenylation mixtures consisted of up to 30 nmol of GFP-trimera, and the amounts of geranylgeranyl pyrophosphate and geranylgeranyltransferase were adjusted accordingly, to conserve the substrate/reagents ratio described for the 5-nmol GFP-trimera unit.
Assessing the Success of Prenylation
This was done by phase separation in Triton X-114, as described in Ref. 28. Briefly, aliquots from the prenylation reaction were supplemented with Triton X-114 to a final concentration of 1% and kept at 4 °C for 30 min, with occasional mixing. Next, the mixture was incubated at 37 °C for 10 min in a Thermomix Comfort heating block, without mixing, followed by centrifugation at 11,600 × g for 5 min at room temperature in an Eppendorf fixed angle microcentrifuge. The prenylated trimera partitioned into the lower Triton X-114 layer (10% of the total volume), and the nonprenylated trimera was present in the upper aqueous layer (90% of the total volume). The contents of the two layers were brought to an equal volume and subjected to SDS-PAGE, and the extent of prenylation was estimated from the relative intensities of the bands present in the two layers.
Preparation of Prenylated GFP-Trimera Free of Nonprenylated Trimera
Prenylation of GFP-trimeras was never complete, and the need arose for the removal of the nonprenylated material to ensure that the experimental results were due exclusively to the prenylated trimera. This was achieved by gel filtration on a Superdex 200 Hiload 16/60 FPLC column, based on the observation that the prenylated GFP-trimera eluted as a large molecular weight peak well ahead of and clearly separated from the nonprenylated fraction. 2-mg amounts of prenylated GFP-trimera (=18.5 nmol) were injected into the column, and chromatography was performed with the Tris-HCl buffer also used for the purification of the nonprenylated trimera, supplemented with 10 mm octyl glucoside and 10% v/v glycerol on an HPLC system (Waters), at a flow rate of 1 ml/min. 1-ml fractions were collected, and those corresponding to the center of the large molecular weight peak were pooled and used in experiments in which prenylated GFP-trimera, free of nonprenylated material, was required.
Determination of Protein Concentration
The concentration of recombinant proteins was estimated by the method of Bradford (29), modified for use with 96-well microplates, using the Bio-Rad protein assay dye reagent concentrate and bovine γ-globulin as a standard.
Preparation of Macrophage Membrane Liposomes
Phagocyte membranes were prepared from guinea pig macrophages obtained by injection of mineral oil into the peritoneal cavity, as described previously (30). The membranes were solubilized in 40 mm octyl glucoside and reconstituted into liposomes by dialysis against detergent-free buffer, as described previously (31). The cytochrome b559 heme content of membrane liposomes was measured by the difference spectrum of sodium dithionite-reduced minus oxidized samples (32).
Preparation of Membrane Liposomes Supplemented with Exogenous Phospholipids
Native macrophage membranes were supplemented with exogenous phospholipids to generate membrane liposomes with an artificially modified electrical charge (anionic or neutral). For this purpose, we first dissolved 1,2-dioleoyl-sn-glycero-3-phosphocholine or 1,2-dioleoyl-sn-glycero-3-(phospho-rac-(1-glycerol)) at a concentration of 5 mm in the buffer, which was also used to solubilize macrophage membranes (31), according to a procedure described previously (33, 34). The buffer consisted of 120 mm potassium/sodium phosphate buffer, pH 7.4, 1 mm MgCl2, 2 mm NaN3, 1 mm EGTA, 1 mm dithiothreitol, 10 μm FAD, 20% v/v glycerol, and 40 mm octyl glucoside. The phospholipids were added to the solubilized macrophage membranes at a ratio of 4 volumes of phospholipid (5 mm) to 1 volume of membrane (at a concentration equivalent to 1.2 μm cytochrome b559 heme). The membrane/phospholipid mixtures were dialyzed against 100–200 volumes of octyl glucoside-free buffer, for 18 h at 4 °C, which led to the formation of phospholipid-enriched membrane liposomes. The concentration of cytochrome b559 in the modified liposomes was measured and was typically found to be close to 0.24 μm cytochrome b559 heme. The theoretical final concentration of exogenous phospholipids (1,2-dioleoyl-sn-glycero-3-phosphocholine and 1,2-dioleoyl-sn-glycero-3-(phospho-rac-(1-glycerol))) was 4 mm. The total concentration of phospholipids in the native unmodified membranes was determined as described previously (33).
Preparation of Membrane Liposomes Supplemented with Exogenous Phospholipid Deprived of Cytochrome b559
Cytochrome b559 was removed from membrane preparations based on its ability to bind to heparin (35). To 1.5 ml of solubilized membrane containing 40 mm octyl glucoside (1.2 μm cytochrome b559 heme) were added 0.4 ml of packed heparin-agarose gel (type I, 750–1000 μg/ml, Sigma, product number H-6508), and the mixture was kept for 14–16 h at 4 °C with continuous rotary mixing. The mixture was centrifuged for 2 min at 600 × g, and the cytochrome b559 content of the supernatant was measured. If the cytochrome b559 content was above 10% of the original amount, the supernatant was subjected to yet another cycle of absorption by heparin-agarose by exposure to the 0.4 ml of heparin gel for 3 h at 4 °C. Two rounds of absorption were sufficient for the removal of more than 95% of the cytochrome b559 from the membrane. The cytochrome b559-depleted supernatant was brought to the volume of the original solubilized membrane (1.5 ml) and used for enrichment with exogenous 1,2-dioleoyl-sn-glycero-3-(phospho-rac-(1-glycerol)), followed by preparation of liposomes, as described above.
Assessment of the Capability of GFP-Trimeras to Bind to Membrane Liposomes of Defined Composition by In-line Fluorescence Recording
In these experiments, membrane liposomes of defined composition, containing the equivalent of 100 pmol of cytochrome b559 heme, were mixed with 500 pmol of either nonprenylated or prenylated GFP-trimeras (or mutants of these), and the total volume was brought to 0.5 ml. The following types of liposomes were used: native membrane liposomes; membrane liposomes enriched in neutral phospholipid (phosphatidylcholine (PC)); membrane liposomes enriched in anionic phospholipid (phosphatidylglycerol (PG)), and membrane liposomes enriched in anionic phospholipid (PG), deprived of cytochrome b559. The phospholipid content of liposomes consisting of native membrane, corresponding to 100 pmol of cytochrome b559 heme, was 0.26–0.29 μmol. The phospholipid content of membrane liposomes enriched in anionic or neutral phospholipids, at a ratio of 4 volumes of phospholipid (5 mm) to 1 volume of membrane (1.2 μm cytochrome b559 heme), was 1.92–1.95 μmol per 100 pmol of cytochrome b559 heme, of which 1.66 μmol was exogenous phospholipid. The mixtures were incubated for 5 min at 25 °C on an orbital mixer and were injected into a Superose 6 10/300 FPLC gel filtration column. Chromatography was performed with a buffer consisting of 50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, and 150 mm NaCl on an HPLC system at a flow rate of 0.2 ml/min at 4 °C, and 0.6-ml fractions were collected. Both absorbance (at 280 and 413 nm) and the fluorescent signal (excitation = 488 nm, emission = 511 nm) were measured continuously as described under “Further Purification of GFP-Trimeras.” The areas of the fluorescent peaks, representing free and liposome-bound trimeras, were measured by the Spectra Manager software (Jasco). Absorbance at 413 nm was an indicator of the presence of heme and served as proof for the presence of and as a rough measure of the amount of liposome-associated cytochrome b559.
Cell-free NADPH Oxidase Activation Assay
Activation of the oxidase in vitro was assessed by measuring the NADPH-dependent O2⨪ production in a semi-recombinant cell-free system, as described previously (36). Two cell-free systems were utilized as follows: (a) an amphiphile-dependent system, consisting of membrane liposomes (equivalent to 5 nm cytochrome b559 heme) and GFP-trimera, in the presence of 100 μm lithium dodecyl sulfate (LiDS), and (b) an amphiphile-independent system consisting of membrane liposomes (5 nm cytochrome b559 heme) and GFP-trimera, in the absence of LiDS. Nonprenylated GFP-trimeras were tested in both amphiphile-dependent and -independent systems. Prenylated GFP-trimeras (not freed of the nonprenylated fraction) were tested only in the amphiphile-independent system, in which nonprenylated trimeras are incapable of activation (25). All trimeras were tested at concentrations ranging from 0 to 320 nm in 96-well microplates in a total volume of 200 μl of assay buffer (37) per well, by incubation for 90 s, in the presence or absence of LiDS, before the addition of 240 μm NADPH, to initiate O2⨪ production. This was quantified by the kinetics of cytochrome c reduction, as described previously (37).
Curve Plotting
Plotting of dose-response curves was performed using GraphPad Prism version 5.00.
Assessment of the Ability of Membrane Liposome-GFP-Trimera Complexes to Produce O2⨪ in a Cell-free System
40-μl aliquots of fractions (individual or pooled) collected from Superose 6 gel filtration columns, on which membrane liposome/GFP-trimera mixtures were separated, were added to 160 μl of assay buffer in 96-well microplates. O2⨪ production was initiated by the addition of 240 μm NADPH and was quantified as described above. No LiDS was added, whether the fractions originated from the separation of membrane liposome complexes with nonprenylated or prenylated trimeras.
Immunodetection of GFP-Trimera in Complex with Membrane Liposome following Isolation by Gel Filtration
Aliquots from four 0.6-ml fractions, corresponding to the void volume (6–8.4 ml) of gel filtration experiments on Superose 6 columns in which membrane liposome/GFP-trimera mixtures were separated, were assayed for the presence of GFP-trimera bound to liposomes. Quantification was by a kinetic ELISA. 100-μl volumes from each of the four fractions were added to 96-well microplates (Immulon 4 HBX, ultra-high binding polystyrene, part number 3855, Thermo Labsystems). The proteins were allowed to adhere to the wells for 18 h at 4 °C with continuous mixing on an orbital shaker, and the unattached protein was removed by washing the wells with phosphate-buffered saline, consisting of 5.7 mm sodium/potassium buffer, pH 7.3, 137 mm NaCl, and 2.7 mm KCl, supplemented with 0.05% (v/v) Tween 20 (Sigma). Next, the wells were filled with 300 μl of blocking buffer (phosphate-buffered saline supplemented with 0.1% (v/v) Tween 20 and 1% (w/v) casein sodium salt (Sigma)), and the plates were kept for 1 h at 25 °C. Because all the GFP-trimeras possessed a His6 tag, quantification of the GFP-trimera present in the fractions was performed by using a mouse monoclonal anti-polyhistidine antibody conjugated with horseradish peroxidase (Sigma, product number A7058). This was added to the wells at a dilution of 1:4000 in blocking buffer, and the plates were kept for 1 h at 25 °C with continuous mixing on an orbital shaker. The bound peroxidase-conjugated antibody was quantified by the addition of 3,3′,5,5′- tetramethylbenzidine + substrate chromogen (Dako Cytomation, product number S1599), and the linear section of the increase in absorbance at 650 nm was measured in a Spectramax 190 microplate reader in the kinetic mode (Molecular Devices). Results were expressed as absorbance at 650 nm × 1000/min.
RESULTS
Rationale for the Design of Tripartite p47phox-p67phox-Rac1 Chimeras with an Intrinsic Fluorescent Tag and a Potential for Prenylation
In an earlier study, we described the generation of a single molecule that served as an oxidase activator, represented by a tripartite p47phox-p67phox-Rac1 chimera (25). The basic construct, which was termed “prototype trimera,” was composed of the PX domain and the two SH3 domains of p47phox (comprising residues 1–286), the tetratricopeptide repeat and activation domains of p67phox (comprising residues 1–212), and full-length Rac1 (residues 1–192). The trimera was found to activate the oxidase in its natural membrane habitat in a cell-free system in the presence of an anionic amphiphile, but activation became amphiphile-independent upon enrichment of membranes with anionic phospholipids. Mutants were generated to ascertain the importance of particular regions or residues in protein-protein and protein-phospholipid interactions, essential for the assembly of a functional oxidase complex. The results of this earlier study pointed to a key role of the membrane phospholipid environment in oxidase activation. In this study, we focused on the design of a variant of the prototype trimera by fusing it with a fluorescent protein (GFP), via its N terminus, and by adding an isoprenyl group to the C terminus of the Rac1 part. These modifications had two important consequences as follows: first, binding to the membrane could be readily followed and quantified, and second, the presence of the isoprenyl group ensured that in vitro experiments resemble the in vivo reality, in which Rac exists exclusively in the prenylated form. GFP was fused to the N terminus of the prototype trimera by means of a 10-residue linker (Fig. 1). A second 10-residue linker was present between the p47phox and p67phox parts of the trimera, as in the original construct. A further change from the original construct was the introduction of a Q61L mutation in the Rac1 part of the trimera, ensuring that this was constitutively in the GTP-bound form. This modification was intended to ensure that an intramolecular bond was established between the Rac1 and p67phox parts of the trimera, a requirement for oxidase activating ability of both tripartite chimeras (25) and p67phox-Rac1 chimeras (10, 20, 22). A significant characteristic “inherited” from the parent trimera was the truncation of the p47phox part at residue 286, which removes the C-terminal region containing the polybasic segment, involved in an auto-inhibitory loop with the SH3 tandem (11–13), and recently also shown to participate in the prevention of PX domain-phospholipid interaction (38). This is likely to make the trimera more reactive with p22phox and the PX target phosphatidylinositol 3,4-bisphosphate.
All GFP-trimeras had an N-terminal His6 tag, preceding the GFP part, allowing easy purification by metal affinity chromatography. In addition to the prototype GFP-trimera, GFP-p47phox-(1- 286)-p67phox-(1–212)-Rac1(Q61L),with an Mr of 108,530, five variants were constructed, modeled on the trimeras described before (25). This group included proteins in which the following mutations, deletions, or sequence exchanges were performed: 1) deletion of residues 1–150 from the p47phox part, resulting in removal of the PX domain (Mr = 90,838); 2) introducing a W193R mutation in the p47phox part (Mr = 108,500); 3) deletion of the “insert domain” (residues 124–135) in the Rac1 part (Mr = 107,103); 4) exchanging residues 183–188 in the Rac1 part for the corresponding residues in Rac2 (Mr = 108,473); and 5) replacing residues 183–188 in the Rac1 part by six glutamines (Mr = 108,474). All these recombinant proteins were successfully produced in E. coli and purified to above 90% homogeneity. Their purity and molecular mass were confirmed by SDS-PAGE analysis and found to correspond to the theoretical values listed above (results not shown). As will be described below and in Fig. 4A, the native molecular dimensions of the prototype and mutant trimeras, in the nonprenylated form, were determined by gel filtration on a Superose 6 FPLC column and found to correspond closely to the expected values, proving that the proteins behaved as monomers in solution.
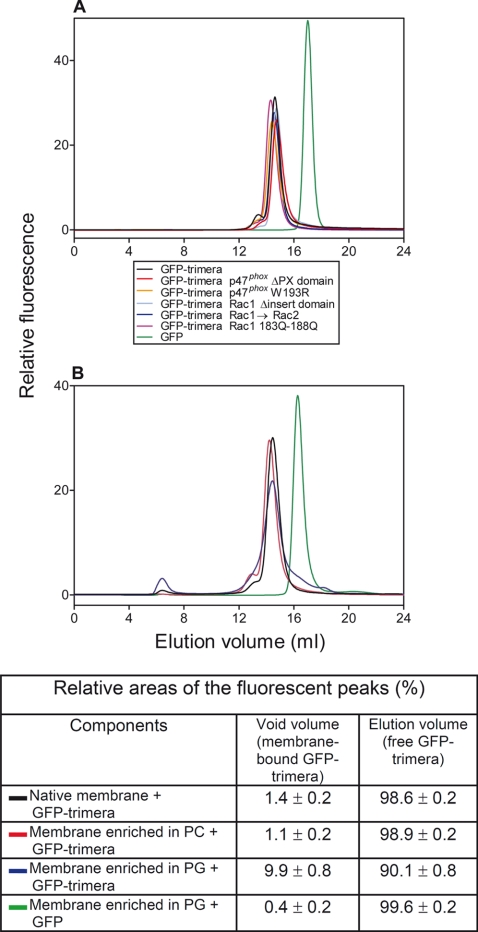
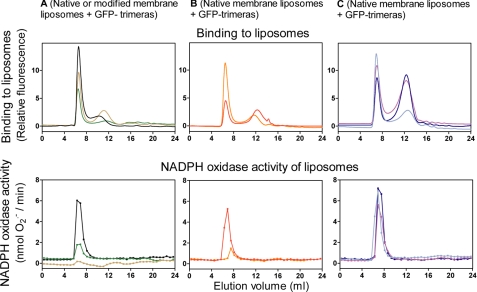
FIGURE 4.
Quantification of nonprenylated GFP-trimeras in solution and bound to macrophage membrane liposomes of various compositions by gel filtration and in-line detection by a fluorescence detector. A, 500 pmol of GFP-trimera or GFP (in a volume of 0.5 ml) were subjected to gel filtration on a Superose 6 column, and the eluates were monitored in-line for the fluorescent signal (excitation = 488 nm; emission = 511 nm). The following proteins were examined: GFP-trimera (black line); GFP-trimera p47phox ΔPX domain (red line); GFP-trimera p47phox W193R (orange line); GFP-trimera Rac1 Δinsert domain (light blue line); GFP-trimera Rac1 → Rac2 (blue line); GFP-trimera Rac1 Gln183–Gln188 (purple line), and GFP (green line). B, membrane liposomes of various lipid compositions, at an amount equivalent to 100 pmol of cytochrome b559 heme, were mixed with 500 pmol of prototype GFP-trimera or GFP, in a total volume of 0.5 ml, and preincubated for 5 min at 25 °C. The following mixtures were then subjected to gel filtration on a Superose 6 FPLC column: native membrane + GFP-trimera (black line); membrane enriched in PC + GFP-trimera (red line); membrane enriched in PG + GFP-trimera (blue line), and membrane enriched in PG + GFP (green line). The eluates were monitored in-line for the fluorescent signal. The 1st peak (void volume) represents GFP-trimeras or GFP bound to membrane liposomes, and the 2nd peak represents free GFP-trimeras or GFP. Calculations were based on the integration of the areas of the peaks. A and B illustrate one characteristic group of experiments each out of three performed. The table below B shows the numerical results of the type of experiment illustrated in B, representing the means ± S.E. of 3–5 experiments, for each combination of membrane liposomes and GFP-trimera or GFP.
GFP-Trimeras Support NADPH Oxidase Activation in the Presence of Amphiphile in the Same Manner as Their Non-GFP-fused Counterparts
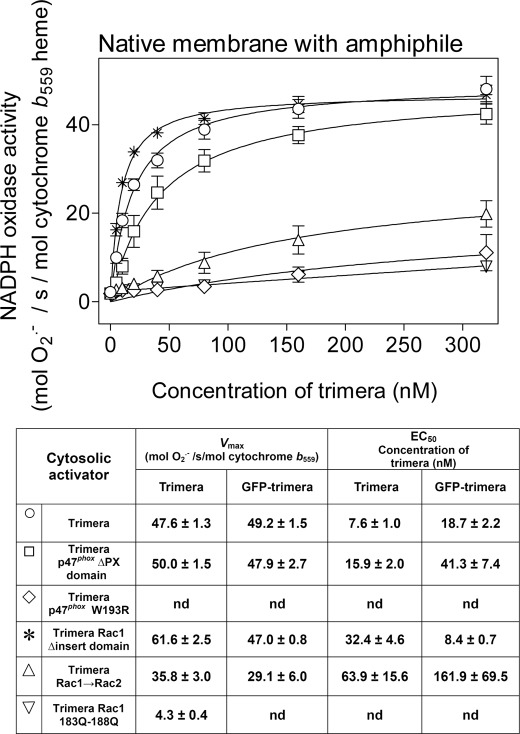
A precondition for using the GFP-fused trimeras as model compounds was to ascertain that they are functionally comparable with trimeras lacking the GFP part. It should be noted that the prototype GFP-trimera differs from the trimera lacking GFP not only by molecular mass (108.5 versus 81.5 kDa) but also by pI (6.41 versus 7.24). Also, the GFP-trimera was designed to be constitutively in the GTP-bound form, by virtue of the Q61L mutation in the Rac1 part, whereas the original trimera had to be exchanged to a nonhydrolyzable GTP analog for optimal activity. We have shown that the p47phox-(1–286)-p67phox-(1–212)-Rac1-(1–192) trimera was an efficient oxidase activator in the canonical amphiphile-dependent cell-free system, when added to phagocyte membrane liposomes (25). Using the same experimental conditions, the prototype GFP-trimera possessed an oxidase activating ability that was similar to that of the trimera lacking GFP, as evident by the comparison of the kinetic parameters, Vmax and EC50 (Fig. 2). Exchanging Rac1 residues 183–188 for the corresponding Rac2 residues, resulting in a reduction from six to three positively charged residues, and the exchange of all six positively charged residues to neutral glutamines led to a marked reduction in oxidase activating ability, more pronounced with the Rac1 Gln183–Gln188 mutant (Fig. 2). An equally pronounced reduction in activating ability was caused by a W193R mutation in the N-terminal SH3 domain of the p47phox part. Deletion of the p47phox PX domain and of the Rac1 insert domain had no significant influence on the Vmax, and deletion of the Rac1 insert domain even resulted in a moderate enhancing effect, expressed in a modest lowering of the EC50 (Fig. 2). These results show that the kinetic attributes of the mutant GFP-trimeras were similar to those of their counterparts lacking GFP, providing a secure basis for utilizing GFP-trimeras throughout this investigation.
FIGURE 2.
Amphiphile-dependent NADPH oxidase activation by nonprenylated GFP-trimeras. The ability of prototype GFP-trimera and its mutants to activate the NADPH oxidase in the presence of an anionic amphiphile was assayed in a cell-free system consisting of native membrane, at a concentration equivalent to 5 nm cytochrome b559 heme and 100 μm LiDS. Each one of the following GFP-trimeras (all at concentrations varying from 0 to 320 nm) was added to the reaction as follows: prototype GFP-trimera (○); GFP-trimera p47phox ΔPX domain (□); GFP-trimera p47phox W193R (◇); GFP-trimera Rac1 Δinsert domain (*); GFP-trimera Rac1 → Rac2 (▵), and GFP-trimera Rac1 Gln183–Gln188 (▿). O2⨪ production was initiated by the addition of NADPH (240 μm) and measured as described under “Experimental Procedures.” The table below the graph offers a comparison of the kinetic parameters of NADPH oxidase activation by the prototype and mutant nonprenylated GFP-trimeras with that of the corresponding nonprenylated trimeras (lacking GFP), subjected to nucleotide exchange to GMPPNP, as described in Ref. 25 (nd = not determinable). All trimeras were assayed on native membrane in the presence of LiDS. The results represent the means ± S.E. of 3–6 experiments, for each type of chimera.
In the Absence of Amphiphile, Enrichment of Membranes in Anionic Phospholipids Is Mandatory for NADPH Oxidase Activation by Nonprenylated GFP-Trimera
As expected from work done with trimeras lacking GFP, the prototype GFP-trimera and its mutants were incapable of activating the oxidase, as represented by native membrane liposomes, in the absence of amphiphile (Fig. 3A). Also, in accordance with earlier work, enriching the native membrane with exogenous neutral phospholipid (represented by PC) did not enable amphiphile-free activation (Fig. 3B). Enrichment of the membrane with the anionic phospholipid PG resulted in a dose-dependent activation of the oxidase by the prototype GFP-trimera in the absence of amphiphile, as found before with the trimera lacking GFP (Fig. 3C). GFP-trimeras Rac1 Gln183–Gln188, p47phox W193R, Rac1 → Rac2, and p47phox ΔPX domain exhibited deficient activating ability, in decreasing order of severity. A comparison of these results with those of amphiphile-dependent oxidase activation using native membrane (Fig. 2) clearly indicated the pronounced similarity of the two activating mechanisms, with the negative charge of the membrane acting as an alternative for the exogenous anionic amphiphile. In both situations, a paramount element required for successful oxidase activation was an intact Rac1 polybasic domain at the C terminus of the trimera, emphasizing the centrality of electrostatic protein-lipid interaction between the negatively charged membrane and the positively charged C terminus of the cytosolic activator. Another shared element was the requirement for an intact Trp193 in the N-terminal SH3 domain of the p47phox part, involved in direct protein-protein interaction with the p22phox subunit of cytochrome b559 in the membrane (13).
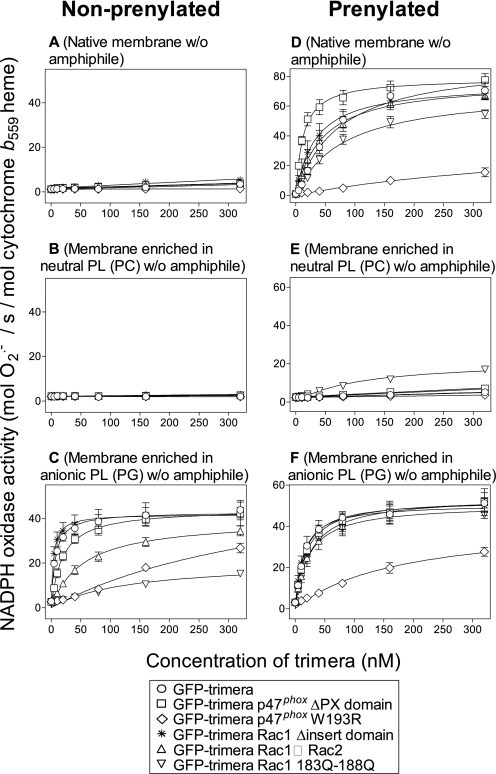
FIGURE 3.
Comparison of NADPH oxidase activating ability of nonprenylated versus prenylated GFP-trimeras in an amphiphile-independent cell-free system including membrane liposomes of various phospholipid composition. The ability to support NADPH oxidase activation by nonprenylated or prenylated prototype GFP-trimera and mutants was assayed in a cell-free system containing one of the following types of membrane liposomes: native membrane (A and D), membrane supplemented with PC (B and E), or membrane supplemented with PG (C and F). To each of these were added the following GFP-trimeras: prototype GFP-trimera (○); GFP-trimera p47phox ΔPX domain (□); GFP-trimera p47phox W193R (◇); GFP-trimera Rac1 Δinsert domain (*); GFP-trimera Rac1 → Rac2 (▵), and GFP-trimera Rac1 Gln183–Gln188 (▿). O2⨪ production was assessed in the absence of amphiphile as described under “Experimental Procedures.” The prenylated trimera preparations were not freed of nonprenylated material. The results represent the means ± S.E. of 3–10 experiments for each combination of membrane liposomes and GFP-trimera (prototype or mutant).
Prenylation of GFP-Trimera Endows It with the Ability to Activate the NADPH Oxidase in the Native Membrane Environment and in the Absence of an Amphiphilic Activator and Diminishes the Differences in Activating Abilities between Native and Mutant Trimeras
One of the drawbacks of studying oxidase assembly in the cell-free system is the uncertainty about the applicability of the findings to the in vivo reality. This can be remedied in part by the introduction of oxidase components in a form similar to that present in the intact cell, as exemplified by membrane liposomes of controllable lipid composition (31) and by the use of prenylated Rac (33, 34, 39–41) and prenylated p67phox-Rac1 chimeras (10, 16) as activators. The availability of recombinant geranylgeranyltransferase I (27) and the introduction of a simple method of in vitro prenylation (42) greatly facilitated the production of prenylated Rac and its application to fusion proteins comprising Rac in a C-terminal location. In this work, we applied this method to prenylate the prototype GFP-trimera and the mutants derived from it. As described under “Experimental Procedures,” prenylation of GFP-trimeras required some changes from the technique used for Rac1 and for p67phox-Rac1 chimeras (16); these consisted of prolonging the incubation with geranylgeranyltransferase I from 90 min to 6 h and reducing the temperature from 37 to 25 °C. The prenylated GFP-trimeras were found to have the expected sizes, when assessed under denaturing conditions (SDS-PAGE; results not shown). As will be described below, prenylation of GFP-trimera was never complete, and preparations usually contained a certain amount of nonprenylated trimera. For experiments involving quantification of binding of prenylated trimera to membrane liposomes, we prepared prenylated trimera free of the nonprenylated contaminant, as described under “Experimental Procedures.”
We examined the ability of prenylated GFP-trimeras to activate the oxidase in the absence of an amphiphilic activator. Because the trimera, in the nonprenylated form, was incapable of activating the oxidase under these conditions, the experiments described in this section were performed with prenylated trimera preparations not freed of nonprenylated material. As seen in Fig. 3D, the prenylated GFP-trimera activated the oxidase in liposomes of native membrane in the absence of amphiphile, in sharp contrast with the inability of the nonprenylated trimera to do the same (Fig. 3A). Enriching membrane liposomes with exogenous neutral phospholipid (PC) abrogated oxidase activation by prenylated trimera (Fig. 3E). On the other hand, membrane liposomes supplemented with exogenous anionic phospholipid (PG) served as a suitable, albeit not superior to the native, environment for oxidase activation by prenylated trimera (Fig. 3F). Prenylation also overcame partially (native membranes) or completely (anionic phospholipid-enriched membrane) the negative effect of mutations on oxidase activating ability by the trimera, with the exception of mutation W193R in the p47phox part. Thus, comparing the dose-response curves illustrated in Fig. 3F (prenylated trimeras; membrane enriched in PG) with those in Fig. 3C (nonprenylated trimeras; membrane enriched in PG) revealed that only the W193R mutation reduced oxidase activation by the prenylated trimera, whereas four out of five mutants exhibited various degrees of reduced activating ability by the nonprenylated trimera. A compensatory effect of prenylation for loss of trimera function, due to the same mutations, was also apparent with native membranes (Fig. 3D compared with Fig. 2). In summary, prenylation was found capable of partially or fully overcoming inadequate protein-membrane lipid interactions, based on charge, but not failures in protein-protein interactions such as that resulting from the p47phox W193R mutation, the target of which is p22phox.
Fusion of GFP to Trimera Enables In-line Detection and Quantification by Gel Filtration
The results described above point to an important role for membrane attachment of the trimeras in oxidase activation. The GFP-trimeras were designed to enable us to separate and quantify free and membrane liposome-bound trimera. In earlier work, we utilized for this purpose 2′-(or 3′)-O-(N-methylanthraniloyl) (mant)-labeled guanine nucleotides (mant-GTP or mant-GDP) to prepare fluorescent Rac (33, 34, 39, 40) or Rac-containing chimeras (20). Fusion of GFP has the advantages of the fluorescent label being an intrinsic part of the protein and possessing high intensity fluorescent emission. The prototype nonprenylated GFP-trimera, its five mutants, and isolated GFP protein were subjected to gel filtration on a Superose 6 column, and the eluate was monitored in-line for fluorescence and absorbance. As apparent in Fig. 4A, all GFP-trimeras eluted as sharp peaks, at positions corresponding to their molecular mass. Free GFP eluted as a symmetrical peak, corresponding to its molecular mass of 30 kDa. Membrane liposomes eluted within the void volume of the column (6–8.4 ml), indicating a Mr equal to or in excess of 4 × 107. We next mixed nonprenylated GFP-trimera with three types of membrane liposomes (native, enriched in PC, and enriched in PG), at the ratios described under “Experimental Procedures,” and subjected the mixtures to gel filtration. A mixture of PG-enriched membrane and GFP served as a negative control. As apparent in Fig. 4B and the accompanying table, less than 1.5% of the prototype GFP-trimera was bound to native membrane liposomes, an amount that was further decreased by enriching the membrane with neutral phospholipid (PC). Enrichment of membrane with anionic phospholipid (PG) led to a marked increase in binding, reaching 10%. There was no significant binding of GFP to membrane liposomes. The numerical values presented here should be considered in relation to the relative quantities of trimera (0.5 nmol) and membrane liposomes (0.1 nmol of cytochrome b559 heme and 0.26–0.29 μmol of endogenous membrane phospholipid ± 1.66 μmol of exogenous neutral or anionic phospholipid). The quantitative predominance of exogenous phospholipid explains its pronounced effect on both binding of nonprenylated trimera and on the ability of the trimera to activate the oxidase (see enhancement by anionic phospholipid; Fig. 3C). These findings demonstrate that, in the absence of prenylation, electrostatic attraction of the trimera to the membrane is well correlated with oxidase activation.
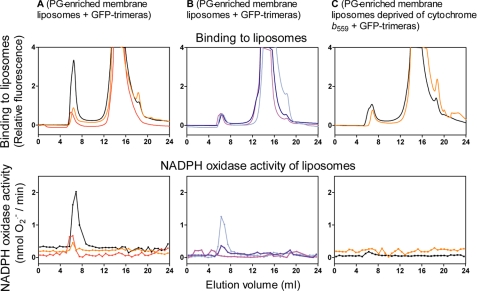
Nonprenylated GFP-Trimeras Form Complexes with Membrane Liposomes Enriched in Anionic Phospholipid That Exhibit Activator-independent NADPH Oxidase Activity
In these experiments, we assessed the binding of nonprenylated trimeras to membrane liposomes enriched in PG under the conditions used in the experiments illustrated in Fig. 4B. This time, however, accent was placed on the effect of mutations in the trimera, found to affect oxidase activation, as described earlier for trimeras lacking GFP (25) and for GFP-trimeras, in this report (Fig. 3C), on binding to PG-enriched membrane liposomes. In addition, we examined the ability of isolated membrane liposome-trimera complexes to function as stimulus-independent, NADPH-dependent, O2⨪-generating entities. We found that most mutations in the GFP-trimera, whether affecting the Rac part (Rac1 → Rac2; Rac1 Gln183–Gln188; Δ insert domain) or the p47phox part (ΔPX domain) reduced membrane attachment by about 80% (Fig. 5, upper panels, and Table 1). A more moderate reduction in binding to the membrane was caused by the W193R mutation in the p47phox part (Fig. 5, upper panel, A, and Table 1). Depleting the membrane of cytochrome b559 reduced binding by about 50% and effaced the difference between the binding of the prototype trimera and the W193R mutant, indicating that Trp193 participates in the binding of the trimera to the membrane by its affinity for the p22PHOX subunit of cytochrome b559 (Fig. 5, upper panel, C, and Table 1).
FIGURE 5.
Binding of nonprenylated GFP-trimeras to macrophage membrane liposomes enriched in anionic phospholipids quantified by gel filtration followed by assessment of NADPH oxidase activity of membrane liposomes with bound GFP-trimera. A, membrane liposomes enriched in PG were mixed with the following: prototype GFP-trimera (black line); GFP-trimera p47phox ΔPX domain (red line), or GFP-trimera p47PHOX W193R (orange line). B, membrane liposomes enriched in PG were mixed with: GFP-trimera Rac1 Δinsert domain (light blue line); GFP-trimera Rac1 → Rac2 (blue line), or GFP-trimera Rac1 Gln183–Gln188 (purple line). C, membrane liposomes enriched in PG but depleted of cytochrome b559 were mixed with GFP-trimera (black line) or GFP-trimera p47phox W193R (orange line). The mixtures were subjected to gel filtration on a Superose 6 column, and the eluates were monitored in-line for the fluorescent signal (upper three panels). The ability of fractions collected from the column eluate to express activator-independent NADPH oxidase activity is shown in the lower three panels. Aliquots of each fraction were added to a cell-free assay, in the absence of amphiphile and any other NADPH oxidase component, and O2⨪ production was measured upon addition of NADPH only (see “Experimental Procedures”). The upper and lower panels, A and B, and upper panel, C, illustrate one characteristic group of experiments, out of three performed for each group. The lower panel, C, illustrates a single experiment. Numerical results expressing the amounts of liposome-bound trimera, as percentage of the total, and the NADPH oxidase activities of liposome-bound trimera appear in Table 1.
TABLE 1.
Binding of nonprenylated and purified prenylated GFP-trimeras to native and modified macrophage membrane liposomes quantified by gel filtration followed by assessment of NADPH oxidase activity of membrane liposomes with bound GFP-trimera
The table summarizes the numerical results of the binding of nonprenylated and purified prenylated GFP-trimeras to membrane liposomes, illustrated in Figs. 5 and 7, respectively. The prenylated trimeras were freed of nonprenylated material by gel filtration as described under “Experimental Procedures.” The results represent means ± S.E. of 3–6 experiments, for each combination of membrane liposomes and GFP-trimera, with the exception of the results of NADPH oxidase activity of liposomes of PG-enriched membranes deprived of cytochrome b559, which are those of a single experiment.
| Type of GFP-trimera | Nonprenylated GFP-trimeras |
Prenylated GFP-trimeras |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Binding to membrane liposomes (% of trimera bound to liposomes)a |
NADPH oxidase activity of liposomes with bound trimera (nmol of O2⨪/min)b |
Binding to membrane liposomes (% of trimera bound to liposomes)a |
NADPH oxidase activity of liposomes with bound trimera (nmol of O2⨪/min)b |
|||||||
| PG-enriched | PG-enriched deprived of cytochrome b559 | PG-enriched | PG-enriched deprived of cytochrome b559 | Native | PG-enriched | PC-enriched | Native | PG-enriched | PC-enriched | |
| Prototype trimera | 10.4 ± 0.8 | 5.5 ± 0.2 | 1.64 ± 0.20 | 0.15 | 73.4 ± 0.5 | 69.0 ± 1.7 | 55.5 ± 3.1 | 5.67 ± 0.67 | 1.41 ± 0.37 | 0.43 ± 0.16 |
| Trimera p47phox ΔPX domain | 2.4 ± 0.3 | 0.69 ± 0.05 | 43.3 ± 3.5 | 4.97 ± 0.60 | ||||||
| Trimera p47phox W193R | 3.5 ± 0.4 | 4.2 ± 1.4 | 0.28 ± 0.01 | 0.26 | 59.9 ± 4.2 | 1.13 ± 0.13 | ||||
| Trimera Rac1 Δinsert domain | 2.1 ± 0.6 | 0.98 ± 0.14 | 57.6 ± 4.7 | 5.12 ± 0.60 | ||||||
| Trimera Rac1 → Rac | 2.1 ± 0.3 | 0.51 ± 0.18 | 32.0 ± 0.6 | 6.56 ± 0.54 | ||||||
| Trimera Rac1 183Q-188Q | 2.0 ± 0.2 | 0.00 ± 0.05 | 39.1 ± 0.5 | 4.60 ± 0.44 | ||||||
a Numbers represent the integrated areas of the fluorescent signal of the void volume peak (representing liposome-bound trimera) as percentage of the total fluorescent signal recorded (sum of the areas of free and liposome-bound trimera).
b Numbers represent the NADPH oxidase activities of 40-μl aliquots of the pooled void volume (4 fractions, eluting between 6 and 8.4 ml) assayed in the absence of amphiphile and of any other NADPH oxidase component, as described under “Experimental Procedures”.
The membrane-bound trimera was found to form a stable complex that could be separated from unbound trimera by gel filtration and that was capable of generating O2⨪ upon the mere addition of NADPH. The oxidase activities of the membrane-mutant trimera complexes reflected quite closely the differences in oxidase activities between native and mutant trimeras as measured in the amphiphile-independent cell-free system with PG-enriched membranes (Fig. 3C). By assuming that the totality of membrane liposomes (containing an equivalent of 100 pmol of cytochrome b559) was recovered in the void volume upon gel filtration, we calculated a turnover of 16.46 ± 2.00 mol of O2⨪/s/mol of cytochrome b559 heme for the oxidase activity of the prototype nonprenylated trimera in complex with PG-enriched membrane liposomes. There was a general but not a close correlation between the ability of the mutant trimeras to bind to liposomes and to confer oxidase activity (Fig. 5, lower panels, and Table 1). Thus, the complex with the prototype trimera was the most active, followed in decreasing order by mutants Rac1 Δinsert domain, p47phox ΔPX domain, Rac1 → Rac2, p47phox W193R, and Rac1 Gln183–Gln188. Cytochrome b559-depleted liposomes, which were found to bind about half of the amount of prototype trimera bound by native liposomes, expressed close to zero oxidase activity (Fig. 5, lower panel, C, and Table 1). The most likely explanation for the only partial correlation between binding and activity is that measuring binding to membrane liposomes does not provide information on the precise location of the trimeras within the membrane microenvironment and, most importantly, on their chance to interact with cytochrome b559 and with Nox2, in particular.
Prenylated GFP-Trimeras Form Complexes with Native Membrane Liposomes That Exhibit Activator-independent NADPH Oxidase Activity
In the previous section, we described the binding of nonprenylated trimeras to membrane liposomes enriched with the anionic phospholipid, PG, which represents an only partial rendering of the in vivo reality. Consequently, we repeated the binding experiments using prenylated trimeras and native membrane liposomes, a situation closely resembling that occurring in the intact cell.
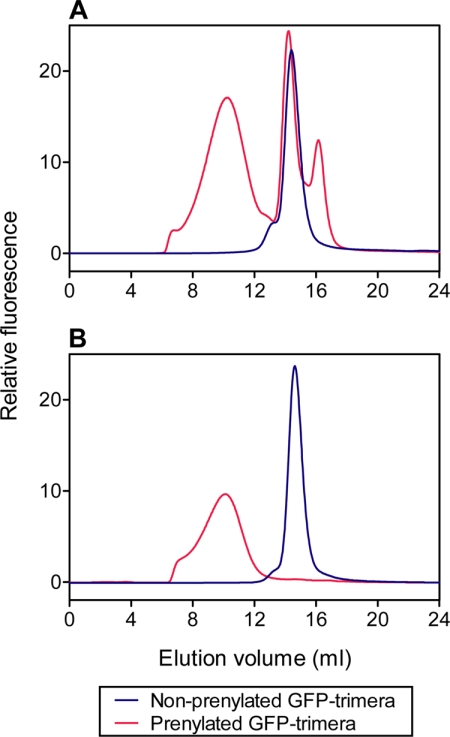
These experiments were performed with prenylated trimeras freed of nonprenylated material by preparative gel filtration on Superdex 200 column, as described under “Experimental Procedures.” We first compared the characteristics of nonpurified prenylated GFP-trimeras with that of the same trimera freed on nonprenylated material by subjecting both to gel filtration on a Superose 6 column. As seen in the characteristic experiment shown in Fig. 6A, the nonpurified prenylated trimera eluted as a wide peak, the center of which corresponded to Mr of 2 × 106, followed by a sharp peak corresponding to Mr close to 108,000, and a small third peak corresponding to Mr of 37,000. As also apparent in Fig. 6A, on the same column, nonprenylated trimera eluted as a single sharp peak overlapping the second peak in the elution pattern of the nonpurified prenylated trimera. Analysis of column fractions for the relative amounts of prenylated and nonprenylated trimera revealed that the major first peak consisted of prenylated trimera, and the second sharp peak consisted of nonprenylated trimera, indicating that prenylation was not complete. The identity of the third peak could not be established. As seen in Fig. 6B, under the same conditions, the prenylated trimera purified on Superdex 200 eluted as a single peak, identical to the first peak in the elution pattern of the nonpurified prenylated trimera, and free of nonprenylated trimera and of the “third peak” contaminant. To make a clear distinction between the binding characteristics of the nonprenylated trimeras, described above, and those of the prenylated trimeras, the present experiments were performed with purified prenylated trimeras, free of contamination by nonprenylated material.
FIGURE 6.
Elution patterns of nonpurified and purified prenylated GFP-trimeras on a Superose 6 gel filtration column. A, 2000 pmol of nonpurified prenylated GFP-trimera (red line) or 500 pmol of nonprenylated GFP-trimera (blue line) were subjected to gel filtration on a Superose 6 column. B, 500 pmol of purified prenylated GFP-trimera (freed of nonprenylated material by gel filtration as described under “Experimental Procedures”) (red line) or 500 pmol of nonprenylated GFP-trimera (blue line) were subjected to gel filtration on a Superose 6 column. Tracings illustrate the in-line recording of the fluorescent signal (excitation = 488 nm; emission = 511 nm). The results illustrated are those of characteristic individual gel filtration experiments out of three performed for each pair of chimeras.
Purified prenylated prototype trimera and its mutants were mixed with native or modified membrane liposomes and the mixtures subjected to separation on a Superose 6 column, under conditions identical to those used with nonprenylated trimeras. The amount of liposome-bound trimera was calculated in relation to the total amount of purified prenylated trimera. As apparent in Fig. 7 and Table 1, the extent of binding of all prenylated trimeras vastly exceeded that of their nonprenylated counterparts (shown in Fig. 5 and Table 1). Thus, about 50 times more prenylated prototype trimera was bound to native membrane liposomes and seven times more to PG-enriched membrane liposomes than the nonprenylated equivalent. Modifying the charge of membrane phospholipids had only a limited effect on the amount of bound trimera. Thus, supplementation with PC moderately decreased binding of the trimera, whereas supplementation with PG did not affect binding (Fig. 7, upper panel, A, and Table 1). Binding was reduced by 50–60% by the Gln183–Gln188 and Rac1 → Rac2 mutations in the Rac1 part, by 40% by the deletion of the PX domain in the p47phox part, and by only 20% by the W193R mutation in the p47phox part and the deletion of the insert domain in the Rac1 part (Fig. 7, upper panels, B and C, and Table 1).
FIGURE 7.
Binding of purified prenylated GFP-trimeras to native macrophage membrane liposomes quantified by gel filtration followed by assessment of NADPH oxidase activity of membrane liposomes with bound GFP-trimera. A, native membrane liposomes (black line) or membrane liposomes supplemented with the neutral phospholipid PC (gold line) or the anionic phospholipid PG (green line) were mixed with prenylated prototype GFP-trimera. B, native membrane liposomes were mixed with the following: prenylated GFP-trimera p47phox ΔPX domain (red line) or prenylated GFP-trimera p47PHOX W193R (orange line). C, native membrane liposomes were mixed with the following: prenylated GFP-trimera Rac1 Δinsert domain (light blue line), prenylated GFP-trimera Rac1 → Rac2 (blue line), or prenylated GFP-trimera Rac1 Gln183–Gln188 (purple line). The mixtures were subjected to gel filtration on a Superose 6 column. The eluates were monitored in-line for the fluorescent signal (upper three panels). The ability of the fractions collected from the column eluate to express activator-independent NADPH oxidase activity is shown in the lower three panels and was assayed as described in the legend of Fig. 5. The upper and lower panels illustrate characteristic groups of experiments, out of three performed, for each group. The prenylated trimeras were freed of nonprenylated material by gel filtration as described under “Experimental Procedures.” Numerical results expressing the amounts of liposome-bound trimera, as percentage of the total, and the NADPH oxidase activities of liposome-bound trimera appear in Table 1.
As also found with its nonprenylated counterpart, the membrane-bound prenylated trimera formed a stable complex that could be separated from unbound trimera by gel filtration and that was capable of generating O2⨪ upon the mere addition of NADPH. The oxidase activities of the prenylated membrane-trimera complexes were much higher than those found with nonprenylated trimeras (Fig. 7, lower panels, A–C, and Table 1). By assuming that the totality of membrane liposomes (containing an equivalent of 100 pmol cytochrome b559) was recovered in the void volume upon gel filtration, we calculated a turnover of 56.92 ± 6.72 mol of O2⨪/s/mol of cytochrome b559 heme for the oxidase activity of prototype-prenylated trimera in complex with native membrane liposomes. This value is close to the maximal turnover values measured in the amphiphile-independent cell-free system with prenylated trimera and native membranes (Fig. 3D). It is 3.5 times higher than the turnover value of nonprenylated trimera-PG-enriched membrane liposome complexes (see above), reflecting the seven times higher binding of the prenylated trimera to membrane liposomes. Enrichment of the membrane with PC caused a marked fall in the recovered oxidase activity, which was well correlated with the inability of prenylated trimera to activate the oxidase in an amphiphile-independent cell-free system using PC-enriched membranes (Fig. 3E). Paradoxically, enriching the membrane with the anionic phospholipid PG also resulted in the recovery of a complex with reduced oxidase activity. We have no explanation for this finding, which contrasts with the marked difference in the ability of the prenylated trimera to activate the oxidase in an amphiphile-independent cell-free system containing PG-enriched (activates) versus PC-enriched (does not activate) membranes (see Fig. 3, E and F). Unlike the case with nonprenylated trimeras, mutations in the prenylated trimera had a much lesser effect on the recovery of active complexes (0–20% reduction). The only mutation that markedly decreased the oxidase activity of the complex (by 80%) was W193R in the p47phox part (Fig. 7, lower panel, B, and Table 1). This closely resembles the effect of mutations on oxidase activation by prenylated trimera in an amphiphile-independent cell-free system using native (Fig. 3D) or PG-enriched (Fig. 3F) membranes. The message derived from these experiments is that prenylation endows the trimera with a most powerful membrane-localizing mechanism that supersedes, at least partially, the role of intrinsic protein domains with membrane affinity (polybasic Rac1 C terminus, Rac1 insert domain, and p47phox PX domain). The only bond not compensated for by prenylation, as far as oxidase activation is concerned, is that between Trp193 in the p47phox part of the trimera and the p22phox subunit of cytochrome b559.
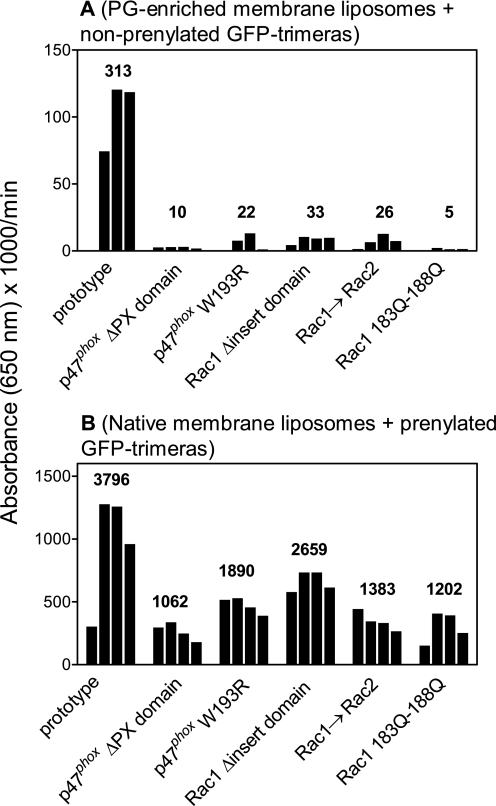
Binding of GFP-Trimeras to Membrane Liposomes Assessed by ELISA
To confirm that the fluorescent signal associated with material eluted in the void volume of mixtures of membrane liposomes and GFP-trimeras subjected to gel filtration was indeed liposome-bound trimera, we also analyzed these fractions by ELISA. The fractions were derived from mixtures of PG-enriched membrane liposomes and nonprenylated GFP-trimeras (as summarized in Fig. 5) and mixtures of native membrane liposomes and purified prenylated GFP-trimeras (as summarized in Fig. 7).
The ELISA-based results faithfully reflected the much higher binding of the prenylated trimera to membrane liposomes when compared with that of nonprenylated trimera, even if the binding of the latter to PG-enriched membrane liposomes served as the basis for comparison. As seen in Fig. 8A, there was a close correlation between the amounts of bound nonprenylated GFP-trimera and mutants, detected by ELISA, and the levels of fluorescence detected in the void volume fractions (Fig. 5, upper panels, A and B, and Table 1). The ELISA-based results confirmed that mutations and deletions in all three parts of the nonprenylated trimera had a pronounced, although rather nonspecific, effect on the binding of nonprenylated trimeras to membrane liposomes. A fair but not perfect correlation was also found between the amounts of membrane-bound prenylated GFP-trimera and mutants, as detected by ELISA, and the levels of fluorescence detected in the void volume fractions (Fig. 7, upper panels, A–C, and Table 1). Mutations affecting the C terminus of the Rac1 part and the PX domain in the p47phox part caused the most pronounced decrease in the amount of membrane-bound prenylated trimera, as detected by ELISA, demonstrating yet again the participation of charge in what was essentially a hydrophobic interaction.
FIGURE 8.
Quantification of nonprenylated (A) and purified prenylated (B) GFP-trimeras bound to membrane liposomes by immunodetection. Void volume fractions (four 0.6-ml fractions, eluting between 6 and 8.4 ml) were collected in gel filtration experiments in which mixtures of nonprenylated GFP-trimeras were mixed with PG-enriched membrane liposomes (panel A, also see Fig. 5, A and B) or purified prenylated GFP-trimeras were mixed with native membrane liposomes (B, also see Fig. 7, A–C) and injected in a Superose 6 column. 100-μl aliquots of these fractions were used to coat the wells of microplates, and the GFP-trimera present in these fractions, representing liposome-bound material, was detected by a kinetic ELISA, using anti-polyhistidine antibody, as described under “Experimental Procedures.” The prenylated trimeras were freed of nonprenylated material by gel filtration as described under “Experimental Procedures.” The numbers on the top of the column fraction clusters represent the sum of the kinetic absorbance values measured with the individual aliquots from the four fractions. A and B illustrate characteristic groups of experiments, out of three performed, for each group.
DISCUSSION
We describe the construction of a recombinant chimera with the structure GFP-p47phox-(1–286)-p67phox-(1–212)-Rac1-(1–192). It encompasses functionally indispensable regions of the three cytosolic oxidase components and also contains N-terminally fused GFP, to serve as a fluorescent tag in studies involving attachment to phagocyte membranes. The chimera does not include the fourth cytosolic component, p40phox, because this is not required for oxidase activation in the in vitro model utilized in these studies. The construct possesses a number of characteristics enabling its use as a flexible and modular instrument for the study of oxidase activation as follows. 1) The protein is of a reasonably small molecular mass (108 kDa), soluble, monomeric, and stable in an aqueous environment. 2) The GFP tag endows it with high emission fluorescent properties and reasonable photostability, ideal for in vitro binding studies of relatively long duration. 3) Two 10-residue spacers within the trimera ensure flexibility. 4) The Q61L mutation in the Rac1 part leads to a constitutive GTP-bound state and to the consequent intramolecular bond between the Rac1 and p67phox parts of the trimera, found essential for oxidase activation. 5) The trimera includes the complete p47phox SH3 tandem in the absence of the polybasic auto-inhibitory region, enabling the trimera to interact spontaneously (in the absence of an exogenous activator) with the PRR region of p22phox. A recently revealed consequence of this is a secondary induction of a conformational change in Nox2 (43). 6) A sequence (residues 151–158) serving as a linker between the PX and the N-terminal SH3 domains, present in the p47phox part of the prototype trimera and of the p47phox ΔPX mutant, was found to be essential for oxidase activation (44–46). 7) The free Rac1 C terminus offers the option of attaching a geranylgeranyl group. 8) A number of mutations, sequence replacements, and deletions in the three parts of the trimera serve as useful indicators of the roles played by specific domains in the absence and presence of prenylation. The most significant property of this novel chimeric construct is the ability to be prenylated in vitro, bringing it closer to the in vivo situation. The importance of using prenylated Rac in in vitro models of oxidase activation has been emphasized repeatedly (15, 16, 34, 36, 40, 47).
At the membrane level, we replaced the routinely used sedimented membrane fragment preparations by liposomes obtained by solubilizing the membrane with octyl glucoside followed by the removal of the detergent by dialysis. The resulting liposomes eluted on gel filtration as a sharp peak in the void volume, clearly separated from free prenylated and nonprenylated trimera, and could thus be used as an accurate and easy means to measure binding of trimera to the membrane. Membrane liposomes could be readily supplemented with exogenous PC or PG, resulting in preparations enriched in either neutral or anionic phospholipid.
The GFP-tagged trimera possessed oxidase activating ability undistinguishable from that of its untagged precedent (25). In the nonprenylated form, its ability to activate the oxidase in membrane liposome preparations was absolutely dependent on either the presence of a soluble exogenous anionic amphiphile or the enrichment of the liposomes with anionic phospholipids, revealing a key role for electrostatic forces. In the prenylated form, likely to mimic the in vivo situation, the trimera activated the oxidase in native unmodified membrane liposomes independently of an exogenous activator. To the best of our knowledge, this is the first description of a single molecule oxidase activator acting on native phagocyte membranes in the absence of any additional factor, which includes functionally significant segments of all three cytosolic components involved in oxidase assembly under physiological conditions.
Binding to the membrane experiments with the two forms of the trimera revealed the basic mechanisms involved in what we defined earlier as “second stage” (cytosolic components-membrane) interactions. Thus, results obtained with the nonprenylated trimera demonstrated the dominance of charge-dependent interactions, as evident in the binding to membranes enriched in anionic phospholipids but not to native membranes and by the negative effect of mutations affecting domains in the trimera participating in electrostatic interactions, most notably the Rac C terminus and the p47phox N terminus. In contrast to this, the prenylated trimera showed high binding to native membranes, which was not enhanced by supplementation with anionic phospholipids, suggesting a dominant role for hydrophobic forces. However, this interaction also was not entirely independent of charge, as shown by the moderate inhibition of binding to membranes by mutations that reduced the positive charge of the Rac1 C terminus, or by removing the cationic PX domain at the p47phox N terminus, and by the fact that the Rac1 Gln183–Gln188 mutant had somewhat reduced activating ability in a cell-free system with native membranes but exhibited normal activity with membranes enriched in anionic phospholipids. The membrane “partners” for such electrostatic interactions are negatively charged phospholipids on the cytosolic aspect of the membrane. An increase in negative charge in this location is due to generation of polyphosphoinositides and phosphatidylserine flipping, which function as signal transducers from membrane receptors to the oxidase in phagocytosing leukocytes (reviewed in Refs. 48, 49).
Physiological second stage interactions between cytosolic proteins and membrane phospholipids are likely to be essentially nonspecific. Their function is to bring p67phox to the vicinity of Nox2, but a more focused juxtaposition is required for actual binding to Nox2. This latter function is performed by the SH3 tandem of p47phox (predominantly via residue Trp193) by binding to p22phox in the membrane. Mutating Trp193 to Arg had an inhibitory effect on the binding of nonprenylated trimera to “acidified” membranes but did not significantly affect binding of prenylated trimera to native membranes. The W193R mutants of both nonprenylated and prenylated trimeras were, however, severely impaired in their ability to activate the oxidase in the cell-free system with both native and acidified membranes. Also, although complexes of nonprenylated and prenylated W193R mutant trimeras and membrane liposomes could be recovered by gel filtration, these lacked significant oxidase activity. These data suggest that the interaction of the SH3 tandem of p47phox with p22phox plays no role in the binding of p47phox to the membrane under physiological conditions, but it is essential for facilitating its specific interaction with Nox2. No evidence was found for a similar role for the insert domain of Rac, which was reported to be involved in the binding of Rac to Nox2 (17, 18). This domain was found to participate in the binding of nonprenylated trimera to PG-enriched membrane (probably due to its positive charge) but not in that of the prenylated trimera to native membranes. Thus, prenylated trimera, lacking the Rac insert domain, had an unchanged oxidase activating capacity in the cell-free system and formed stable complexes with membrane liposomes with almost intact oxidase activity, corroborating other reports of the lack of involvement of the Rac insert domain in oxidase assembly (16, 20, 21).
A legitimate question, which cannot be answered, is whether geranylgeranylation at the C terminus of the Rac part of the trimera is the only post-translational modification capable of promoting membrane tropism. Possible alternatives are farnesylation, which could be made possible by switching the X residue in the CAAX box (where AA indicate aliphatic amino acids) at the C terminus of the Rac part, or myristoylation of the GFP part at the N terminus of the trimera. It is conceivable that both acyl chains would promote some degree of binding to the membrane, but this is likely to be inferior to that found with the geranylgeranylated trimera because of the fact that both farnesyl (15-carbon) and myristoyl (14-carbon) are less hydrophobic than the 20-carbon geranylgeranyl chain (50). Our choice of a geranylgeranylated trimera was principally motivated by the fact that this represents the physiological modification of Rac and is thus the closest approximation of the in vivo reality.
Our study does not include an analysis of the third and last stage of assembly, namely the interaction of p67phox with Nox2. The reason for this is the paucity of data on the identity of the binding site(s) for Nox2 in p67PHOX, the only information available at present being the involvement of the activation domain (residues 199–210) in p67phox in oxidase activation (51). We also did not address directly the important issue of the mechanism of the termination of oxidase activity (52). We have shown before (25) that a nonprenylated p47phox-p67phox-Rac1 trimera forms a much more stable complex with membrane liposomes in vitro than the combined individual cytosolic components, suggesting that the reversibility of first stage interactions might be responsible for the termination of the oxidative burst. Although no comparable measurements were done with the prenylated equivalent, one can fairly assume that the presence of the isoprenyl tail of the Rac part would lead to an even more stable complex and that Rac dynamics might determine the longevity of the assembled oxidase complex. The present findings might also be relevant to other Noxes, particularly to those for which there is evidence for the involvement of Rac in their regulation, such as Nox1 and Nox3 (53).
We trust that the prenylated trimera will become a useful instrument for the high throughput screening in vitro of modulators of oxidase activation, particularly inhibitors acting directly on the components of the complex, to serve as drug candidates. There also exists a potential for the use of GFP-trimera cDNA in cell transfection experiments.
This work was supported, in whole or in part, by National Institutes of Health Grant GM46372 (to P. J. C.). This work was also supported by Israel Science Foundation Grants 19/05 and 49/09, the David and Natalie Roberts Chair in Immunopharmacology, the Julius Friedrich Cohnheim-Minerva Center for Phagocyte Research, and the Ela Kodesz Institute of Host Defense against Infectious Diseases (to E. P.).
- O2⨪
- superoxide
- PRR
- proline-rich region
- SH3
- Src homology 3
- PX
- phox homology
- PC
- phosphatidylcholine
- PG
- phosphatidylglycerol
- GFP
- green fluorescent protein
- FPLC
- fast protein liquid chromatography
- HPLC
- high pressure liquid chromatography
- octyl glucoside
- octyl-β-d-glucopyranoside
- LiDS
- lithium dodecyl sulfate
- mant
- 2′-(or 3′)-O-(N-methylanthraniloyl)
- GMPPNP
- guanylyl-imidodiphosphate
- ELISA
- enzyme-linked immunosorbent assay.
REFERENCES
- 1.Nauseef W. M. (2007) Immunol. Rev. 219, 88–102 [DOI] [PubMed] [Google Scholar]
- 2.Cross A. R., Segal A. W. (2004) Biochim. Biophys. Acta 1657, 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quinn M. T., Gauss K. A. (2004) J. Leukocyte Biol. 76, 760–781 [DOI] [PubMed] [Google Scholar]
- 4.Nauseef W. M. (2004) Histochem. Cell Biol. 122, 277–291 [DOI] [PubMed] [Google Scholar]
- 5.Groemping Y., Rittinger K. (2005) Biochem. J. 386, 401–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sumimoto H. (2008) FEBS J. 275, 3249–3277 [DOI] [PubMed] [Google Scholar]
- 7.de Mendez I., Adams A. G., Sokolic R. A., Malech H. L., Leto T. L. (1996) EMBO J. 15, 1211–1220 [PMC free article] [PubMed] [Google Scholar]
- 8.Koga H., Terasawa H., Nunoi H., Takeshige K., Inagaki F., Sumimoto H. (1999) J. Biol. Chem. 274, 25051–25060 [DOI] [PubMed] [Google Scholar]
- 9.Lapouge K., Smith S. J., Walker P. A., Gamblin S. J., Smerdon S. J., Rittinger K. (2000) Mol. Cell 6, 899–907 [DOI] [PubMed] [Google Scholar]
- 10.Sarfstein R., Gorzalczany Y., Mizrahi A., Berdichevsky Y., Molshanski-Mor S., Weinbaum C., Hirshberg M., Dagher M. C., Pick E. (2004) J. Biol. Chem. 279, 16007–16016 [DOI] [PubMed] [Google Scholar]
- 11.Sumimoto H., Kage Y., Nunoi H., Sasaki H., Nose T., Fukumaki Y., Ohno M., Minakami S., Takeshige K. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 5345–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leto T. L., Adams A. G., de Mendez I. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 10650–10654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groemping Y., Lapouge K., Smerdon S. J., Rittinger K. (2003) Cell 113, 343–355 [DOI] [PubMed] [Google Scholar]
- 14.Kanai F., Liu H., Field S. J., Akbary H., Matsuo T., Brown G. E., Cantley L. C., Yaffe M. B. (2001) Nat. Cell Biol. 3, 675–678 [DOI] [PubMed] [Google Scholar]
- 15.Kreck M. L., Freeman J. L., Abo A., Lambeth J. D. (1996) Biochemistry 35, 15683–15692 [DOI] [PubMed] [Google Scholar]
- 16.Gorzalczany Y., Alloul N., Sigal N., Weinbaum C., Pick E. (2002) J. Biol. Chem. 277, 18605–18610 [DOI] [PubMed] [Google Scholar]
- 17.Diebold B. A., Bokoch G. M. (2001) Nat. Immunol. 2, 211–215 [DOI] [PubMed] [Google Scholar]
- 18.Diebold B. A., Fowler B., Lu J., Dinauer M. C., Bokoch G. M. (2004) J. Biol. Chem. 279, 28136–28142 [DOI] [PubMed] [Google Scholar]
- 19.Kao Y. Y., Gianni D., Bohl B., Taylor R. M., Bokoch G. M. (2008) J. Biol. Chem. 283, 12736–12746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alloul N., Gorzalczany Y., Itan M., Sigal N., Pick E. (2001) Biochemistry 40, 14557–14566 [DOI] [PubMed] [Google Scholar]
- 21.Miyano K., Koga H., Minakami R., Sumimoto H. (2009) Biochem. J. 422, 373–382 [DOI] [PubMed] [Google Scholar]
- 22.Mizrahi A., Berdichevsky Y., Ugolev Y., Molshanski-Mor S., Nakash Y., Dahan I., Alloul N., Gorzalczany Y., Sarfstein R., Hirshberg M., Pick E. (2006) J. Leukocyte Biol. 79, 881–895 [DOI] [PubMed] [Google Scholar]
- 23.Ebisu K., Nagasawa T., Watanabe K., Kakinuma K., Miyano K., Tamura M. (2001) J. Biol. Chem. 276, 24498–24505 [DOI] [PubMed] [Google Scholar]
- 24.Miyano K., Ogasawara S., Han C. H., Fukuda H., Tamura M. (2001) Biochemistry 40, 14089–14097 [DOI] [PubMed] [Google Scholar]
- 25.Berdichevsky Y., Mizrahi A., Ugolev Y., Molshanski-Mor S., Pick E. (2007) J. Biol. Chem. 282, 22122–22139 [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27.Zhang F. L., Moomaw J. F., Casey P. J. (1994) J. Biol. Chem. 269, 23465–23470 [PubMed] [Google Scholar]
- 28.Gutierrez L., Magee A. I., Marshall C. J., Hancock J. F. (1989) EMBO J. 8, 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 30.Bromberg Y., Pick E. (1984) Cell. Immunol. 88, 213–221 [DOI] [PubMed] [Google Scholar]
- 31.Shpungin S., Dotan I., Abo A., Pick E. (1989) J. Biol. Chem. 264, 9195–9203 [PubMed] [Google Scholar]
- 32.Pick E., Bromberg Y., Shpungin S., Gadba R. (1987) J. Biol. Chem. 262, 16476–16483 [PubMed] [Google Scholar]
- 33.Ugolev Y., Molshanski-Mor S., Weinbaum C., Pick E. (2006) J. Biol. Chem. 281, 19204–19219 [DOI] [PubMed] [Google Scholar]
- 34.Sigal N., Gorzalczany Y., Pick E. (2003) Inflammation 27, 147–159 [DOI] [PubMed] [Google Scholar]
- 35.Harper A. M., Dunne M. J., Segal A. W. (1984) Biochem. J. 219, 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molshanski-Mor S., Mizrahi A., Ugolev Y., Dahan I., Berdichevsky Y., Pick E. (2007) in Neutrophil Methods and Protocols (Quinn M. T., DeLeo F. R., Bokoch G. M.) pp. 385–428, Humana Press Inc., Totowa, NJ [Google Scholar]
- 37.Toporik A., Gorzalczany Y., Hirshberg M., Pick E., Lotan O. (1998) Biochemistry 37, 7147–7156 [DOI] [PubMed] [Google Scholar]
- 38.Marcoux J., Man P., Castellan M., Vivès C., Forest E., Fieschi F. (2009) FEBS Lett. 583, 835–840 [DOI] [PubMed] [Google Scholar]
- 39.Ugolev Y., Berdichevsky Y., Weinbaum C., Pick E. (2008) J. Biol. Chem. 283, 22257–22271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gorzalczany Y., Sigal N., Itan M., Lotan O., Pick E. (2000) J. Biol. Chem. 275, 40073–40081 [DOI] [PubMed] [Google Scholar]
- 41.Mizrahi A., Molshanski-Mor S., Weinbaum C., Zheng Y., Hirshberg M., Pick E. (2005) J. Biol. Chem. 280, 3802–3811 [DOI] [PubMed] [Google Scholar]
- 42.Casey P. J., Thissen J. A., Moomaw J. F. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 8631–8635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor R. M., Lord C. I., Riesselman M. H., Gripentrog J. M., Leto T. L., McPhail L. C., Berdichevsky Y., Pick E., Jesaitis A. J. (2007) Biochemistry 46, 14291–14304 [DOI] [PubMed] [Google Scholar]
- 44.Morozov I., Lotan O., Joseph G., Gorzalczany Y., Pick E. (1998) J. Biol. Chem. 273, 15435–15444 [DOI] [PubMed] [Google Scholar]
- 45.Shen K., Sergeant S., Hantgan R. R., McPhail L. C., Horita D. A. (2008) Biochemistry 47, 8855–8865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taura M., Miyano K., Minakami R., Kamakura S., Takeya R., Sumimoto H. (2009) Biochem. J. 419, 329–338 [DOI] [PubMed] [Google Scholar]
- 47.Heyworth P. G., Knaus U. G., Xu X., Uhlinger D. J., Conroy L., Bokoch G. M., Curnutte J. T. (1993) Mol. Biol. Cell 4, 261–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeung T., Grinstein S. (2007) Immunol. Rev. 219, 17–36 [DOI] [PubMed] [Google Scholar]
- 49.Pick E. (2010) J. Leukocyte Biol. 87, 537–540 [DOI] [PubMed] [Google Scholar]
- 50.Hancock J. F., Cadwallader K., Paterson H., Marshall C. J. (1991) EMBO J. 10, 4033–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han C. H., Freeman J. L., Lee T., Motalebi S. A., Lambeth J. D. (1998) J. Biol. Chem. 273, 16663–16668 [DOI] [PubMed] [Google Scholar]
- 52.Decoursey T. E., Ligeti E. (2005) Cell. Mol. Life Sci. 62, 2173–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bedard K., Krause K. H. (2007) Physiol. Rev. 87, 245–313 [DOI] [PubMed] [Google Scholar]