Abstract
Recent studies have demonstrated that embryonic stem cell-like induced pluripotent stem (iPS) cells can be generated by enforced expression of defined transcription factors. The fact that cell fate change is accompanied by changes in epigenetic modifications prompted us to investigate whether chemicals known to modulate epigenetic regulators are capable of enhancing the efficiency of iPS cell generation. Here, we report that butyrate, a natural small fatty acid and histone deacetylase inhibitor, significantly increases the efficiency of mouse iPS cell generation using the transcription factors Oct4, Sox2, Klf4, and c-Myc. We show that butyrate not only changes the reprogramming dynamics, but also increases the ratio of iPS cell colonies to total colonies by reducing the frequency of partially reprogrammed cells and transformed cells. Detailed analysis reveals that the effect of butyrate on reprogramming appears to be mediated by c-Myc and occurs during an early stage of reprogramming. Genome-wide gene expression analysis reveals up-regulation of ES cell-enriched genes when mouse embryonic fibroblasts are treated with butyrate during reprogramming. Thus, our study identifies butyrate as a chemical factor capable of promoting iPS cell generation.
Keywords: Embryonic Stem Cell, Enzyme Inhibitors, Epigenetics, Histone Deacetylase, Induced Pluripotent Stem (iPS) Cell, Butyrate
Introduction
Reprogramming from somatic cells to induced pluripotent stem (iPS)2 cells can be achieved by retroviral expression of transcription factors Oct4, Sox2, Klf4, and c-Myc (1–4). However, the slow reprogramming process and low reprogramming efficiency impede detailed mechanistic studies and potential applications of this technology. One solution to overcome these problems is to identify small molecules that can enhance the reprogramming efficiency. Indeed, recent studies have demonstrated that several chemicals with the capacity to modulate epigenetic enzymes exhibit positive effects on iPS cell generation. For example, valproic acid (VPA), a histone deacetylase inhibitor, has been shown to improve both the kinetics and efficiency of mouse and human iPS cell generation (5, 6). In addition, BIX-01294, an inhibitor for the histone methyltransferase G9a, and RG108, a DNA methyltransferase inhibitor, have been reported to enhance the efficiency of iPS cell generation (7, 8). Furthermore, another DNA methyltransferase inhibitor, 5-aza-cytidine, has been shown to facilitate the conversion of partially reprogrammed cells to fully reprogrammed iPS cells (9).
Recently, butyrate, a naturally occurring short chain fatty acid and histone deacetylase inhibitor, has been shown to support self-renewal of both human and mouse embryonic stem (ES) cells in a range of relatively low concentrations (10). However, butyrate has also been reported to induce differentiation when applied at higher concentrations (11). Therefore, whether butyrate has an effect on iPS cell generation is an intriguing question. In this report, we sought to examine the effect of butyrate on iPS cell generation. We found that butyrate facilitates iPS cell generation in the range of 0.5–1 mm. This effect appears to be mediated through one of the reprogramming factors c-Myc. In addition, butyrate is able to increase the percentage of fully reprogrammed iPS cells by reducing partially and/or unsuccessfully reprogrammed cells. Genome-wide gene expression analysis indicates that butyrate can specifically increase the expression of some ES cell-enriched genes in fibroblasts in the presence of exogenous c-Myc. Thus, our studies uncover another chemical capable of facilitating iPS cell generation, contributing to the iPS cell tool box.
EXPERIMENTAL PROCEDURES
Mouse Embryonic Fibroblast (MEF) Derivation and iPS Cell Culture
MEFs were derived from day 13.5 embryos (E13.5) of Sox2-GFP/Rosa26-M2rtTA double knock-in mice. MEFs were cultured in rich fibroblast growth medium (Dulbecco's modified Eagle's medium supplemented with 15% fetal bovine serum, nonessential amino acid, GlutaMax, and penicillin/streptomycin) for no more than two passages before retroviral transduction. iPS cells were cultured in mouse ES cell medium (Dulbecco's modified Eagle's medium with 15% fetal bovine serum, nonessential amino acid, GlutaMax, sodium pyruvate, β-mercaptoethanol, penicillin/streptomycin, and 1,000 units/ml leukemia inhibitory factors) with mitomycin C-treated STO cells as feeder cells or on 0.1% gelatin-coated plates.
Retrovirus Preparation and Infection
Retroviral plasmids pMXs expressing murine Oct4, Sox2, Klf4, and c-Myc (2) were transfected respectively into 293T cells with packaging plasmids pGag-pol and pVSVG. Virus-containing supernatants were harvested at 48 and 72 h after transfection, filtered through 0.45-μm filter membrane, and concentrated by spin column before being used in MEF transduction in the presence of Polybrene (5 μg/ml).
Generation of iPS Cells and Calculation of Reprogramming Efficiency
MEFs were seeded in 6-well plates with 1 × 105 cells/well, 16 h before the first infection. Concentrated viruses were applied to MEFs twice within 48 h. The day when the viral supernatant was removed was defined as day 0 after infection. Transduced fibroblasts were then cultured in mouse ES cell medium in the presence or absence of butyrate for 12–16 days. The concentration of butyrate used was 1 mm, except stated otherwise. In most cases, reprogramming efficiency is represented by the Sox2-GFP+ colony number derived from 1 × 105 MEFs; in some case, relative reprogramming efficiency is also used, which is the fold change of Sox2-GFP+ colony number with butyrate treatment compared with that without treatment. Alkaline phosphatase (AP) staining was performed with alkaline phosphatase detection kit (Millipore).
Teratoma Formation and Analysis
Teratomas were induced by subcutaneously injecting 1 × 106 iPS cells into Rag2−/−::γC−/− immunodeficient mice. Xenografted tumor samples were isolated from mice in 4–6 weeks, fixed by 4% paraformaldehyde, embedded in paraffin, and processed for hematoxyin and eosin staining, using standard protocols.
Quantitative and Semiquantitative RT-PCR
Total RNAs were harvested using RNeasy kit (Qiagen). Primers for quantitative and semiquantitative RT-PCR are listed in supplemental Table 1. Quantitative PCRs were performed with SYBR GreenER mix (Invitrogen). Relative gene expression levels were normalized to Gapdh mRNA.
Genome-wide Expression Analysis
2 μg of total RNA was reverse-transcribed into cDNA with a T7-(dT)24 primer from a custom kit (Invitrogen). Biotinylated cRNA was then generated from the cDNA reaction using the BioArray High Yield RNA Transcript kit. The cRNA was then fragmented in fragmentation buffer (40 mm Tris acetate, pH 8.1, 100 mm KOAc, and 150 mm MgOAc) at 94 °C for 35 min before microarray hybridization. 15 μg of fragmented cRNA was then added to a hybridization mixture (0.05 μg/μl fragmented cRNA, 50 pm control oligonucleotide B2, BioB, BioC, BioD, and Cre hybridization controls, 0.1 mg/ml herring sperm DNA, 0.5 mg/ml acetylated bovine serum albumin, 100 mm MES, 1 m [Na+], 20 mm EDTA, 0.01% Tween 20). 10 μg of cRNA was used for hybridization to Affymetrix GeneChip Mouse Genome 430A 2.0 Array. Hybridization was carried out at 45 °C for 16 h. The array was then washed and stained with R-phycoerythrin streptavidin before scanning. Washing, scanning, and basic analysis were carried out using Affymetrix GeneChip Microarray Suite 5.0 software. For details of Western blotting, see supplemental Methods.
RESULTS
Butyrate Promotes iPS Cell Generation
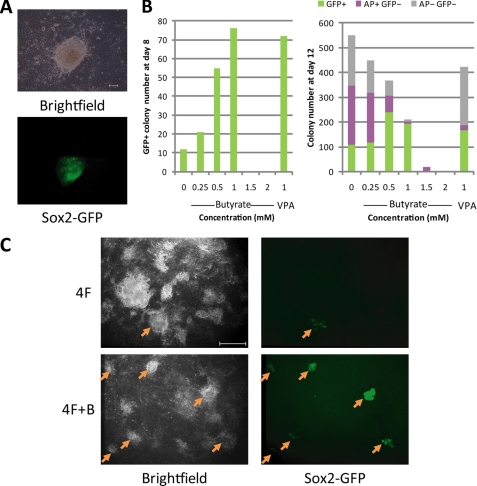
A recent study indicated that butyrate, a small fatty acid, supports self-renewal in mouse and human embryonic stem cells (10). To determine whether butyrate has an effect on iPS cell generation, we transduced 1 × 105 MEFs derived from hemizygote a Sox2-GFP mouse with retroviruses expressing Oct4, Sox2, Klf4, and c-Myc in the presence of varying concentrations of butyrate. The effect of butyrate on reprogramming was monitored for a period of 12 days after infection. In the presence of butyrate, GFP+ colonies with ES cell-like morphology were observed at day 6 after infection (Fig. 1A). At day 8, we observed a dose-dependent enhancement of reprogramming efficiency when butyrate was used at concentrations between 0.25 and 1 mm. A maximum 7-fold increase was observed when butyrate was applied at 1 mm. However, at higher concentrations (1.5 and 2 mm), butyrate becomes cell-toxic, and no GFP+ colonies were observed (Fig. 1B). On day 12, treatment of butyrate at 0.5 and 1 mm still showed an approximate 2-fold increase in the number of GFP+ colonies (Fig. 1B, right panel, green bars). The effect of butyrate on promoting the generation of GFP+ iPS cell colonies is comparable with that of VPA (Fig. 1B).
FIGURE 1.
Butyrate promotes iPS cell generation. A, representative Sox2-GFP+ iPS cell colony generated in the presence of butyrate at postinfection day 6. Scale bar, 100 μm. MEFs from hemizygote Sox2-GFP mouse were transduced with Oct4, Sox2, Klf4, and c-Myc (4F) and treated with butyrate. B, GFP+ colony numbers counted at posttransduction day 8 (left panel) of 1 × 105 MEFs with the four reprogramming factors in the absence or the presence of various concentrations (0.25, 0.5, 1, 1.5, and 2 mm) of butyrate or VPA (1 mm). Presented in the right panel are the numbers of GFP+ and total colony counted at day 12 after transduction. Subsequently, cells were stained for AP. The number of GFP+ colonies (green), AP+GFP− colonies (purple), and AP−GFP− colonies (gray) are shown in the chart. C, representative fluorescent microscopic pictures taken at day 12 after transduction by 4F and 4F in the presence of 0.5 mm butyrate (4F+B). Arrows indicate Sox2-GFP+ colonies. Scale bar, 1 mm.
In addition to counting the GFP+ iPS cell colony numbers, we also counted the total and the AP-positive colony numbers on day 12. Interestingly, at lower concentrations of butyrate (0.25–1 mm), both the AP+ and total colony numbers show a concentration-dependent reduction (Fig. 1B, right panel), although more GFP+ colonies were observed. This leads to exceptionally high GFP+/AP+ and GFP+/total colony ratios under butyrate treatment, which is distinct from the effect of VPA (Fig. 1B, right panel). For example, in the absence of butyrate, only 20% of the total colonies are GFP+ and 63% are AP+; at 0.5 mm, 66% are GFP+ and 83% are AP+; at 1 mm, these ratios are further increased to 92 and 96%, respectively (Fig. 1B, right panel, and Fig. 1C). Because Sox2-GFP is a more stringent pluripotency marker than AP, the reduction of GFP− colonies in total population and AP+ population suggests that butyrate is capable of suppressing the formation of transformed cells or partially reprogrammed cells that were not destined to the pluripotent cell fate. This observation is consistent with the well characterized effect of butyrate on limiting the cell growth of cancerous cells (12).
Butyrate Accelerates iPS Cell Generation, and Its Effect Is c-Myc-dependent
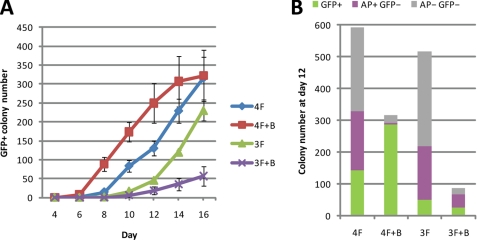
To characterize further the effect of butyrate on iPS cell generation, we monitored the effect of butyrate (1 mm) on the kinetics of reprogramming by introducing the four transcription factors (4F; Oct4, Sox2, Klf4, and c-Myc) and three factors (3F; Oct4, Sox2, and Klf4). In the case of 4F reprogramming, butyrate accelerated formation of GFP+ colonies by 2–3 days until postinfection day 16 when the effect of butyrate on the GFP+ colony number becomes unnoticeable (Fig. 2A). However, when the reprogramming was performed using three factors, butyrate appears to have a negative effect on reprogramming efficiency (Fig. 2A). This suggests that the enhancement effect of butyrate on reprogramming is dependent on exogenous c-Myc under our experimental condition. We also monitored the number of AP+ colony and total colony at postinfection day 12. In both 4F and 3F reprogramming, butyrate strongly reduces the total colony number and GFP− colony number (Fig. 2B), indicating that butyrate is capable of limiting the formation of partially reprogrammed cells or transformed cells regardless of whether three factors or four factors were used for reprogramming. In addition, we also tested the potential effect of butyrate on reprogramming by withdrawing other factors from the 4F combination; however, we did not observe any positive effect of butyrate on reprogramming under these conditions (data not shown).
FIGURE 2.
Butyrate improves the reprogramming kinetics in a c-Myc-dependent manner. A, kinetics of reprogramming in the presence of butyrate. Sox2-GFP MEFs transduced with 4F and 3F were treated with butyrate (1 mm). The number of GFP+ colonies for each treatment was counted at different days until day 16 after transduction. B, GFP+ colonies and total colonies at day 12. AP+ colonies were also counted, after cells were stained for AP.
Sox2-GFP-positive Colonies Are Pluripotent
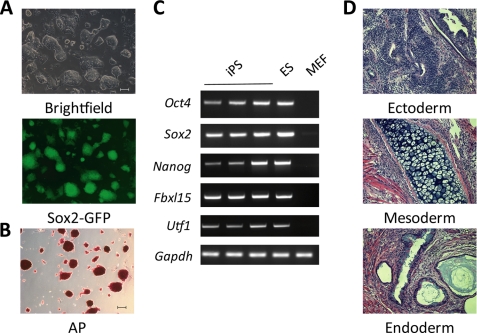
Next, we set out to characterize the GFP+ colonies generated in the presence of butyrate. To this end, individual GFP+ colonies were picked up and propagated in standard ES cell culture medium in the absence of butyrate. All of the GFP+ colonies derived in the presence of butyrate exhibit ES-like morphology (Fig. 3A) and high AP activity (Fig. 3B). We randomly picked three colonies for further characterization. RT-PCR analysis demonstrated that all three lines expressed endogenous genes encoding stem cell factors Oct4, Sox2, Nanog, as well as other pluripotency-related genes, such as Fbxl15 and Utf1 (Fig. 3C). When these iPS cells were injected into immunodeficient mice, all lines were able to form complex-structured teratoma containing tissues from the three germ layers (Fig. 3D). Collectively, these results suggest that Sox2-GFP+ cells generated in the presence of butyrate are pluripotent.
FIGURE 3.
iPS cells generated in the presence of butyrate are pluripotent. A, representative pictures of an iPS cell line derived in the presence of butyrate. iPS cell lines derived from 4F reprogramming in the presence of butyrate were cultured in ES cell medium on gelatin-coated plates without feeder cells. Scale bar, 100 μm. B, AP activity of an iPS cell line derived in the presence of butyrate. Scale bar, 100 μm. C, RT-PCR demonstrating that three randomly picked iPS cell lines derived in the presence of butyrate express pluripotent marker genes. D, representative pictures of teratoma derived from butyrate-assisted iPS cell lines comprising cell types from all three germ layers (endoderm, mesoderm, and ectoderm). Scale bar, 100 μm. All three randomly picked iPS cell lines showed a similar capacity in generating teratomas harboring cells belonging to the three germ layers.
Butyrate Facilitates iPS Cell Generation in an Early Time Window
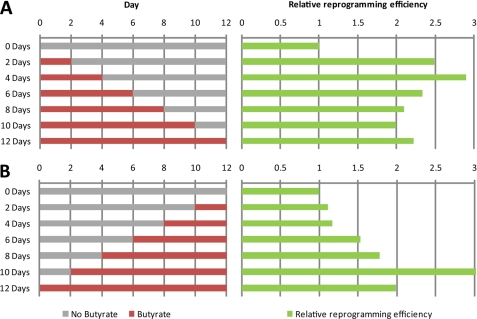
To shed light on the role of butyrate in promoting iPS generation, we sought to determine the time window during which butyrate exerts its effect. First, butyrate was applied to the culture medium immediately after infection and was withdrawn from the medium at different time points during the 12-day reprogramming process (Fig. 4A, left panel). Successfully reprogrammed GFP+ colonies were counted at day 12, and the effect of butyrate on reprogramming was determined by comparison with GFP+ colony numbers in the absence of butyrate. Results shown in Fig. 4A (right panel) demonstrate that the exposure to butyrate for only 2–4 days following transduction has an effect similar to that of continued exposure during the reprogramming process. Next, we determined whether the 2–4 days of butyrate treatment needed to be performed at a particular time point during the reprogramming process. To this end, butyrate was added to the culture medium at different time points during the reprogramming process (Fig. 4B, left panel), and the effect of butyrate on reprogramming was determined in a way similar to that described above. Interestingly, we found that exposure to butyrate from day 2 after transduction has the maximum positive effect on the efficiency of reprogramming. Based on the above experiments, we conclude that butyrate exhibits the maximum effect on iPS generation at the initial 2–4 days of reprogramming, suggesting that butyrate functions early during the reprogramming process. Our findings that butyrate exerts its effect in a c-Myc-dependent manner (Fig. 2) and that this occurs early during the reprogramming process are consistent with previous studies demonstrating that Myc mainly contributes to reprogramming at an early stage (13).
FIGURE 4.
Butyrate facilitates iPS cell generation at an early time window during reprogramming. A, Sox2-GFP MEFs transduced with 4F were treated with butyrate immediately after transduction with 4F, and butyrate was removed from the culture medium at different time points. GFP+ colonies were counted at day 12. The reprogramming efficiencies of various treatments were compared with that without the butyrate treatment and are presented in the right panel. B, Sox2-GFP MEFs transduced with 4F were treated with butyrate at different times after transduction with 4F, and butyrate was maintained in the culture media until day 12 when the GFP+ colonies were counted. The reprogramming efficiencies of various treatments were compared with that without the butyrate treatment and are presented in the right panel.
Butyrate Up-regulates a Set of ES Cell-enriched Genes in c-Myc- mediated Reprogramming
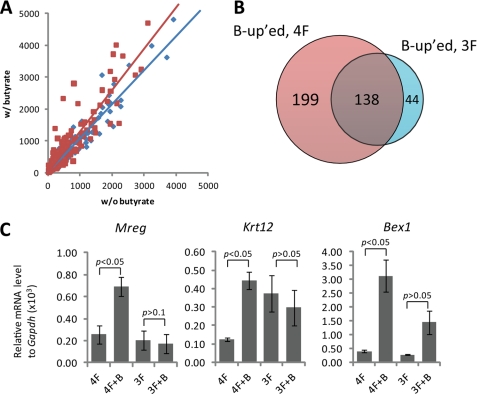
To gain insight into the molecular mechanism of butyrate enhanced reprogramming, we carried out gene expression studies using cDNA microarrays with the following four samples: (i) 4F transduction, (ii) 4F transduction with butyrate treatment (4F+B), (iii) 3F transduction, and (iv) 3F transduction with butyrate treatment (3F+B). Because the effective time window for butyrate is 2–4 days after infection and because we are mainly interested in a primary effect, we treated the transduced MEFs with butyrate for a period of 48 h prior to harvesting RNA. We first focused on the genes whose expression is at least 8-fold higher in ES cells than in MEFs (9). We plotted the expression level of these ES cell-enriched genes in 4F (Fig. 5A, red dots) and 3F (blue dots)-transduced MEFs at day 2 on a scatter chart, in which the x and y axis, respectively, represent the expression level in the absence or presence of butyrate (Fig. 5A). Linear regression of the scatter plot shows that the slope of the 4F regression line (1.3137) is significantly higher than that of the 3F regression line (1.0918), indicating that the expression of these ES cell-enriched genes as a whole is increased by butyrate treatment in a c-Myc-dependent manner (Fig. 5A). Similarly, we have also analyzed the expression of MEF-enriched genes in response to butyrate treatment in reprogramming. However, a significant down-regulation of these genes due to the treatment of butyrate was not noticed, regardless of whether 4F or 3F was used in reprogramming (data not shown).
FIGURE 5.
Butyrate enhances the expression of a set of ES cell-enriched genes in a c-Myc-dependent manner. A, global gene expression was analyzed by microarray (Affymetrix) using total RNA samples harvested 2 days after mock or butyrate treatment. The expression levels of ES cell-enriched genes from samples with butyrate treatment (y axis) are plotted against those without butyrate treatment (x axis). The expression from cells transduced with 4F and 3F are, respectively, shown in red squares and blue diamonds. The linear regression line for 4F (red, y = 1.3137x − 10.675, R2 = 0.8705) is significantly different (p < 0.05) from that of 3F (blue, y = 1.0918x + 2.5027, R2 = 0.9653). B, Venn diagram depicts probes of genes that have 2-fold up-regulation in response to butyrate treatment in 4F (B-up'ed, 4F) or 3F (B-up'ed, 3F) reprogramming. 199 probes that are up-regulated by butyrate in 4F, but not 3F, reprogramming were further analyzed. C, RT-quantitative PCR verification of three randomly picked ES cell-enriched genes listed in supplemental Table 2, Mreg, Krt12, and Bex1, is shown. Data presented are normalized to Gapdh.
Further analysis of the microarray data indicates that a total of 337 probes were at least 2-fold up-regulated by the treatment of butyrate in the 4F reprogramming, whereas only 182 probes were up-regulated at least 2-fold in 3F reprogramming (Fig. 5B). Interestingly, 199 of the 337 probes up-regulated in the 4F reprogramming appear to be c-Myc-dependent as butyrate treatment failed to significantly up-regulate them in the 3F reprogramming. Among the 199 probes, 21 probes correspond to 19 known ES cell-enriched genes (supplemental Table 2). RT-quantitative PCR analysis of randomly selected genes out of the 19 ES cell-enriched genes confirmed that their expression is significantly up-regulated by the treatment of butyrate in the 4F reprogramming, but not in the 3F reprogramming (Fig. 5C). The selected genes, Bex1, Mreg, and Krt12 are most up-regulated by butyrate at the concentration of 1 mm (supplemental Fig. 1), which is consistent with the observed maximum effect of butyrate on iPS cell generation at this concentration. How up-regulation of these genes contributes to the iPS cell generation process remains to be determined.
DISCUSSION
In this study, we demonstrate that butyrate, a small fatty acid and histone deacetylase inhibitor, promotes mouse iPS cell generation by Oct4, Sox2, Klf4, and c-Myc. Under the optimal concentration (0.5–1 mm), butyrate can enhance the generation of Sox2-GFP+ iPS cells by 7-fold when the four factors were used in reprogramming (Fig. 1B). Butyrate facilitates iPS cell generation mainly by shifting the kinetic of 4F reprogramming 2–3 days forward (Fig. 2A). A recent study indicated that ES cell self-renewal is facilitated by the presence of butyrate at a lower concentration (around 0.2 mm) (10). Given that butyrate can relax the chromatin structure by functioning as a histone deacetylase inhibitor, a higher (0.5–1 mm) optimal concentration in 4F reprogramming might indicate that establishment of pluripotency may need more accessible chromatin structure compared with that required for maintenance of ES cell status. Although butyrate is widely used as a differentiation reagent when applied at a higher concentration (≥1 mm) (11), we did not notice differentiation of the fully reprogrammed Sox2-GFP+ colonies during the time of butyrate treatment. Neither did we observe any negative effect of butyrate on the quality of iPS cells derived in the presence of butyrate. The apparent conflicting roles that butyrate displays in differentiation and iPS cell generation suggest that this epigenetic modulator may generally facilitate cell fate changes by increasing the flexibility of chromatin. In this regard, it will be interesting to test whether butyrate can facilitate transdifferentiation between different cell types, for example from fibroblasts to muscle cells (14) and from B cells to macrophage (15).
In addition to facilitating iPS cell generation, we also noticed that butyrate can reduce the GFP− colony numbers, regardless of their AP activity status. This effect results in a significant increase in the ratio of GFP+ colonies, representing authentically reprogrammed iPS cells (Fig. 1, B and C). The AP−GFP− colonies are probably cells that failed to express all four reprogramming factors. These cells usually exhibit properties of transformed cells, such as granulated or cobblestone morphology with fast cell growth (2). Elimination of these AP−GFP− colonies can be attributed to the well characterized anti-cancer effect of butyrate (12), including induction of p21 and Arf (supplemental Fig. 2). On the other hand, AP+GFP− colonies most likely represent partially reprogrammed cells that somehow have not achieved the pluripotent cell fate (9, 13). With the assistance of butyrate, these partially reprogrammed cells may gain full pluripotency and express Sox2-GFP contributing to the observed higher reprogramming efficiency. We note that although VPA, another histone deacetylase inhibitor, can also increase the GFP+ colony number, it does not seem to have the same capacity to suppress the total colony numbers (Fig. 1B). Another epigenetic modulator, 5-aza-cytidine, has been shown capable of facilitating conversion of partially reprogrammed cells to fully reprogrammed cells (9). It will be interesting to test whether butyrate has a similar property. Given that butyrate functions at an early stage of reprogramming, whereas 5-aza-cytidine facilitates conversion of partially reprogrammed cells to fully reprogrammed cells, it will be interesting to test whether they can function synergistically to facilitate the reprogramming process.
Because reprogramming is a process with multiple steps (16, 17), we determined the functioning time window of butyrate to be at an early stage of the reprogramming process (Fig. 4). This is consistent with our finding that butyrate facilitates reprogramming only in the presence of exogenous c-Myc (Fig. 2) as c-Myc has been suggested to contribute to the early events of reprogramming (13). To explore the effect of butyrate in the early reprogramming process at the molecular level, we analyzed the genome-wide expression profiles of cells undergoing reprogramming in this time window. We observed a trend of up-regulation for ES cell-enriched genes in response to butyrate (Fig. 5A), but did not detect genome-wide down-regulation of MEF-enriched genes (data not shown). This observation is consistent with the role of butyrate in gene activation as a histone deacetylase inhibitor. Furthermore, we identified 19 ES cell-enriched genes that are specifically up-regulated by butyrate only when c-Myc is included as a reprogramming factor (supplemental Table 2). It remains to be determined whether up-regulation of these genes mediates the effect of butyrate. Given that reprogramming efficiency can be increased by suppression of the p53-p21 pathway (18–21) as well as elimination of the senescence barrier imposed by Ink4a and/or Arf (22, 23), we also analyzed whether butyrate can suppress the expression of p53, p21, Ink4a, and Arf. Instead of down-regulation, we indeed observed a slight up-regulation of some of these genes (p21 and Arf) by the butyrate treatment (supplemental Fig. 2), largely excluding the involvement of p53-p21 pathway and Arf in mediating the butyrate effects.
During the preparation of this paper, Mali et al. (24) reported that butyrate greatly facilitates human iPS cell generation. In their report, reprogramming efficiency is increased remarkably by butyrate even in the absence of c-Myc and Klf4. Another difference between these two studies is the timing at which butyrate exhibits its effect. Although it exerts an effect at an early stage during reprogramming in mouse cell reprogramming, Mali et al. reported a later effect in human cells (24). Furthermore, although we noticed an inhibitory effect for transformed cells and partially reprogrammed cells, it is not clear whether a similar effect was seen in human cells. Whether these differences are due to the endogenous c-Myc levels, the different durations required to achieve reprogramming for human and mouse MEFs, or other technical aspects remains to be determined. Nonetheless, the demonstration that butyrate, an histone deacetylase inhibitor, is capable of facilitating iPS cell generation suggests that alteration of epigenetic status is an important step for the establishment of pluripotency.
Supplementary Material
Acknowledgments
We thank Larysa Pevny for the Sox2-GFP transgenic mice, Jin He for technical help, and Anh Tram Nguyen for critical reading of the manuscript.
This work was supported, in whole or in part, by a National Institutes of Health Beta Cell Biology Consortium project through the NIDDK.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2, Tables 1 and 2, and Methods.
- iPS
- induced pluripotent stem
- AP
- alkaline phosphatase
- ES
- embryonic stem
- GFP
- green fluorescent protein
- MEF
- mouse embryonic fibroblast
- MES
- 4-morpholineethanesulfonic acid
- RT
- reverse transcription
- VPA
- valproic acid.
REFERENCES
- 1.Maherali N., Sridharan R., Xie W., Utikal J., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R., Plath K., Hochedlinger K. (2007) Cell Stem Cell 1, 55–70 [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 3.Okita K., Ichisaka T., Yamanaka S. (2007) Nature 448, 313–317 [DOI] [PubMed] [Google Scholar]
- 4.Wernig M., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B. E., Jaenisch R. (2007) Nature 448, 318–324 [DOI] [PubMed] [Google Scholar]
- 5.Huangfu D., Osafune K., Maehr R., Guo W., Eijkelenboom A., Chen S., Muhlestein W., Melton D. A. (2008) Nat. Biotechnol. 26, 1269–1275 [DOI] [PubMed] [Google Scholar]
- 6.Huangfu D., Maehr R., Guo W., Eijkelenboom A., Snitow M., Chen A. E., Melton D. A. (2008) Nat. Biotechnol. 26, 795–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Y., Desponts C., Do J. T., Hahm H. S., Schöler H. R., Ding S. (2008) Cell Stem Cell 3, 568–574 [DOI] [PubMed] [Google Scholar]
- 8.Shi Y., Do J. T., Desponts C., Hahm H. S., Schöler H. R., Ding S. (2008) Cell Stem Cell 2, 525–528 [DOI] [PubMed] [Google Scholar]
- 9.Mikkelsen T. S., Ku M., Jaffe D. B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T. K., Koche R. P., Lee W., Mendenhall E., O'Donovan A., Presser A., Russ C., Xie X., Meissner A., Wernig M., Jaenisch R., Nusbaum C., Lander E. S., Bernstein B. E. (2007) Nature 448, 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ware C. B., Wang L., Mecham B. H., Shen L., Nelson A. M., Bar M., Lamba D. A., Dauphin D. S., Buckingham B., Askari B., Lim R., Tewari M., Gartler S. M., Issa J. P., Pavlidis P., Duan Z., Blau C. A. (2009) Cell Stem Cell 4, 359–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newmark H. L., Lupton J. R., Young C. W. (1994) Cancer Lett. 78, 1–5 [DOI] [PubMed] [Google Scholar]
- 12.Bolden J. E., Peart M. J., Johnstone R. W. (2006) Nat. Rev. Drug Discov. 5, 769–784 [DOI] [PubMed] [Google Scholar]
- 13.Sridharan R., Tchieu J., Mason M. J., Yachechko R., Kuoy E., Horvath S., Zhou Q., Plath K. (2009) Cell 136, 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis R. L., Weintraub H., Lassar A. B. (1987) Cell 51, 987–1000 [DOI] [PubMed] [Google Scholar]
- 15.Xie H., Ye M., Feng R., Graf T. (2004) Cell 117, 663–676 [DOI] [PubMed] [Google Scholar]
- 16.Stadtfeld M., Maherali N., Breault D. T., Hochedlinger K. (2008) Cell Stem Cell 2, 230–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brambrink T., Foreman R., Welstead G. G., Lengner C. J., Wernig M., Suh H., Jaenisch R. (2008) Cell Stem Cell 2, 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong H., Takahashi K., Ichisaka T., Aoi T., Kanagawa O., Nakagawa M., Okita K., Yamanaka S. (2009) Nature 460, 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawamura T., Suzuki J., Wang Y. V., Menendez S., Morera L. B., Raya A., Wahl G. M., Belmonte J. C. (2009) Nature 460, 1140–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanna J., Saha K., Pando B., van Zon J., Lengner C. J., Creyghton M. P., van Oudenaarden A., Jaenisch R. (2009) Nature 462, 595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marión R. M., Strati K., Li H., Murga M., Blanco R., Ortega S., Fernandez-Capetillo O., Serrano M., Blasco M. A. (2009) Nature 460, 1149–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Utikal J., Polo J. M., Stadtfeld M., Maherali N., Kulalert W., Walsh R. M., Khalil A., Rheinwald J. G., Hochedlinger K. (2009) Nature 460, 1145–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H., Collado M., Villasante A., Strati K., Ortega S., Cañamero M., Blasco M. A., Serrano M. (2009) Nature 460, 1136–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mali P., Chou B. K., Yen J., Ye Z., Zou J., Dowey S., Brodsky R. A., Ohm J. E., Yu W., Baylin S. B., Yusa K., Bradley A., Meyers D. J., Mukherjee C., Cole P. A., Cheng L. (2010) Stem Cells 28, 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.