Abstract
Trypanosoma cruzi causes Chagas disease (American trypanosomiasis), which threatens the lives of millions of people and remains incurable in its chronic stage. The antifungal drug posaconazole that blocks sterol biosynthesis in the parasite is the only compound entering clinical trials for the chronic form of this infection. Crystal structures of the drug target enzyme, Trypanosoma cruzi sterol 14α-demethylase (CYP51), complexed with posaconazole, another antifungal agent fluconazole and an experimental inhibitor, (R)-4′-chloro-N-(1-(2,4-dichlorophenyl)-2-(1H-imid-azol-1-yl)ethyl)biphenyl-4-carboxamide (VNF), allow prediction of important chemical features that enhance the drug potencies. Combined with comparative analysis of inhibitor binding parameters, influence on the catalytic activity of the trypanosomal enzyme and its human counterpart, and their cellular effects at different stages of the Trypanosoma cruzi life cycle, the structural data provide a molecular background to CYP51 inhibition and azole resistance and enlighten the path for directed design of new, more potent and selective drugs to develop an efficient treatment for Chagas disease.
Keywords: Crystal Structure, Cytochrome P450, Enzyme Inhibitors, Membrane Lipids, Parasitology, Protein Structure, Chagas Disease, Trypanosoma Cruzi, Sterol 14alpha-Demethylase (CYP51)
Introduction
Chagas disease is a deadly protozoan infection endemic in Central and South America.2 With human migration and HIV co-infections, it also is now found in all other parts of the globe (2–5). Worldwide, an estimated 16 to 18 million people are infected with Trypanosoma cruzi (TC), and ∼50,000 people die each year (3). The parasite is transmitted to humans and >100 mammalian species that form a reservoir by Triatomine insect vector (kissing bugs), bites, feces, and food contamination. The infection also spreads from mother to child, congenitally and by breastfeeding, via organ transplantation, and blood transfusion. In 2007, most blood banks in the U.S. began screening for Chagas disease (3, 6). After a nonspecific, frequently flu-like acute stage of infection, which sometimes passes unnoticed, TC enters the host cells, and the chronic stage develops in about one-third of patients 5 to 15 years later. Most often, it affects the heart (cardiomegaly, arrhythmias, cardiomyopathy, consequently causing heart failure and death) and the gastrointestinal tract (megaesophagus and megacolon), though the parasites can be found in other organs and tissues.
For more than a century, Chagas disease has remained one of the most neglected and deadly human infections with only two available clinical drugs, benznidazole and nifurtimox. Toxic and nonspecific, they are effective only for the acute stage but do not cure the chronic stage of Chagas. Corticosteroids, pacemakers, and medications for treating cardiac arrhythmias are aimed only to manage the symptoms. Even heart transplantation or recently suggested development of stem cell therapy (7) cannot assure a cure because the parasite from other tissues often re-enters the heart (3). Current approaches for development of new therapies for Chagas disease include blind screening for compounds that produce antiparasitic effects in cellular experiments, searches for new drug targets in the parasite genome, as well as searches for effective compounds to act on potential drug targets known to be essential for parasite biology (8–10).
Similar to fungi and yeasts, TC is strictly dependent on endogenously produced sterols. Ergosterol-like molecules are required for parasite membranes, growth, development, and division (11, 12). Sterol 14α-demethylase (14DM,3 CYP51 gene family), the membrane-bound cytochrome P450 catalyzing oxidative removal of the 14α-methyl group from the sterol core, is a highly drug-targetable enzyme in sterol biosynthetic pathways (13). 14DM is strongly inhibited by heterocyclic compounds with a basic atom that coordinates to the heme iron affecting substrate binding and catalysis. 14DM inhibitors (imidazole and triazole derivatives; see Fig. 1a) are among the most efficient current antifungal drugs (14, 15). In addition to blocking ergosterol production, antifungal azoles cause accumulation of toxic methylated sterol precursors, leading to pathogen growth arrest and cell death (16). The antiparasitic effect of antifungal azoles on TC has been observed by several investigators (17–22); yet, only recently, the antifungal drug posaconazole, proven to be capable of producing a parasitological cure in a mouse model of the chronic stage of Chagas disease, has been reported to be entering clinical trials in June 2010 (23).
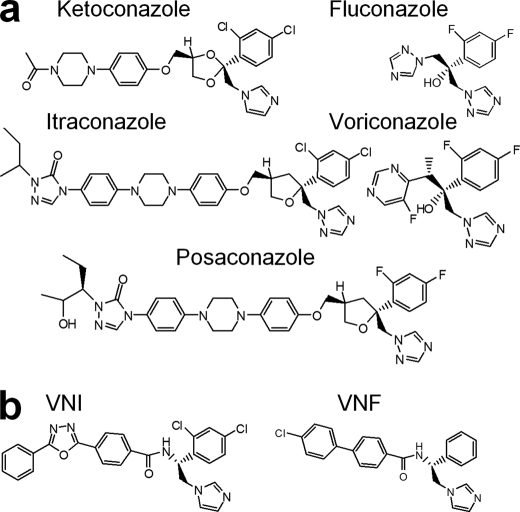
FIGURE 1.
14DM inhibitors. a, five azoles currently available for systemic use as clinical antifungal drugs. b, VNI and VNF represent a novel experimentally identified inhibitory scaffold selective for trypanosomal 14DMs.
Posaconazole is a highly potent broad spectrum antifungal drug, approved by the U.S. Food and Drug Administration in 2006 as salvage therapy for invasive fungal infections in immunocompromised patients (15, 17, 24). The disadvantages of this drug include its cost and severe adverse side effects, especially gastrointestinal problems, because the drug is produced only in an orally available form. To our knowledge, the direct inhibitory effect of posaconazole on 14DM catalysis has not yet been tested.
Fluconazole is a cheap first generation antifungal agent (15). Water soluble and well tolerated, it easily penetrates the blood-brain barrier and currently is used for treatment of yeast infections but has much lower efficiency for filamentous fungi.
VNF ((R)-4′-chloro-N-(1-(2,4-dichlorohenyl)-2-(1H-imidazol-1-yl)ethyl)biphenyl-4-carboxamide) (Fig. 1b) is an experimental inhibitor (12). Highly potent for trypanosomal 14DMs, this compound displays low general cytotoxicity and does not inhibit the human 14DM ortholog, which makes this carboxamide-containing inhibitory scaffold potentially advantageous in the cases of long term treatment.
Acquired azole resistance, the major problem in current fungal therapy (25), is another reason to search for new 14DM inhibitors. Activation of the efflux pumps that help to expel azoles from the fungal cells has been proven for ketoconazole, voriconazole, and fluconazole (15, 16, 24). Moreover, easily induced resistance to fluconazole was demonstrated in the laboratory strains of TC (26). Posaconazole has been reported not to induce the efflux pump mechanism (24); however, sparse cases of resistance associated with point mutations in the fungal 14DM have been observed (27).
Here, we describe the structures of TC14DM, in complex with posaconazole, fluconazole, and VNF. Accompanied by comparative analysis of the inhibitor binding parameters, inhibitory potencies in the reconstituted enzyme reaction in vitro and antiparasitic effects in TC cells, the structural data not only elucidate inhibitory details but uncover important general aspects in 14DM inhibition, which we are showing to be likely applicable for the fungal 14DMs as well. Interestingly, despite the differences in the inhibitor structures, molecular volumes, and binding poses, they do not cause large scale conformational rearrangements in the enzyme, filling the available active site cavity without significantly changing its shape and topology. This characterizes 14DM as possibly one of the most structurally rigid members of the cytochrome P450 superfamily (28) and provides an excellent basis for structure-directed design of new, highly potent and ideally pathogen-specific drugs. Analysis of the 14DM expression in TC confirms that the gene is regulated developmentally and essential at all stages of the parasite life cycle. In contrast to posaconazole and fluconazole, VNF does not induce an obvious increase in the 14DM gene expression. Low doses of VNF produce a steady antiparasitic effect over time, which distinguishes this inhibitory scaffold as a very promising lead for antitrypanosomal therapy.
EXPERIMENTAL PROCEDURES
TC14DM Gene Cloning and Modifications
The gene-encoding 14DM was amplified from T. cruzi genomic DNA with the addition of a His6 tag at the protein C terminus and cloned into a pCW expression plasmid as described previously (29). For crystallization purposes, the N-terminal transmembrane domain upstream of Pro32 was replaced with MAKKTSSKGKL- (28) in the construct used for co-crystallization with fluconazole and VNF (construct 1) and with MAKKT- (5′-ATGGTCAAGAAAACG-3′) in the complex with posaconazole (construct 2). All gene modifications were carried out in pET17b (Novagen) and verified by DNA sequencing.
Purification and Crystallization of TC14DM
Expression and purification of the full-length TC and human 14DMs was reported elsewhere (29, 30). The N-terminal truncation resulted in the increased expression levels of TC14DM, up to 0.7 and 1.5 μmol/liter for constructs 1 and 2, respectively. Truncated protein was purified following the same steps as the full-length TC14DM (29), including nickel-nitrilotriacetic acid (Qiagen) and ion-exchange chromatography (Q-Sepharose followed by SP-Sepharose, Amersham Biosciences), except that Triton X-100 was replaced by 0.048 mm n-tridecyl-β-d-maltoside (Anatrace). The initial screening of crystallization conditions was performed using Hampton Research crystallization kits. Crystals were obtained in hanging drops at 25 °C from equal mixtures of 260 μm cytochrome P450 solution in 20 mm potassium phosphate buffer, pH 7.2, containing 200 mm NaCl, 0.1 mm EDTA, 10% glycerol, and 0.048 mm n-tridecyl-β-d-maltoside preincubated with 1.2-fold molar excess of posaconazole or VNF and 2-fold molar excess of fluconazole against a well solution containing 0.2 m potassium formate, pH 7.2, or 0.2 m sodium formate, pH 7.4, in the case of VNF, and 15% (w/v) polyethylene glycol 3550.
Data Collection, Structure Determination, and Analysis
Crystals were soaked briefly in a 40% glycerol cryo-buffer and flash-cooled in liquid nitrogen. Data were collected at the Advanced Photon Source, Argonne National Laboratory, Life Sciences Collaborative Access Team, beamline 21ID-F (posaconazole complex), and Southwest Regional Collaborative Access Team, beamlines 22ID and 22BM for fluconazole and VNF complexes. The data were processed with the HKL2000 software package (31). The posaconazole-bound structure was solved by molecular replacement using ligand-free Trypanosoma brucei (Tbb) 14DM (3g1q) as a search ensemble in PhaserMR (CCP4 suite; 32). Model building and refinement were performed with COOT (33) and REFMAC5 (CCP4 suite), respectively. Structures of the TC14DM complexes with fluconazole and VNF were solved using the coordinates of TC14DM complexed with posaconazole as a search ensemble. Supplemental Table S1 summarizes the diffraction and refinement data statistics. The coordinates and structure factors have been deposited at the Protein Data Bank under codes 3K1O, 3KHM, and 3KSW, for posaconazole, fluconazole, and VNF-bound TC14DM, respectively. The codes of other structures related to this work are 3G1Q and 3GW9 (Tbb14DM) and 3LD6 (most recent release for human 14DM). Structure superpositions were done in LSQkab of the CCP4 suite. Molecular volumes were calculated in Accelrys Discovery Studio Visualizer 2.5 (probe radius 1.4 Å). Figures were prepared with Accelrys, Chimera, and PyMOL.
Spectrophotometric Studies and Catalytic Assays
UV-visible spectra were recorded on a dual-beam spectrophotometer (Shimadzu UV-2401PC) in 1-cm optical path length quartz cuvettes. P450 concentration was quantified from the Soret band absolute absorbance of the ligand-free ferric form of the protein and confirmed by carbon monoxide difference spectra using extinction coefficients ϵ418 nm and Δϵ(490–448) nm of 117 and 91 mm−1 cm−1, respectively (29). Titrations with the inhibitors were carried out at cytochrome P450 concentration of 2 μm and difference spectra were recorded between 350 and 500 nm. The apparent dissociation constants were calculated by plotting P450 spectral response (ΔA420–390) against free ligand concentration as described previously (30). Difference CO-binding spectra were recorded between 400 and 500 nm, 14DM was reduced with sodium dithionite, and CO gas was bubbled through the sample cuvette. Influence of the inhibitors on 14DM enzymatic activity was assayed at 1 μm P450 as described (12) using 24-methylenedihydrolanosterol and lanosterol as substrates (50 μm) for TC and human 14DMs, respectively.
Immunoblot Analysis of 14DM Expression in T. cruzi
To evaluate expression of 14DM in the three developmental stages of T. cruzi, 5 μg of whole extracts of trypomastigote (12), epimastigote (12), and amastigote (34) forms of the highly invasive 20A clone of the Tulahuen strain (35) were solubilized for 10 min at 96 °C in a nonreducing SDS mixture (2% SDS, 10% glycerol, 0.005% bromphenol blue, and 80 mm Tris-HCL, pH 6.8), subjected to SDS-PAGE, transferred to nitrocellulose membranes, and probed with a 1:2000 dilution of mouse anti-Tbb14DM antiserum (28) or with the same dilution of preimmune serum. Bound antibodies were detected by chemiluminescence (catalog no. 34077, Thermo Fisher Scientific).
Immunogold Localization of 14DM within T. cruzi Cells
T. cruzi epimastigotes (108 organisms) were fixed with 2.5% paraformaldehyde and embedded in low acryl resin as described (36). Ultrathin cell sections were probed with mouse anti-Tbb14DM antiserum or with mouse preimmune serum, followed by incubation with anti-mouse IgG conjugated with 10-nm gold particles (catalog no. RPN431V, GE Healthcare), stained with 0.25% phosphotungstic acid as described (36) and observed with a Phillips CM-12 electron microscope. The sections probed with preimmune serum did not show reactivity.
Cellular Infection Assays
T. cruzi-expressing GFP were generated by transfection with pTRex-GFP plasmid (kindly provided by Dr. P. Guevara) (37), followed by clonal selection with G418. A single clone of epimastigotes with high GFP expression was used to generate GFP-transgenic trypomastigotes by metacyclogenesis in low pH buffer. For inhibition of T. cruzi infection within cardiomyoblasts by 14DM inhibitors, GFP-expressing trypomastigotes (8 × 104 organisms) were pretreated for 30 min with several concentrations of the inhibitors dissolved in dimethyl sulfoxide/Dulbecco's modified Eagle's medium free of phenol red and exposed in triplicates to cardiomyocyte monolayers (each at 4 × 103 cells) in 96-well tissue culture plates for 12 h. Unbound trypomastigotes were removed by washing with Dulbecco's modified Eagle's medium. Infected monolayers were incubated for 72 h for parasite multiplication and washed with phosphate-buffered saline, and the infection was quantified using a Synergy HT fluorometer (Biotek Instruments). For fluorescence microscopic observation of the inhibition of T. cruzi multiplication within cardiomyocytes, the monolayers were fixed with 2.5% paraformaldehyde and stained with 4′,6-diamidino-2-phenylindole to visualize DNA. T. cruzi epimastigotes (106 cells/ml) were exposed to different concentrations of 14DM inhibitors and mock-treated in triplicates, and the kinetics of growth in brain infusion medium was evaluated microscopically during 6 days.
RESULTS
Binding and Inhibition of TC14DM with Posaconazole, Fluconazole, and VNF
Although the apparent Kds determined by spectral titration are useful for primary identification of new binding ligands, they do not always correlate with inhibition of 14DM activity (12). Thus, we found that several imidazole derivatives that reveal nanomolar Kds were easily replaced in the enzyme reaction by substrate, whereas VNF derivatives were not (12). On the other hand, some pyridine derivatives, which induce moderate spectral responses (the apparent Kds ∼ 0.2–0.5 μm), turned out to be rather strong inhibitors of TC14DM (38). This contradiction might be explained by the fact that because 14DM spectral responses to azoles (an example is shown in supplemental Fig. S1a) only reflect displacement of a weaker ligand (water) from the sixth iron coordination sphere (28), they might be enhanced by basicity of the iron-coordinating atom (imidazole > triazole > pyridine (39)), whereas the effects of additional lipophilic interactions with the protein residues may sometimes remain unrecognized. Therefore, to estimate the inhibitors relative affinities to 14DM, we also have monitored their influence on the formation of the reduced P450-CO complexes (supplemental Fig. S1b). This approach appears to be rather sensitive, because it reflects the ability of CO to replace the inhibitors in the iron coordination sphere while the enzyme is in the reduced, reactive state. Based on the binding parameters and inhibitory effects on enzyme activity (Table 1), fluconazole is the weakest ligand/inhibitor for TC14DM, whereas the potencies of posaconazole and VNF are comparable; they both completely inhibit TC14DM activity at 1:1 molar ratio inhibitor/enzyme, yet VNF reveals higher selectivity (no influence on the host (human) 14DM catalysis at up to 200-fold molar excess).
TABLE 1.
Binding parameters and inhibition of 14DM activity by posaconazole, fluconazole, and VNF
| TC14DM | No azole | Posaconazole | Fluconazole | VNF |
|---|---|---|---|---|
| Azole binding: spectral titrations of ferric form | ||||
| Soret band maximum (nm) | 418.5 | 422.5 | 422.5 | 424.5 |
| Apparent Kd (μm) | 0.06 ± 0.01 (1.9 ± 0.12)a | 0.44 ± 0.03 (40 ± 2.11) | 0.07 ± 0.01 (0.3 ± 0.04) | |
| Influence on CO complex formation: ferrous form | ||||
| Rate (%/min) | 80 | ndd | 22 | ndd |
| Total (%) | 100 | <5 | 70 | <5 |
| Inhibitory effect on 14DM catalysis | ||||
| I/E2b | <1 (40) | 1.8 (>200) | <1 (>200) | |
| Activity at 1:1 molar ratio I/E (%)c | ||||
| Initial rate | 0 | 70 | 0 | |
| Substrate conversion after a 1-h reaction | 0 | 100 | 0 | |
a Data in parentheses are for the human 14DM ortholog.
b This refers to the molar ratio inhibitor/enzyme that causes a 2-fold decrease in the initial rate of catalysis (12).
c The P450 concentration in the reaction was 1 μm (see “Experimental Procedures”).
d nd, not determined.
Structural Characterization of the Enzyme-Inhibitor Complexes
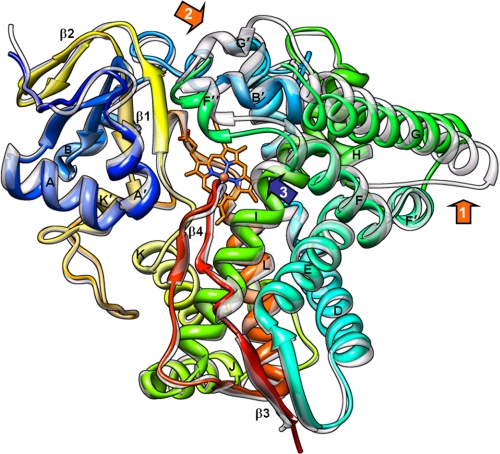
Binding of any of the three structurally different inhibitors does not cause large scale conformational rearrangements in TC14DM. Overall, the structures are similar to that of ligand-free 14DM from another pathogen, Trypanosoma brucei (P1 space group) (Fig. 2), which we have published recently (28). Only two loop regions, FG and GH, that have the highest B-factor (elevated flexibility) in Tbb14DM (1.9 Å) in the TC14DM structures clearly display different conformations and are partially disordered (Fig. 3, left panel). Similar to the inhibitor-bound Tbb14DM, the middle portion of the I helix (Ala291 region) in the TC14DM-inhibitor complexes is pushed ∼1.5 Å away from the heme iron (Fig. 2), releasing additional space for azole ring coordination, thus supporting our previous suggestion (28) that close location of the I helix to the heme might be a structural signature of the CYP51 family. Such proximity may provide the basis for the 14DMs elevated susceptibility to azole inhibitors and consequent high drug targetability.
FIGURE 2.
Overall 14DM structure. Posaconazole-bound TC14DM (rainbow color, from blue N terminus to red C terminus) is superimposed with the ligand-free Tbb14DM (semitransparent gray); root mean square deviation for all Cα atoms is 0.91 Å. Major P450 structural elements are marked. The heme is shown in stick representation; posaconazole was deleted for clarity. The most flexible areas (GH loop and FG loop regions) are marked with orange arrows, 1 and 2, respectively. Repositioning the rigid I helix that is observed in all inhibitor bound trypanosomal 14DMs is marked with a blue arrow 3.
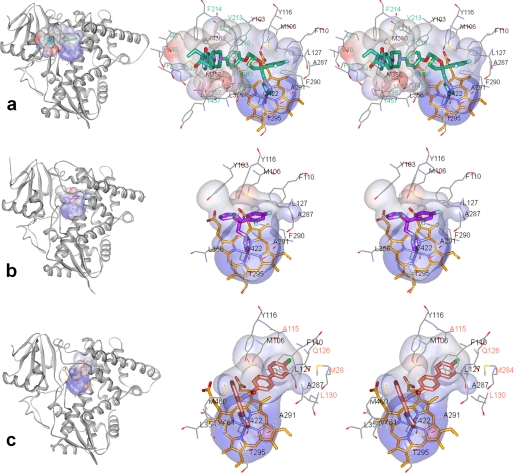
FIGURE 3.
Location of the inhibitors in the TC14DM structures (left) and stereoview of the active site (right) in complexes with: posaconazole (a), fluconazole (b), and VNF (c). The protein is in the same orientation shown from the distal view; the backbone is displayed as gray ribbons. The active site surfaces (Accelrys) are colored by interpolating charge; the inhibitors and the heme are shown in stick representation; the residues located within Van der Waals bond distances (4.5 Å) are depicted by lines. For posaconazole, fluconazole, and VNF (throughout all figures) the carbons are colored in green, violet, and salmon, respectively; the heme carbons are orange. Nitrogens are blue, oxygens are red, chlorines are gray, fluorines are light blue, sulfurs are orange; protein atoms follow the same scheme except carbons are gray. Residue labels are color-coded according to their specificity, with green labels for the posaconazole and salmon labels for VNF-specific residues. The list of inhibitor-contacting residues in the three TC14DM structures and in the VNI-bound Tbb14DM is provided as supplemental Table S2. The electron density maps for the inhibitors are presented in supplemental Fig. S2.
TC14DM-Posaconazole
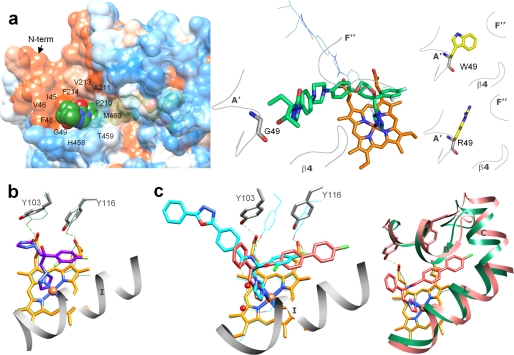
The profound inhibitory potency of posaconazole appears to result from multiple interactions that tightly bind and position the inhibitor within the TC14DM active site geometry (Fig. 3a). Posaconazole is the largest among the three inhibitors, with a molecular volume of 673 Å3 (Accelrys). Within Van der Waals bond distances (<4.5 Å), it is surrounded by 25 residues; 13 of them are located in the active site cavity, and 12 others are from the access channel. The deep hydrophobic active site cavity buries the inhibitor triazole ring, fluorinated aromatic ring and the proximal part of its long arm. The active site cavity residues, Tyr103, Met106, Phe110, and Tyr116 are contributed by the B′ helix/B′C loop; Leu127 is contributed by helix C; Ala297, Phe290, Ala291, and Thr295 are provided by helix I; and Leu356, Leu357, Met358, and Met360 are from the αK–β1–4 loop. The access channel accommodates the middle portion of the posaconazole arm, the interactions being formed around the entrance, 20–25 Å from the heme iron, by helices A′ (Ile45, Val46, Phe48, and Gly49), F″ (Pro210, Ala211, Val213, and Phe214), and the β4 hairpin (Tyr457, His458, Thr459, and Met460). These contacts with the inhibitor appear to have a stabilizing effect on the F″ helix, because only in the posaconazole-bound structure, the density for the helical structure is clearly defined. The distal part of the posaconazole arm extends outside of the protein molecule and is completely exposed to the surface (Fig. 4a, left panel). The arm is bent in comparison with its position in the energy-minimized solution model of posaconazole (Fig. 4a, right panel) accommodating the available shape within the enzyme binding cleft and potentially contributing to its inhibitory potency. Location of posaconazole in the TC14DM structure provides a possible explanation to posaconazole resistance observed in some strains (clinical isolates) of a pathogenic fungus Aspergillus fumigatus (27), where single point mutations resulted in substitutions of Gly54 (aligns with Gly49 in TC14DM) to arginine or tryptophan have been detected. Assuming that the fungal 14DM structure is similar to that of TC14DM, the introduction of a bulky and less flexible residue in this position might narrow the channel entrance and affect the surface interactions (Fig. 4a, right panel), thus decreasing the posaconazole inhibitory potency.
FIGURE 4.
Specific features in the binding of inhibitors. a, the long arm of posaconazole is extended above the protein surface forming 12 additional interactions around the access channel entrance (left panel). The surface is colored by hydrophobicity (from blue for the most polar to white to orange red for the most hydrophobic residues; the surfaces for the three TC14DM structures can be seen as supplemental Fig. S3). Right panel, posaconazole in the TC14DM structure (stick representation) superimposed with its energy-minimized model (line representation). Location of Gly49 in the structure and models of its mutations to the residues found in the posaconazole-resistant A. fumigatus 14DM are shown. b, fluconazole alters positions of conserved Tyr103 and Tyr116 affecting their hydrogen bonding with the heme propionates (rings A and D). The corresponding tyrosines and the heme in the superimposed posaconazole bound TC14DM structure are shown as green lines; the hydrogen bonds are depicted as dotted lines. c, VNF (salmon) binds to TC14DM in the orientation opposite to that of its scaffold analog VNI (cyan) in the Tbb14DM structure. Two water molecules (Tbb14DM) are displayed as red spheres (left panel). Right panel, the longer arm of VNF is intercalated between the B′ helix/B′C loop, helix C, and N-terminal (N-term) part of helix I, slightly broadening the active site cavity. The corresponding structural elements in the superimposed posaconazole-bound TC14DM are colored in green.
TC14DM-Fluconazole
The smallest of the three inhibitors, fluconazole (molecular volume of 348 Å3) is bound completely inside the active site cavity, contacting 10 of the 13 residues that interact with posaconazole (Fig. 3b). The significantly weaker inhibitory effect of fluconazole on TC14DM activity supports the importance of the entrance channel interactions in the complex of TC14DM with posaconazole. On the other hand, binding of the fluorinated aromatic ring of fluconazole between helices I and B′ alters the side chain positions of Tyr103 and Tyr116 and disrupts their hydrogen bonds with the heme propionates, rings A and D (Fig. 4b). (An electron density map for this region can be seen in supplemental Fig. S2b.) Disruption of this heme support might allow fluconazole to inhibit TC14DM activity selectively regardless of its small molecular volume. (The inhibitory effect of fluconazole on the Tbb14DM ortholog is ∼7-fold weaker (12).) It is conceivable that in Tbb14DM, the I helix ”pushing” effect on fluconazole can be neutralized by bulky Phe105 in the B′ helix (Ile105 in the TC14DM (29)).
TC14DM-VNF
VNF was an intriguing molecule for co-crystallization with TC14DM because this carboxamide-containing compound represents a novel, potent, and highly selective to the inhibitory scaffold of protozoan 14DMs. Closely related to VNI (Fig. 1b), whose structure we have determined recently in complex with Tbb14DM (28), even though smaller (molecular volumes 522 and 625 Å3, respectively), VNF produces the same, functionally irreversible effect on trypanosomal 14DMs (12), completely inhibiting their activity at a 1:1 molar ratio to the enzyme (Table 1).
Interestingly, in the TC14DM structure, we found VNF bound to the enzyme in the orientation opposite to that of VNI in Tbb14DM (Fig. 4c) including 180° rotation of its carboxamide fragment around the chiral carbon atom (supplemental Fig. S4). While the longer, three-ring arm of VNI lies in the access channel of Tbb14DM (like the posaconazole arm in TC14DM), the two-ring arm of VNF is intercalated between the C-terminal part of helix B′, B′C loop, helix C, and the N-terminal portion of helix I. As a result, in addition to the 10 posaconazole-contacting residues, the VNF complex with TC14DM involves four specific Van der Waals contacts, with Ala115 (B′C loop), Gln126, Leu127 (helix C), and Met284 (helix I) (Fig. 3c). To accommodate the two-ring arm of the inhibitor molecule, the N-terminal part of the I helix is pushed ∼1.4 Å away from the inhibitor, the B′ helical structure shortens (Val110 versus Gly111), and the elongated B′C loop expands ∼2Å above the corresponding loop in the complex with posaconazole. The N-terminal part of the C helix moves away, extending the proximal surface of the protein molecule and making additional space inside the globule. Reflecting these rearrangements within the active site cavity, the molecular volume of VNF-bound TC14DM also slightly increases (from ∼61,500 to ∼61, 900 A3).
Similar to VNI, which disrupts the hydrogen bond between Tyr103 and the heme ring A propionate in the Tbb14DM structure (28), VNF breaks the hydrogen bond between the TC14DM heme ring D propionate and Tyr116 (Fig. 3c). Observed in complexes of trypanosomal 14DMs with three different inhibitors, repositioning of these conserved and functionally important heme-supporting tyrosines (28) might be another general feature (related to the I helix proximity to the heme) that elevates susceptibility of 14DMs to heme-coordinating inhibitors. Finally, in the Tbb14DM structure, the interaction with VNI is strengthened by the hydrogen bond network formed between the amide group fragment specific for this inhibitory scaffold and two functionally essential protein regions, helices B′ (via Tyr103) and I (via Ala291) (28). The same can be true for VNF (Tyr116), although at medium resolution water molecules cannot be seen clearly in the TC14DM-VNF complex density map.
14DM and Effects of the Inhibitors in the Parasite
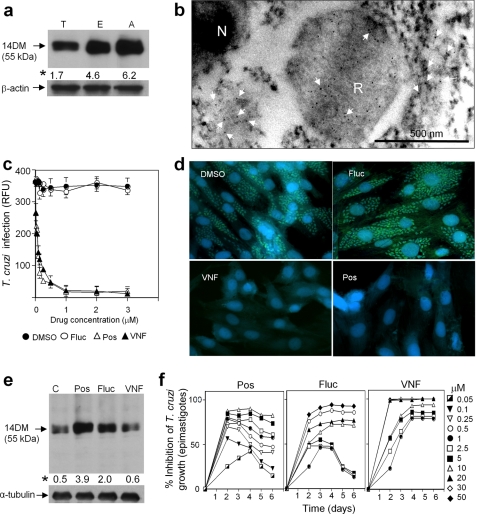
Like many other trypanosomes, TC has a complex life cycle, including insect (proliferating epimastigotes and infective metacyclic trypomastigotes) and mammalian (infective bloodstream trypomastigotes and proliferating intracellular amastigotes) forms. Contrary to Tbb, where CYP51 gene expression is >30-fold lower in the mammalian stages (28), it is expressed ubiquitously in TC, the protein amount increasing in multiplying amastigotes and epimastigotes (Fig. 5a). This confirms a crucial role of endogenous sterol production for all stages of the parasite life cycle. Subcellular distribution of TC14DM demonstrates that in addition to its expected localization in the cytoplasm (endoplasmic reticulum), a large amount of the enzyme is accumulated in reservosomes (Fig. 5b). These organelles are specific for trypanosomes. Surrounded by multiple vesicles and enriched in the lipid content, they are believed to represent the endocytic system in the parasite (40). It has been reported that the sterol biosynthetic pathway in Trypanosomatidae might have multiorganelle distribution, which in addition to ER, includes the mitochondrion and glycosomes (41–43). Localization of other enzymes from the sterol biosynthetic pathway in TC cells can be helpful to investigate this; however, it is not excluded that 14DM enters reservosomes on its way to other cellular compartments.
FIGURE 5.
14DM expression, cellular localization, and inhibition in T. cruzi. a, immunoblot analysis of 14DM expression in different life stages of the parasite. Equal concentrations of whole cell extracts of trypomastigotes (T), epimastigotes (E), and amastigotes (A) were separated by SDS-PAGE, blotted onto nitrocellulose, probed with mouse anti-Tbb14DM antiserum, and densitometrically scanned. The values (indicated by an asterisk) correspond to relative intensities normalized to the β-actin loading control. b, localization of 14DM in T. cruzi by immunogold electron microscopy. 14DM (arrows) is seen in the cytoplasm and reservosomes (R). N, nucleus. c, inhibition of the ability of trypomastigotes to infect cardiomyocytes by fluconazole (Fluc), posaconazole (Pos), and VNF. GFP-expressing trypomastigotes were pretreated with 14DM inhibitors and exposed to cardiomyocytes monolayers, and infected monolayers were incubated for 72 h to assay for parasite multiplication. The infection was evaluated by GFP fluorescence (RFU). The data represent the mean ± S.D. of the results from triplicate samples. d, fluorescence microscopic observation of the inhibition of T. cruzi multiplication by 1 μm 14DM inhibitors within cardiomyocytes. GFP-expressing amastigotes are seen as small green spots inside cardiomyocytes, and cardiomyocytes nuclei are blue. e, 14DM expression upon exposure of T. cruzi to azole inhibitors. Epimastigotes (106 organisms/ml) were allowed to grow for 5 days after exposure to inhibitors. 14DM expression was analyzed as in a, the values (indicated by an asterisk) were normalized to α-tubulin. f, dose-dependent inhibition of T. cruzi epimastigotes multiplication by 14DM inhibitors. The data represent the mean ± S.D. of the results from triplicate samples.
Both posaconazole and VNF inhibit the ability of TC trypomastigotes (the form prevalent in the acute stage of Chagas disease) to infect cardiomyocytes (Fig. 5c) at nanomolar concentrations, much stronger than does fluconazole (EC50 ∼ 40 μm, data not shown). At 1 μm, they completely destroy the parasite amastigotes (prevalent in the chronic stage of the disease), clearing the pathogen from cardiomyocytes (Fig. 5d). Quite remarkably, although treatment with posaconazole and fluconazole clearly induces an increase in TC14DM expression (which can be a warning sign for possible development of resistance, e.g. gene amplification), no such increase is seen in the case of VNF (Fig. 5e). Furthermore, we observed that while increasing concentrations of posaconazole and fluconazole are required to maintain the parasite growth inhibition, the effect of VNF remains steady over time (Fig. 5f). Though a more extended period of time is required to study possible induced resistance in the parasite, the results presented here strongly suggest that development of 14DM inhibitors with alternative binding modes can be highly beneficial.
DISCUSSION
It generally has been accepted for years that extremely wide distribution of cytochromes P450 in nature (more than 11,000 gene family members known to date),4 fulfilling multiple functions in living organisms and metabolizing an enormous variety of chemical structures has evolved largely due to high plasticity of the P450 structural fold (44). Particularly, the active site pockets of several drug-metabolizing P450s have been found to change their shape significantly to accommodate structurally different compounds (1, 45, 46); an example is shown in supplemental Fig. S6.
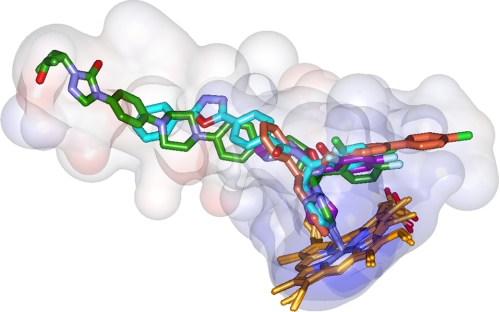
14DM (CYP51) is the only P450 family found in all biological kingdoms. At amino acid identities <25% across the kingdoms, these enzymes completely lack substrate promiscuity, catalyzing only one, strictly specific three-step reaction on the sterol core to remove the 14α-methyl group. Crystal structures of TC14DM support our suggestion (28) that it must be spatial rigidity of the active site cavity that provides the basis for the strict 14DM functional conservation. Contrary to other CYP families, in the 14DM structures, the inhibitors and not the enzyme reveal their flexibility, acquiring the space available inside the cleft in the protein. Superimposition of the 14DMs complexed with four different inhibitors depicts the 14DM active site cavity composition (Fig. 6), providing the first structure-based pharmacophore that outlines possible directions for further development. Thus, modifying the VNI structure by adding a posaconazole-like surface interaction subsite can further enhance its antiparasitic efficiency, while putting stronger accent on the development of the B′/C helices-bound portion of VNF might provide a helpful alternative for the cases of 14DM mutation-derived resistance.
FIGURE 6.
14DM structure-based inhibitory pharmacophore. Posaconazole is green, fluconazole is violet, VNF is salmon (TC14DM), and VNI (Tbb14DM) is cyan. Semitransparent (4-Å radius) surfaces are shown. Stereoview of the active site including contacting residues from all four structures can be seen in supplemental Fig. S5.
High selectivity of 14DM inhibitors toward parasitic targets with minimal potential harm to the human body is one of the critical issues in drug development. All three inhibitors reported here have a stronger inhibitory effect on TC14DM than on its human ortholog, with VNF being the most selective among them (Table 1). Superimposition of TC14DM with the recently reported structure of the human 14DM ortholog, confirms high structural similarity between the enzyme from the host and the parasite (supplemental Fig. S7); at 26% amino acid identity between the proteins, the structures reveal an only 1.7 Å root mean square deviation for the Cα atoms. However, contrary to the four trypanosomal 14DM structures, where the I helix is whole, in human 14DM, a low energy loop-like region is intercalated into the I helix in the closest to the heme iron area (supplemental Fig. S8). This suggests that relatively weaker susceptibility to many azole inhibitors can be an intrinsic feature of the human enzyme, possibly connected with higher flexibility of the central portion of its I helix (the P450 fold skeleton).
Altogether, the 14DM structures open an excellent opportunity for structure-directed, rational design of novel, highly potent, and selective inhibitors. Instead of blind screening, they allow concentration on the pharmacologically important features, such as toxicity, cellular permeability, solubility, volume of distribution, and lifetime in the human body. With multiple 14DM inhibitors available, diseases like Chagas and opportunistic fungal infections in immunocompromised patients may become more easily treatable, with the adverse side effects significantly minimized, and the potential problem of resistance may be resolved by using inhibitors with alternative binding modes.
Supplementary Material
Acknowledgments
We thank M. Gelb and F. Buckner (University of Washington) for posaconazole.
The work was supported, in whole or in part, by National Institutes of Health Grants GM 067871 (to M. R. W. and G. I. L.) and AI 080580 (to F. V.). This work was also supported by American Heart Association Grant 0535121N (to G. I. L.), and use of the Advanced Photon Source and Life Sciences Collaborative Access Team Sector 21 was supported by Grants DE-AC02-06CH11357 and 085P1000817.
The atomic coordinates and structure factors (codes 3K1O, 3KHM, and 3KSW) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S8.
Chagas disease, Pan American Health Organization.
D. R. Nelson, personal communication.
- 14DM
- sterol 14α-demethylase (CYP51)
- CYP
- cytochrome P450
- GFP
- green fluorescent protein
- Tbb
- Trypanosoma brucei
- TC
- Trypanosoma cruzi
- VNF
- (R)-4′-chloro-N-(1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl)biphenyl-4-carboxamide.
REFERENCES
- 1.Ekroos M., Sjogren T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13682–13687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moncayo A., Silveira A. C. (2009) Mem. Inst. Oswaldo Cruz. 104, 17–30 [DOI] [PubMed] [Google Scholar]
- 3.Neva F. A. (2007) Cecil Medicine:Expert Consult, Ch. 368, 23rd Ed., Saunders Elsevier, Philadelphia [Google Scholar]
- 4.Schmunis G. A. (2007) Mem. Inst. Oswaldo Cruz. 102, 75–85 [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez-Granado L. I., Rojo-Conejo P., Ruiz-Contreras J., Gonzalez-Tomé M. I. (2009) Lancet 373, 2025. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention (2007) MMWR Morb. Mortal. Wkly. Rep. 56, 141–14317318113 [Google Scholar]
- 7.Soares M. B., Santos R. R. (2009) Mem. Inst .Oswaldo. Cruz. 104, 325–332 [DOI] [PubMed] [Google Scholar]
- 8.Paulino M., Iribarne F., Dubin M., Aguilera-Morales S., Tapia O., Stoppani A. O. (2005) miniRev. Med. Chem. 5, 499–519 [DOI] [PubMed] [Google Scholar]
- 9.Moreira D. R., Leite A. C., dos Santos R. R., Soares M. B. (2009) Curr. Drug. Targets. 10, 212–231 [DOI] [PubMed] [Google Scholar]
- 10.Soeiro M. N., de Castro S. L. (2009) Expert .Opin. Ther. Targets 13, 105–121 [DOI] [PubMed] [Google Scholar]
- 11.Roberts C. W., McLeod R., Rice D. W., Ginger M., Chance M. L., Goad L. J. (2003) Mol. Biochem. Parasitol. 126, 129–142 [DOI] [PubMed] [Google Scholar]
- 12.Lepesheva G. I., Ott R. D., Hargrove T. Y., Kleshchenko Y. Y., Schuster I., Nes W. D., Hill G. C., Villalta F., Waterman M. R. (2007) Chem. Biol. 14, 1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lepesheva G. I., Waterman M. R. (2007) Biochim. Biophys. Acta. 1770, 467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petrikkos G., Skiada A. (2007) Int. J. Antimicrob. Agents. 30, 108–117 [DOI] [PubMed] [Google Scholar]
- 15.Zonios D. I., Bennett J. E. (2008) Semin. Respir. Crit. Care Med. 29, 198–210 [DOI] [PubMed] [Google Scholar]
- 16.Nagappan V., Deresinski S. (2007) Clin. Infect. Dis. 45, 1610–1617 [DOI] [PubMed] [Google Scholar]
- 17.Docampo R., Moreno S. N., Turrens J. F., Katzin A. M., Gonzalez-Cappa S. M., Stoppani A. O. (1981) Mol. Biochem. Parasitol. 3, 169–180 [DOI] [PubMed] [Google Scholar]
- 18.Beach D. H., Goad L. J., Holz G. G., Jr. (1986) Biochem. Biophys. Res. Commun. 136, 851–856 [DOI] [PubMed] [Google Scholar]
- 19.Urbina J. A., Lazardi K., Aguirre T., Piras M. M., Piras R. (1988) Antimicrob. Agents. Chemother. 32, 1237–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Apt W., Aguilera X., Arribada A., Pérez C., Miranda C., Sánchez G., Zulantay I., Cortés P., Rodriguez J., Juri D. (1998) Am. J. Trop. Med. Hyg. 59, 133–138 [DOI] [PubMed] [Google Scholar]
- 21.Araújo M. S., Martins-Filho O. A., Pereira M. E., Brener Z. J. (2000) Antimicrob. Chemother. 45, 819–824 [DOI] [PubMed] [Google Scholar]
- 22.Molina J., Martins-Filho O., Brener Z., Romanha A. J., Loebenberg D., Urbina J. A. (2000) Antimicrob. Agents. Chemother. 44, 150–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Urbina J. A. (2009) Mem. Inst. Oswaldo Cruz. 104, 311–318 [DOI] [PubMed] [Google Scholar]
- 24.Schiller D. S., Fung H. B. (2007) Clin. Ther. 29, 1862–1886 [DOI] [PubMed] [Google Scholar]
- 25.Kanafani Z. A., Perfect J. R. (2008) Clin. Infect. Dis. 46, 120–128 [DOI] [PubMed] [Google Scholar]
- 26.Buckner F. S., Wilson A. J., White T. C., Van Voorhis W. C. (1998) Antimicrob. Agents. Chemother. 42, 3245–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mann P. A., Parmegiani R. M., Wei S. Q., Mendrick C. A., Li X., Loebenberg D., DiDomenico B., Hare R. S., Walker S. S., McNicholas P. M. (2003) Antimicrob. Agents. Chemother. 47, 577–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lepesheva G. I., Park H. W., Hargrove T. Y., Vanhollebeke B., Wawrzak Z., Harp J. M., Sundaramoorthy M., Nes W. D., Pays E., Chaudhuri M., Villalta F., Waterman M. R. (2010) J. Biol. Chem. 285, 1773–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lepesheva G. I., Zaitseva N. G., Nes W. D., Zhou W., Arase M., Liu J., Hill G. C., Waterman M. R. (2006) J. Biol. Chem. 281, 3577–3585 [DOI] [PubMed] [Google Scholar]
- 30.Lepesheva G. I., Virus C., Waterman M. R. (2003) Biochemistry 42, 9091–9101 [DOI] [PubMed] [Google Scholar]
- 31.Otninowski Z., Minor W. (1995) HKL Manual, Yale University [Google Scholar]
- 32.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D. Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 33.Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34.Villalta F., Kierszenbaum F. (1982) J. Protozool. 29, 570–576 [DOI] [PubMed] [Google Scholar]
- 35.Lima M. F., Villalta F. (1989) Mol. Biochem. Parasitol. 33, 159–170 [DOI] [PubMed] [Google Scholar]
- 36.Madison M. N., Kleshchenko Y. Y., Nde P. N., Simmons K. J., Lima M. F., Villalta F. (2007) Infect. Immun. 75, 4780–4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guevara P., Dias M., Rojas A., Crisante G., Abreu-Blanco M. T., Umezawa E., Vazquez M., Levin M., Añez N., Ramirez J. L. (2005) J. Med. Entomol. 42, 48–56 [DOI] [PubMed] [Google Scholar]
- 38.Konkle M. E., Hargrove T. Y., Kleshchenko Y. Y., von Kries J. P., Ridenour W., Uddin M. J., Caprioli R. M., Marnett L. J., Nes W. D., Villalta F., Waterman M. R., Lepesheva G. I. (2009) J. Med. Chem. 52, 2846–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghomari M., Mokhlisse R., Laurence C., Le Questel J. Y., Bertelot M. (1997) J. Phys. Org. Chem. 10, 669–674 [Google Scholar]
- 40.Figueiredo R. C., Rosa D. S., Gomes Y. M., Nakasawa M., Soares M. J. (2004) Parasitology 129, 431–438 [DOI] [PubMed] [Google Scholar]
- 41.Quinones W., Urbina J. A., Dubourdieu M., Concepcion J. L. (2004) Exp. Parasitol. 106, 135–149 [DOI] [PubMed] [Google Scholar]
- 42.Peña-Diaz J., Montalvetti A., Flores C. L., Constán A., Hurtado-Guerrero R., De Souza W., Gancedo C., Ruiz-Perez L. M., Gonzalez-Pacanowska D. (2004) Mol. Biol. Cell 15, 1356–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Souza W., Rodrigues J. C. (2009) Interdiscip. Perspect. Infect. Dis. 2009, 642502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poulos T. L., Johnson E. F. (2005) Cytochrome P450: Structure, Mechanism, and Biochemistry, 3rd Ed., Kluwer Academic/Plenum Publishers, NY [Google Scholar]
- 45.Williams P. A., Cosme J., Vinkovic D. M., Ward A., Angove H. C., Day P. J., Vonrhein C., Tickle I. J., Jhoti H. (2004) Science 305, 683–686 [DOI] [PubMed] [Google Scholar]
- 46.Scott E. E., He Y. A., Wester M. R., White M. A., Chin C. C., Halpert J. R., Johnson E. F., Stout C. D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13196–13201 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.