Abstract
Hepatitis C virus (HCV) relies on many interactions with host cell proteins for propagation. Successful HCV infection also requires enzymatic activity of host cell enzymes for key post-translational modifications. To identify such enzymes, we have applied activity-based protein profiling to examine the activity of serine hydrolases during HCV replication. Profiling of hydrolases in Huh7 cells replicating HCV identified CES1 (carboxylesterase 1) as a differentially active enzyme. CES1 is an endogenous liver protein involved in processing of triglycerides and cholesterol. We observe that CES1 expression and activity were altered in the presence of HCV. The knockdown of CES1 with siRNA resulted in lower levels of HCV replication, and up-regulation of CES1 was observed to favor HCV propagation, implying an important role for this host cell protein. Experiments in HCV JFH1-infected cells suggest that CES1 facilitates HCV release because less intracellular HCV core protein was observed, whereas HCV titers remained high. CES1 activity was observed to increase the size and density of lipid droplets, which are necessary for the maturation of very low density lipoproteins, one of the likely vehicles for HCV release. In transgenic mice containing human-mouse chimeric livers, HCV infection also correlates with higher levels of endogenous CES1, providing further evidence that CES1 has an important role in HCV propagation.
Keywords: Biophysics, Lipid Droplet, Liver, Low Density Lipoprotein (LDL), Proteomics, Viral Replication, Virus Assembly, CARS Microscopy
Introduction
HCV2 is a growing global health problem, with ∼3% of the world population chronically infected, no vaccines available, and limited clinical treatments that are only successful in a small subset of patients (1). HCV is a positive strand RNA virus of the Flaviviridae family with a ∼9.6-kb genome (see Fig. 1A), which encodes a ∼3,000-amino acid polyprotein that is processed into 10 mature viral proteins (1–3). HCV is known to induce changes to lipid metabolism (4–7) and causes the formation of endoplasmic reticulum (ER)-derived membranous webs on which HCV replicates (3). A genome-wide siRNA screen identified 96 human genes that support HCV replication, with a significant number having a role in vesicle organization and biogenesis as well as metabolism- and membrane-related genes that may affect the membranous web (8). HCV also induces the accumulation of LDs on which certain HCV proteins are known to reside (9, 10). The accumulation of LDs leads to hepatic steatosis, which is defined as an elevated fat content in the liver that is accounted for by increases in triglyceride (TG) levels.
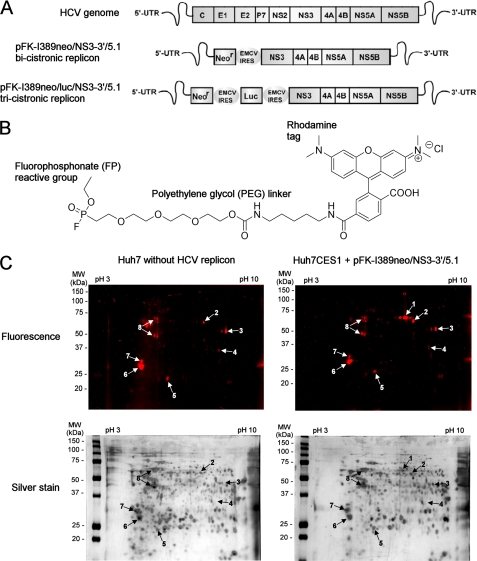
FIGURE 1.
Activity specific labeling of serine hydrolases with the FP-PEG-rhodamine probe in hepatoma cells replicating HCV identifies CES1 as a differentially active enzyme. A, schematic representations of the HCV genome and the bicistronic (pFK-I389neo/NS3-3′/5.1) and tricistronic (pFK-I389neo/luc/NS3-3′/5.1) subgenomic replicon variants used in this study (NS, non-structural proteins). B, chemical structure of the FP-PEG-rhodamine probe. C, the active proteome isolated from naive Huh7 cells or a unique Huh7CES1 hepatoma cell line stably expressing the pFK-I389neo/NS3-3′/5.1 replicon was labeled in vitro for 1 h at 37 °C with 1 nm FP-PEG-rhodamine probe (n = 3). After separation of the proteomes by two-dimensional gel electrophoresis, the fluorescence-tagged proteins, indicated with arrows, were identified by liquid chromatography-MS/MS (supplemental Table S1). UTR, untranslated region.
LDs are involved in the production of infectious virus particles (11). HCV capsid protein (core) recruits non-structural (NS) proteins and replication complexes to LD-associated membranes, which is critical for producing infectious viruses (11). Lipid droplets are composed mostly of neutral lipids belonging to TG and cholesterol esters (CE) encapsulated by a monolayer of phospholipids originating from the ER during lipid budding (12). The phospholipid monolayer is also associated with various lipid droplet proteins, mostly from the PAT family of proteins, which are involved in cell trafficking and signaling, and may play a role in modulating host-pathogen interactions (13, 14). Because the LDs are surrounded by a phospholipid monolayer (13, 14) and HCV replication complexes are likely enclosed by lipid bilayer membranes (15), the replication complexes are not likely to be directly associated with the membranes of LDs (11). Supporting this, NS5A has been shown to mainly localize near HCV core, resulting in a doughnut-shaped signal around LDs with a diameter slightly larger than that of core protein (11). Thus, virion assembly proceeds around the LD-associated membrane (11). Recently, HCV assembly and maturation was shown to occur in the ER and post-ER compartments with a dependence on apolipoprotein B similar to very low density lipoprotein (VLDL) production and secretion (16). This suggests that HCV uses LD components and associated membranes, such as the ER-derived membrane, which are part of the VLDL assembly and secretory machinery in order to propagate and disseminate. These dynamic and transient interactions encompass host molecule-HCV molecule interactions, requisite enzyme activity, and intracellular trafficking. Novel experimental approaches are needed to discover these interactions and determine their importance in HCV pathogenesis and persistence.
High throughput techniques, including gene expression profiling and proteomic approaches, have led to the identification of a number of important host-virus interactions in cell culture models of HCV replication and infection (5, 17). However, these methods provide virtually no information on the dynamic changes in the activity of enzymes and their zymogens. Activity-based protein profiling (ABPP) is a technique that offers direct insight into changes in catalytic activity of enzyme classes in complex proteomes (18, 19) and can be used to functionally annotate the enzyme function of proteins (20). ABPP has been successfully applied to a number of interesting systems, including cancer tumor growth and metastasis, identifying proteomic signatures that are associated with changes in cancer biology (21), to the study of metalloproteins of the extracellular matrix (22), and to the study of histone deacetylation (23), which is a cornerstone of epigenetics. Serine hydrolases comprise ∼1% of all predicted expressed human genes (24), and HCV itself encodes a serine hydrolase within the NS3/4A protein (3). Herein we have examined the activity profile of serine hydrolases during HCV replication to identify enzyme activity needed for HCV propagation and identified a new Huh7 cell line, Huh7CES1, that retains the ability to express CES1.
EXPERIMENTAL PROCEDURES
Proteome Extraction for Reaction with Fluorophosphonate (FP)-PEG-rhodamine
Subconfluent cells were washed and pooled with ice-cold 10 mm sodium phosphate buffer, pH 7. The cells were then subjected to Dounce homogenization at 30% power (T8 Ika-Werke Homogenizer, GMBH (Staufen, Germany)) and sonicated (20 pulses, 50% duty cycle; Sonifier 250, Branson Ultrasonic (Danbury, CT)) in ice-cold sodium phosphate buffer supplemented with 1% Triton X-100. The crude proteome extract was cleared by ultracentrifugation at 100,000 × g at 4 °C for 45 min, quantified with the BCA protein assay (Pierce), and diluted with sodium phosphate buffer to a final protein concentration of 1 mg/ml.
Active Proteome Labeling with FP-PEG-rhodamine
The active proteome labeling conditions were optimized as follows. The proteome extract (1 mg/ml in sodium phosphate buffer) was incubated in the dark with 1 nm FP-PEG-rhodamine (from a 0.5 μm stock in DMSO, final DMSO concentration of 0.2%) for 1 h at 37 °C, with occasional mixing. After 1 h, the reaction was quenched by precipitating the proteome with five volumes of ice-cold acetone to remove salts and unreacted probe.
Two-dimensional Separation and Identification of FP-labeled Proteins
Precipitated protein pellets (200 μg) were subjected to two-dimensional gel electrophoresis and identified by liquid chromatography-MS/MS against the National Centre for Biotechnology Information non-redundant data base (October 16, 2006; 2,879,860 sequence entries; 1,012,985,077 residues) as described previously (25, 26).
Transient Transfection with pFK-I389neo/luc/NS3-3′/5.1 Replicon
pFK-I389neo/luc/NS3-3′/5.1 HCV and Renilla luciferase RNAs were generated and transiently transfected as described previously (27). Replication and translation of the transiently transfected HCV RNA was evaluated by using the dual luciferase reporter assay (Promega, Madison, WI).
Infection with JFH1 HCV
Huh7.5 and Huh7CES1 cells were seeded 24 h before infection at a density of 1.2 × 105 cells/well in a 12-well plate. Cells were infected with 10 μl of serially passaged JFH1 virus (8 × 107 copies/ml, filtered through a 0.22-μm filter). Four days after infection, the HCV titers were quantified as described previously (28), and the cells were visualized by immunofluorescence as described below.
Immunofluorescence Analysis of HCV-infected Chimeric Human-Mouse Liver
All mice were treated according to the University of Alberta Health Research Ethics Board and the Canadian Council on Animal Care guidelines. SCID-beige/Alb-uPA mice were transplanted with human primary hepatocytes and then infected with human serum containing HCV (genotype 2a) as described previously (29, 30). Formaldehyde-fixed paraffin-embedded sections (4 μm) were prepared as described previously (28) and subjected to immunofluorescence staining as described below.
Statistical Analysis
Individual experiments in this study were performed in triplicate in order to confirm the reproducibility of the results. Values are represented as means ± S.D. The statistical significance of differences between two or more means was evaluated by using analysis of variance; p values of less than 0.05 (indicated by asterisks) were considered to be statistically significant.
RESULTS
CES1 Is Differentially Active in Huh7 Cells Highly Replicating HCV
We examined the serine hydrolase ABPP in hepatoma cells replicating a genotype 1b HCV subgenomic replicon RNA (31) (Fig. 1A), using a FP-rhodamine ABPP probe (Fig. 1B) that has been shown to react with serine hydrolases in an activity-dependent manner (19). The FP probe sensitivity was confirmed using the serine hydrolase trypsin (detection limit of 100 ng of active trypsin) (supplemental Fig. S1A), and its optimal concentration for Huh7 ABPP was observed to be 1 nm. At this concentration, the heat-denatured proteome showed minimal nonspecific labeling (supplemental Fig. S1B). To identify host cell serine hydrolase activity required for HCV replication, the active proteomes of naive Huh7 cells (bearing no replicons) and of a Huh7-derived hepatoma cell line capable of high levels of pFK-I389neo/NS3-3′/5.1 HCV RNA replication (supplemental Fig. S2, A and B) were treated with the FP probe. After two-dimensional gel separation of the proteome (Fig. 1C), the proteins that showed specific activity-based labeling by the probe were then identified by liquid chromatography-MS/MS (supplemental Table S1). Interestingly, the HCV NS3/4A serine protease was not detected in Huh7-derived cells stably replicating pFK-I389neo/NS3-3′/5.1. By using purified recombinant NS3/4A, the enzyme was showed to have minimal reactivity toward several ABPP probes (26), which could be attributed to inherent reactivity. In the cellular environment, additional limiting factors could include the low abundance of NS3/4A relative to other host cell hydrolases or product inhibition of NS3/4A (32). However, three known serine hydrolases were identified by the FP ABPP: CES1, acyl-CoA thioesterase 1, and monoglyceride lipase (Fig. 1C and supplemental Table S1). From these, only spot number 1 appeared to be unique to the Huh7-derived cells (Huh7CES1 cells) when compared with naive cells. CES1 was identified unambiguously from spot 1 (Fig. 1C) because the amino acid sequences of 19 peptides determined by MS/MS covered 51% of the CES1 sequence (supplemental Table S1) (herein we refer to the clonal cell line derived from Huh7 cells as Huh7CES1 cells because they have endogenous CES1 expression, whereas Huh7 and Huh7.5 do not; see below). Proteomes from naive Huh7 and Huh7.5 cells as well as from Huh7 stably replicating pFK-I389neo/luc/NS3-3′/5.1 did not show any appreciable CES1 activity as measured by the FP probe, which correlated with the low abundance of CES1 measured by Northern and Western blotting (supplemental Fig. S2A). Because CES1 displayed the greatest differential activity between the pFK-I389neo/NS3-3′/5.1 replicating in Huh7CES1 cells and the other cell lines, naive Huh7, Huh7.5, and Huh7 replicating pFK-I389neo/luc/NS3-3′/5.1 (supplemental Fig. S2, A and B), we sought to understand if cell line-specific differences in hepatoma cells gave rise to differences in CES1 expression or if these differences could also be attributed to HCV pFK-I389neo/NS3-3′/5.1 replication.
HCV Modulates CES1 Abundance
To determine if the high CES1 expression observed was attributed to HCV, we attempted to cure the pFK-I389neo/NS3-3′/5.1 replicon-bearing cells. This was accomplished by treating the cells for 2 weeks with NS5B-6367 siRNA (33), followed by a 2-month IFNγ treatment (200 units/ml). We identified a number of cured clones, including Huh7CES1 cells, all of which eventually displayed greatly reduced CES1 RNA and protein levels (Fig. 2A) but retained the ability to express CES1 when stimulated with HCV replicon RNA. The loss of CES1 expression, after curing cells of HCV replicon RNA, displays slow kinetics and takes days to weeks to reach minimal levels. Immunofluorescence and FACS analysis of the cured Huh7CES1 cells also confirmed these results because they showed significantly reduced CES1 staining, which was not completely abolished, unlike naive Huh7 and Huh7.5, which displayed no detectable CES1 expression (Fig. 2B). A basal CES1 expression remains in about 15% cured Huh7CES1 cells after 60 days of interferon treatment. The presence of over 40% of cells with moderate CES1 expression following a 96-h IFNγ treatment indicates that the decrease of CES1 expression appears to be gradual over a significant period of time and seems to follow the trend of HCV NS5A levels (whose expression was also reduced to about 40% of the cells at this point) (Fig. 2B).
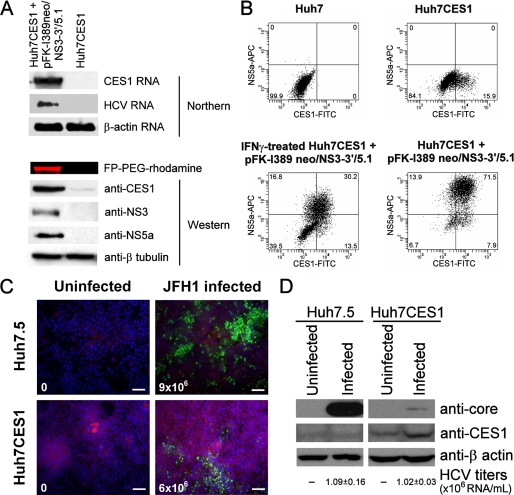
FIGURE 2.
CES1 modulates JFH1 HCV infection in cured Huh7CES1 cells. A, pFK-I389neo/NS3-3′/5.1-bearing Huh7CES1 cells were cured with NS5B-6367 siRNA (33) for 2 weeks, followed by IFNγ treatment (200 units/ml) for 2 months, leading to the isolation of a clonal cell line referred to as Huh7CES1 cells. The cellular proteome was analyzed by Northern blotting against HCV and β-actin as well as Western blotting using anti-CES1, anti-NS3, anti-NS5A, and anti β-tubulin antibodies. B, representative flow cytometry analysis used to monitor CES1 and NS5A expression in various cell lines. The Huh7.5 and pFK-I389neo/luc/NS3-3′/5.1 replicon cell FACS profiles (not shown) were identical to that of Huh7 cells. The Huh7CES1 cells bearing pFK-I389 neo/NS3-3′/5.1 were cured by a 2-week HCV siRNA exposure followed by a 60-day IFNγ treatment (200 units/ml). The IFNγ-treated Huh7CES1 cells bearing pFK-I389 neo/NS3-3′/5.1 were subjected to a 96-h IFNγ treatment prior to FACS analysis. The numbers indicate the percentage of positive cells in each quadrant. C, Huh7.5 and cured Huh7CES1 cells were infected with 8 × 104 JFH1 particles for 96 h. Immunofluorescence was subsequently performed to visualize CES1 (red) and HCV core (green) intracellular abundance. HCV titers are indicated in the lower left of each panel (scale bars, 40 μm). D, Huh7.5 and Huh7CES1 cells were infected with 8 × 104 JFH1 particles for 96 h, after which intracellular core and CES1 levels were measured by Western blotting and HCV titers in the cellular media were quantified by quantitative PCR.
HCV Replication and Infection Are Influenced by CES1 Levels
The importance of CES1 activity for HCV replication was assessed using siRNA knockdown and overexpression experiments on Huh7CES1 cells harboring the pFK-I389neo/NS3-3′/5.1 HCV replicon. Overexpression of active CES1 increased HCV replication and translation because both HCV RNA and proteins correlated with the higher levels of CES1 from 48 h post-transfection (Fig. 3, A and B).
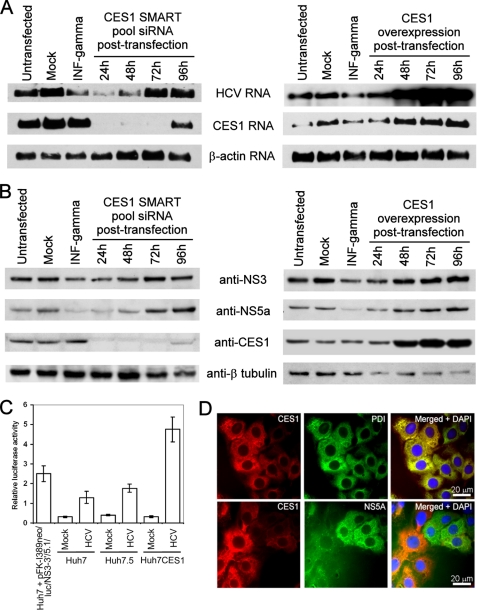
FIGURE 3.
The abundance of CES1 modulates HCV replication levels in Huh7 cells. A,B, pFK-I389neo/NS3-3′/5.1 stably replicating Huh7CES1 cells were treated with non-targeting siRNA or empty overexpression vector (mock), INFγ (48h treatment), CES1 SMART pool siRNA or CES1 overexpression plasmid. Their effects on replication level were measured at different time points post-transfection by northern blotting against HCV, CES1 and β-actin (A); and by Western blotting using anti-NS3, anti-NS5A, anti-CES1 and anti β-tubulin antibodies (B). The untreated and mock control showed are representative of the RNA and protein abundance at the four time point studied during these experiments (supplemental Fig. S3, G and H). C, the replication and translation of HCV RNA was measured in the indicated cell lines co-transfected with pFK-I389neo/luc/NS3-3′/5.1 HCV replicon and Renilla luciferase mRNA. Luciferase activity was measured 24 h post-transfection and normalized with Renilla luciferase activity. D, immunofluorescence detection of CES1 (red), PDI (green), and NS5A (green) proteins in pFK-I389neo/NS3-3′/5.1 replicon-containing Huh7CES1 cells.
In order to confirm the importance of CES1 expression, we performed transient transfections of HCV replicon RNA into the cured Huh7CES1 cells. Cells were transiently transfected with the subgenomic replicon pFK-I389/luc/NS3-3′/5.1 RNA containing a luciferase reporter gene. The luciferase reporter assay revealed that HCV replication levels were 3 and 2 times higher in Huh7CES1 cells when compared with Huh7 and Huh7.5, respectively (Fig. 3C).
Although CES1 levels correlated with HCV replication on a collective basis (i.e. within a cell population) (Fig. 3, A and B, and supplemental Fig. S3, A–D), we do observe some cells that show higher expression levels of either CES1 or HCV nonstructural proteins in about 20% of pFK-I389neo/NS3-3′/5.1 HCV replicon-containing Huh7CES1 cells, as observed by FACS analysis and fluorescence microscopy (Figs. 2B and 3D). These data suggest that the HCV-mediated mechanism for regulation of CES1 expression and activity is complex and probably involves other regulatory elements within the hepatoma cell line. These results are consistent with results from the JFH1 infectious model, where although CES1 and core proteins are simultaneously present at high levels within a given cell, they also appear in subpopulations of cells that show either high levels of CES1 expression or high levels of HCV core expression but not both (Fig. 2C). This implies that CES1 activity/function assists HCV propagation rather than there being a requirement for a direct interaction of HCV with CES1. This was further supported with FLAG-CES1 pull-down experiments, where CES1 was not observed to interact with any of the HCV non-structural proteins (data not shown).
To further evaluate the importance of CES1 for HCV replication, we used siRNA knockdown approaches. siRNAs targeting CES1 reduced CES1 RNA and protein levels by 90 and 40%, respectively, at 48 h post-transfection (Fig. 3, A and B, and supplemental Fig. S3, A, C, and F) when compared with non-targeting and mismatched CES1 siRNA controls. The CES1 knockdown and overexpression did not have any cytotoxic effect on the Huh7CES1 cells (supplemental Fig. S3E). The reduction of CES1 levels after CES1 siRNA treatments also led to a reduction in HCV RNA and NS3 and NS5A proteins by 80, 50, and 75%, respectively, up to 72 h post-transfection. Interestingly, we observe that HCV RNA and protein levels recover to nearly normal levels after transient CES1 knockdown (Fig. 3, A and B, and supplemental S3) and that this recovery appears to happen more rapidly than for the recovery of CES1 expression. Because we do not know the minimum level of CES1 expression that is required for HCV replication to occur efficiently in this cell line, it is possible that there is enough of a rebound of CES1 levels to cause HCV to begin to recover but remains below the detection limits of our Western or Northern blots. Also, we have observed that there can be differential cellular expression of CES1 and HCV (Figs. 2B and 3D) within a cell population, as discussed above, and that this could explain the different kinetics observed for recovery of HCV and CES1 levels.
The effects of CES1 knockdown on HCV RNA and protein levels were observed to be similar to that of the antiviral cytokine IFNγ. The recovery of HCV and CES1 levels at 96 h post-siRNA transfection was due to the short half-life of transiently transfected siRNA (Fig. 3, A and B). These results suggest that CES1 can influence HCV replication and that HCV also seemed to promote CES1 expression in the pFK-I389neo/NS3-3′/5.1 Huh7CES1 cell line. This cell line will be useful for identifying the mechanisms of CES1 function in the liver and how this function influences HCV.
Next, we conducted HCV infection experiments on the cured Huh7CES1 cells using the JFH1 genotype 2a infectious model (34, 35) to determine the effects of basal CES1 levels on HCV infection. Huh7.5 cells were used as a control because they are ideal for supporting infection by the JFH1 HCV strain. When exposed to three genome equivalents of infectious JFH1 HCV virion particles for 96 h, we observed a 3-fold reduction of cells with high intracellular core staining (Fig. 2C) and a 20-fold decrease of intracellular core protein levels (Fig. 2D) in the Huh7CES1 cells when compared with Huh7.5. Interestingly, HCV titer levels of secreted virus were very similar between the two cell lines. These results suggest that CES1 expression in Huh7CES1 cells (Fig. 2, A–D) probably plays a role in the export of HCV virions because core does not accumulate intracellularly as much as in the CES1-negative Huh7.5 cells, although similar viral titers between the two cell lines were observed.
CES1 Increases TG and CE Levels and Apolipoprotein B (ApoB) Secretion
To further study the potential role of CES1 in HCV secretion, the role of CES1 in intracellular lipid modulation was analyzed. CES1 is a luminal ER protein (36, 37) (Fig. 3D, top panels) that is highly expressed in primary human hepatocytes (38) and is the most abundant hydrolase in the human liver (39, 40). CES1 has been shown to catalyze the hydrolysis of ester, thioester, and amide bonds and modulates neutral lipid homeostasis by displaying cholesteryl ester hydrolase (CEH) (41), acyl-CoA:acyl transferase (ACAT) (42), and triacylglycerol hydrolase (TGH) (43) activities. In Homo sapiens, the CES1 enzyme not only has TGH, ACAT, and CEH activity, but these abbreviations are also used as alternative names for the same enzyme. It is also involved in the transport of neutral lipids (i.e. CE and TGs) across the ER for loading in LDs and thus participates in the assembly and maturation of CE- and TG-rich VLDL (43).
CES1 displays CEH, ACAT, and TGH activities (41–44) to maintain constant levels of free lipids in the cell that may be important for HCV assembly and budding (11, 16, 45). To delineate the role of CES1 in HCV replication, we examined the cellular function of CES1 in modulating neutral lipid homeostasis by measuring the intracellular levels of TG, cholesterol, and apoB. In untransfected cells, TG, cholesterol, and secreted apoB levels were found to directly correlate with the endogenous abundance of CES1 (Fig. 4, A–C, and supplemental Fig. S2A). In pFK-I389neo/NS3-3′/5.1 replicating Huh7CES1 cells with high endogenous CES1 expression, cholesterol, CE, TGs, and secreted apoB levels were 11-, 3-, 3-, and 2-fold higher, respectively, compared with the other cell lines in which CES1 levels were low or undetectable (Fig. 4, A–C). Overexpression of CES1 in Huh7CES1 replicating pFK-I389neo/NS3-3′/5.1 was shown to increase cholesterol, CE, TGs, and secreted apoB levels by 10-, 3-, 2-, and 2-fold, respectively. Our observations are consistent with the known role of CES1 in CE- and TG-rich VLDL assembly and secretion (43).
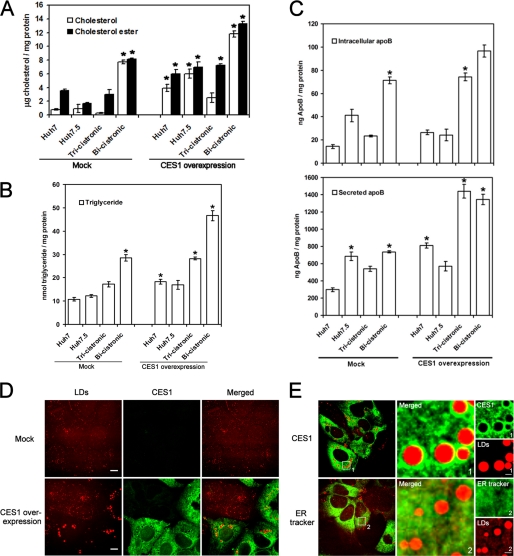
FIGURE 4.
CES1 surrounds LDs and increases the storage of inert lipids in LDs. A–C, cholesterol (A), triglyceride (B), and apoB (C) levels measured before and after overexpressing CES1 for 48 h in naive Huh7 and Huh7.5, as well as in cells stably expressing the subgenomic bi- and tricistronic replicons. Statistically significant differences of three independent experiments were determined using analysis of variance, with probabilities p < 0.05 (asterisk). D, Huh7 cells with basal or overexpressed CES1 levels were analyzed 48 h post-transfection with CARS and two-photon immunofluorescence microscopy to visualize lipid droplets (red) and CES1 protein (green) (scale bar, 10 μm). E, after overexpression of CES1 for 48 h in Huh7 cells, two-photon immunofluorescence microscopy was used to visualize CES1 (green) or ER tracker (green) in parallel with LD analysis (red) by CARS microscopy. Higher magnification images of areas 1 and 2 are shown in the middle and right panels (scale bar, 2 μm). Error bars, S.D.
CES1 Does Not Influence the Mevalonate Pathway
The role of CES1 in intracellular cholesterol homeostasis was further investigated through the mevalonate pathway because it was previously demonstrated that an increased activity of this pathway promotes protein geranylgeranylation (such as FBL2) that facilitates HCV replication (7). To determine whether CES1 influences the HCV replication through FBL2 geranylgeranylation, the mRNA levels of HMG-CoA synthase, one of the most tightly regulated enzymes in the mevalonate pathway, were monitored by real-time PCR in pFK-I389neo/NS3-3′/5.1 replicating Huh7CES1 cells. After knockdown and overexpression of CES1, no significant differences were observed among HMG-CoA synthase mRNA levels when compared with the benzamide control (supplemental Table S2). Benzamide was previously shown to decrease HMG-CoA synthase expression (31). It is therefore unlikely that CES1 modulates HCV levels through geranylgeranylation of FBL2 from up-regulation of HMG-CoA synthase (46).
CES1 Increases LD Size and Abundance
We have shown that CES1 activity alters levels of TG and CE, which are the main components of cellular LDs (Fig. 4). We hypothesized that CES1 may affect the size and number of LDs. The accumulation of LDs leads to hepatic steatosis, or fatty liver, which results from increases in TGs (47). LDs are associated with hepatic steatosis and are also involved in the production of infectious HCV particles (11). We tested whether there is a correlation between CES1 activity and LD abundance using coherent anti-Stokes Raman scattering (CARS) and two-photon fluorescence microscopy, a modern technique previously applied to the visualization of lipids in living cells (48–54) and host-HCV interactions (31, 55). Endogenous LD levels in the four hepatoma-derived cell lines (supplemental Fig. S4, A and B) were found to be proportional to CES1 levels (supplemental Fig. S2A), for which the Huh7CES1 cells had a 25% stronger CARS lipid signal per cell (n = 40). The increased abundance of TGs and CE measured during CES1 overexpression (Fig. 4, A and B) resulted in a 35% increase of cells (n = 100) with larger and more abundant LDs (Fig. 4D), consistent with observations in CHO cells (42).
Given that components of the HCV replication complex localize to ER membranes near LDs, resulting in doughnut-shaped immunohistochemical staining patterns surrounding LDs that have a diameter slightly larger than that of HCV core (11), the boundary between the ER and LDs appears critical to the HCV life cycle. Virion assembly also occurs around the LD-associated membrane (11). Using immunofluorescence analysis, CES1 was found to concentrate around LDs by encircling them when visualized by CARS microscopy (Fig. 4E) and Oil red-O staining (supplemental Fig. S4C). The absence of a fluorescent ring around LDs from the ER tracker control (Fig. 4E) supports the observations that the CES1 protein occupies LD-associated ER membranes, reminiscent of HCV localization (11). Although CES1 is mainly found in the ER, its enrichment around the LDs and the continuity between the ER and LDs make lipid delivery possible (12). CES1 was identified from LD proteomic analyses (56), suggesting that it has a strong association with LDs. This is consistent with the enrichment of CES1 around LDs observed by CARS and two photon fluorescence microscopy (Fig. 4E). These observations implicate a possible role for CES1 in CE and TG loading of LDs (56). CEH (another acronym for CES1) was shown to relocate at the surface of LDs upon lipid loading in human macrophages, resulting in a weak cellular ER background fluorescence with a stronger signal in a ring around the LDs (57), which is similar to our CARS and two photon fluorescence imaging of Huh7CES1 cells (Fig. 4E). Therefore, CES1 activity is probably creating more LDs and more LD-associated ER membranes that are needed for HCV replication and release (11).
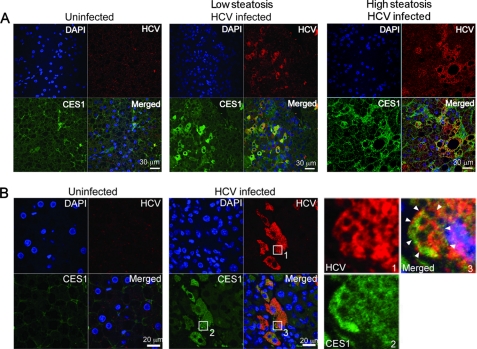
CES1 Abundance in Albumin/uPA-SCID Mice Harboring Primary Human Hepatocytes Correlates with HCV Levels
We sought to validate the importance of CES1 function in animal models representing in vivo HCV infection. We employed the small animal model for HCV infection, where primary human hepatocytes are transplanted into SCID mice carrying a plasminogen activator transgene (Alb-uPA) that have been shown to support HCV infection (supplemental Fig. S5A) without stimulating an adaptive immune response in the mice (30). In these in vivo models, HCV genomic RNA and proteins are present at biologically relevant levels, and infectious virion particles are assembled and released. We used chimeric mice that were infected with HCV JFH1 (genotype 2a, serum titer 6 × 104) (34, 58). In the chimeric liver, endogenous CES1 was detected from transplanted human hepatocytes in the uninfected controls (Fig. 5), confirming that CES1 is an abundant hydrolase in the human liver (39, 40). Upon HCV infection, immunohistochemical analysis revealed a good correlation between HCV NS3/NS4 and CES1 expression within the primary human hepatocytes, which suggests an HCV-induced CES1 expression under physiological conditions. This JFH1-induced CES1 expression in chimeric mice supports our previous finding with HCV replication and JFH1 infection in the Huh7CES1 cell line (Fig. 2). Similar results were obtained with mice containing hepatocytes from a different donor, serum titer 2.5 × 105 of JFH1 (Fig. 5B and supplemental Fig. S5B). A closer look at higher magnification reveals that both CES1 and NS3/NS4A proteins appear to be partially localized around lipid droplets in infected cells, even in a low steatosis area (Fig. 5B, arrowheads). Similar results were also obtained for chimeric mice infected with patient-derived HCV genotype 1a for 4 weeks, serum titer 3.6 × 107 (supplemental Fig. S5B) (30). Interestingly, HCV-induced CES1 expression was observed both in low and high steatosis regions of the chimeric livers (Fig. 5 and supplemental Fig. S5B). The lack of direct association between CES1- and HCV-mediated steatosis suggests that additional factors besides CES1 might be involved in triggering steatosis. Perhaps the combination of HCV-induced fatty acid synthesis and CES1 up-regulation could trigger enough CES1 to store the excessively produced TG in LDs, which would eventually lead to steatosis. Although CES1 was shown to increase the number and size of LDs in vitro (Fig. 4, D and E, and supplemental Fig. S4A), these LDs were not of the magnitude of the LDs seen in the high steatosis region (Fig. 5A and supplemental Fig. S5B).
FIGURE 5.
HCV up-regulates CES1 expression in JFH1-infected chimeric mouse-human livers. A and B, immunofluorescence co-localization of HCV NS3/NS4A proteins (red) and CES1 (green) within JFH1 HCV-infected primary human hepatocytes transplanted in SCID Alb-uPA mice from donor A (A), and analogously for another hepatocyte donor B (B). The arrowheads in the magnified image (B), box 3, delineate HCV and CES1 proteins occupying distinct areas surrounding LDs. Only human hepatocytes are present in this figure. DAPI, 4′,6-diamidino-2-phenylindole.
In summary, our data suggest that CES1 supports HCV replication and virion release and that HCV can induce CES1 expression to modulate the cellular environment to its advantage. Moreover, there is a strong link between HCV pathogenesis and CES1, and this holds true across different patient-derived primary hepatocytes, Huh7 cell lines, HCV genotypes (1b and 2a), and cellular environment (murine liver and in vitro cellular media).
DISCUSSION
ABPP is a powerful screening tool that allows for the profiling of cells and tissues based on protein activity, giving insight into the functional state of the protein rather than merely its abundance (59–62). Here, we demonstrate the application of ABPP on HCV replication to aid in the identification of host factors in the functional proteome that are involved and facilitate HCV propagation. The serine hydrolase activity profile obtained during HCV replication identified CES1 as a differentially active enzyme in the novel Huh7CES1 cell line (derived from Huh7 cells stably replicating pFK-I389neo/NS3-3′/5.1) capable of high levels of HCV replication. CES1 is abundantly expressed by primary hepatocytes (38) and is the most abundant hydrolase in the human liver (39, 40). These results demonstrate the ability of ABPP to screen for clonal variations in the hepatocytes and to identify and study the role of catalytically active enzymes, such as CES1, during HCV propagation.
HCV replication and budding occur at altered ER membranes called membraneous webs (3, 11) that are in close association with LDs (11). However, little is known about how HCV induces the formation of the membranous web or how these altered ER-LD structures might assist the virus to propagate (3, 11). We showed that both HCV genotypes 1b and 2a were influenced by CES1 expression in the Huh7CES1 cell line. Furthermore, we observed a higher abundance of large LDs in this cell line. Consistent with its known CEH, ACAT, and TGH functions (41–44), we showed that high levels of active CES1 promote both LD formation and growth through TG and CE loading into LDs. Because CES1 localizes in the luminal ER (36, 37) and the majority of cytosolic LDs are believed to be in permanent association with the ER leaflet (63), there is probably an increase in ER-LD associations in Huh7CES1 cells and in the primary hepatocytes studied, based simply on an increase in surface area of the larger LDs. Because HCV replication and infection influences CES1 levels, we postulate that the virus modulates CES1 activity to create a favorable environment around the LDs used for its efficient propagation.
We observe that CES1 displayed the greatest differential activity between naive Huh7 cells and Huh7CES1 cells stably replicating the pFK-I389neo/luc/NS3-3′/5.1 replicon. Huh7CES1 cells containing stably replicating pFK-I389neo/NS3-3′/5.1 HCV replicon showed high levels of both CES1 and HCV proteins (supplemental Figs. S2B and 3C). Transient transfection experiments did show HCV-mediated increases in CES1 expression but suggested that the kinetics of CES1 expression change and that its subsequent effects on HCV replication follow slow kinetics and may involve other changes to the host cell beyond just CES1 activity.
CARS and two-photon fluorescence cellular imaging were used to study changes in LD size and shape while simultaneously probing levels of either CES1 or HCV non-structural and core proteins. Although CARS experiments revealed dramatic differences in LD size and number during CES1 overexpression, simultaneous two-photon fluorescence imaging of CES1 confirmed its known localization on the luminal ER and surrounding LDs (36, 37). However, CES1 was found not to directly interact with HCV proteins, as supported by CES1 pull-down proteomics experiments utilizing FLAG-tagged active CES1. Therefore, CES1 function rather than protein-protein interactions seem to result in a favorable host cell environment that subsequently aids HCV pathogenesis.
It is possible that the reduced intracellular accumulation of core protein observed during JFH1 infection in Huh7CES1 cells might be attributable to an increase of production of TG- and CE-rich ApoB VLDLs created by CES1 activity. Several studies have shown that HCV secretion is dependent on VLDL secretion (45, 64), and our data show that CES1 may play a regulatory role in this process. When insufficient loading of nascent ApoB particles with neutral lipids (i.e. TG and CE stored in LDs) occurs, these poorly lipidated pre-VLDL particles are targeted for intracellular degradation (65). Many hepatoma cells lines, such as McArdle RH7777, HepG2, Huh7, and Huh7.5, poorly mobilize TG essential for proper VLDL assembly secretion (64, 66), which could be attributed to a lack in TGH (i.e. CES1) activity (66) (this study). Furthermore, specific TGH inhibitors were shown to reduce intracellular lipolysis and decreased apoB and TG secretion in both rat hepatocytes and hepatoma cells (67). CES1 therefore appears to have a regulatory role in ApoB/VLDL secretion, supporting our finding that high CES1 activity in Huh7CES1 cells increases intracellular TG and CE stores in LDs, which results in an augmented lipidation and secretion of VLDL particles. The increased VLDL secretion observed in Huh7CES1 cells could favor JFH1 HCV release because these cells showed less intracellular core staining during JFH1 infection but display similar HCV titers as observed with Huh7.5.
In summary, we have identified a unique relationship between CES1, a host ER protein, and HCV that assists and regulates HCV propagation. HCV-host interactions of this type represent novel targets for development of antivirals because it is more difficult for HCV to develop escape mutations against therapeutics that target host cell factors. Our findings may lead to novel antiviral strategies and help facilitate further studies of this important cellular enzyme and its influence on HCV pathogenesis.
Supplementary Material
Acknowledgments
We thank Dr. Takaji Wakita for JFH1 and associated materials, Dr. Ralf Bartenschlager for materials for HCV mouse models and replicon generation, Dr. John Kelly for mass spectrometry, Shuqiong Lin for assistance with probe synthesis, and Sylvie Belanger and Kasia Kieliszkiewicz for technical assistance. The anti-CES1 serum was a kind gift from Dr. Lance Pohl.
This work was supported by the Canadian Liver Foundation, the Canadian Institutes for Health Research, and the National Research Council Genomics and Health Initiative.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” “Results,” Tables S1 and S2, and Figs. S1–S5.
- HCV
- hepatitis C virus
- ABPP
- activity-based protein profiling
- ACAT
- acyl-CoA:acyl transferase
- apoB
- apolipoprotein B
- CARS
- coherent anti-Stokes Raman scattering
- CE
- cholesterol ester(s)
- CEH
- cholesteryl ester hydrolase
- ER
- endoplasmic reticulum
- FP
- fluorophosphonate
- LD
- lipid droplet
- PEG
- polyethylene glycol
- TG
- triacylglycerol
- TGH
- triacylglycerol hydrolase
- VLDL
- very low density lipoprotein
- MS
- mass spectrometry
- IFN
- interferon
- siRNA
- small interfering RNA
- FACS
- fluorescence-activated cell sorting.
REFERENCES
- 1.Lindenbach B. D., Rice C. M. (2005) Nature 436, 933–938 [DOI] [PubMed] [Google Scholar]
- 2.Chisari F. V. (2005) Nature 436, 930–932 [DOI] [PubMed] [Google Scholar]
- 3.Moradpour D., Penin F., Rice C. M. (2007) Nat. Rev. Microbiol. 5, 453–463 [DOI] [PubMed] [Google Scholar]
- 4.Sagan S. M., Rouleau Y., Leggiadro C., Supekova L., Schultz P. G., Su A. I., Pezacki J. P. (2006) Biochem. Cell Biol. 84, 67–79 [DOI] [PubMed] [Google Scholar]
- 5.Pezacki J. P., Singaravelu R., Lyn R. K. (2010) Mol. Biosys. 6, 1131–1142 [DOI] [PubMed] [Google Scholar]
- 6.Adinolfi L. E., Gambardella M., Andreana A., Tripodi M. F., Utili R., Ruggiero G. (2001) Hepatology 33, 1358–1364 [DOI] [PubMed] [Google Scholar]
- 7.Kapadia S. B., Chisari F. V. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2561–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tai A. W., Benita Y., Peng L. F., Kim S. S., Sakamoto N., Xavier R. J., Chung R. T. (2009) Cell Host Microbe 5, 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moriya K., Fujie H., Shintani Y., Yotsuyanagi H., Tsutsumi T., Ishibashi K., Matsuura Y., Kimura S., Miyamura T., Koike K. (1998) Nat. Med. 4, 1065–1067 [DOI] [PubMed] [Google Scholar]
- 10.Shi S. T., Polyak S. J., Tu H., Taylor D. R., Gretch D. R., Lai M. M. (2002) Virology 292, 198–210 [DOI] [PubMed] [Google Scholar]
- 11.Miyanari Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M., Bartenschlager R., Wakita T., Hijikata M., Shimotohno K. (2007) Nat. Cell Biol. 9, 1089–1097 [DOI] [PubMed] [Google Scholar]
- 12.Martin S., Parton R. G. (2006) Nat. Rev. Mol. Cell Biol. 7, 373–378 [DOI] [PubMed] [Google Scholar]
- 13.Beckman M. (2006) Science 311, 1232–1234 [DOI] [PubMed] [Google Scholar]
- 14.Listenberger L. L., Brown D. A. (2008) Curr. Biol. 18, R237–R238 [DOI] [PubMed] [Google Scholar]
- 15.Boulant S., Douglas M. W., Moody L., Budkowska A., Targett-Adams P., McLauchlan J. (2008) Traffic 9, 1268–1282 [DOI] [PubMed] [Google Scholar]
- 16.Gastaminza P., Cheng G., Wieland S., Zhong J., Liao W., Chisari F. V. (2008) J. Virol. 82, 2120–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su A. I., Pezacki J. P., Wodicka L., Brideau A. D., Supekova L., Thimme R., Wieland S., Bukh J., Purcell R. H., Schultz P. G., Chisari F. V. (2002) Proc. Natl. Acad. Sci. U. S. A. 99, 15669–15674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans M. J., Cravatt B. F. (2006) Chem. Rev. 106, 3279–3301 [DOI] [PubMed] [Google Scholar]
- 19.Liu Y., Patricelli M. P., Cravatt B. F. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14694–14699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barglow K. T., Cravatt B. F. (2007) Nat. Methods 4, 822–827 [DOI] [PubMed] [Google Scholar]
- 21.Jessani N., Humphrey M., McDonald W. H., Niessen S., Masuda K., Gangadharan B., Yates J. R., 3rd, Mueller B. M., Cravatt B. F. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13756–13761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saghatelian A., Jessani N., Joseph A., Humphrey M., Cravatt B. F. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10000–10005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salisbury C. M., Cravatt B. F. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1171–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Funke R., Gage D., Harris K., Heaford A., Howland J., Kann L., Lehoczky J., LeVine R., McEwan P., McKernan K., Meldrim J., Mesirov J. P., Miranda C., Morris W., Naylor J., Raymond C., Rosetti M., Santos R., Sheridan A., Sougnez C., Stange-Thomann N., Stojanovic N., Subramanian A., Wyman D., Rogers J., Sulston J., Ainscough R., Beck S., Bentley D., Burton J., Clee C., Carter N., Coulson A., Deadman R., Deloukas P., Dunham A., Dunham I., Durbin R., French L., Grafham D., Gregory S., Hubbard T., Humphray S., Hunt A., Jones M., Lloyd C., McMurray A., Matthews L., Mercer S., Milne S., Mullikin J. C., Mungall A., Plumb R., Ross M., Shownkeen R., Sims S., Waterston R. H., Wilson R. K., Hillier L. W., McPherson J. D., Marra M. A., Mardis E. R., Fulton L. A., Chinwalla A. T., Pepin K. H., Gish W. R., Chissoe S. L., Wendl M. C., Delehaunty K. D., Miner T. L., Delehaunty A., Kramer J. B., Cook L. L., Fulton R. S., Johnson D. L., Minx P. J., Clifton S. W., Hawkins T., Branscomb E., Predki P., Richardson P., Wenning S., Slezak T., Doggett N., Cheng J. F., Olsen A., Lucas S., Elkin C., Uberbacher E., Frazier M., Gibbs R. A., Muzny D. M., Scherer S. E., Bouck J. B., Sodergren E. J., Worley K. C., Rives C. M., Gorrell J. H., Metzker M. L., Naylor S. L., Kucherlapati R. S., Nelson D. L., Weinstock G. M., Sakaki Y., Fujiyama A., Hattori M., Yada T., Toyoda A., Itoh T., Kawagoe C., Watanabe H., Totoki Y., Taylor T., Weissenbach J., Heilig R., Saurin W., Artiguenave F., Brottier P., Bruls T., Pelletier E., Robert C., Wincker P., Smith D. R., Doucette-Stamm L., Rubenfield M., Weinstock K., Lee H. M., Dubois J., Rosenthal A., Platzer M., Nyakatura G., Taudien S., Rump A., Yang H., Yu J., Wang J., Huang G., Gu J., Hood L., Rowen L., Madan A., Qin S., Davis R. W., Federspiel N. A., Abola A. P., Proctor M. J., Myers R. M., Schmutz J., Dickson M., Grimwood J., Cox D. R., Olson M. V., Kaul R., Raymond C., Shimizu N., Kawasaki K., Minoshima S., Evans G. A., Athanasiou M., Schultz R., Roe B. A., Chen F., Pan H., Ramser J., Lehrach H., Reinhardt R., McCombie W. R., de la Bastide M., Dedhia N., Blöcker H., Hornischer K., Nordsiek G., Agarwala R., Aravind L., Bailey J. A., Bateman A., Batzoglou S., Birney E., Bork P., Brown D. G., Burge C. B., Cerutti L., Chen H. C., Church D., Clamp M., Copley R. R., Doerks T., Eddy S. R., Eichler E. E., Furey T. S., Galagan J., Gilbert J. G., Harmon C., Hayashizaki Y., Haussler D., Hermjakob H., Hokamp K., Jang W., Johnson L. S., Jones T. A., Kasif S., Kaspryzk A., Kennedy S., Kent W. J., Kitts P., Koonin E. V., Korf I., Kulp D., Lancet D., Lowe T. M., McLysaght A., Mikkelsen T., Moran J. V., Mulder N., Pollara V. J., Ponting C. P., Schuler G., Schultz J., Slater G., Smit A. F., Stupka E., Szustakowski J., Thierry-Mieg D., Thierry-Mieg J., Wagner L., Wallis J., Wheeler R., Williams A., Wolf Y. I., Wolfe K. H., Yang S. P., Yeh R. F., Collins F., Guyer M. S., Peterson J., Felsenfeld A., Wetterstrand K. A., Patrinos A., Morgan M. J., de Jong P., Catanese J. J., Osoegawa K., Shizuya H., Choi S., Chen Y. J., Szustakowki J. (2001) Nature 409, 860–921 [DOI] [PubMed] [Google Scholar]
- 25.Singaravelu R., Blais D. R., McKay C. S., Pezacki J. P. (2010) Proteome Sci. 8, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blais D. R., Brûlotte M., Qian Y., Bélanger S., Yao S. Q., Pezacki J. P. (2010) J. Proteome Res. 9, 912–923 [DOI] [PubMed] [Google Scholar]
- 27.Noestheden M., Hu Q., Tonary A. M., Tay L. L., Pezacki J. P. (2007) Org. Biomol. Chem. 5, 2380–2389 [DOI] [PubMed] [Google Scholar]
- 28.Joyce M. A., Walters K. A., Lamb S. E., Yeh M. M., Zhu L. F., Kneteman N., Doyle J. S., Katze M. G., Tyrrell D. L. (2009) PLoS Pathog. 5, e1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mercer D. F., Schiller D. E., Elliott J. F., Douglas D. N., Hao C., Rinfret A., Addison W. R., Fischer K. P., Churchill T. A., Lakey J. R., Tyrrell D. L., Kneteman N. M. (2001) Nat. Med. 7, 927–933 [DOI] [PubMed] [Google Scholar]
- 30.Walters K. A., Joyce M. A., Thompson J. C., Smith M. W., Yeh M. M., Proll S., Zhu L. F., Gao T. J., Kneteman N. M., Tyrrell D. L., Katze M. G. (2006) PLoS Pathog. 2, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rakic B., Sagan S. M., Noestheden M., Bélanger S., Nan X., Evans C. L., Xie X. S., Pezacki J. P. (2006) Chem. Biol. 13, 23–30 [DOI] [PubMed] [Google Scholar]
- 32.Steinkühler C., Biasiol G., Brunetti M., Urbani A., Koch U., Cortese R., Pessi A., De Francesco R. (1998) Biochemistry 37, 8899–8905 [DOI] [PubMed] [Google Scholar]
- 33.Wilson J. A., Jayasena S., Khvorova A., Sabatinos S., Rodrigue-Gervais I. G., Arya S., Sarangi F., Harris-Brandts M., Beaulieu S., Richardson C. D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2783–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindenbach B. D., Meuleman P., Ploss A., Vanwolleghem T., Syder A. J., McKeating J. A., Lanford R. E., Feinstone S. M., Major M. E., Leroux-Roels G., Rice C. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3805–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong J., Gastaminza P., Cheng G., Kapadia S., Kato T., Burton D. R., Wieland S. F., Uprichard S. L., Wakita T., Chisari F. V. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9294–9299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robbi M., Beaufay H. (1991) J. Biol. Chem. 266, 20498–20503 [PubMed] [Google Scholar]
- 37.Potter P. M., Wolverton J. S., Morton C. L., Wierdl M., Danks M. K. (1998) Cancer Res. 58, 3627–3632 [PubMed] [Google Scholar]
- 38.Zhao B., Natarajan R., Ghosh S. (2005) Physiol. Genomics 23, 304–310 [DOI] [PubMed] [Google Scholar]
- 39.Satoh T., Taylor P., Bosron W. F., Sanghani S. P., Hosokawa M., La Du B. N. (2002) Drug Metab. Dispos. 30, 488–493 [DOI] [PubMed] [Google Scholar]
- 40.Taketani M., Shii M., Ohura K., Ninomiya S., Imai T. (2007) Life Sci. 81, 924–932 [DOI] [PubMed] [Google Scholar]
- 41.Ghosh S. (2000) Physiol. Genomics 2, 1–8 [DOI] [PubMed] [Google Scholar]
- 42.Becker A., Böttcher A., Lackner K. J., Fehringer P., Notka F., Aslanidis C., Schmitz G. (1994) Arterioscler. Thromb. 14, 1346–1355 [DOI] [PubMed] [Google Scholar]
- 43.Gilham D., Alam M., Gao W., Vance D. E., Lehner R. (2005) Mol. Biol. Cell 16, 984–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei E., Alam M., Sun F., Agellon L. B., Vance D. E., Lehner R. (2007) J. Lipid Res. 48, 2597–2606 [DOI] [PubMed] [Google Scholar]
- 45.Huang H., Sun F., Owen D. M., Li W., Chen Y., Gale M., Jr., Ye J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5848–5853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang C., Gale M., Jr., Keller B. C., Huang H., Brown M. S., Goldstein J. L., Ye J. (2005) Mol. Cell 18, 425–434 [DOI] [PubMed] [Google Scholar]
- 47.Patton H. M., Patel K., Behling C., Bylund D., Blatt L. M., Vallée M., Heaton S., Conrad A., Pockros P. J., McHutchison J. G. (2004) J. Hepatol. 40, 484–490 [DOI] [PubMed] [Google Scholar]
- 48.Pegoraro A. F., Ridsdale A., Moffatt D. J., Jia Y., Pezacki J. P., Stolow A. (2009) Opt. Express 17, 2984–2996 [DOI] [PubMed] [Google Scholar]
- 49.Cheng J. X., Jia Y. K., Zheng G., Xie X. S. (2002) Biophys. J. 83, 502–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans C. L., Potma E. O., Puoris'haag M., Côté D., Lin C. P., Xie X. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16807–16812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Evans C. L., Xie X. S. (2008) Annu. Rev. Anal. Chem. 1, 883–909 [DOI] [PubMed] [Google Scholar]
- 52.Freudiger C. W., Min W., Saar B. G., Lu S., Holtom G. R., He C., Tsai J. C., Kang J. X., Xie X. S. (2008) Science 322, 1857–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Potma E. O., Xie X. S. (2003) J. Raman Spectrosc. 34, 642–650 [Google Scholar]
- 54.Lyn R. K., Kennedy D. C., Sagan S. M., Blais D. R., Rouleau Y., Pegoraro A. F., Xie X. S., Stolow A., Pezacki J. P. (2009) Virology 394, 130–142 [DOI] [PubMed] [Google Scholar]
- 55.Nan X., Tonary A. M., Stolow A., Xie X. S., Pezacki J. P. (2006) Chembiochem 7, 1895–1897 [DOI] [PubMed] [Google Scholar]
- 56.Wang H., Gilham D., Lehner R. (2007) J. Biol. Chem. 282, 33218–33226 [DOI] [PubMed] [Google Scholar]
- 57.Zhao B., Fisher B. J., St Clair R. W., Rudel L. L., Ghosh S. (2005) J. Lipid Res. 46, 2114–2121 [DOI] [PubMed] [Google Scholar]
- 58.Kato T., Date T., Murayama A., Morikawa K., Akazawa D., Wakita T. (2006) Nat. Protoc. 1, 2334–2339 [DOI] [PubMed] [Google Scholar]
- 59.Yang Z., Fonović M., Verhelst S. H., Blum G., Bogyo M. (2009) Bioorg. Med. Chem. 17, 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pungercar J. R., Caglic D., Sajid M., Dolinar M., Vasiljeva O., Pozgan U., Turk D., Bogyo M., Turk V., Turk B. (2009) FEBS J. 276, 660–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin B. R., Cravatt B. F. (2009) Nat. Methods 6, 135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barglow K. T., Saikatendu K. S., Bracey M. H., Huey R., Morris G. M., Olson A. J., Stevens R. C., Cravatt B. F. (2008) Biochemistry 47, 13514–13523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Targett-Adams P., Chambers D., Gledhill S., Hope R. G., Coy J. F., Girod A., McLauchlan J. (2003) J. Biol. Chem. 278, 15998–16007 [DOI] [PubMed] [Google Scholar]
- 64.Icard V., Diaz O., Scholtes C., Perrin-Cocon L., Ramière C., Bartenschlager R., Penin F., Lotteau V., André P. (2009) PLoS One 4, e4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fisher E. A., Ginsberg H. N. (2002) J. Biol. Chem. 277, 17377–17380 [DOI] [PubMed] [Google Scholar]
- 66.Lehner R., Cui Z., Vance D. E. (1999) Biochem. J. 338, 761–768 [PMC free article] [PubMed] [Google Scholar]
- 67.Gilham D., Ho S., Rasouli M., Martres P., Vance D. E., Lehner R. (2003) FASEB J. 17, 1685–1687 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.